Abstract
Although chronic kidney disease (CKD) is common, only a fraction of CKD patients progress to end-stage renal disease. Molecular predictors to stratify CKD populations according to their risk of progression remain undiscovered. Here we applied transcriptional profiling of kidneys from transforming growth factor-β1 transgenic (Tg) mice, characterized by heterogeneity of kidney disease progression, to identify 43 genes that discriminate kidneys by severity of glomerular apoptosis before the onset of tubulointerstitial fibrosis in 2-week-old animals. Among the genes examined, 19 showed significant correlation between mRNA expression in uninephrectomized left kidneys at 2 weeks of age and renal disease severity in right kidneys of Tg mice at 4 weeks of age. Gene expression profiles of human orthologs of the 43 genes in kidney biopsies were highly significantly related (R2 = 0.53; P < 0.001) to the estimated glomerular filtration rates in patients with CKD stages I to V, and discriminated groups of CKD stages I/II and III/IV/V with positive and negative predictive values of 0.8 and 0.83, respectively. Protein expression patterns for selected genes were successfully validated by immunohistochemistry in kidneys of Tg mice and kidney biopsies of patients with IgA nephropathy and CKD stages I to V, respectively. In conclusion, we developed novel mRNA and protein expression signatures that predict progressive renal fibrosis in mice and may be useful molecular predictors of CKD progression in humans.
Awareness of chronic kidney disease (CKD) and its consequences has increased enormously during the past decade as a result of the worldwide adoption of a uniform classification system developed by the Kidney Disease Outcomes Quality Initiative (K/DOQI).1 Recent epidemiological studies indicate that 16.8% of the US population may be affected with CKD,2 suggesting that CKD is far more prevalent in the general population than previously thought. Importantly, cardiovascular disease morbidity and mortality are strongly associated with advanced CKD and proteinuria before end-stage renal disease, suggesting that some underlying pathomechanisms may be shared, or interactive, between cardiovascular disease and CKD.3,4,5
Progression of CKD to end-stage renal disease occurs only in a minority of CKD patients, suggesting considerable heterogeneity in the risk of progressive decline of renal function in CKD. Because the K/DOQI definition of CKD stages is based on GFR alone, the relative risk of progression of patients within each stage is not characterized. As a simple method of risk assessment, family history of advanced CKD or end-stage renal disease and the extent of proteinuria are currently the best predictors of the risk to develop progressive CKD.6,7 However the accuracy of these clinical markers is currently not sufficient to predict CKD progression risk reliably, or to guide preventive interventions. Thus, one of the most important unmet needs in renal medicine is the identification and validation of predictive markers of CKD progression that facilitate targeted treatment of those at high risk, while avoiding unnecessary treatment and the attendant financial costs in low-risk patients.8 However, in contrast with other disorders that are characterized by heterogeneity in progression, in particular malignancies,9,10 the development of predictive molecular markers in CKD is limited by the paucity of tissue-based diagnostic procedures in clinical renal medicine.
Among candidate pathways mediating CKD progression, the transforming growth factor-β (TGF-β) pathway is prominent because it controls principle pathobiological processes associated with CKD, including fibrogenesis, apoptosis, epithelial-to-mesenchymal transition, and inflammation.8,11 Indeed, TGF-β and its receptors are increased in most forms of CKD in humans and experimental animal models.12,13,14 In addition, DNA polymorphisms of codon 10 in the TGF-β gene have been associated with progressive CKD.15,16 Consistent with numerous clinical studies, overexpression of TGF-β1 in transgenic (Tg) mice can cause glomerulosclerosis and tubulointerstitial fibrosis.17 Similar to the heterogeneity of CKD progression observed in humans, a fraction of TGF-β1 Tg mice develop progressive glomerulosclerosis and tubulointerstitial fibrosis leading to marked proteinuria, uremia, and death, whereas the majority of animals manifest moderate, nonprogressive renal fibrosis with mild proteinuria and normal lifespan.17,18 The heterogeneity of renal disease manifestations in TGF-β1 Tg mice is dependent on their mixed genetic background, mimicking the familial clustering and importance of genetic susceptibility observed in CKD progression in humans.6
In this study, we combined unique experimental animal and clinical research resources to identify and characterize molecular markers as predictors of estimated glomerular filtration rate (eGFR) and clinical stage of CKD in human cohorts. First, we used gene expression profiling in cross-sectional and prospective study designs to identify and validate genes that predict heterogeneity in renal disease progression in TGF-β1 Tg mice. In a second step, expression values of human orthologs of these genes, available in a unique database of human kidney biopsies,19,20 were shown to be highly-significantly related to linear GFR or K/DOQI CKD stages in human cohorts affected with hypertensive nephrosclerosis, IgA nephropathy, minimal change disease, or thin basement membrane disease. Finally, immunohistochemical protein expression patterns for several markers were associated with renal lesions in patients with CKD and IgA nephropathy.
Materials and Methods
Mouse Models
Albumin/Tgfb1 Tg mice18 in C57BL/6J × CBA background were maintained at the Animal Resource Center of Mount Sinai School of Medicine. Experiments were performed according to an approved protocol of the institutional animal care and use committee.
Kidney Total RNA Isolation
Harvested mouse kidneys were homogenized in Trizol reagent (Invitrogen, Carlsbad, CA) for 40 seconds using PowerGen125 (Fisher Scientific, Pittsburgh, PA) at maximum speed. Total RNA was isolated according to the manufacturer’s protocol. Quality and quantity of total RNA was checked by Bio-analyzer (Agilent, Santa Clara, CA).
Histological Analysis
Harvested kidneys were embedded in paraffin after fixing in 10% formalin overnight and then sectioned at 4-μm thickness. Periodic acid-Schiff (PAS)-stained sections were examined for glomerulosclerosis, mesangial expansion, tubular atrophy, and interstitial inflammation. For in situ detection of macrophage, anti-Mac-3 (BD Biosciences Pharmingen, San Jose, CA) was used. Anti-collagen I (Biogenesis, Mill Creek, WA) antibody was used to detect collagen accumulation. Apoptotic nuclei were detected by terminal dUTP nick-end labeling (TUNEL) assay (Chemicon, Temecula, CA), as described previously.21
At least 50 glomeruli per kidney were evaluated by two renal pathologists in a blinded manner. Glomerulosclerosis was scored as proportion of glomeruli with sclerosis relative to all glomeruli examined per mouse. Ten ×40 fields were scored for tubular atrophy/dilation/casts and for interstitial or perivascular inflammation for each mouse on the following scale: 0 = none, 1 = <25% of tubules/vessels with tubular features or adjacent mononuclear cells, 3 ≥50% and 4 ≥ 75% of tubules/vessels with those features or with adjacent mononuclear cells. Mean value was used for tubular and interstitial score.
Other semiquantitative histopathological scores include: Mac-3 positive cells per tubular interstitial high-power field, Col1a1-positive cells per total glomerular area (%), TUNEL-positive cells were counted as podocytes when residing on the outer aspect of PAS-positive basement membrane. Podocyte apoptotic score was defined as apoptotic podocyte/100 glomeruli. Cells were counted as nonpodocyte glomerular cells when residing inside the outer aspect of PAS-positive basement membrane. Nonpodocyte glomerular apoptotic score was defined as apoptotic nonpodocyte glomerular cells/100 glomeruli. Tubular interstitial apoptotic cell was defined as apoptotic cells/per tubular interstitial high-power field. All of these methods have been previously reported from this laboratory.22,23
Uninephrectomy
Two-week-old mice were anesthetized with isoflurane. After removing hair from the left flank, an incision was made and left kidney was decapsulated, ligated with silk suture, and excised. The area was cleansed with antimicrobial agent Amerse (ConvaTec, St. Louis, MO), and the flank incision was sutured closed. Total RNA was isolated from the left kidneys and qrt-polymerase chain reaction (PCR) was performed for gene expression analysis. At 4 weeks of age, these mice were euthanized and the right kidneys were harvested. A half of the right kidney was snap-frozen for RNA isolation, and the other half was fixed in 10% normal-buffered formalin for histopathological studies.
Quantitative Real-Time (qrt) PCR
One μg of kidney total RNA was reversely transcribed into single strand cDNA. The qrt-PCR was performed as described previously.24 Expression of Gapdh and β-actin was used to normalize the sample amount.
cDNA Microarray
Mouse cDNA arrays (9M series) were obtained from the Albert Einstein College of Medicine cDNA Microarray Facility (www.aecom.yu.edu/home/molgen/facilities.html). Each slide contained an unbiased, random collection of 8976 cDNA probe elements derived from the sequence-verified GEM1 clone set (Incyte Genomics, Palo Alto, CA). Microarray procedures were performed as previously described.25 For each hybridization cDNA was prepared from RNA samples obtained from individual kidneys from Tg or Wt mouse (Cy3-labeled) and co-hybridized with a standard reference cDNA prepared from age-matched, pooled RNA obtained from wild-type mouse kidneys (Cy5-labeled).
Data Processing of Murine Data
Preprocessing and Normalization
In the preprocessing step, the signal intensities were not background subtracted because of the local nature in which background intensity was calculated. All spots flagged during the scanning and quantification steps were removed from further analysis. The array had a scarcity of negative controls preventing an is expressed threshold from being set. The data were normalized using a within-slide scaled loess normalization that corrects for print-tip effects.
Tg (Progressive) versus Tg (Nonprogressive) Comparisons
The strength of the correlation (using Spearman coefficients) among phenotypic traits in the Tg group was used to determine informative traits. The Tg mice were then clustered using hierarchical sampling based on a phenotypic similarity metric for podocyte apoptosis. Rank order statistics and permutation tests were used to compare expression patterns among the different Tg animals. Finally, bi-clustering was done to identify subsets of genes and samples that when one is used to cluster the other, stable and significant partitions emerge. This allows utilization of both the phenotypic and expression matrices.
Clustering Analysis
Hierarchical cluster dendrograms were generated with TIGR Multiexperiment Viewer software (The Institute for Genomics Research, Rockville, MD) by using Manhattan distance metrics and bootstrapping protocols for resampling.
Immunohistochemistry Staining
Immunohistochemistry staining was performed using Vectastain ABC systems (Vector Laboratories, Burlingame, CA) and anti-Ncf2 (Santa Cruz Biotechnology Inc., Santa Cruz, CA), anti-Itgb5 (Abcam, Cambridge, MA), anti-Bgn (Abcam), anti-Col6a1 (Santa Cruz), anti-S100a6 (Santa Cruz), anti-Dkk3 (Santa Cruz), anti-Slc13a3 (Abcam), and anti-Mpv17l (affinity-purified in our laboratory26) antibodies. Images were generated by Zeiss (Thornwood, NY) Axioskop microscope in Mount Sinai Medical Center.
Gene Expression Analysis of Human Renal Biopsies
Microdissection and RNA Isolation
After renal biopsy, the tissue was transferred to RNase inhibitor and microdissected into glomerular and tubular fragments. Total RNA was isolated from microdissected tubulointerstitial and glomerular tissue as previously described.27
Target Preparation
A total of 300 to 800 ng of total RNA was reverse-transcribed and linearly amplified according to a protocol previously reported.28 The fragmentation, hybridization, staining, and imaging were performed according the Affymetrix (Santa Clara, CA) Expression Analysis Technical Manual.
Image files were initially obtained through Affymetrix GeneChip software. Subsequently, robust multichip analysis was performed using RMAexpress. Robust multichip analysis is an R-based technique using the Affymetrix microarray image file and is comprised of three steps: background adjustment, quartile normalization, and summarization. The expression values for the Affymetrix probesets are reported as log2 transformed.
Affymetrix-based gene expression profiling was performed as described in detail.28 In brief, human renal biopsy specimens were procured in an international multicenter study, the European Renal cDNA Bank-Kroener-Fresenius biopsy bank (see the Acknowledgments for participating centers). Biopsies were obtained from patients after informed consent and with approval of the local ethics committees.
Regression and Prediction of GFR at Time of Biopsy
Ridge regression was chosen to improve the predictive potential of the model in the presence of collinearities among the markers.29 To choose the regularization parameter with the lowest prediction error we used from the available methods a leave-one-out cross validation. Additionally we calculated the Pearson correlation between the expression values and the MDRD GFR of the patient for each probeset. To estimate significance of the correlation coefficients, the false discovery rate was determined using a permutation approach.
Results
Identification of a Gene Expression Signature of Advanced Glomerular Apoptosis Activity in Kidneys of Tgfb1 Tg Mice
We reported previously that kidneys from 2-week-old Tg mice were characterized by severe podocyte apoptosis and mild mesangial expansion in some, but not all animals, while the tubulointerstitial compartment was normal.23 To identify gene expression patterns that are associated with quantitative measures of apoptosis, extracellular matrix accumulation, or inflammatory cell infiltrates at the early stage of progressive renal disease in this model, respectively, we performed microarray and detailed quantitative phenotype analysis in wild-type and Tgfb1 Tg mice. Matrix accumulation and tubulointerstitial inflammation were assessed by quantitative digital analysis of α-1-collagen 1 and Mac3 immunohistochemistry, respectively. Glomerular and tubulointerstitial cell apoptosis rates were quantitated by TUNEL assay. With exception of glomerular apoptosis rates, the quantitative phenotype markers were either not significantly different among Tg mice (glomerular anti-Col1a1 labeling), and/or not significantly different when compared with wild-type mice (tubulointerstitial apoptosis and anti-Mac3 labeling) (Table 1). In contrast, glomerular apoptosis rates (GA) were sufficiently variable to separate Tgfb1 Tg mice into two groups as defined by GA less than threefold (Tg2, Tg5, Tg6), or more than threefold (Tg1, Tg3, Tg4, Tg7) of maximum value observed in wild-type control mice, respectively (Table 1). To identify the genes that are differentially expressed in the two groups, we performed linear discriminant analysis30 on expression patterns of 9000 genes. Linear discriminant analysis is a statistical method usually used to find the linear combination of features that best separate two or more classes of objects or events. Linear discriminant analysis revealed 43 genes with significantly different expression patterns between Tg mice with GA less than threefold and more than threefold of wild-type control (see Supplementary Table S1 at http://ajp.amipathol.org). Quantitative assays for these 43 genes were developed to quantitate their mRNA levels in three additional studies and to evaluate their potential utility as molecular classifiers and/or predictors of progressive renal disease.
Table 1.
Quantitative Analysis of Extracellular Matrix Accumulation, Inflammation, and Apoptosis in Wild-Type (Wt) and TGF-β1 Transgenic (Tg) Mice
| Parameters | Tg6 | Tg5 | Tg2 | Tg3 | Tg7 | Tg1 | Tg4 | Wt1 | Wt2 | Wt3 | Wt4 | Wt5 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Col1a1 | 0.20 | 0.22 | 0.34 | 0.21 | 0.37 | 0.25 | 0.20 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| Mac-3 | 0.00 | 0.00 | 2.00 | 1.00 | 1.00 | 0.00 | 1.00 | 0.00 | 0.00 | 0.56 | 0.56 | 0.56 |
| GA | 0.00 | 2.22 | 4.77 | 10.00 | 13.16 | 16.67 | 22.73 | 0.00 | 2.03 | 0.36 | 0.91 | 0.00 |
| TIA | 1.60 | 1.42 | 1.40 | 1.15 | 3.05 | 2.67 | 1.09 | 0.17 | 0.61 | 1.21 | 0.48 | 0.22 |
Col1a1, collagen I α-1-positive area, presented as fraction of Col1a1-positive area per total glomerular area; Mac-3, presented as Mac-3-positive cells per high-power field; GA, glomerular cell apoptosis was scored as apoptotic glomerular cells/100 glomeruli; TIA, tubular interstitial apoptosis was defined as apoptotic cells/per tubular interstitial high-power field based on TUNEL assay.
Validation of 43-Gene Expression Signature to Classify Tubulointerstitial Disease Heterogeneity in Older Tgfb1 Tg Mice
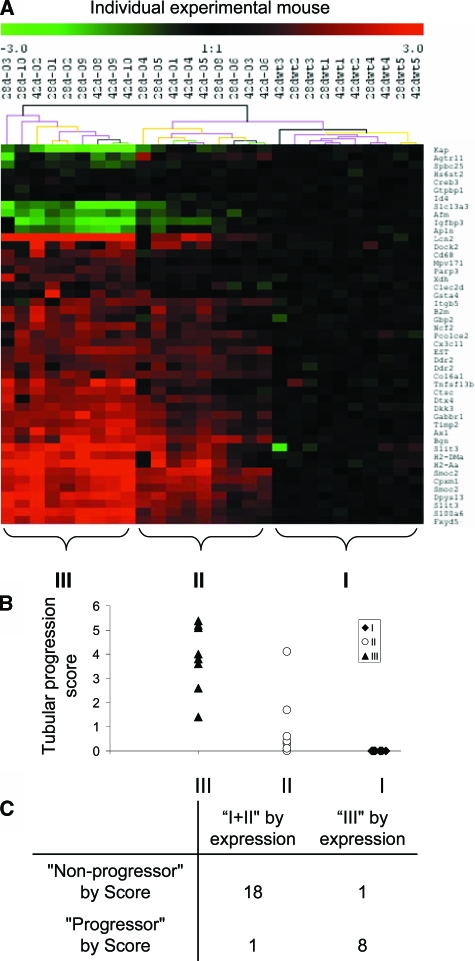
By 4 or 6 weeks of age, histopathological manifestations of progressive renal disease, including tubular atrophy and interstitial inflammation, were established and highly variable in Tgfb1 Tg mice (see Supplementary Table S2 at http://ajp.amipathol.org). Next, we asked the question if the expression patterns of the 43-geneset in 4- and 6-week-old mice is able to classify the mice by progression of histopathological manifestations in the kidney. We chose to use an interrelated two-way (that is, genes against mice) hierarchical clustering method using unsupervised approach. The goal of clustering is to find important gene patterns and perform cluster discovery on experimental mice. The advantage of this approach is that we can dynamically use the relationships between the groups of genes and mice while iteratively clustering through both gene dimension and experimental mouse dimension. We applied unsupervised two-way hierarchical clustering, based on expression profiles of the 43-geneset, to an independent set of 4- and 6-week-old wild-type (10 mice) and Tgfb1 Tg (18 mice). To show the reliability of the clusters, we performed bootstrapped cluster analysis (the bootstrap is widely accepted as a method to assess the reliability of reconstructed phylogenetic trees31 using the TMEV (TIGR Multiple Experiment Viewer) program32 and we could demonstrate that expression profiles of 43-geneset reliably identified a highly-significant (100% reproducibility) cluster of 9 animals (cluster III) among 18 Tgfb1 Tg mice and 10 wild-type mice (Figure 1A).
Figure 1.
A: Bootstrapped hierarchical clustering analysis of 10 wild-type and 18 Tg mice based on expression values of the 43 genes at 4 and 6 weeks of age. Gene expression values were acquired from cDNA microarray data. Transcripts are annotated with gene symbol. The individual experimental mouse was labeled as “age-Mouse ID.” Black line of the supporting tree indicates the cluster is 100% reproducible. B: Scatter plot graph of the histopathological score (evaluated by level of tubular atrophy and interstitial inflammation based on PAS staining) of individual mouse in groups I, II, and III, as clustered by bootstrapped hierarchical clustering analysis in A. Detailed scoring method is described in Materials and Methods. C: Distribution of experimental mice of group I + II and group III as defined by gene expression profile in nonprogressor or progressor as defined by histopathological scores (mouse is classified as progressor if the semiquantitative histopathological score is more than 2, otherwise is considered as nonprogressor; nonprogressor subgroup includes wild-type mice).
Semiquantitative scores for tubular atrophy (TA) and interstitial-perivascular inflammation (IPI) were tightly correlated in 4- to 6-week-old Tgfb1 Tg mice and combined to calculate a composite tubular progression score (TA+IPI) in all animals of this set (see Supplementary Table S2 at http://ajp.amipathol.org). The composite tubular progression score was significantly higher in cluster III (median, 4.0) compared with cluster II Tgfb1 Tg mice (median, 0.4) (P = 0.00015) and cluster I (wild-type mice) (Figure 1B). A cut-off value of 2.0 of the composite tubular progression score classified Tgfb1 Tg and wild-type mice into progressive (cluster III) and nonprogressive (clusters I, II) groups with 88.9% and 95% sensitivity and specificity, respectively (Figure 1C). These findings indicate that the 43-geneset classified advanced versus mild tubulointerstitial progression of renal disease with high sensitivity and specificity in an independent set of older Tgfb1 Tg mice.
Identification of Prospective Predictor Expression Signatures for Progressive Renal Fibrosis in Tgfb1 Tg Mice
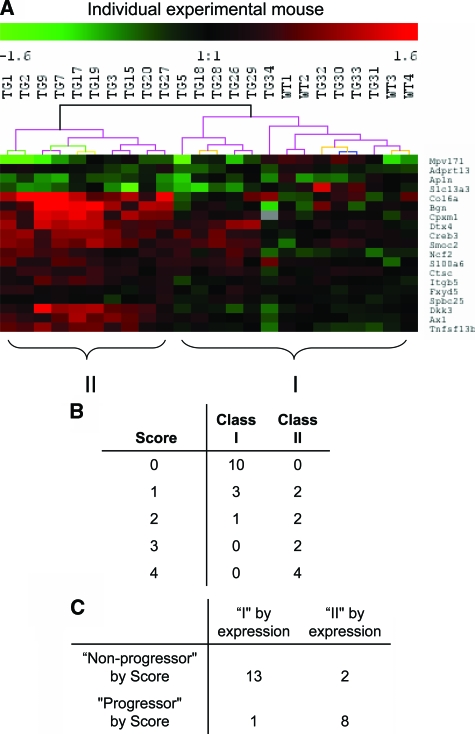
Next we devised a longitudinal study to examine the correlation and predictive values of gene expression profiles obtained among the 43-geneset in left kidneys removed by uninephrectomy at 2 weeks of age, with the histopathological manifestations in right kidneys of the same animal at 4 weeks of age. Expression levels of the 43-geneset were determined by qrt-PCR analysis of total RNA extracted from whole kidney of 24 experimental mice (20 Tgfb1 Tg mice and 4 wild-type mice) at 2 weeks of age and histopathological scoring of the remaining right kidney was performed on PAS-stained sections by three independent investigators in a blinded manner at 4 weeks of age. F-test statistics identified 19 genes among the 43-geneset, which grouped animals prospectively according to the severity of histopathological scores with statistical significance of P < 0.05 (see Supplementary Table S3 at http://ajp.amipathol.org). Bootstrapped unsupervised clustering demonstrated that expression profiles of 19-geneset, determined by qrt-PCR of left kidney RNA at 2 weeks of age, reliably identified a highly-significant (100% reproducibility) cluster of 10 animals (cluster II) among 20 Tgfb1 Tg mice and 4 wild-type mice (Figure 2A). Histopathology scores were recorded for each animal on a scale of 0 (normal) to 4 (global glomerulosclerosis and tubulointerstitial fibrosis). Scores were consistent across all three investigators and the median score for each animal was used for statistical analysis. The median of all median histopathology scores among animals grouped in cluster II (Tgfb1 Tg mice) was 3.0, compared with 0 among all animals in cluster I (10 Tgfb1 Tg and 4 wild-type mice) (P = 0.000078)(Figure 2B). By assigning an optimal cut-off value of the histopathology score between 1 and 2 for classification, the gene expression profiles for 19-geneset, as determined at 2 weeks of age, prospectively predicted the severity of renal disease progression as assessed by semiquantitative scoring in Tgfb1 Tg mice with 87.5%, 88.9%, and 86.6% accuracy, sensitivity, and specificity, respectively (Figure 2C). Thus, results obtained from the prospective, longitudinal validation study in an independent cohort of Tgfb1 Tg and wild-type mice demonstrated that the 19-geneset predicted advanced versus mild progression of renal disease with high accuracy, sensitivity, and specificity. Of note, renal TGF-β1 mRNA levels were not significantly different between the advanced and mild progression groups identified by 19-geneset expression signature (see Supplementary Figure S1 at http://ajp.amipathol.org), indicating that the separation of mild and advanced progression groups is not associated with differences in renal TGF-β1 levels between both groups.
Figure 2.
A: Bootstrapped hierarchical clustering analysis of uninephrectomized wild-type (n = 4) and Tg mice (n = 20) at 4 weeks of age based on the expression value of 19 predictive genes in their 2-week-old kidneys. Gene expression values were acquired from 2-week-old kidneys by qrt-PCR. Phenotypic lesions of the remaining kidneys were scored at 4 weeks of age. B: Semiquantitative histopathological score (0 to 4) of the experimental mice in groups I and II (clustered by expression value). C: Distribution of experimental mice according to group I or II as defined by gene expression profile and progressor and nonprogressor by histopathological scores (mouse is classified as progressor if the score is more than 2, otherwise is considered as nonprogressor; nonprogressor subgroup includes wild-type mice).
Development of a Renal Gene Expression Signature Related to Continuous eGFR and CKD Stages in Human CKD Cohorts
Because the 43-geneset and a subset of 19 genes were validated as classifier and/or predictive markers of advanced renal histopathology in two independent cohorts of Tgfb1 Tg mice, using a cross-sectional and a longitudinal study design, respectively, we devised a third validation study using cohorts of humans with various stages of CKD from the European Renal cDNA Bank-Kroener-Fresenius Biopsy Bank (ERCB).19 Consented ERCB participants were screened for cases with i) diagnostic kidney biopsies; ii) quality-controlled, high-quality RNA/cDNA from microdissected tubular interstitial and glomerular compartments; iii) high-quality genome-wide expression microarray data for both, glomerular and tubulointerstitial compartments;, and iv) clinical information on medical treatments and K/DOQI stage classification of kidney function. This screen of the ERCB database identified patients with hypertensive nephropathy (HTN) (n = 19), IgA nephropathy (IgAN) (n = 21), minimal change disease (n = 1), thin membrane disease (n = 6), and unaffected, normal renal tissue from tumor nephrectomies (n = 3) (see Supplementary Table S4 at http://ajp.amipathol.org).
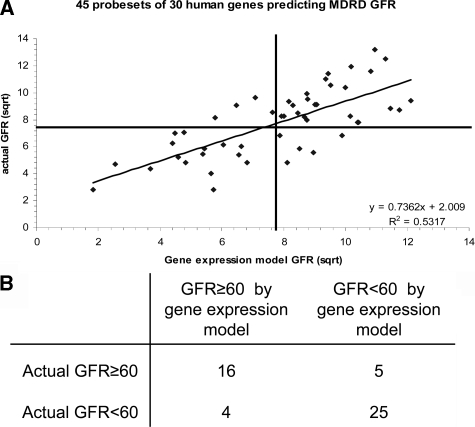
Among the 43 murine geneset, unequivocal human orthologs were identified for 33 genes, which are represented by 59 corresponding probesets on Affymetrix HGU133A GeneChips. 14 probesets (probing three genes) did not pass the quality control threshold for expression values when applied to the tubular compartment samples and were excluded from further analysis. Similarly, nine probesets (measuring two genes) did not pass quality control threshold when applied to the glomerular compartment. Although the severity of progressive renal disease is typically assessed in murine models by histopathological, but not functional, parameters, stages of human CKD are well-defined and assessed by measured or eGFR.33 The eGFR at time of kidney biopsy was calculated using a modified MDRD formula for all patients eligible for this study.34 eGFR ranged from 7.58 ml/minute/1.73m2 to 157.30 ml/minute/1.73m2 across the entire cohort, including tumor nephrectomy patients (n = 3) and patients with stage I (n = 8), stage II (n = 18), stage III (n = 11), stage IV (n = 8), and stage V (n = 2) as defined by K/DOQI CKD staging criteria.1,33,35 To test whether the continuous actual eGFR is statistically related to the expression values of the ortholog geneset in glomerular and/or tubular interstitial compartments, we applied regression analysis using a ridge regression model and leave-one-out cross validation as previously described.29,36 The glomerular expression dataset did not achieve statistically significant relationship results of the actual MDRD eGFR. In contrast, the tubular interstitial expression levels for 30 human orthologs assessed by 45 probesets provided a highly-significant relationship of continuous actual eGFR with a cross-validated R2 of 0.53 (r = 0.74, P < 0.001) (Figure 3A). Clinical cohort studies demonstrate that patients with CKD stages III, IV, and V (GFR < 60 ml/minute/1.73m2) have a high likelihood of progressive CKD.8 In contrast, progression of CKD in patients with CKD stages I and II (GFR ≥ 60 ml/minute/1.73m2) remains poorly defined. By assigning the clinically relevant eGFR threshold of 60 ml/minute/1.73m2 between stages II and III as a cutoff, tubulointerstitial expression signature of the 45 human probeset classified patients into stage I/II or stage III/IV/V groups with 83%, 80%, 86.2%, and 76.2% positive predictive value (PPV), negative predictive value (NPV), sensitivity, and specificity, respectively (Figure 3B).
Figure 3.
A: Statistical analysis of functional relationship between tubulointerstitial compartment gene expression and renal function in 50 CKD patients. Ridge regression analysis of tubulointerstitial compartment gene expression values (45 probeset corresponding to 30 human orthologs of the 43 mouse genes) and continuous actual eGFR (ml/minute/1.73m2) shows significant relationship with a cross-validated R2 = 0.53, P < 0.001. B: Performance of ridge regression model as classifier of two groups of CKD patients with measured eGFR higher or lower than 60 ml/minute/1.73m2, respectively.
Comparative Characterization of Marker Protein Expression Patterns in Kidneys of Tgfb1 Tg Mice and Patients with IgA Nephropathy
Individual tubular expression profiles for 16 genes were significantly correlated with continuous actual eGFR by univariate statistical analysis (false discovery rate < 0.01) (see Supplementary Table S5 at http://ajp.amipathol.org). Because RNA profiling of diagnostic kidney biopsies is currently limited to research applications, we initiated studies to examine the protein expression profiles of gene products from these 16 genes in Tgfb1 Tg mice. Antibodies were obtained from commercial or academic sources where available. Their utility in immunohistochemistry of kidney sections from wild-type and Tgfb1 Tg mice with moderate (histopathology score 1) or advanced (score 3) renal disease at 4 weeks of age was evaluated for specificity and quality, providing satisfactory results for the following antibodies: neutrophil cytosolic factor 2 (Ncf2), biglycan (Bgn), integrin β 5 (Itgb5), collagen type VI, α 1 (Col6A1), S100 calcium-binding protein A6 (S100a6), solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 3 (Slc13a3). and dickkopf 3 (Dkk3). In addition, we used a new antibody for Mpv17-like protein (Mpv17l), recently generated by our group.26 Results are presented in Supplementary Figure S2 (see Supplementary Figure S2 at http://ajp.amipathol.org) and summarized in Table 2.
Table 2.
Protein Expression Signatures in Glomeruli (Glom.), Tubules (Tub.), and Interstitial Cells (Int.) as Assessed by Immunolabeling of Sections from Wild-Type (Wt) Mice and Tg Mice with Nonprogressive or Progressive Kidney Disease (n ≥ Five Animals per Group)
| Protein | WT
|
Nonprogressive
|
Progressive
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Glom. | Tub. | Int. | Glom. | Tub. | Int. | Glom. | Tub. | Int. | |
| Ncf2 | − | − | − | − | − | − | ++ | ++ | − |
| Mpv17l | − | ++ | − | − | + | − | − | − | − |
| S100a6 | − | − | − | − | − | + | + | − | ++ |
| Bgn | − | +/− | − | + | ++ | − | ++ | ++ | − |
| Col6a1 | − | + | − | − | ++ | − | − | ++ | − |
| Itgb5 | + | + | − | ++ | ++ | − | ++ | ++ | − |
| Slc13a3 | − | + | − | − | + | − | + | + | − |
| Dkk3 | − | − | − | + | − | − | ++ | − | − |
−, Absent staining; +, minimal/moderate staining; ++, advanced staining.
Staining for Ncf2, a 67-kDa cytosolic subunit of the multiprotein complex of NADPH oxidase in neutrophils, was modest and comparable in tubules of wild-type and score 1 Tgf1b Tg mice, but strongly increased in tubules of score 3 Tgfb1 Tg mice, where de novo expression was also observed in glomerular cells, representing a podocyte-like pattern. Biglycan is a member of the small leucine repeat proteoglycan family (SLRP) that is barely detectable in tubules of wild-type kidney, but strongly increased in tubules, parietal epithelial cells, and glomerular cells with a podocyte pattern in score 1, and especially in score 3 Tgfb1 Tg mice. Itgb5 is strongly expressed in kidneys from wild-type mice at the corticomedullary junction and moderately expressed in glomerular cells, whereas it is expressed with increasing intensity in cortical tubules and glomerular cells with a podocyte pattern of score 1 and score 3 Tgfb1 Tg mice. Col6a1 was expressed in tubules of wild-type mice and expression became increased in tubules of Tgfb1 Tg mice and intensity is correlated with disease severity. Mpv17l is a member of the Mpv17/PMP22 protein family that was strongly expressed in tubuli at the cortical and corticomedullary junction tubuli in kidneys of wild-type mice. Cortical tubular expression was greatly reduced in score 1 Tgf1 Tg mice, and both, cortical and corticomedullary tubular expression was lost in score 3 Tgfb1 Tg mice. S100a6, also called calcyclin, is a 10.5-kDa calcium-binding protein that is not detectable in kidneys of wild-type mice. S100a6 was detected in cortical interstitial cells in score 1 and score 3 Tgfb1 Tg mice, and in podocytes of score 3 Tgfb1 Tg animals. Slc13a3 is one of the Na+-dependent dicarboxylate transporters that were encoded by Slc13 gene family members. It is highly expressed in cortical and corticomedullary junction tubuli in kidneys of WT and Tgf1 Tg mice. High-level expression of Slc13a3 was also observed exclusively in glomerular cells with a podocyte pattern in score 3 Tgfb1 Tg mice. De novo expression of Dkk3 proteins was detected in podocytes of Tgfb1 Tg mice with nonprogressive disease (as indicted in Supplementary Figure S2 at http://ajp.amipathol.org) by double-immunofluorescence staining with podocyte-specific marker synaptopodin). The expression of Dkk3 was further significantly increased in kidneys with progressive kidney disease. Taken together, these observations delineate novel marker protein expression patterns that distinguish normal kidneys and kidneys with nonprogressive and progressive disease, including de novo podocyte expression of Bgn and Dkk3, de novo cortical interstitial S100a6 expression, gradual increase in tubular Bgn, Col6a1, and Itgb5. In contrast, de novo expression of S100a6, Ncf2, and Slc13a3 in glomerular cells with a podocyte pattern (Figure 4A) and global loss of tubular Mpv17l expression (Figure 4B) was a characteristic protein expression pattern that distinguished advanced kidney disease from nonprogressive kidney disease in Tgfb1 Tg mice.
Figure 4.
Immunohistochemical analysis of protein expression patterns in kidney sections of wild-type, Tg mice with nonprogressive and progressive kidney lesions at 4 weeks of age (n ≥ 5 for each group). A: Single glomerulus staining for Ncf2, S100a6, and Slc13a3. B: Cortex-medulla staining for Mpv17l and Ncf2 with insets of cortex-medulla images. Original magnifications: ×63 (A); {times]10 (B); ×40 (B, insets).
Because no additional tissue was available for confirmatory immunohistochemistry from the ERCB renal biopsy tissue used for expression profiling studies, we obtained sections of routine clinical biopsies from 20 patients with IgA nephropathy and eGFRs between 7 to 134 ml/minute/1.73 m2. Four of the eight antibodies evaluated in Tgfb1 Tg mice provided specific and consistent staining patterns when applied to human kidney biopsy sections (Figure 5). Protein expression patterns of S100A6, NCF2, SLC13A3, and BGN were strongly increased in kidney biopsies of patients with stage III/IV CKD (Figure 5, A and B; group 3), compared with patients with stage I/II CKD (Figure 5, A and B; groups 1 and 2), and similar to the expression patterns detected in Tgfb1 Tg mice (Figure 4A). Interestingly, in the biopsies from patients with stage I/II CKD (eGFR higher 60 ml/minute/1.73 m2), staining for all of these proteins was either present or absent, ie, these four proteins appeared closely co-expressed. It will be of interest to examine in the future a their expression in stage I/II CKD may turn out to be prognostic indicator for further disease progression.
Figure 5.
Immunohistochemical analysis of protein expression patterns in kidney biopsies of patients with various stages of CKD and IgA nephropathy. A: Representative glomerular staining for NCF2, S100A6, and SLC13A3. Groups 1 and 2, patients with CKD I/II, GFR value greater than 60 ml/minute/1.73 m2; group 3, patients with CKD III/IV, GFR value is less than 60 ml/minute/1.73 m2. B: Tubular staining for NCF2 and BGN. Groups 1, 2, and 3, as described in A. Original magnifications, ×40.
Discussion
During the past decade, the application of gene expression profiling in cancer research has resulted in development of new therapeutic targets, and of prognostic profiling assays that are now in phase III clinical trials designed to evaluate their contribution to therapeutic decision making.37 The impressive progress and successes of genomic profiling in oncology have been facilitated by an abundance of surgical tumor tissues and samples from hematological malignancies. CKD is emerging as a major public health problem, estimated to affect ∼1 in 10 Americans.2 However, only a fraction of CKD patients progress to end-stage kidney disease. Thus, the development of new prognostic and therapeutic approaches to assess and treat the risk of progression of CKD is a major unmet need in clinical nephrology. In contrast with the success of gene expression profiling in oncology, several challenges have severely limited the application of genomic profiling in nonmalignant kidney diseases. First, tissue availability is limited because diagnostic kidney biopsies or nonmalignant nephrectomies are performed relatively infrequently. Second, the composition of kidney tissue cores is inherently heterogeneous contributing to sampling error,38 which renders standardized, quantitative gene expression profiling across large series of kidney biopsies technically challenging.20
To begin to overcome these challenges, hampering development of urgently needed predictive markers of CKD progression, we report a two-step comparative genomics approach, combining unique resources focused on systematic molecular analysis of murine renal models (Bottinger group39) and human kidney biopsy (Kretzler group40), respectively. The initial development of predictive expression signatures for CKD progression was accomplished by applying microarray analysis of whole mouse kidney obtained from the Tgfb1 Tg mouse model, which recapitulates key pathomechanisms and heterogeneity of progression of CKD. This information was then applied to interrogate a human kidney gene expression database to develop and validate a model algorithm for prediction of estimated GFR in humans with various stages of CKD (I to V). A similar approach has recently been reported, in which mouse to human metagene profiles were identified and developed in peripheral blood mononuclear cells of mouse to predict radiation response in humans.41
Although the classification of progressive CKD in our study was based on distinct endpoints and methods, namely advanced histopathological scores in mouse versus estimated GFR on a continuum of stage I to V in human cohorts, the results of our comparative analysis were remarkably consistent (see Supplementary Table S1 at http://ajp.amipathol.org), supporting the validity and robustness of our approach. For example, the intersection between the 19 murine gene expression profiles that were significantly correlated with advanced histopathology scores in Tgfb1 Tg mice, and the 16 human gene expression profiles that were significantly correlated with eGFR in humans, were comprised of nine genes, including Axl, Bgn, Col6a1, Creb3, Dkk3, Itgb5, Ncf2, S100a6, and Slc13a3. Only 4 of 19 murine expression profiles, Ctsc, Dtx4, Fxyd5, and Adprtl3 were correlated with progressive disease in our longitudinal validation study in Tgfb1 Tg mice, but were not correlated with eGFR in human CKD, whereas Apln, Cpxm1, Mpv17l, Smoc2, Spc25, and Tnfsf13b were correlated in mice, but expression data for their human orthologs was not available in the human gene expression database (either the probesets were not present in Affymetrix HGU133A or the data did not pass quality control as described above). Because previous reports demonstrated that the deletion of the Mpv17 gene, a close homolog of Mpv17l, caused nephrotic syndrome and progressive glomerulosclerosis in mice,42 we used qrt-PCR to confirm the correlation of MPV17L expression with CKD progression independently (data not shown). Thus, 10 molecular markers were correlated with CKD progression based on histopathological and functional (eGFR) parameters in murine and human kidney disease, respectively. Among these, only S100a6 (calcyclin) has previously been reported as potential marker for acute ischemic tubular injury.43,44
Although the primary focus of this report is the development of novel markers for CKD progression, our findings also imply novel pathways that support an important role for imbalances in stress response and cell survival/cell death signaling in CKD progression. Several of the 10 genes have previously been linked with multiple pathways controlling apoptosis, inflammation, and organization of extracellular matrix, including inducers and modulators of receptor for advanced glycation end products (RAGE), TGF-β, and Akt signaling (S100a6, Bgn, Axl, Dkk3), endoplasmatic reticulum (ER) stress (Creb3), mitochondrial dysfunction (Col6a1, Mpv17l), innate immune response (Bgn), and fibrillar collagen network formation (Bgn). For example, Axl is a receptor tyrosine kinase and activator of Akt survival signaling that has previously been implicated in early diabetic nephropathy and experimental glomerulonephritis models, respectively,45,46 whereas S100a6 can modulate cell survival by interacting with distinct RAGE immunglobulin domains.47 S100a6 is also involved in the processing of apoptosis by modulating the transcriptional regulation of caspase-3.48 Dkk3 may regulate TGF-β signaling in Xenopus,49 and its overexpression in cancer cells induced apoptosis.50 Mpv17l is an inner mitochondrial membrane protein of proximal tubular cells that protects mitochondria against superoxide generation, apoptosis, and mitochondrial dysfunction.26 Supramolecular assemblies of collagen VI microfibrils provide scaffolds for the formation of the structurally critical fibrillar collagen networks through connection with the small proteoglycans decorin and biglycan.51 Although biglycan itself functions as an extracellular matrix organizer, it can modulate stress signaling directly by binding extracellular TGF-β, or toll-like receptor 4 (TLR4), which has recently been implicated as innate immune response mediator in hepatic fibrosis and ischemia reperfusion injury models.52,53 The leucine zipper transcription factor Creb3 regulates transcription of mediators of endoplasmic reticulum (ER) stress response.54 Only the Slc13a3 gene belongs to the family of renal sodium-dicarboxylate co-transporters involved in the handling of citrate by the kidney and has not been implicated in cell stress or apoptosis to date. Together, the functional roles in their respective pathways of the CKD progression gene and protein profiles reported here support the emerging concept of glomerular and tubular epithelial cell injury and apoptosis as initiating mechanisms in TGF-β-mediated nephron loss and renal fibrosis, considered central pathomechanisms in progression of CKD.11,55 Finally, these results are also consistent with the validity of our initial screening strategy of selecting gene expression profiles for development of markers of CKD progression based on their ability to discriminate the extent of epithelial apoptosis in kidneys of Tgfb1 Tg mice before the emergence of histopathological lesions.
In routine diagnostic kidney biopsy procedures, typically two tissue cores are obtained by needle biopsy and processed for histopathology, immunofluorescence, or electron microscopy methods. However, microdissection or fractionation of glomerular and tubular tissue compartments, prerequisite steps for standardized, quantitative molecular analysis, are currently only performed in research settings, but not in routine diagnostic biopsy procedures. Thus, we reasoned that molecular markers that can be applied by in situ immunodetection methods using routine kidney biopsy tissue blocks would likely be of broader clinical utility. To enhance the potential clinical utility of our study, we began to verify the feasibility of using routine immunodetection methods on kidney sections by interrogating the in situ protein expression profiles of the 10 putative markers of CKD progression that we identified in mouse and human kidney disease. From these, we developed a panel of eight informative molecular markers for CKD progression (see Table 2), whereas antibodies for Axl and Creb3, were either not available, or did not provide specific protein detection, respectively (data not shown). As characteristic protein expression signatures of advanced disease, de novo expression of Ncf2, S100a6, and Slc13a3 proteins was exclusively detected in glomerular cells with a podocyte pattern of Tgfb1 Tg mice with progressive disease (Figure 4A), in addition to de novo expression of Ncf2 and significant loss of Mpv17l protein expression in corticomedullary tubules (Figure 4B). In contrast, de novo expression of Bgn and Dkk3 proteins in glomerular cells with a podocyte pattern, and of Itgb5 in subcortical tubules, was already detected in Tgfb1 Tg mice with nonprogressive disease, and their expression was markedly increased in kidneys manifesting progressive disease compared with nonprogressive disease (see Supplementary Figure S2 at http://ajp.amipathol.org). Thus, we have developed protein expression signatures, on the basis of gene expression signatures, that distinguish progressive and nonprogressive kidney disease in Tgfb1 Tg mice.
In addition, our initial observations with immunohistochemistry on human kidney biopsies indicate that a subset of these protein expression signatures, including S100A6, SLC13A3, BGN, and NCF2 may be applied to archival and prospective human kidney biopsy collections obtained through routine diagnostic biopsy protocols. However, further clinical development and validation of the protein expression signatures for CKD progression identified here will require longitudinal studies of extended cohorts of human CKD patients with diagnostic kidney biopsies that are beyond the scope of our current study and will require collaborative study networks involving multiple centers.
Supplementary Material
Acknowledgments
We thank Dan Liang for technical support; Dr. Almut Nitsche and Dr. Bodo Brunner (Sanofi-Aventis Deutschland GmbH, Frankfurt, Germany) for DNA chip hybridizations and delivery of the human gene expression data set as part of the EU-FP V framework program on chronic renal disease; and the members of the European Renal cDNA Bank-Kroener-Fresenius biopsy bank at the time of the expression profiling: Clemens. D. Cohen, Michael Fischereder, Holger Schmid, Peter J. Nelson, Matthias Kretzler, Detlef Schlondorff, University of Munich, Munich, Germany; Jean-Daniel Sraer, Pierre Ronco, Paris, France; Maria P. Rastaldi, Gennaro D’Amico, San Carlo Borromeo Hospital, Milano, Italy; Francisco Mampaso, Hospital Universitario Ramon y Cajal, Madrid, Spain; Peter Doran, Hugh R. Brady, University College Dublin, Dublin, Ireland; Detlev Monks, Christoph Wanner, Medizinische Klinik, Würzburg, Germany; Andy J. Rees, Paul Brown, Institute of Medical Sciences, Aberdeen Scotland; Frank Strutz, Gerhard Mueller, Zentrum Innere Medizin, Goettingen Germany; Peter Mertens, Juergen Floege, Medizinische Klinik II, Aachen, Germany; Norbert Braun, Teut Risler, Universität Tübingen, Tuebingen; Loreto Gesualdo, Francesco P. Schena, Institute of Nephrology, Bari, Italy; Jens Gerth, Gunter Wolf, Medizinische Klinik IV, Nephrologie, Jena, Germany; Rainer Oberbauer, Dontscho Kerjaschki, AKH Universitätsklinik, Vienna, Austria; Brigitte Banas, Bernhard K. Kraemer, Medizinische Klinik der Universität Regensburg, Regensburg, Germany; Walter Samtleben, University of Munich, Munich, Germany; Harm Peters, Hans-Hellmut Neumayer, Berlin; Katrin Ivens, Bernd Grabensee, Klinik der Heinrich-Heine-Universität, Duesseldorf, Germany; Rudolf P. Wuethrich, University of Zurich, Zurich, Switzerland; and Vladimir Tesar, Charles University, Prague, Czech Republic.
Footnotes
Address reprint requests to Erwin P. Böttinger, Dept. of Medicine, Mount Sinai Medical Center, One Gustave L. Levy Pl., Box 1118, New York, NY 10029. E-mail: erwin.bottinger@mssm, or Matthias Kretzler, Internal Medicine-Nephrology, University of Michigan, Medical Science Research Building II 1150 West Medical Center DriveRoom 1560 Ann Arbor, MI 48109-5676. E-mail: kretzler@med.umich.edu.
Supported by the National Institutes of Health (grants 5R01DK056077-09 to E.P.B., 5R01DK060043-07 to E.P.B., 5R01DK073960-02 to E.P.B., 5U01DK060995-08 to E.P.B., R01 DK079912-01 to M.K. and R21 DK079441-01 M.K.), the Else-Kroener Fresenius Foundation (to C.D.C., D.S., and M.K.), the American Heart Association (research fellowship to W.J.), and the National Kidney Foundation (to M.B. and C.B).
Supplemental material for this article can be found on http://ajp.amipathol.org.
References
- K/DOQI clinical practice guidelines for chronic kidney disease: evaluation, classification, and stratification. Am J Kidney Dis. 2002;39:S1–S266. [PubMed] [Google Scholar]
- Prevalence of chronic kidney disease and associated risk factors–United States, 1999–2004. MMWR Morb Mortal Wkly Rep. 2007;56:161–165. [PubMed] [Google Scholar]
- Freedman BI, Bowden DW, Sale MM, Langefeld CD, Rich SS. Genetic susceptibility contributes to renal and cardiovascular complications of type 2 diabetes mellitus. Hypertension. 2006;48:8–13. doi: 10.1161/01.HYP.0000227047.26988.3e. [DOI] [PubMed] [Google Scholar]
- Garg JP, Bakris GL. Microalbuminuria: marker of vascular dysfunction, risk factor for cardiovascular disease. Vasc Med. 2002;7:35–43. doi: 10.1191/1358863x02vm412ra. [DOI] [PubMed] [Google Scholar]
- Schieppati A, Remuzzi G. Chronic renal diseases as a public health problem: epidemiology, social, and economic implications. Kidney Int Suppl. 2005;98:S7–S10. doi: 10.1111/j.1523-1755.2005.09801.x. [DOI] [PubMed] [Google Scholar]
- Satko SG, Sedor JR, Iyengar SK, Freedman BI. Familial clustering of chronic kidney disease. Semin Dial. 2007;20:229–236. doi: 10.1111/j.1525-139X.2007.00282.x. [DOI] [PubMed] [Google Scholar]
- Taal MW, Brenner BM. Renal risk scores: progress and prospects. Kidney Int. 2008;73:1216–1219. doi: 10.1038/ki.2008.36. [DOI] [PubMed] [Google Scholar]
- Taal MW, Brenner BM. Predicting initiation and progression of chronic kidney disease: developing renal risk scores. Kidney Int. 2006;70:1694–1705. doi: 10.1038/sj.ki.5001794. [DOI] [PubMed] [Google Scholar]
- Beer DG, Kardia SL, Huang CC, Giordano TJ, Levin AM, Misek DE, Lin L, Chen G, Gharib TG, Thomas DG, Lizyness ML, Kuick R, Hayasaka S, Taylor JM, Iannettoni MD, Orringer MB, Hanash S. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat Med. 2002;8:816–824. doi: 10.1038/nm733. [DOI] [PubMed] [Google Scholar]
- Ye QH, Qin LX, Forgues M, He P, Kim JW, Peng AC, Simon R, Li Y, Robles AI, Chen Y, Ma ZC, Wu ZQ, Ye SL, Liu YK, Tang ZY, Wang XW. Predicting hepatitis B virus-positive metastatic hepatocellular carcinomas using gene expression profiling and supervised machine learning. Nat Med. 2003;9:416–423. doi: 10.1038/nm843. [DOI] [PubMed] [Google Scholar]
- Bottinger EP. TGF-beta in renal injury and disease. Semin Nephrol. 2007;27:309–320. doi: 10.1016/j.semnephrol.2007.02.009. [DOI] [PubMed] [Google Scholar]
- Border WA, Noble NA. Transforming growth factor beta in tissue fibrosis. N Engl J Med. 1994;331:1286–1292. doi: 10.1056/NEJM199411103311907. [DOI] [PubMed] [Google Scholar]
- Bitzer M, Sterzel RB, Bottinger EP. Transforming growth factor-beta in renal disease. Kidney Blood Press Res. 1998;21:1–12. doi: 10.1159/000025837. [DOI] [PubMed] [Google Scholar]
- Liu Y. Renal fibrosis: new insights into the pathogenesis and therapeutics. Kidney Int. 2006;69:213–217. doi: 10.1038/sj.ki.5000054. [DOI] [PubMed] [Google Scholar]
- Patel A, Scott WR, Lympany PA, Rippin JD, Gill GV, Barnett AH, Bain SC. The TGF-beta 1 gene codon 10 polymorphism contributes to the genetic predisposition to nephropathy in type 1 diabetes. Diabet Med. 2005;22:69–73. doi: 10.1111/j.1464-5491.2005.01376.x. [DOI] [PubMed] [Google Scholar]
- August P, Suthanthiran M. Transforming growth factor beta and progression of renal disease. Kidney Int Suppl. 2003;87:S99–S104. doi: 10.1046/j.1523-1755.64.s87.15.x. [DOI] [PubMed] [Google Scholar]
- Sanderson N, Factor V, Nagy P, Kopp J, Kondaiah P, Wakefield L, Roberts AB, Sporn MB, Thorgeirsson SS. Hepatic expression of mature transforming growth factor beta 1 in transgenic mice results in multiple tissue lesions. Proc Natl Acad Sci USA. 1995;92:2572–2576. doi: 10.1073/pnas.92.7.2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp JB, Factor VM, Mozes M, Nagy P, Sanderson N, Bottinger EP, Klotman PE, Thorgeirsson SS. Transgenic mice with increased plasma levels of TGF-beta 1 develop progressive renal disease. Lab Invest. 1996;74:991–1003. [PubMed] [Google Scholar]
- Schmid H, Cohen CD, Henger A, Schlondorff D, Kretzler M. Gene expression analysis in renal biopsies. Nephrol Dial Transplant. 2004;19:1347–1351. doi: 10.1093/ndt/gfh181. [DOI] [PubMed] [Google Scholar]
- Yasuda Y, Cohen CD, Henger A, Kretzler M. Gene expression profiling analysis in nephrology: towards molecular definition of renal disease. Clin Exp Nephrol. 2006;10:91–98. doi: 10.1007/s10157-006-0421-z. [DOI] [PubMed] [Google Scholar]
- Schiffer M, Mundel P, Shaw AS, Bottinger EP. A novel role for the adaptor molecule CD2-associated protein in transforming growth factor-beta-induced apoptosis. J Biol Chem. 2004;279:37004–37012. doi: 10.1074/jbc.M403534200. [DOI] [PubMed] [Google Scholar]
- Bitzer M, von Gersdorff G, Liang D, Dominguez-Rosales A, Beg AA, Rojkind M, Bottinger EP. A mechanism of suppression of TGF-beta/SMAD signaling by NF-kappa B/RelA. Genes Dev. 2000;14:187–197. [PMC free article] [PubMed] [Google Scholar]
- Schiffer M, Bitzer M, Roberts IS, Kopp JB, ten Dijke P, Mundel P, Bottinger EP. Apoptosis in podocytes induced by TGF-beta and Smad7. J Clin Invest. 2001;108:807–816. doi: 10.1172/JCI12367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju W, Ogawa A, Heyer J, Nierhof D, Yu L, Kucherlapati R, Shafritz DA, Bottinger EP. Deletion of Smad2 in mouse liver reveals novel functions in hepatocyte growth and differentiation. Mol Cell Biol. 2006;26:654–667. doi: 10.1128/MCB.26.2.654-667.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YC, Piek E, Zavadil J, Liang D, Xie D, Heyer J, Pavlidis P, Kucherlapati R, Roberts AB, Bottinger EP. Hierarchical model of gene regulation by transforming growth factor beta. Proc Natl Acad Sci USA. 2003;100:10269–10274. doi: 10.1073/pnas.1834070100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krick S, Shi S, Ju W, Faul C, Tsai SY, Mundel P, Bottinger EP. Mpv17l protects against mitochondrial oxidative stress and apoptosis by activation of Omi/HtrA2 protease. Proc Natl Acad Sci USA. 2008;105:14106–14111. doi: 10.1073/pnas.0801146105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen CD, Kretzler M. Gene expression analysis in microdissected renal tissue. Current challenges and strategies. Nephron. 2002;92:522–528. doi: 10.1159/000064099. [DOI] [PubMed] [Google Scholar]
- Schmid H, Boucherot A, Yasuda Y, Henger A, Brunner B, Eichinger F, Nitsche A, Kiss E, Bleich M, Grone HJ, Nelson PJ, Schlondorff D, Cohen CD, Kretzler M. Modular activation of nuclear factor-kappaB transcriptional programs in human diabetic nephropathy. Diabetes. 2006;55:2993–3003. doi: 10.2337/db06-0477. [DOI] [PubMed] [Google Scholar]
- Hoerl AE, Kennard RW. Ridge regression: biased estimation for nonorthogonal problems. Technometrics. 2007;12:55–67. [Google Scholar]
- Brabender J, Marjoram P, Salonga D, Metzger R, Schneider PM, Park JM, Schneider S, Holscher AH, Yin J, Meltzer SJ, Danenberg KD, Danenberg PV, Lord RV. A multigene expression panel for the molecular diagnosis of Barrett’s esophagus and Barrett’s adenocarcinoma of the esophagus. Oncogene. 2004;23:4780–4788. doi: 10.1038/sj.onc.1207663. [DOI] [PubMed] [Google Scholar]
- Kerr MK, Churchill GA. Bootstrapping cluster analysis: assessing the reliability of conclusions from microarray experiments. Proc Natl Acad Sci USA. 2001;98:8961–8965. doi: 10.1073/pnas.161273698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, Sturn A, Snuffin M, Rezantsev A, Popov D, Ryltsov A, Kostukovich E, Borisovsky I, Liu Z, Vinsavich A, Trush V, Quackenbush J. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- Snyder S, Pendergraph B. Detection and evaluation of chronic kidney disease. Am Fam Physician. 2005;72:1723–1732. [PubMed] [Google Scholar]
- Levey AS, Bosch JP, Lewis JB, Greene T, Rogers N, Roth D. A more accurate method to estimate glomerular filtration rate from serum creatinine: a new prediction equation. Modification of Diet in Renal Disease Study Group. Ann Intern Med. 1999;130:461–470. doi: 10.7326/0003-4819-130-6-199903160-00002. [DOI] [PubMed] [Google Scholar]
- Bauer C, Melamed ML, Hostetter TH. Staging of chronic kidney disease: time for a course correction. J Am Soc Nephrol. 2008;19:844–846. doi: 10.1681/ASN.2008010110. [DOI] [PubMed] [Google Scholar]
- Bovelstad HM, Nygard S, Storvold HL, Aldrin M, Borgan O, Frigessi A, Lingjaerde OC. Predicting survival from microarray data—a comparative study. Bioinformatics. 2007;23:2080–2087. doi: 10.1093/bioinformatics/btm305. [DOI] [PubMed] [Google Scholar]
- Morris SR, Carey LA. Gene expression profiling in breast cancer. Curr Opin Oncol. 2007;19:547–551. doi: 10.1097/CCO.0b013e3282f0ada3. [DOI] [PubMed] [Google Scholar]
- Corwin HL, Schwartz MM, Lewis EJ. The importance of sample size in the interpretation of the renal biopsy. Am J Nephrol. 1988;8:85–89. doi: 10.1159/000167563. [DOI] [PubMed] [Google Scholar]
- Bottinger EP, Ju W, Zavadil J. Applications for microarrays in renal biology and medicine. Exp Nephrol. 2002;10:93–101. doi: 10.1159/000049904. [DOI] [PubMed] [Google Scholar]
- Cohen CD, Frach K, Schlondorff D, Kretzler M. Quantitative gene expression analysis in renal biopsies: a novel protocol for a high-throughput multicenter application. Kidney Int. 2002;61:133–140. doi: 10.1046/j.1523-1755.2002.00113.x. [DOI] [PubMed] [Google Scholar]
- Dressman HK, Muramoto GG, Chao NJ, Meadows S, Marshall D, Ginsburg GS, Nevins JR, Chute JP. Gene expression signatures that predict radiation exposure in mice and humans. PLoS Med. 2007;4:e106. doi: 10.1371/journal.pmed.0040106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiher H, Noda T, Gray DA, Sharpe AH, Jaenisch R. Transgenic mouse model of kidney disease: insertional inactivation of ubiquitously expressed gene leads to nephrotic syndrome. Cell. 1990;62:425–434. doi: 10.1016/0092-8674(90)90008-3. [DOI] [PubMed] [Google Scholar]
- Lewington AJ, Padanilam BJ, Hammerman MR. Induction of calcyclin after ischemic injury to rat kidney. Am J Physiol. 1997;273:F380–F385. doi: 10.1152/ajprenal.1997.273.3.F380. [DOI] [PubMed] [Google Scholar]
- Cheng CW, Rifai A, Ka SM, Shui HA, Lin YF, Lee WH, Chen A. Calcium-binding proteins annexin A2 and S100A6 are sensors of tubular injury and recovery in acute renal failure. Kidney Int. 2005;68:2694–2703. doi: 10.1111/j.1523-1755.2005.00740.x. [DOI] [PubMed] [Google Scholar]
- Yanagita M, Ishimoto Y, Arai H, Nagai K, Ito T, Nakano T, Salant DJ, Fukatsu A, Doi T, Kita T. Essential role of Gas6 for glomerular injury in nephrotoxic nephritis. J Clin Invest. 2002;110:239–246. doi: 10.1172/JCI14861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai K, Matsubara T, Mima A, Sumi E, Kanamori H, Iehara N, Fukatsu A, Yanagita M, Nakano T, Ishimoto Y, Kita T, Doi T, Arai H. Gas6 induces Akt/mTOR-mediated mesangial hypertrophy in diabetic nephropathy. Kidney Int. 2005;68:552–561. doi: 10.1111/j.1523-1755.2005.00433.x. [DOI] [PubMed] [Google Scholar]
- Leclerc E, Fritz G, Weibel M, Heizmann CW, Galichet A. S100B and S100A6 differentially modulate cell survival by interacting with distinct RAGE (receptor for advanced glycation end products) immunoglobulin domains. J Biol Chem. 2007;282:31317–31331. doi: 10.1074/jbc.M703951200. [DOI] [PubMed] [Google Scholar]
- Joo JH, Yoon SY, Kim JH, Paik SG, Min SR, Lim JS, Choe IS, Choi I, Kim JW. S100A6 (calcyclin) enhances the sensitivity to apoptosis via the upregulation of caspase-3 activity in Hep3B cells. J Cell Biochem. 2008;103:1183–1197. doi: 10.1002/jcb.21496. [DOI] [PubMed] [Google Scholar]
- Pinho S, Niehrs C. Dkk3 is required for TGF-beta signaling during Xenopus mesoderm induction. Differentiation. 2007;75:957–967. doi: 10.1111/j.1432-0436.2007.00185.x. [DOI] [PubMed] [Google Scholar]
- Yue W, Sun Q, Dacic S, Landreneau RJ, Siegfried JM, Yu J, Zhang L. Downregulation of Dkk3 activates beta-catenin/TCF-4 signaling in lung cancer. Carcinogenesis. 2008;29:84–92. doi: 10.1093/carcin/bgm267. [DOI] [PubMed] [Google Scholar]
- Lampe AK, Bushby KM. Collagen VI related muscle disorders. J Med Genet. 2005;42:673–685. doi: 10.1136/jmg.2002.002311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki E, De Minicis S, Osterreicher CH, Kluwe J, Osawa Y, Brenner DA, Schwabe RF. TLR4 enhances TGF-beta signaling and hepatic fibrosis. Nat Med. 2007;13:1324–1332. doi: 10.1038/nm1663. [DOI] [PubMed] [Google Scholar]
- Wu H, Chen G, Wyburn KR, Yin J, Bertolino P, Eris JM, Alexander SI, Sharland AF, Chadban SJ. TLR4 activation mediates kidney ischemia/reperfusion injury. J Clin Invest. 2007;117:2847–2859. doi: 10.1172/JCI31008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang G, Audas TE, Li Y, Cockram GP, Dean JD, Martyn AC, Kokame K, Lu R. Luman/CREB3 induces transcription of the endoplasmic reticulum (ER) stress response protein Herp through an ER stress response element. Mol Cell Biol. 2006;26:7999–8010. doi: 10.1128/MCB.01046-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottinger EP. TGF-beta and fibrosis. Derynck R, Miyazono K, editors. Cold Spring Harbor: Cold Spring Harbor Laboratory Press,; The TGF-Beta Family. 2008:pp 989–1022. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.