Abstract
X box-binding protein 1 (Xbp-1) is a transcription factor that is required for the terminal differentiation of B lymphocytes into plasma cells. The Xbp-1 gene is activated in response to endoplasmic reticulum stress signals, which generate a 50-kDa nuclear protein that acts as a potent transactivator and regulates the expression of genes related to the unfolded protein response. Activated Xbp-1 is essential for cell survival in plasma-cell tumors but its role in B-cell lymphomas is unknown. We analyzed the expression of activated Xbp-1 in reactive lymphoid tissues, 411 lymphomas and plasma-cell neoplasms, and 24 B-cell lines. In reactive tissues, Xbp-1 was only found in nuclear extracts. Nuclear expression of Xbp-1 was observed in occasional reactive plasma cells and in a subpopulation of Irf-4+/Bcl-6−/Pax-5− B cells in the light zones of reactive germinal centers, probably representing cells committed to plasma-cell differentiation. None of the low-grade lymphomas showed evidence of Xbp-1 activation; however, Xbp-1 activation was found in 28% of diffuse large B-cell lymphomas, independent of germinal or postgerminal center phenotype, as well as in 48% of plasmablastic lymphomas and 69% of plasma-cell neoplasms. Diffuse large B-cell lymphomas with nuclear Xbp-1 expression had a significantly worse response to therapy and shorter overall survival compared with negative tumors. These findings suggest that Xbp-1 activation may play a role in the pathogenesis of aggressive B-cell lymphomas.
The endoplasmic reticulum is a dynamic compartment of the cell where folding and quality control of nascent proteins occurs.1 The homeostasis of the endoplasmic reticulum is regulated by a complex adaptive pathway, known as the unfolded protein response, that coordinates the translation and transcription rates to the increased demands of protein folding in the endoplasmic reticulum.2,3 One of the main modulators of this response is the X-box binding protein-1 (Xbp-1, also TREB5),4,5,6 a potent transcription factor regulating the expression of genes that increase the capacity of the protein folding system or the degradation of misfolded proteins. The endoplasmic reticulum stress generated by the accumulation of unfolded proteins activates Xbp-1 by a specific enzymatic mechanism named frame switch splicing that modifies the coding sequence of the Xbp-1 mRNA transcript. The Xbp-1 full-length mRNA transcript encodes for an inactive p33 Xbp-1 protein. Under endoplasmic reticulum stress a small nucleotide fragment of the Xbp-1 transcript is spliced modifying its open reading frame to encode for a larger p50 Xbp-1 protein containing a potent transcription activator domain.7,8
Xbp-1 is expressed in several mammalian tissues including exocrine glands, bone remodeling cells, and hepatocytes.9,10 The expression and distribution of this transcription factor in hematopoietic cells is not well known, but experimental studies have shown that Xbp-1 activation is required for B-cells to switch the differentiation program to immunoglobulin secreting plasma-cells. In vitro and animal model experiments have shown that Xbp-1 deficient lymphocytes are able to form normal germinal centers, but have a severe defect in the generation of plasma-cells and immunoglobulin production emphasizing the essential role of this transcription factor in terminal B-cell differentiation.11,12,13,14 Although the role of Xbp-1 in the generation of plasma-cells has been well established in these experimental models, its expression and activation in human lymphoid tissues is not well known.
In addition to the physiological participation of Xbp-1 in normal plasma cell differentiation, this transcription factor is highly expressed in multiple myeloma (MM) cells, and evidence suggests that it may play a role in the pathogenesis of this tumor.15 For example, forced expression of the Xbp-1 transcript coding for the active form of the protein drives the development of MM in mice. On the other hand, disruption of Xbp-1 activation mediated by proteasome inhibitors promotes apoptosis of MM cells suggesting that Xbp-1 may play a role in sustaining the viability and perpetuation of these tumor cells.7
Multiple myeloma represents the most terminally differentiated B-cell neoplasm. However, different grades of plasma cell differentiation may occur in virtually all types of B-cell neoplasms from small B-cell to aggressive large B-cell lymphomas. In low-grade lymphomas, the presence of plasma cell differentiation does not affect the tumor behavior. However, large cell lymphomas with plasmablastic differentiation are clinically very aggressive with poor response to therapy and short survival of the patients. The distribution and activation of Xbp-1 in human lymphomas, its role in their pathogenesis and the potential clinical impact have not been previously investigated.
The aims of this study were to examine the expression and activation of Xbp-1 in human reactive lymphoid tissues, particularly in relation to the B-cell differentiation process, and determine the potential role of this transcription factor in the pathogenesis of human B-cell neoplasms and its clinical significance, particularly in aggressive lymphomas.
Materials and Methods
Samples and Patients
Human reactive lymphoid tissues, including tonsil, lymph node, spleen, and Peyer’s patches were studied. Tumor specimens from 411 patients were obtained from the files of the Laboratory of Pathology of the Hospital Clinic, Barcelona, Spain; Hospital Germans Trias i Pujol, Badalona, Spain; and Memorial Sloan-Kettering Cancer Center, Sloan-Kettering Institute, New York, with appropriate consent. All cases were reviewed by three pathologists (O.B., A.M., and E.C.) and re-classified according to the criteria of the World Health Organization Classification. Moreover, diffuse large B-cell lymphomas (DLBCL) were further classified as germinal center and non-germinal center subtypes according to the algorithm proposed by Hans et al.16
The clinical histories of the 103 patients with DLBCL and 13 patients with plasmablastic lymphoma diagnosed between 1986 and 2002 were available for review. These patients had a median age of 60 years, 56% were male and 44% female. 53% were in advanced stage, 63% had extranodal involvement, including bone marrow in 12%, and 58% high serum low-density lipoprotein. The distribution according to the International Prognostic Index (IPI) was as follows: low-risk, 36%; low/intermediate, 16%; high/intermediate, 19%; high-risk, 29%. Staging and re-staging maneuvers were the standard. Treatment consisted of adriamycin-containing regimens (in most cases cyclosphosphamide, hydroxydaunorubicin (adriamycin), oncovin (vincristine), prednisone/prednisolone (CHOP)) in 83% of the cases whereas the remainder 17% of cases received polychemotherapy without Adriamycin. Among 89 patients with assessable response, 56 (63%) achieved a complete response. After a median follow-up of 6.9 years for surviving patients, 61 had died, with a 5-year overall survival (OS) of 47% (95% confidence interval: 37% to 57%).
Cell Lines, Culture Conditions, and Treatments
Twenty-three human B-cell lines representing different types of lymphoid neoplasms were included in the study. These cell lines were derived from hairy cell leukemia (Eskol), mantle cell lymphoma (MCL) (REC-1, Granta-519, Jeko-1, and NCEB), DLBCL (Ly-8, SUDHL-4, 6, 7, and 10), Burkitt lymphoma (Thomas, Ramos 2G6.4C10, BJAB, and JD38) multiple myeloma (RPMI 8226, KMM-1, KMS-11,12-BM, 1,2-PE, and OPM-2), and primary effusion lymphoma (BC1, BC-2, BC-3). All of the cell lines (0.3 to 0.5 × 106 cells/ml) were grown in RPMI 1640 supplemented with 10% fetal bovine serum, 2 mmol/L glutamine, and 50 μg/ml penicillin-streptomycin (GIBCO BRL, Gaithersburg, Maryland) at 37°C in a humidified atmosphere containing 5% carbon dioxide. Dulbecco’s Modified Eagles’ Medium (DMEM) culture media (GIBCO BRL) was used instead of RPMI 1640 for Granta-519 cell line. All cultures were routinely tested for mycoplasma contamination by PCR.
Immunohistochemistry
Immunohistochemical studies were performed on formalin-fixed, paraffin-embedded tissue sections. A rabbit polyclonal antibody raised against the amino acids 73 to 263 of mouse Xbp-1 (Xbp-1-M186, Santa Cruz Biotechnology, Santa Cruz, CA) was used to recognize both spliced and unspliced forms of human Xbp-1 as previously described.17 Briefly, paraffin sections on silane-coated slides were developed in a fully automated immunostainer (Bond Max, Vision Biosystems, Mount Waverley, Australia). Low pH retrieval in Bond ER1 Buffer solution (Vision Biosystems) was done for 20 minutes followed by 2 hours incubation with primary antibody (1:100) at room temperature and 30 minutes of Bond Refine Polymer (Vision Biosystems). 5′, 3′-diaminobenzidine was used for 10 minutes as a chromogen. A minimum of 10 high power field (HPF) were analyzed. Most tumors were clearly negative or positive for nuclear staining with a homogeneous diffuse staining pattern. When needed, a cutoff of 30% of nuclear positivity was used.
Double immunostainings were performed in a automated immunostainer (BondMax) in two sequential detections with a peroxidase linked secondary antibody and 5′, 3′-diaminobenzidine (brown color) for the first detection and Barjoran Purple Chromogen kit (purple color, Biocare Medical, Concord, CA) or 3-amino-9-ethylcarbazole (AEC) (red color, Dako) for the second.
Immunofluorescence and Confocal Microscopy
Double immunofluorescence staining for Xbp-1/Pax-5, Xbp-1/Bcl-6, and Xbp-1/Irf-4 were performed on formalin-fixed, paraffin-embedded tissue tonsil sections approximately 3 to 4 microns in thickness. Table 1 summarizes antibodies used, source, and dilution. Streptavidin-conjugated quantum dots (585 and 655 nm) (Quantum Dot Corp., Hayward, CA) (2 mmol/L stock solution) were used at a final concentration of 6 nmol/L in PBS with 2% albumin. Generally, after deparaffinization, slides were placed in a microwavable pressure cooker (Nordic Ware, Minneapolis, MN) containing 1.5 l of 1× ‘Target Retrieval Solution’ high or low pH (Dako, Carpinteria, CA), and heated in a 1100W microwave for 4 (high pH) or 8 minutes (low pH)(hot start). After retrieval, slides were incubated in Tris-buffered saline (pH 7.6) containing 3% goat serum for 15 minutes. Primary antibodies were incubated for 1 hour (PAX-5, Bcl-6, Irf-4) and overnight (Xbp-1) at room temperature, then rinsed in Tris-buffered saline (pH 7.6) for 15 minutes (three times for 5 minutes each). A cocktail of biotinylated secondary anti-mouse/anti-rabbit/anti-goat antibody (Dako) was applied and incubated for 30 minutes at room temperature. Slides were washed in Tris-buffered saline (pH 7.6) for 15 minutes (3 × 5 minutes) and then incubated with individual streptavidin-conjugated quantum dots for 30 minutes at room temperature. After rinsing in Tris-buffered saline (pH 7.6) for 15 minutes (3 × 5 minutes), the slides were air dried and mounted in aqueous mounting media with 406-4,6-diamidino-2-phenylindole (Vector Laboratories, Inc, Burlingame, CA). For the sequential double staining, an extra avidin–biotin block was included. Avidin block was applied for 10 minutes, followed by a rinse in Tris buffered saline (pH 7.6) and 10 minutes incubation with biotin block (Dako). This extra blocking step was performed between the end of the first full detection and the beginning of the second primary.
Table 1.
Antibodies and Conditions of Use for Immunohistochemistry and Immunofluorescence
| Antibody | Clone | Source* | Dilution |
|---|---|---|---|
| Xbp-1 | Polyclonal | Santa Cruz | 1:50 |
| Pax-5 | 24 | BD Biosciences, Pharmingen | 1:50 |
| Bcl-6 | PG-B6p | DAKO | 1:2 |
| Irf-4 | MUM-1p | DAKO | 1:200 |
| CD20 | L26 | DAKO | 1:100 |
| CD3 | F7.2.38 | DAKO | 1:50 |
| Ki67 | Mib1 | DAKO | 1:25 |
Santa Cruz, Santa Cruz, CA; BD Biosciences, Pharmingen, San Jose, CA; DAKO, Glostrup, Denmark.
Images were acquired using a Leica TCS SP2 (Leica Lasertechnik GmbH, Mannheim, Germany) confocal system adapted to an inverted Leitz DM IRBE microscope and a ×40 (NA 1.25, oil) Leitz Plan Apochromatic objective. Excitation of streptavidin-conjugated quantum dots was performed using the 351 nm and 364 nm lines from an Ar-UV laser. Fluorescence of streptavidin-conjugated quantum dots was sequentially collected using two photo multiplier tubes as follows: 565 to 606 nm for streptavidin-conjugated quantum dot 585 and 615 to 670 nm for streptavidin-conjugated quantum dot 655. All confocal images were acquired with a frame size of 1024 × 1024 pixels and were averaged eight times. Average projection images were obtained from several optical sections9,10,11 at 400 Hz (z-step = 0.5 μm).
Western Blot
Exponentially growing cells (about 106 cells/ml) and frozen tumor samples were lysed in a non-denaturing detergent in 25 mmol/L bicine, 150 mmol/L NaCl at pH 7,6 buffer (M-PER and T-PER, Pierce, Rockford, IL) in ice-cold PBS buffer containing protease inhibitors (Complete Mini, Roche, Manheim, Germany) and phosphatase inhibitors (Cocktails 1 and 2, Sigma, Saint Louis, MO). Nuclear and cytoplasmic extracts were performed by using the NE-PER kit (Pierce) following manufacturer’s instructions. Polyadenosine diphosphate ribose polymerase (Roche, Mannheim, Germany) was used to asses the purity of the cytoplasmic extracts. The protein content was determined using a BCA Protein Assay kit (Pierce), according to the manufacturer’s instructions. Identical amounts (7 μg of total extracted protein) were heated 10 minutes at 70°C in NuPAGE LDS Sample buffer and separated by electrophoresis on 4 to 12% (w/v) polyacrylamide gradient gels (Novex NuPAGE, Invitrogen, Carlsbad, CA) with a 2-(N-morpholino) ethanesulfonic acid. After transfer to a 0.45-μm pore size nitrocellulose membrane (Bio-Rad Laboratories, Hercules, CA) the immunoblotting was performed as follows. The membrane was blocked two hours at room temperature in TTBS (50 mmol/L Tris-buffered saline, pH 7.6, with 0.05% Tween-20) containing 5% nonfat dry milk. The nitrocellulose membranes were incubated with a rabbit anti-Xbp-1(SC-7160, Santa Cruz Biotechnologies) and mouse anti-tubulin (Sigma, Saint Louis, MO) as a loading control. Binding was detected using a secondary antibody conjugated to horseradish peroxidase (Amersham, Buckinghamshire, UK) and an enhanced chemiluminiscence Supersignal WestPico detection kit (Pierce). Visualization and image analysis was done in a LAS-3000 cooled CCD camera system (Fuji Photo Film, Minato-Ku, Tokyo, Japan) and Multi GAUGE V2.0 software (Fuji Photo Film).
RNA Analysis
Total RNA was extracted from frozen samples of tumors and reactive tissues by using the RNeasy Mini Kit (Qiagen, GmbH, Germany). For reverse transcription 1 μg of total RNA of each sample was used with the Quantitect reverse transcription kit following the manufacturer recommendations. PCR amplification was performed using 200 nmol/L dNTPs, 2 mmol/L MgCl2, 1.75U of Taq polymerase Expand high fidelity (Roche) and 1 nmol/L each of a pair of primers labeled with TAMRA (Operon Biotechnologies, Cologne, Germany) and corresponding to nucleotides 412 to 431 (5′-CCTTGTAGTTGAGAACCAGG-3′) and 834 to 853 (5′-GGGGCTTGGTATATATGTGG-3′) of Xbp-1 cDNA.18 Amplification consisted of 35 cycles of 1 minute 94°C, 1 minute 60°C, and 1 minute 72°C. Reverse transcription-PCR products were analyzed using 310 Gene Scan 3.1 (Applied Biosystems, Foster City, CA). The ratio of the two mRNA species was analyzed measuring the area under each of the peaks corresponding to the 411bp and 437bp products (Genotyper 3.7NT, Applied Biosystems).
Statistical Analysis
The main initial and evolution variables of patients with DLBCL were recorded and analyzed for the prognostic significance. Overall survival was defined as the period of time from diagnosis to the death or last follow-up. Categorical data were compared using Chi-Square or Fisher’s exact test, two-sided P value, whereas for ordinal data non-parametric tests were used. The actuarial survival analysis was performed according to the method described by Kaplan and Meier and the curves compared by the log-rank test.
Results
Activation of Xbp-1 in Reactive Lymphoid Tissues
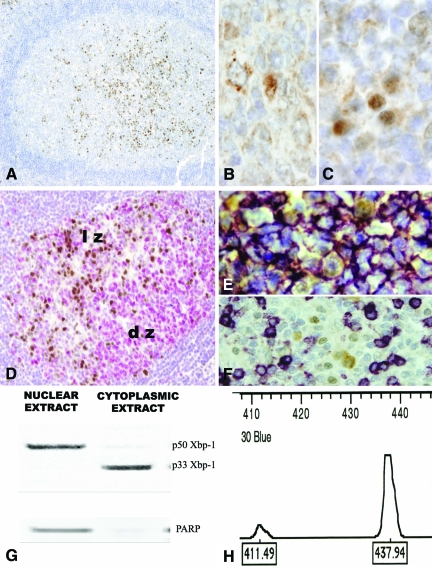
We first characterized the distribution of the Xbp-1 expression in reactive lymphoid tissues by immunohistochemistry. Nuclear Xbp-1 expression was found in lymphoid cells of the light zones of the secondary follicles (Figure 1, A and D), a non-proliferative compartment of the germinal center, where cells committed with a postgerminal center differentiation program tend to accumulate. Moreover, Xbp-1 was ubiquitously expressed in the cytoplasm of all germinal center cells with a granular and perinuclear pattern (Figure 1B). Interestingly, the majority of plasma cells lacked nuclear Xbp-1 expression although it was seen occasionally in some of these cells in intrafollicular, interfollicular, and subepithelial areas of the tonsils (Figure 1C). Double immunostaining demonstrated that all cells with nuclear Xbp-1 expression were positive for CD20 (Figure 1E) and negative for CD3 (Figure 1F).
Figure 1.
Xbp-1 Expression in reactive lymphoid tissue. Human tonsil. A: In the light zones of secondary lymphoid follicles clusters of nuclear Xbp-1 positive cells can be easily identified (×10). B: The cytoplasmic Xbp-1 is ubiquitously expressed in the cytoplasm of B-cells with a granular and paranuclear pattern (×60 under oil). C: Some cells with plasmacytic features exhibit a strong nuclear positivity of Xbp-1 (×60 under oil). D: Ki-67 positive cells (red) are found in the dark zone of the germinal centers. Ki-67 negative cells in the light zone are positive for nuclear Xbp-1(brown) (×20). E: Double staining for CD20 (purple) and Xbp-1(brown) showed that cells with nuclear Xbp-1 expression are B-cells. F: T cells stained with CD3 (purple) are negative for nuclear Xbp-1(brown). G: Western-blot on compartmental protein extracts showed the unspliced p33 in the cytoplasmic extracts and the p50 protein in the nuclear extracts. A nuclear protein, PAARP was used to assed the quality of the extracts. H: Reverse transcription-PCR flanking the splicing region reveals a two peaks pattern in the Gene Scan. A main 437bp unspliced expression is in agreement with the immunohistochemical results. lz: light zone of the germinal center; dz: dark zone of the germinal center.
To confirm that the nuclear expression observed immunohistochemically corresponded to the activated form of Xbp-1, we performed a Western blot analysis of nuclear and cytoplasmic extracts of the same reactive lymphoid tissues. The 33-kDa band of the inactive Xbp1 protein was only seen in cytoplasmic extracts, whereas the 50-kDa band corresponding to the active form was detected in the nuclear extracts (Figure 1G), in agreement with the distribution observed by immunohistochemistry.
RNA extracted from the same tissues was examined by reverse transcription-PCR. Two peaks corresponding to the 437-bp product representing the unspliced mRNA transcript coding for the inactive form of Xbp-1 and the 411-bp product representing the spliced mRNA transcript coding for the active form of Xbp-1 were identified (Figure 1H).
Activated Xbp-1 and Other Transcription Factors Involved in B-Cell Differentiation
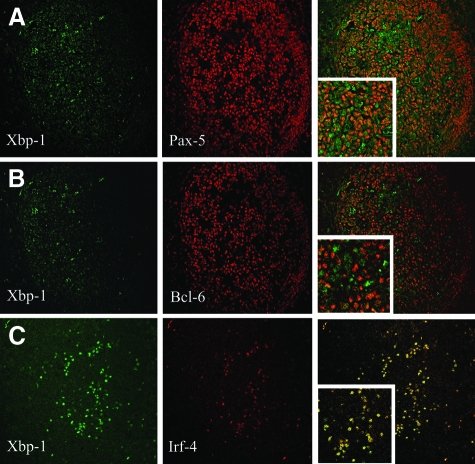
We have used immunofluorescence and confocal microscopy to assess the relationship between the nuclear expression of Xbp-1 and other transcription factors important for B-cell differentiation. As expected, Pax-5 was expressed in virtually all B cells (Figure 2A). However, a small fraction of centrocytes in the light zone of the germinal centers were negative for Pax-5 but expressed nuclear Xbp-1. This pattern is consistent with the repressor function of Pax-5 on the transcription of Xbp-1. A mutually exclusive expression pattern was also observed between nuclear Xbp-1 and Bcl-6 (Figure 2B). The plasma cell associated transcription factor Irf-4 was co-expressed in all Xbp-1 nuclear positive cells, in agreement with its role in plasma cell differentiation (Figure 2C).
Figure 2.
Relation of Xbp-1 with other transcription B cell transcription factors. Human tonsil. A: Pax-5 is expressed in virtually all B cells, only in the light zone of the germinal centers a small fraction of cells lack Pax-5 and express nuclear Xbp-1, in a mutually exclusive pattern (×20). B: Bcl-6 expression is restricted to germinal centers and only Bcl-6 negative centrocytes expressed nuclear Xbp-1 (×20). C: The plasma cell associated transcription factor Irf-4 is coexpressed in all nuclear Xbp-1 positive cells (×20).
Activation of Xbp-1 in Human B-NHL Cell Lines
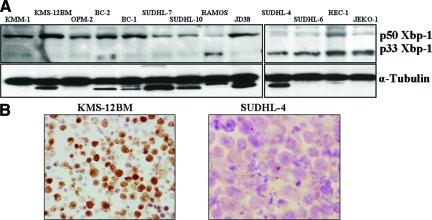
We analyzed the Xbp-1 expression in 23 human B-cell lines representing the spectrum of B-cell neoplasms in different stages of B-cell differentiation, including MCL, hairy cell leukemia, DLBCL, Burkitt lymphoma, primary effusion lymphoma, and multiple myeloma. Western blot analysis showed that the immunoglobulin secreting multiple myeloma cell lines KMM-1, KMS-11, OPM2, and RPMI-822619,20 had a strong expression of the activated p50 Xbp-1 form whereas the inactive p33 was weak (Figure 3A). The non-secretory myeloma cell lines (KMS-12PE and KMS-12BM20) and the primary effusion lymphoma cell lines (BC-1, BC-2, and BC-3), characterized by the lack of immunoglobulin secretion,21 exhibited the same strong expression of activated Xbp-1 and weak p33 (Figure 3A). Burkitt lymphoma cell lines lacking a plasma cell phenotype (JD38, Thomas, Ramos, and BJAB) had levels of activated p50 Xbp-1 expression similar to those found in multiple myeloma cell lines. These results suggest that Xbp-1 activation in these cell lines is independent of immunoglobulin secretion and plasma cell differentiation. The DLBCL cell lines showed a heterogeneous pattern of Xbp-1 activation. Ly-8, SUDHL-7 and 10 had stronger expression of the active Xbp-1 p50 form whereas the cell lines bearing the t(14,18) (SUDHL-4,6) had higher levels of the inactive p33 Xbp-1 form. In the MCL and hairy cell leukemia cell lines the predominant Xbp-1 protein was the inactive p33 form (Figure 3A).
Figure 3.
Xbp-1 Activation in plasma cell and lymphoma cell lines. A: Western blotting. All cell lines have some extent of Xbp-1 activation. Myeloma cell lines had the highest expression of the active form of Xbp-1(p50 Xbp-1) with a very weak band, corresponding to the cytoplasmic form of the protein (p33 Xbp-1) in the immunoglobulin secreting cell lines (KMM-1and OPM2) and in the non-secreting cell lines (KMS-12BM). Ramos 2G6.4C10, Jeko-1 and SUHL-4,6 cell lines show the weakest Xbp-1 activation with a prominent p33Xbp-1 expression. B: Immunocytochemistry. All KMS-12BM cells expressed nuclear Xbp-1 concordantly with the high expression of p50 Xbp-1 in the blots. SUDHL-4 exhibits a homogeneous cytoplasmic localization of the protein, according with the higher expression of p33 Xbp-1 (× 60 under oil).
Immunocytochemical staining of representative cells lines demonstrated a good correlation between the nuclear localization of Xbp-1 and the strong expression of p50 form by Western blot (KMS12, KMM1, OPM2, BC-1, BC-2 SUCHL-7, 10, and JD38), whereas cell lines with a predominant expression of the inactive p33 Xbp-1 protein by Western blot showed only a cytoplasmic expression in the immunohistochemical staining (SUDHL-4,6, REC-1, JEKO, and Ramos) (Figure 3B).
Activation of Xbp-1 in B-Cell Neoplasms
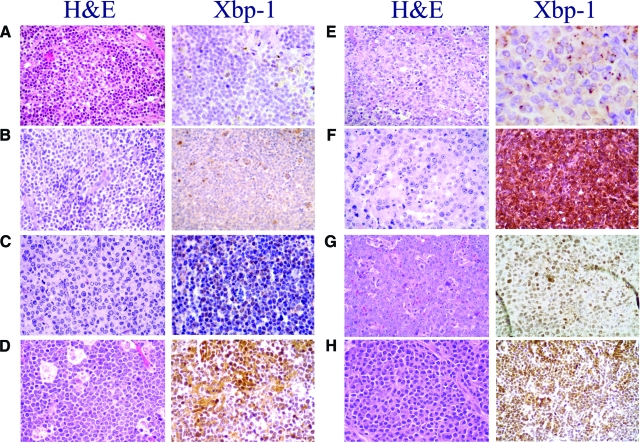
Activation of Xbp-1 was further evaluated in 411 lymphoid and plasma-cell neoplasms assessing the nuclear expression of the protein by immunohistochemistry. The results are summarized in Table 2. In low grade lymphomas, no nuclear Xbp-1 was found regardless of morphological evidence of plasmacytic differentiation in some of these tumors. Scattered positive cells were identified in the proliferation centers of 4 out 34 (11%) chronic lymphocytic leukemia, whereas the small lymphocytic component was consistently negative in all cases (Figure 4A). All follicular, marginal zone, Hodgkin, and peripheral T-cell lymphomas were consistently negative for nuclear Xbp-1 although scattered positive plasma cells and endothelial cells could be found (Figure 4, B and D).
Table 2.
Xbp-1 Activation in B-Cell and Plasma Cell Tumors
| Diagnosis | Nuclear Xbp-1 IHQ | XBP1s/XBP1u mRNA |
|---|---|---|
| Chronic lymphocytic leukemia | 0/17* | .011 |
| Follicular lymphoma | 0/12 | .012 |
| Mantle cell lymphoma | 4/25 (16%) | .040 |
| Splenic marginal zone lymphoma | 0/4 | |
| Marginal zone lymphoma | 0/5† | .036 |
| Burkitt lymphoma | 3/15 (20%) | .092 |
| DLBCL | 37/126 (29%) | |
| GCB | 15/53 (28%) | .105 |
| Non-GCB | 22/73 (30%) | .143 |
| Plasmablastic lymphoma | 25/54 (46%) | .122 |
| PEL | 4/6 (67%) | |
| Plasmacytoma | 27/39 (69%) | .301 |
| Classical Hodgkin lymphoma | 0/40 | |
| Peripheral T-cell lymphoma | 0/11 |
ND: Not done; DLBCL: Diffuse large B-cell lymphoma; GCB: Germinal center type DLBCL; PEL: Primary effusion lymphoma.
Scattered cells positive in the proliferative growth centers.
Scattered plasma cells positive.
Figure 4.
Xbp-1 activation in B-NHL and plasma cell tumors. A: In chronic lymphocytic leukemia, only scattered positive cells expressed nuclear Xbp-1 in the proliferation centers. The small lymphocytic component was consistently negative (×40). B: Marginal zone lymphoma contains some mature plasma cells that expressed nuclear Xbp-1, while the monocytoid neoplastic cells are negative (×40). C: Nuclear positivity for Xbp-1 in a typical mantle cell lymphoma with a high proliferative index (×40). D: Burkitt lymphoma with plasma cell differentiation has more than 20% of tumor cells with nuclear positivity for Xbp-1. E: Diffuse large B-cell lymphoma, germinal center type with a cytoplasmic expression of Xbp-1 (×40). F: Intense nuclear positivity for Xbp-1 in a diffuse large B-cell lymphoma non-germinal center type (×40).G: Plasmablastic lymphoma (×40) and (H) plasmacytoma exhibit a strong and diffuse nuclear positivity for Xbp-1(×40).
A diffuse nuclear staining was seen in 4 of the 25 (16%) MCL. Interestingly, three of these cases had a blastoid morphology (Figure 4C) and one was a classical variant but had a high proliferation index (2 mitosis/×40 field).
Nuclear Xbp-1 was found in 37 of the 126 (29%) DLBCL. No significant differences were found between tumors with a germinal center (28%) or non- germinal center (30%) phenotype (Figure 4, E and F). Tumors with plasmablastic differentiation, including plasmablastic lymphoma and primary effusion lymphoma, showed nuclear expression of Xbp-1 in 29 of the 60 (48%) cases (P = 0.01, vs. DLBCL) (Figure 4G). Plasma cell neoplasms showed the highest proportion of positive cases with 27 out of 39 tumors (69%) expressing nuclear protein (Figure 4H).
In a subgroup of 22 tumors from which frozen material was available, we examined by reverse transcription-PCR the expression of the two spliced and unspliced (XBP-1s/XBP-1u) mRNA transcripts encoding for the active and inactive Xbp-1 protein, respectively. The ratio between these two transcripts was calculated. The chronic lymphocytic leukemia, follicular lymphomas, marginal zone lymphomas and mantle cell lymphomas lacking nuclear Xbp-1 expression had a concordant predominant expression of the unspliced transcript with XBP-1s/XBP-1u ratios of 0.011 to 0.040 (Table 2). The expression of the spliced Xbp-1 transcript was significantly higher in DLBCL (0.105), plasmablastic lymphomas (0.143) and, particularly, plasma cell neoplasms (0.301) (Table 2). These findings are concordant with the nuclear protein expression observed in these tumors.
Clinical Correlation and Prognostic Value of Activated Xbp-1 in DLBCL
The relationship between activated nuclear Xbp-1 expression and the clinico-pathological features of the patients was evaluated in 116 aggressive lymphomas, 103 DLBCL, and 13 plasmablastic lymphomas in which the clinical data were available. No correlation was observed between the nuclear expression and the characteristics of the patients at diagnosis, including age, sex, bulky disease, stage, low-density lipoprotein, beta2-microglobulin, and IPI, either in plasmablastic and non-plasmablastic DLBCL.
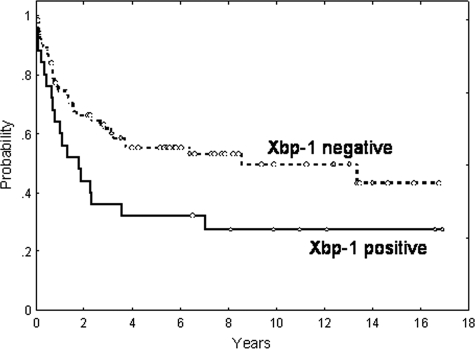
The complete remission rate was significantly poorer in patients with activated Xbp-1 than in the remainder (68 vs. 41%, respectively; P = 0.03). Patients showing activated Xbp-1 had shorter OS than the others (5-year OS: 29 vs. 53%, respectively; P = 0.01). In the 103 non-plasmablastic DLBCL patients Xbp-1 maintained its prognostic value (5-year OS: 32 vs. 55% for patients with activated or not-activated Xbp-1, respectively; P = 0.03) (Figure 5). A multivariate analysis was performed including IPI (low versus intermediate versus high-risk), histology (non-plasmablastic DLBCL versus plasmablastic), and Xbp-1 (non-activated versus activated). In the final model, with 95 patients, IPI (relative risk [RR]: 2.5; P < 0.001), histology (RR: 7.0; P = 0.04) and Xbp-1 activation (RR: 2.03; P = 0.032) maintained the independent prognostic value for OS.
Figure 5.
Overall survival of 103 diffuse large B-cell lymphoma patients. Overall survival curves after Kaplan and Meier analysis of 103 diffuse large B-cell lymphoma patients regarding the nuclear expression of active Xbp-1 spliced form by immunohistochemistry.
Discussion
The Unfolded Protein Response is a complex signaling pathway that in lymphoid cells has an essential role in the differentiation process from activated B-cells to immunoglobulin secreting plasma-cells.11,22 The transcription factor Xbp-1 is one of the main regulators of this pathway and experimental studies have shown its selective requirement for the terminal B-cell differentiation to plasma cells. The active form of Xbp-1 is generated by a specific enzymatic splicing of the Xbp-1 transcript that switches the expression of the Xbp-1 protein from a p33 form to a longer p50 form containing a potent transcription domain. In this study we have shown that the active p50 Xbp-1 protein is found only in nuclear extracts of lymphoid tissues whereas the p33 form is restricted to the cytoplasmic fraction. This finding is consistent with the potent transcriptional function of the p50 form and allows the immunohistochemical study of Xbp-1 activation in human reactive tissues and tumors by detecting the nuclear translocation of the Xbp-1 protein.
In non-neoplastic lymphoid tissues the expression of cytoplasmic Xbp-1 was found in virtually all cells. In contrast, Xbp-1 nuclear expression was confined to a small population of B-cells in the light zone of the follicular germinal centers and a subset of intra and extrafollicular plasma cells. Interestingly, nuclear Xbp-1 was found in germinal center cells expressing Irf-4 and it was mutually exclusive with the expression of bcl-6 and Pax-5. Irf-4 is an important transcriptional regulator of postgerminal center B-cell differentiation that is initially expressed in a subset of cells of the light zone of the germinal centers.23,24 Irf-4 expression in follicular germinal centers cells is mutually exclusive with Bcl-6 and it has been suggested that these cells represent and early population of germinal center cells already committed to plasma-cell differentiation.24 Our findings demonstrating for the first time the mutually exclusive expression of nuclear Xbp-1 and bcl-6 and the co-expression of Irf-4 in cells of the light zone of the germinal center are consistent with this idea. Interestingly, we also observed a mutually exclusive expression of Pax-5 and nuclear Xbp-1 in germinal center B-cells. Pax-5 is a transcription factor required for early B-cell differentiation and maintenance of B-cell identity but it is down-regulated in the terminal stages of B-cell differentiation to plasma cells.25 Pax-5 inhibits the differentiation of B-lymphocytes to plasma cells by repressing a series of genes required for the terminal B-cell differentiation. Xbp-1 is one of the master genes under Pax-5 control and its expression is released by Pax-5 down-regulation.26,27,28 The mutually exclusive expression of Pax-5 and nuclear Xbp-1 in a subset of germinal center B-cells observed in our study is consistent with this regulatory model and, together with the expression pattern of bcl-6 and Irf-4, strongly suggest that Xbp-1 activation in reactive lymphoid tissues occurs in a subset of germinal center cells committed to plasma cell differentiation.
The implication of Xbp-1 activation in the normal process of plasma-cell differentiation is relatively well established. However, its potential role in lymphomagenesis and particularly in tumors showing terminal B-cell differentiation features is less known. The relevance of Xbp-1 activation in plasma-cell neoplasms has been highlighted by experimental studies showing that forced expression of the active form promotes the development of MM in mice.15 In addition, active Xbp-1 seems important to sustain the viability and perpetuation of MM tumor cells.7,15,29,30 In our study we have demonstrated that nuclear Xbp-1 was virtually absent in all indolent lymphomas but it was detected in a subset of aggressive lymphomas with poor prognosis suggesting that Xbp-1 activation may play a role in the pathogenesis and clinical behavior of these tumors.
In indolent lymphomas, nuclear Xbp-1 was only observed in occasional large cells of the proliferative centers in chronic lymphocytic leukemia whereas the small lymphocytic component was consistently negative. This observation parallels the prior finding of Irf-4 expression in cells of the proliferation centers and suggests the activation of the plasma-cell differentiation program in this topographic compartment.31,32 In contrast to indolent lymphomas, Xbp-1 activation was observed in 32% of aggressive lymphomas. Thus, we found Xbp-1 nuclear expression in 4 MCL with high proliferative index, three of them with blastoid morphology. Although plasma-cell differentiation has been observed in occasional MCL,33 none of our cases exhibit plasmacytic features. Two Burkitt lymphomas with plasma-cell differentiation expressed also nuclear Xbp-1. Xbp-1 activation has been observed in human Burkitt cell lines induced experimentally to differentiate into plasma cells.34,35 Plasmacytic differentiation may occur in Burkitt lymphoma associated with HIV infection36 but none of our patients had an underlying immunodeficiency.
Plasma cells in reactive lymphoid tissues were only occasionally positive for nuclear Xbp-1. However, our results confirm the high expression of activated Xbp-1 in plasma-cell neoplasms (69%) and also reveal a high incidence of nuclear Xbp-1 in lymphomas with plasmablastic differentiation such as plasmablastic lymphomas and primary effusion lymphoma, where high expression of genes related to the unfolded protein response pathway, including Xbp-1, have been detected by gene expression profiling.37,38 Interestingly, 28% DLBCL had activation of Xbp-1 regardless of the germinal or postgerminal center derivation, suggesting that activation of this pathway in DLBCL may be independent of the activated B-cell phenotype. Interestingly, nuclear expression of Xbp-1 in these lymphomas was associated with poor response to therapy as well as with a shorter OS of the patients. A similar relationship between high expression levels of the active Xbp-1 form and poor prognosis has been observed in patients with breast cancer.39 How the overexpression of the active Xbp-1 form may confer a more aggressive behavior to the tumor cells is not well understood. Experimental studies in MM have suggested that activation of the unfoled protein response pathway promotes the survival of the tumor cells and drugs targeting this pathway are emerging as an attractive approach for the treatment of these patients. Proteasome inhibitors in MM cells induce the apoptosis of the tumor cells. Interestingly, these drugs shift the balance from the active to the inactive Xbp-1 species that in addition act as a dominant-negative inhibitor of the spliced active form preventing the development of an efficient unfoled protein response response. MM cells with a deficient Xbp-1 activation undergo an increased apoptosis in response to endoplasmic reticulum stress. All together, these observations suggest that, similarly to MM, patients with DLBCL expressing active Xbp-1 may benefit from strategic therapies targeting the unfoled protein response response.
In summary, we have investigated the cellular localization and expression pattern of Xbp-1 in human reactive and neoplastic lymphoid tissues. Our findings indicate that the active p50 form of Xbp-1 is expressed in the nucleus of a subset of germinal center B-cells probably committed to plasma-cell differentiation. In malignant lymphomas, nuclear Xbp-1 was expressed in a subset of aggressive lymphomas with plasmablastic features and DLBCL with poor response to therapy and shorter survival, suggesting that Xbp-1 activation may play a role in the pathogenesis and progression of these tumors.
Acknowledgments
The authors acknowledge Dr. Dan Ron, Skirball Institute of Biomolecular Medicine, New York University School of Medicine for his comments, and Dr. Raquel Garcia Olivas, of the Confocal facility University of Barcelona, Spain for her excellent technical assistance.
Footnotes
Address reprint requests to Antonio Martinez, Hematopathology Section, Hospital Clinic, Villarroel 170, 08036-Barcelona, Barcelona, Spain. E-mail: antonmar@clinic.ub.es.
Supported by the Spanish Comisión Interministerial de Ciencia y Tecnología (CICYT) SAF05-5855, Instituto de Salud Carlos III, Red Temática de Cáncer (2039), FIS PI080095, Ajut de la Generalitat de Catalunya (CRSGR0870). O.B. is a fellow supported by the Instituto de Salud Carlos III. LH is a researcher from IDIBAPS and supported by FIS and “programa d’estabilització d’investigadors” of Direcció d’Estrategia i Coordinació del Departament de Salut (Generalitat de Catalunya).
O.B. and A.M. contributed equally to this work.
Specific contributions of authors: O.B. performed research, collected data, analyzed and interpreted data, and drafted the manuscript; A.M. performed research, analyzed data, and drafted the manuscript; D.M. performed research; L.H. performed research; L.C. collected data; J.L.M. collected data; J.T.-F. collected and interpreted data; O.L. collected data; E.C. interpreted data and drafted the manuscript; A.L.G. analyzed and interpreted data; and A.M. designed research, analyzed and interpreted data and drafted the manuscript.
References
- Brodsky JL, McCracken AA. ER protein quality control and proteasome-mediated protein degradation. Sem Cell Dev Biol. 1999;10:507–513. doi: 10.1006/scdb.1999.0321. [DOI] [PubMed] [Google Scholar]
- Rutkowski DT, Kaufman RJ. A trip to the ER: coping with stress. Trends Cell Biol. 2004;14:20–28. doi: 10.1016/j.tcb.2003.11.001. [DOI] [PubMed] [Google Scholar]
- Welihinda AA, Tirasophon W, Kaufman RJ. The cellular response to protein misfolding in the endoplasmic reticulum. Gene Expr. 1999;7:293–300. [PMC free article] [PubMed] [Google Scholar]
- Haze K, Yoshida H, Yanagi H, Yura T, Mori K. Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol Biol Cell. 1999;10:3787–3799. doi: 10.1091/mbc.10.11.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haze K, Okada T, Yoshida H, Yanagi H, Yura T, Negishi M, Mori K. Identification of the G13 (cAMP-response-element-binding protein-related protein) gene product related to activating transcription factor 6 as a transcriptional activator of the mammalian unfolded protein response. Biochem J. 2001;355:19–28. doi: 10.1042/0264-6021:3550019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Shen J, Arenzana N, Tirasophon W, Kaufman RJ, Prywes R. Activation of ATF6 and an ATF6 DNA binding site by the endoplasmic reticulum stress response. J Biol Chem. 2000;275:27013–27020. doi: 10.1074/jbc.M003322200. [DOI] [PubMed] [Google Scholar]
- Lee AH, Iwakoshi NN, Anderson KC, Glimcher LH. Proteasome inhibitors disrupt the unfolded protein response in myeloma cells. Proc Natl Acad Sci USA. 2003;100:9946–9951. doi: 10.1073/pnas.1334037100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Tirasophon W, Shen X, Michalak M, Prywes R, Okada T, Yoshida H, Mori K, Kaufman RJ. IRE1-mediated unconventional mRNA splicing and S2P-mediated ATF6 cleavage merge to regulate XBP1 in signaling the unfolded protein response. Genes Dev. 2002;16:452–466. doi: 10.1101/gad.964702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AH, Chu GC, Iwakoshi NN, Glimcher LH. XBP-1 is required for biogenesis of cellular secretory machinery of exocrine glands. EMBO J. 2005;24:4368–4380. doi: 10.1038/sj.emboj.7600903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimold AM, Etkin A, Clauss I, Perkins A, Friend DS, Zhang J, Horton HF, Scott A, Orkin SH, Byrne MC, Grusby MJ, Glimcher LH. An essential role in liver development for transcription factor XBP-1. Genes Dev. 2000;14:152–157. [PMC free article] [PubMed] [Google Scholar]
- Gunn KE, Gifford NM, Mori K, Brewer JW. A role for the unfolded protein response in optimizing antibody secretion. Mol Immunol. 2004;41:919–927. doi: 10.1016/j.molimm.2004.04.023. [DOI] [PubMed] [Google Scholar]
- Iwakoshi NN, Lee AH, Glimcher LH. The X-box binding protein-1 transcription factor is required for plasma cell differentiation and the unfolded protein response. Immunol Rev. 2003;194:29–38. doi: 10.1034/j.1600-065x.2003.00057.x. [DOI] [PubMed] [Google Scholar]
- Lee AH, Iwakoshi NN, Glimcher LH. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol Cell Biol. 2003;23:7448–7459. doi: 10.1128/MCB.23.21.7448-7459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagliavacca L, Anelli T, Fagioli C, Mezghrani A, Ruffato E, Sitia R. The making of a professional secretory cell: architectural and functional changes in the ER during B lymphocyte plasma cell differentiation. Biol Chem. 2003;384:1273–1277. doi: 10.1515/BC.2003.141. [DOI] [PubMed] [Google Scholar]
- Carrasco DR, Sukhdeo K, Protopopova M, Sinha R, Enos M, Carrasco DE, Zheng M, Mani M, Henderson J, Pinkus GS, Munshi N, Horner J, Ivanova EV, Protopopov A, Anderson KC, Tonon G, DePinho RA. The differentiation and stress response factor XBP-1 drives multiple myeloma pathogenesis. Cancer Cell. 2007;11:349–360. doi: 10.1016/j.ccr.2007.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hans CP, Weisenburger DD, Greiner TC, Gascoyne RD, Delabie J, Ott G, Muller-Hermelink HK, Campo E, Braziel RM, Jaffe ES, Pan Z, Farinha P, Smith LM, Falini B, Banham AH, Rosenwald A, Staudt LM, Connors JM, Armitage JO, Chan WC. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood. 2004;103:275–282. doi: 10.1182/blood-2003-05-1545. [DOI] [PubMed] [Google Scholar]
- Calfon M, Zeng H, Urano F, Till JH, Hubbard SR, Harding HP, Clark SG, Ron D. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415:92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107:881–891. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- Katagiri S, Yonezawa T, Kuyama J, Kanayama Y, Nishida K, Abe T, Tamaki T, Ohnishi M, Tarui S. Two distinct human myeloma cell lines originating from one patient with myeloma. Int J Cancer. 1985;36:241–246. doi: 10.1002/ijc.2910360217. [DOI] [PubMed] [Google Scholar]
- Namba M, Ohtsuki T, Mori M, Togawa A, Wada H, Sugihara T, Yawata Y, Kimoto T. Establishment of five human myeloma cell lines. In Vitro Cell Dev Biol. 1989;25:723–729. doi: 10.1007/BF02623725. [DOI] [PubMed] [Google Scholar]
- Cesarman E, Chang Y, Moore PS, Said JW, Knowles DM. Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med. 1995;332:1186–1191. doi: 10.1056/NEJM199505043321802. [DOI] [PubMed] [Google Scholar]
- Iwakoshi NN, Lee AH, Vallabhajosyula P, Otipoby KL, Rajewsky K, Glimcher LH. Plasma cell differentiation and the unfolded protein response intersect at the transcription factor XBP-1. Nat Immunol. 2003;4:321–329. doi: 10.1038/ni907. [DOI] [PubMed] [Google Scholar]
- Klein U, Casola S, Cattoretti G, Shen Q, Lia M, Mo T, Ludwig T, Rajewsky K, Della-Favera R. Transcription factor IRF4 controls plasma cell differentiation and class-switch recombination. Nat Immunol. 2006;7:773–782. doi: 10.1038/ni1357. [DOI] [PubMed] [Google Scholar]
- Falini B, Fizzotti M, Pucciarini A, Bigerna B, Marafioti T, Gambacorta M, Pacini R, Alunni C, Natali-Tanci L, Ugolini B, Sebastiani C, Cattoretti G, Pileri S, la-Favera R, Stein H. A monoclonal antibody (MUM1p) detects expression of the MUM1/IRF4 protein in a subset of germinal center B cells, plasma cells, and activated T cells. Blood. 2000;95:2084–2092. [PubMed] [Google Scholar]
- Souabni A, Cobaleda C, Schebesta M, Busslinger M. Pax5 promotes B lymphopoiesis and blocks T cell development by repressing Notch1 6. Immunity. 2002;17:781–793. doi: 10.1016/s1074-7613(02)00472-7. [DOI] [PubMed] [Google Scholar]
- Usui T, Wakatsuki Y, Matsunaga Y, Kaneko S, Koseki H, Kita T. Overexpression of B cell-specific activator protein (BSAP/Pax-5) in a late B cell is sufficient to suppress differentiation to an Ig high producer cell with plasma cell phenotype. J Immunol. 1997;158:3197–3204. [PubMed] [Google Scholar]
- Delogu A, Schebesta A, Sun Q, Aschenbrenner K, Perlot T, Busslinger M. Gene repression by Pax5 in B cells is essential for blood cell homeostasis and is reversed in plasma cells. Immunity. 2006;24:269–281. doi: 10.1016/j.immuni.2006.01.012. [DOI] [PubMed] [Google Scholar]
- Reimold AM, Ponath PD, Li YS, Hardy RR, David CS, Strominger JL, Glimcher LH. Transcription factor B cell lineage-specific activator protein regulates the gene for human X-box binding protein 1. J Exp Med. 1996;183:393–401. doi: 10.1084/jem.183.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M, Gotoh T, Okuno Y, Tatetsu H, Sonoki T, Uneda S, Mori M, Mitsuya H, Hata H. Activation of the endoplasmic reticulum stress pathway is associated with survival of myeloma cells. Leuk Lymphoma. 2006;47:531–539. doi: 10.1080/10428190500312196. [DOI] [PubMed] [Google Scholar]
- Zhou X, Jiang H, Hou J. Coordination of upregulated XBP-1 and downregulated c-myc during myeloma cell differentiation induced by 2-methoxyestradiol. Leuk Res. 2007;31:1259–1265. doi: 10.1016/j.leukres.2006.11.023. [DOI] [PubMed] [Google Scholar]
- Soma LA, Craig FE, Swerdlow SH. The proliferation center microenvironment and prognostic markers in chronic lymphocytic leukemia/small lymphocytic lymphoma. Hum Pathol. 2006;37:152–159. doi: 10.1016/j.humpath.2005.09.029. [DOI] [PubMed] [Google Scholar]
- Martinez A, Pittaluga S, Rudelius M, vies-Hill T, Sebasigari D, Fountaine TJ, Hewitt S, Jaffe ES, Raffeld M. Expression of the interferon regulatory factor 8/ICSBP-1 in human reactive lymphoid tissues and B-cell lymphomas: a novel germinal center marker. Am J Surg Pathol. 2008;32:1190–1200. doi: 10.1097/PAS.0b013e318166f46a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young KH, Chan WC, Fu K, Iqbal J, Sanger WG, Ratashak A, Greiner TC, Weisenburger DD. Mantle cell lymphoma with plasma cell differentiation. Am J Surg Pathol. 2006;30:954–961. doi: 10.1097/00000478-200608000-00004. [DOI] [PubMed] [Google Scholar]
- Teng Y, Takahashi Y, Yamada M, Kurosu T, Koyama T, Miura O, Miki T. IRF4 negatively regulates proliferation of germinal center B cell-derived Burkitt’s lymphoma cell lines and induces differentiation toward plasma cells. Eur J Cell Biol. 2007;86:581–589. doi: 10.1016/j.ejcb.2007.05.006. [DOI] [PubMed] [Google Scholar]
- Guedez L, Martinez A, Zhao S, Vivero A, Pittaluga S, Stetler-Stevenson M, Raffeld M, Stetler-Stevenson WG. Tissue inhibitor of metalloproteinase 1 (TIMP-1) promotes plasmablastic differentiation of a Burkitt lymphoma cell line: implications in the pathogenesis of plasmacytic/plasmablastic tumors. Blood. 2005;105:1660–1668. doi: 10.1182/blood-2004-04-1385. [DOI] [PubMed] [Google Scholar]
- Raphael M, Gentilhomme O, Tulliez M, Byron PA, Diebold J. Histopathologic features of high-grade non-Hodgkin’s lymphomas in acquired immunodeficiency syndrome. The French Study Group of Pathology for Human Immunodeficiency Virus-Associated Tumors. Arch Pathol Lab Med. 1991;115:15–20. [PubMed] [Google Scholar]
- Klein U, Gloghini A, Gaidano G, Chadburn A, Cesarman E, Della-Favera R, Carbone A. Gene expression profile analysis of AIDS-related primary effusion lymphoma (PEL) suggests a plasmablastic derivation and identifies PEL-specific transcripts. Blood. 2003;101:4115–4121. doi: 10.1182/blood-2002-10-3090. [DOI] [PubMed] [Google Scholar]
- Jenner RG, Maillard K, Cattini N, Weiss RA, Boshoff C, Wooster R, Kellam P. Kaposi’s sarcoma-associated herpesvirus-infected primary effusion lymphoma has a plasma cell gene expression profile. Proc Natl Acad Sci USA. 2003;100:10399–10404. doi: 10.1073/pnas.1630810100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies MP, Barraclough DL, Stewart C, Joyce KA, Eccles RM, Barraclough R, Rudland PS, Sibson DR. Expression and splicing of the unfolded protein response gene XBP-1 are significantly associated with clinical outcome of endocrine-treated breast cancer. Int J Cancer. 2008;123:85–88. doi: 10.1002/ijc.23479. [DOI] [PubMed] [Google Scholar]