Abstract
Previously, we showed that the presence of high numbers of macrophages correlates with poor prognosis in nongynecological leiomyosarcoma (LMS). In gynecological LMS, a similar trend was noted but did not reach statistical significance. Colony-stimulating factor-1 (CSF1) is a major chemoattractant for macrophages. Here we show that in a subset of LMS cases, CSF1 is expressed by the malignant cells. Previously, we found that CSF1 is translocated and highly expressed in tenosynovial giant cell tumors (TGCTs), and this observation allowed us to identify genes that showed a coordinate expression with CSF1. Here, we evaluated the expression of CSF1 and TGCT-associated proteins in 149 cases of LMS. The coordinate expression of CSF1 and three TGCT-associated proteins (CD163, FCGR3a, and CTSL1) identified cases with poor prognosis in both gynecological LMS (P = 0.00006) and nongynecological LMS (P = 0.03). In gynecological LMS, the coordinate expression of these four markers was the only independent prognosticator in multivariate analysis (hazard ratio, 4.2; 95% CI, 1.12 to 16; P = 0.03). Our findings indicate that CSF1 may play an important role in the clinical behavior of LMS that may open a window for new therapeutic reagents.
In recent years the association of tumor-associated macrophages with clinical outcome in human malignancies has been a subject of intense study.1,2 In several carcinomas (breast, prostate, endometrium, bladder, kidney, and esophagus) and lymphomas the presence of macrophages in the tumor stroma is associated with poor prognosis.3,4,5,6,7,8,9 In contrast, the presence of macrophages in colon, gastric carcinomas, and melanomas is associated with improved clinical outcome.10,11,12 Colony stimulating factor-1 (CSF1) is a cytokine that induces the proliferation and differentiation of macrophages and monocytes and is involved in the recruitment of monocytes into tumors.13,14 CSF1 can be produced by many different cell types including macrophages, fibroblasts, endothelial cells, and tumor cells. Increased CSF1 serum levels have been found in several malignances, including breast carcinoma, ovarian cancer, and endometrial carcinoma.15 The expression of epithelial CSF1 and CSF1R (colony-stimulating factor-1 receptor) in metastases of ovarian carcinoma was shown to be associated with poor clinical outcome.16
Leiomyosarcoma (LMS) is malignant tumor originating from smooth muscle cells. Two major subtypes exist, those that originate in the uterus and those that arise in extra-uterine soft tissues. Surgery is the main mode of treatment and chemotherapeutic approaches have met with little success. Despite their shared origin in smooth muscle cells for the gynecological and nongynecological LMS there are significant differences in the criteria that are used to determine malignancy in smooth muscle tumors from both sites. There are no shared biological prognosticators between the two types of LMS.
We recently reported that there is variable expression of macrophage-specific mRNAs (CD68 and CD163) in LMS but not in several other sarcomas, including synovial sarcoma and gastrointestinal tumors. The number of macrophages in LMS as determined by CD163 staining showed a significant association with poor clinical outcome in nongynecological LMS but not in gynecological LMS.17 In addition to the mRNAs for CD68 and CD163, variable levels of expression for CSF1 and CSF1R mRNA were noted in LMS cases by gene expression profiling. However, gene expression profiling does not allow localization of the source of CSF1 mRNA to a specific cell type within the tumor. In an attempt to improve our ability to predict outcome in not only nongynecological LMS but also gynecological LMS, we decided to examine the role of CSF1 in LMS.
Here we show by in situ hybridization that LMS tumor cells can produce CSF1, suggesting that CSF1 expression by LMS cells is instrumental in attracting and stimulating tumor-associated macrophages. To identify additional genes that are coordinately expressed with CSF1, a set of 896 genes previously identified in tenosynovial giant cell tumor (TGCT) was used. TGCT is a neoplasm driven by a high levels of CSF1 expression caused by a chromosomal translocation involving the CSF1 gene.18,19 The high levels of secreted CSF1 attract many macrophages to the site of the lesion and the gene expression profile of TGCT can therefore be seen to represent to a great extent the a signature of genes responsive to CSF1. The expression of these 896 genes was subsequently analyzed in five independent, previously published breast carcinoma datasets and we identified 112 genes that were consistently and coordinately expressed in a subset of breast cancer cases from each of these five datasets. The expression of the CSF1 response signature in breast cancer was associated with higher tumor grade.20 CSF1 and a subset of 71 genes from these 112 genes could be analyzed on 59 sarcomas, including 16 cases of LMS. The LMS cases, in contrast to the other sarcomas tested, showed variable levels of expression for these genes. We selected three markers from this geneset for which commercial antibodies were available (CD163, FCGR3a, and CTSL1) to study the expression in 149 cases of LMS. Here we show that the coordinate expression of CSF1 and the other three markers is a powerful tool to predict clinical outcome in not only the nongynecological LMS but also in gynecological LMS.
Materials and Methods
Case Material
For gene expression profiling frozen tissue from 59 tumors was used. This included a previously reported set of sarcomas consisting of 8 LMS, 10 TGCTs, 8 gastrointestinal stromal tumors, 8 gastrointestinal stromal tumors, 7 synovial sarcomas, 5 dermatofibrosarcoma protuberans, and 5 solitary fibrous tumors.17 To this set of 51 tumors we added an additional eight cases of LMS, which facilitated the visualization of variable gene expression within LMS. For in situ hybridization and immunohistochemistry studies, we studied 149 LMS cases distributed over two TMAs (TA-121, and TA-201). All cases on the arrays consisted of material obtained at primary diagnosis. The TMAs were constructed using 0.6-mm cores with a manual tissue arrayer from Beecher Instruments, Silver Spring, MD. Survival information with a mean follow-up of 3.1 years (range, 1 month to 5 years) was available for all of the cases. The FNCLCC (Fédération Nationale des Centres de Lutte Contre le Cancer)21 grading system was used for the nongynecological LMS. Gynecological LMS were classified according to tumor differentiation into three categories: well, moderate, and poorly differentiated. The diagnosis of LMS on hematoxylin and eosin-stained paraffin sections was based on morphological features. These findings consisted of intersecting groups of spindle cells with elongated nuclei and eosinophilic cytoplasm and/or immunohistochemistry staining (positive for smooth muscle actin and/or desmin and negative for CD117 and myogenin). Tumors with less obvious histological features of smooth muscle differentiation required, at a minimum, either both focal smooth muscle actin and desmin staining or strong diffuse smooth muscle actin staining to be included in the study.
Gene Expression Profiling and Gene Selection
The Human Exonic Evidence Based Oligonucleotide microarrays (HEEBO, Stanford) used in the study contain 44,544 70-mer probes that were designed using a transcriptome-based annotation of exonic structure for genomic loci. After confirmation of histology and the presence of viable tumor by frozen section, specimens were homogenized in Trizol reagent (Invitrogen, Carlsbad, CA) and total RNA was extracted. The total RNA was reverse-transcribed into cDNA using a mixture of oligo dT (high performance liquid chromatography purified; Operon, Huntsville, AL) and random hexamer (catalog no. 27-2166-01; Amersham, Arlington Heights, IL) primers with incorporation of amino allyl-dUTP (8439; Ambion, Austin, TX). Cy3 and Cy5 dyes (RPN 5661, Amersham) were used for indirect labeling of the cDNA from reference RNA (universal human reference RNA, catalog no. 740000; Stratagen, La Jolla, CA) and cDNA from tumor specimens, respectively. Microarray hybridization and washing was performed using standard procedures.17,22 Microarrays were scanned on a GenePix 4000 microarray scanner (Molecular Devices, Sunnyvale, CA) and fluorescence ratios (tumor/reference) were calculated using GenePix software. The new dataset was filtered to allow only spots with a ratio of signal over background of at least 2.0 in the Cy5 and 2.0 in the Cy3 channel. Gene centering was applied to the expression values for this series of tumors. Only genes with >90% available good data were analyzed.
In contrast to our previous study17 in which we analyzed genes known to be expressed by macrophages and in which we counted the number of macrophages in LMS cases, we examine here CSF1 and a set of CSF1-related genes. We started out with 896 genes that are highly and specifically expressed in TGCTs. These CSF1-associated genes were identified in a previously published set of gene expression profiling data in which TGCTs were compared with a variety of other soft tissue neoplasms.18 The resulting list of 896 genes was used to cluster five publicly available datasets of breast carcinomas and 112 genes with a highly coordinated pattern of expression that showed the ability to distinguish distinct subsets of breast tumors in each of the five datasets20 were selected for analysis on the current 59 sarcoma samples (Supplemental Table 1, see http://ajp.amjpathol.org). This CSF1 response geneset is highly enriched for genes involved in immune function and contains many genes known to be expressed in macrophages. Using the filtered dataset described above, 71 of 112 CSF1-related genes met the criteria on the series of 59 sarcomas used for this study (Supplemental Table 2, see http://ajp.amjpathol.org).
Immunohistochemistry and in Situ Hybridization
Slides were cut from TA 121 and TA 201 at 4 μm, deparaffinized in xylene, and hydrated in a graded series of alcohol. For immunohistochemistry, the primary antibodies used were FCGR3a (CD16) (MCA1816, mouse monoclonal, 1/40; AbD Serotec, Oxford, UK), CD163 (NCL-CD163, mouse monoclonal, 1:100, Novocastra, Newcastle upon Tyne, UK), CTSL1 (MCA2374, mouse monoclonal, 1/25; AbD Serotec); NCAM1(CD56) [18-0152, mouse monoclonal, 1/20; Zymed, South San Francisco, CA (Invitrogen)], and B3GAT1 (CD57) (347391, mouse monoclonal, 1/40; BD Biosciences, San Jose, CA). The antigen retrieval solution for FCGR3a, CD57, and CD56 was citrate, pH 6, and for CD163 was ethylenediaminetetraacetic acid, pH 8. No antigen retrieval was performed for CTSL1. Slides were boiled by microwaving in antigen retrieval solution for 12 minutes. The immunohistochemical reactions were visualized using mouse versions of the EnVision + system (DAKO, Carpinteria, CA) using diaminobenzidine, except for CD163, which was stained on a Benchmark autostainer (Ventana Medical Systems, Tucson, AZ). We performed CSF1 and CSF1R in situ hybridization on TMA sections based on a protocol published previously.23,24,25 CD163/CSF1 double stains were generated by first performing CSF1 in situ hybridization with diaminobenzidine, followed by routine immunohistochemistry with CD163 using VIP (Vector Laboratories, Burlingame, CA). The scoring criteria are shown in Table 1. Scores 1, 2, and 3 were considered positive. For each antibody the scoring criteria were chosen in such a manner that distinct subsets of LMS cases were detected. This was done before analysis of outcome data. Digital images from all stained cores are available through the Stanford tissue microarray database and the accompanying website (http://tma.stanford.edu/tma_portal/LMS_CSF1/).26
Table 1.
Scoring Criteria
| FCGR3a | CD163 | CTSL1 | CSF1 asISH | |
|---|---|---|---|---|
| Score 0 | ≤10 cells/0.6 mm | ≤5 cells/0.6 mm | ≤10 cells/0.6 mm | No paranuclear dotlike stain or >1 paranuclear dotlike stain per cell in less than 5% of the cells |
| Score 1 | >10 to ≤20 cells/0.6 mm | NA* | >10 to ≤20 cells/0.6 mm | NA* |
| Score 2 | >20 to ≤45 cells/0.6 mm | >5 to ≤25 cells/0.6 mm | >20 to ≤45 cells/0.6 mm | One to four paranuclear dotlike stains per cell in at least 5% of the cells |
| Score 3 | >45 cells/0.6 mm | >25 cells/0.6 mm | >45 cells/0.6 mm | More than 5 paranuclear dotlike stains per cell in at least 5% of the cells |
Not applicable.
Statistical Analysis
The Kaplan-Meier method was used to estimate disease-specific survival (DSS) distributions. DSS was calculated from the time of diagnosis to the date of death of disease. Multivariate Cox regression analysis was used to study the relationship between survival and the different variables. The following variables were introduced into the analysis: age, FNLCC grade, cell differentiation, tumor size, presence of necrosis, mitotic count, and co-expression of CSF1, FCGR3a, CTSL1, and CD163 in LMS cases. To create Cox regression models with the subset of features that showed the strongest association with survival, we used forward step-wise variable selection using likelihood ratio. P values ≤0.05 were considered significant. Statistical calculations were performed using SPSS software v14.0 (Chicago, IL). Hierarchical cluster analysis of TMA-immunostaining results were realized using Deconvoluter 6 and TMA-Combiner 7 programs.27,28
Results
Clinicopathological Features of LMS Cases
The clinicopathological features for eight leiomyosarcoma cases previously analyzed by gene expression profiling17 are summarized in Supplementary Table 3 (see http://ajp.amjpathol.org). An additional eight LMS used in this study for gene expression profiling are summarized in Supplementary Table 4 (see http://ajp.amjpathol.org).
The 149 LMS used in the two tissue arrays (TA-121, and TA-201) are described in Supplementary Table 5 (see http://ajp.amjpathol.org) and are the same as used previously.17 Briefly, in the gynecological LMS cases (n = 76), the median age was 51 years (range, 5 to 67 years). The tumor size ranged from 2 to 35 cm (average, 10.1 cm) and the most common location was uterus. The nongynecological LMS (n = 73) presented a median age of 54 years (range, 13 to 81 years). There were more female than male (42 versus 31) patients, and the most common location for the tumor was retroperitoneum followed by limbs and the genitourinary system. The median tumor size was 9.9 cm (range, 1.2 to 34 cm). In both groups, the most common tumor grade was 2, using a three-tier grading scheme.
Gene Expression Profiling
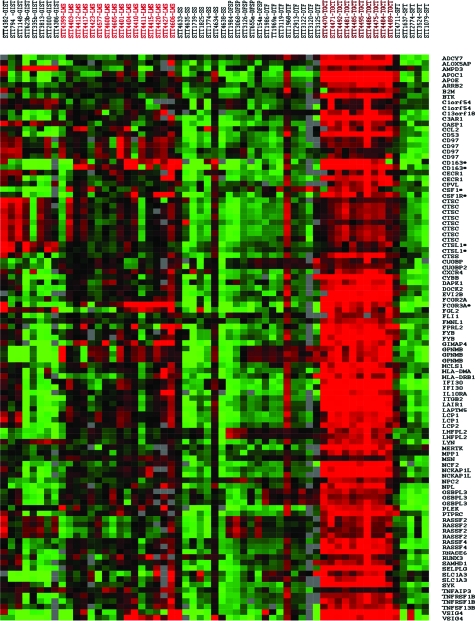
The realization that the presence of tumor-associated macrophages is correlated with poor outcome in nongynecological LMS, and that it shows a trend for statistical significance in gynecological LMS17 led us to investigate the role of CSF1, a major macrophage attractant. We had previously shown that a combination of several prognostic markers in breast carcinoma results in a robust stratification of carcinoma cases that correlates with outcome.29 Rather than focusing on the CSF1 gene alone we hypothesized that coordinated expression of multiple CSF1-associated genes might similarly generate a more potent prognosticator. The identification and generation of the list of 112 CSF1-associated genes is described in the Materials and Methods. The mRNA levels of expression for CSF1, and 71 of the 112 CSF1 response genes that passed the filtering criteria across 16 LMS, 10 TGCTs, 8 gastrointestinal stromal tumors, 8 gastrointestinal stromal tumors, 7 synovial sarcomas, 5 dermatofibrosarcoma protuberans, and 5 solitary fibrous tumors are shown in Figure 1. As expected, the majority of the genes were highly expressed in all of the TGCTs, the tumor used to select the genelist. In contrast to most other sarcomas, the LMS cases showed a variable expression for many of CSF1 response genes, and there is coordinated expression in the moderate to high expression level range of these genes in individual cases.
Figure 1.
Gene expression profiling of 59 tumors with 72 CSF1-related genes. LMS, leiomyosarcomas; GIST, gastrointestinal stromal tumors; DFSP, dermatofibrosarcoma protuberans; DTF, desmoid-type fibromatosis; SFT, solitary fibrous tumor; SS, synovial sarcomas. Asterisks identify the markers selected for immunohistochemistry and in situ hybridization. Green indicates relatively low expression level, red indicates a relatively high level of expression, gray denotes missing data.
To further evaluate this observation and to determine whether the core CSF1 response genes observed in breast carcinoma also show the highest levels of expression in LMS, we performed unsupervised hierarchical clustering of the 59 STTs including 16 LMS with the full set of 459 CSF1-related genes that passed quality-based filtering criteria. This analysis demonstrates a distinct subset of LMS that shows increased expression of a subset of the CSF1-related genes. Interestingly, the genes that are most highly expressed in this subset of LMS cases are the core CSF1 response genes identified in breast cancer as compared with the noncore TGCT-related genes (mean log2 expression ratio for core CSF1 response genes = 0.83 versus 0.16 for noncore TGCT-related genes, P < 0.0001). The expression of core CSF1 response genes in this LMS subset is significantly higher than is seen in other LMS and other STTs (mean log2 expression ratio = 0.83 in LMS with CSF1 response signature, compared with −0.15 in other LMS, and 0.26 in other STTs, P < 0.0001) (Supplemental Figure 1, see http://ajp.amjpathol.org). These findings support our hypothesis that a similar CSF1 response signature to that observed in a subset of breast carcinomas is also seen in a subset of leiomyosarcomas.
Only rare cases of synovial sarcomas and gastrointestinal stromal tumors showed coordinated expression of a significant number of CSF1 response genes, whereas the majority of gastrointestinal stromal tumors showed increased expression for only a few of the CSF1 response genes (especially CTSL1). This pattern of TGCT-related gene expression in LMS but not in others tumors suggests a potential role of this signature in LMS behavior. In general, the gene expression intensity in the LMS cases was weaker than in TGCT most likely attributable to the fact that the latter tumor is composed mostly of macrophages.
We selected markers from the list of 71 core CSF1 response genes for which antibodies were available that react in paraffin-embedded tissue to analyze two LMS TMAs with outcome data. We used commercial antibodies against FCGR3a (CD16), CTSL1, and CD163 for immunohistochemical staining of these tissue arrays; CSF1, and CSF1R were studied by in situ hybridization.
CSF1/CSF1R Expression in LMS
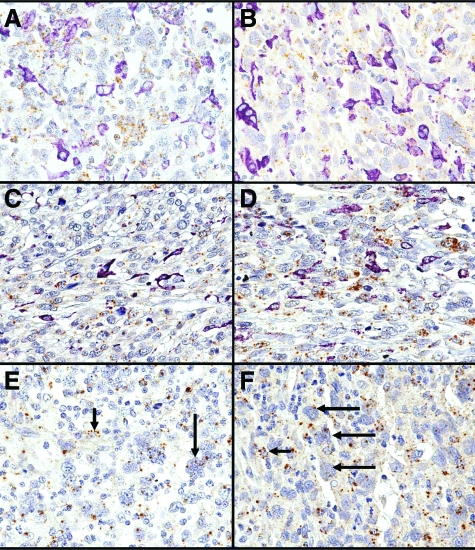
The expression of CSF1 and CSF1R was studied by in situ hybridization. CSF1 reactivity was seen in 98 LMS cases (98 of 144, 68%). In these cases, CSF1 was expressed in the tumor cells and not in the macrophages as identified by morphology and by double staining with CD163 (Figure 2, A–D). In 70 LMS cases (70 of 98, 71%) the staining was weak (1 to 4 paranuclear dot-like stains per cell in at least 5% of the cells), whereas in 28 LMS (28 of 98, 29%) the expression was strong (>5 paranuclear dot-like stains per cell in more than 5% of the cells). The number of CSF1-positive LMS cases in the nongynecological and gynecological group was 53 (53 of 71, 74%), and 45 (45 of 73, 62%) respectively (Table 2).
Figure 2.
A–D: CD163 immunohistochemistry (purple) and CSF1 in situ hybridization (brown) double staining in four different LMS cases. The tumor cells characterized by pleomorphic nuclei express multiple brown dots (CSF1 mRNA). Instead, the majority of the CD163-positive macrophages (purple) do not show CSF1 expression. E and F: CSF1R staining in two different LMS cases. The expression of CSF1R mRNA (brown dots) was seen in macrophages (short arrows), and tumor cells (long arrows). Original magnifications, ×60.
Table 2.
Staining Results from CSF1, FCGR3a, CTSL1, and CD163 in the Gynecological and Nongynecological LMS Groups
| Gynecological LMS | Nongynecological LMS | |
|---|---|---|
| CSF1 | ||
| Score 0 | 28/73 (38%) | 18/71 (25%) |
| Score 1 | NA* | NA* |
| Score 2 | 33/73 (45%) | 37/71 (52%) |
| Score 3 | 12/73 (17%) | 16/71 (23%) |
| FCGR3a | ||
| Score 0 | 25/73 (34%) | 29/72 (40%) |
| Score 1 | 16/73 (22%) | 15/72 (21%) |
| Score 2 | 21/73 (29%) | 14/72 (19%) |
| Score 3 | 11/73 (15%) | 14/72 (19%) |
| CTSL1 | ||
| Score 0 | 34/65 (52%) | 24/64 (38%) |
| Score 1 | 10/65 (15%) | 11/64 (17%) |
| Score 2 | 13/65 (20%) | 18/64 (28%) |
| Score 3 | 8/65 (12%) | 11/64 (17%) |
| CD163 | ||
| Score 0 | 8/76 (10%) | 10/73 (14%) |
| Score 1 | NA* | NA* |
| Score 2 | 34/76 (45%) | 32/73 (44%) |
| Score 3 | 34/76 (45%) | 31/73 (42%) |
Not applicable.
CSF1R could be evaluated in only 89 LMS cases because of the high background in many samples. In contrast to CSF1, CSF1R was expressed both in the tumor cells and macrophages (Figure 2, E and F). CSF1R was expressed in 46 LMS (46 of 89, 52%). In the majority of the CSF1R-positive cases, CSF1 was also expressed (38 of 46, 83%).
FCGR3a Expression in LMS
FCGR3a (CD16) is a receptor for the Fc portion of IgG. It is expressed in macrophages, natural killer cells, and neutrophils.30 The staining pattern was membranous and cytoplasmic (Figure 3). The majority of FCGR3a-positive cells showed typical cytological features of macrophages with abundant (foamy) cytoplasm with a dendritic pattern, and round or oval nuclei with small conspicuous nucleoli. These cells showed no staining for CD56 nor CD57 supporting the interpretation that the FCGR3a-positive cells are truly macrophages instead of natural killer cells (Supplemental Figure 2, see http://ajp.amjpathol.org). FCGR3a was expressed in 91 LMS cases (91 of 145, 63%). In 31 LMS cases (31 of 91, 34%) the numbers of positive cells qualified as score 1, 35 cases (35 of 91, 38%) score 2, and 25 (19 of 91, 19%) LMS cases score 3. The number of FCGR3a-positive cases was similar in the nongynecological and gynecological LMS (43 of 72, 60% versus 48 of 73, 65%) (Table 2).
Figure 3.
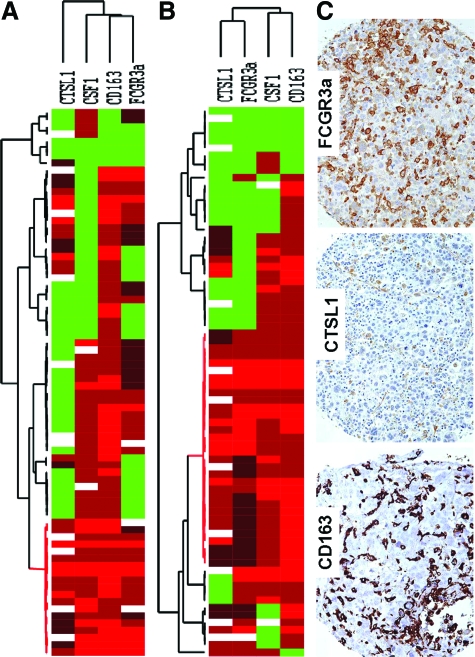
Hierarchical cluster analysis of immunohistochemistry for FCGR3a, CTSL1, CD163, and in situ hybridization for CSF1 in the gynecological (A) and nongynecological (B) LMS. Each column represents a different tumor and each row a different marker. The red segment of the dendrogram indicates a group of cases positive for all markers with interpretable data. C: Stain of a representative case with CD163, CTSL1, and FCGR3a immunohistochemical markers. Green, score 0; black, score1; brown, score 2; red, score 3; and white, missing data.
CTSL1 Expression in LMS
Cathepsin L (CTSL1) is a lysosomal cysteine protease highly expressed in macrophages, where it is responsible for the degradation and turnover of intracellular proteins.31 CTSL1 was also reported to be expressed in a variety of epithelial tumor cells and is thought to be important in tumor progression.32,33,34 In the LMS cases the staining pattern was granular and cytoplasmic and was expressed predominantly in cells with morphological features of macrophages, as described above (Figure 3). In two cases, the tumor cells also expressed CTSL1. For scoring we only considered the CTSL1 expression in the macrophages. Of 129 LMS cases, 71 (55%) were positive for CTSL1. The majority of the positive cases showed a score 2 staining pattern (31 of 71, 44%). In 21 cases (21 of 71, 29%) the score was 1, and in 19 cases (19 of 71, 28%) the score was 3. The LMS cases positive for CTSL1 in the nongynecological and gynecological groups were 40 (40 of 64, 62%) and 31 (31 of 65, 48%), respectively.
CD163 Expression in LMS
As described previously,17 CD163 macrophages were present in the majority of the LMS cases, but different numbers of macrophages were present in different tumors. CD163 was positive in the majority of LMS cases (131 of 149, 88%), and in most the cases more than 25 CD163-positive cells per 0.6 mm tumor core were present (65 of 149, 43%). In the nongynecological LMS group, 63 cases (63 of 73, 86%) were positive [(score 2: 32 of 73 (44%) and score 3: 31 of 73 (42%)] for CD163, whereas in the gynecological LMS group, 68 cases (68 of 76, 89%) were positive [(score 2: 34 of 76 (45%) and score 3: 34 of 76 (45%)] (Table 2).
Co-Expression of CSF1, FCGR3a, CTSL1, and CD163 in LMS
To study the coordinated expression of CSF1- and TGCT-related proteins, we analyzed 125 LMS cases for which stained cores could be evaluated for all four markers studied. Of those 125 LMS cases, 62 cases were gynecological and 63 nongynecologic. Unsupervised hierarchical clustering analysis (average linkage method) with CSF1, FCGR3a, CTSL1, and CD163 identified a distinctive group characterized by co-expression of these four markers in gynecological and nongynecological LMS (Figure 3). Fourteen LMS cases (14 of 62, 23%) in the gynecological group and 28 cases (28 of 63, 44%) in the nongynecological LMS showed a coordinated expression of CSF1, FCGR3a, CTSL1, and CD163.
Association of CSF1, FCGR3a, CTSL1, and CD163 with Prognosis
Kaplan-Meier analysis for individual antibodies and the CSF1 in situ hybridization probe showed variable degrees of correlation with outcome in the two LMS groups (Supplemental Figure 3, see http://ajp.amjpathol.org). All individual markers showed at least a trend for correlation with poor outcome in both gynecological and nongynecological LMS. In the case of CD163, as previously demonstrated,17 only the nongynecological LMS showed a significant association with poor survival.
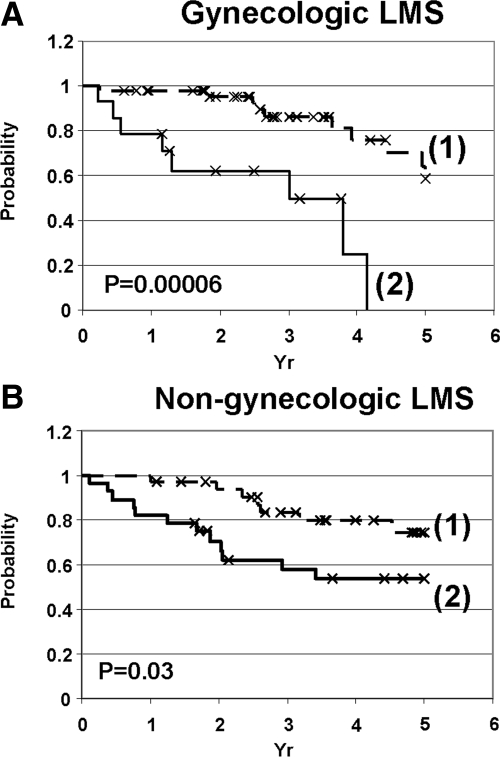
The co-expression of CSF1 and the three TGCT-related markers (FCGR3a, CTSL1, and CD163) identified 14 gynecological leiomyosarcomas and 28 nongynecological leiomyosarcomas. These patients had a worse 5-year DSS than those who did not co-express the four markers. In fact, all 14 patients with gynecological LMS died within 5 years (0% versus 58%; P = 0.00006 in the gynecological group, and 53% versus 74%; P = 0.03 in the nongynecological LMS cases) (Figure 4, A and B).
Figure 4.
Kaplan-Meier analysis for those LMS cases that had interpretable stain for all four markers (FCGR3a, CTSL1, CSF1, and CD163) in the gynecological (A) and nongynecological (B) LMS. 1: LMS with negative stain for one or more markers. 2: LMS-positive for all four markers.
In gynecological LMS, mitotic index and tumor size were also associated with poor survival in a univariate analysis (Table 3). Only the co-expression of CSF1, CTSL1, FCGR3a, and CD163 in the LMS cases was independently associated with a worse DSS in a multivariate analysis (hazard ratio for death, 4.2; 95% CI, 1.12 to 16; P = 0.03). We used a forward stepwise technique of automated model building to create a Cox regression model based on the subset of features most predictive of prognosis. In this analysis in gynecological LMS, the CSF1 response signature (hazard ratio for death, 3.5; 95% CI, 1.1 to 11.0; P = 0.04) and mitotic index (hazard ratio for death, 2.1; 95% CI 1.0 to 4.5; P = 0.05) were selected as the strongest predictors of DSS and the other traditional clinicopathological features were not included in the model.
Table 3.
Univariate Analysis of Prognostic Factors for DSS in the Gynecological and Nongynecological LMS
| Factors | Gynecological LMS (76)
|
P | Nongynecological LMS (73)
|
P |
|---|---|---|---|---|
| 5DSS Rate % (number of cases) | 5DSS Rate % (number of cases) | |||
| Age | ||||
| <50 | 53 (36) | NS (0.8) | 61 (29) | NS (0.8) |
| ≥50 | 56 (40) | 64 (44) | ||
| Tumor size | ||||
| ≤10 cm | 71 (44) | 0.03 | 73 (45) | 0.02 |
| >10 cm | 33 (25) | 44 (24) | ||
| Histological grade* | ||||
| 1 | 37 (20) | NS (0.9) | 83 (19) | NS (0.15) |
| 2 | 62 (38) | 66 (34) | ||
| 3 | 59 (18) | 40 (12) | ||
| Mitotic index | ||||
| 1 | 62 (23) | 0.003 | 82 (24) | NS (0.3) |
| 2 | 95 (21) | 63 (24) | ||
| 3 | 33 (32) | 53 (17) | ||
| Necrosis | ||||
| 0 | 53 (45) | NS (0.7) | 68 (43) | NS (0.7) |
| 1 | 55 (26) | 55 (16) | ||
| 2 | 66 (5) | 75 (6) | ||
| CSF1 signature† | ||||
| Positive | 0 (14) | 6 × 10−5 | 53 (28) | 0.03 |
| Negative | 58 (50) | 74 (35) |
FNCLCC grading system was used to classify nongynecological LMS. We classified the gynecological LMS based on cellular differentiation.
LMS that co-expressed all four markers (CSF1, CTSL1, FCGR3a, and CD163) were considered CSF1 signature-positive. LMS that failed to express at least one or more of the four markers were called CSF1 signature-negative.
5DSS, 5-year disease-specific survival rate in percentage (number of cases); NS, not statistically significant.
In the nongynecological LMS, tumor size, and the coordinated expression of CSF1, CTSL1, FCGR3a, and CD163 were associated with DSS in a univariate analysis. As previously described, in this set of nongynecological LMS, the FNCLCC grading system did not predict clinical outcome (Table 3).17 In the multivariate analysis of nongynecological LMS incorporating age, size, grade, mitotic index, necrosis, cellular differentiation, and the CSF1 response signature, no features in the model were significantly predictive of prognosis. However, using the forward stepwise approach of automated model building, the CSF1 response signature was selected as the feature most predictive of outcome (hazard ratio for death, 3.9; 95% CI 1.2 to 12.6; P = 0.03) and was the only feature included in the Cox regression model. Because of the limited clinical information in our LMS cases we were unable to include tumor stage in the survival analysis.
Finally, to see if a smaller number of CSF1 response markers could predict outcome as well as the CSF1 response signature we performed a Cox regression analysis with forward step-wise variable selection using likelihood ratio. The input variables were: CSF1 response signature, CSF1, FCGR3a, CTSL1, and CD163. This analysis showed that in gynecological LMS, the CSF1 response signature is significantly better at predicting survival than any of the markers alone (or a smaller combination) (hazard ratio for death, 6.4; 95% CI 2.3 to 18.1; P = 0.0005). In the multivariate model for nongynecological LMS, only CSF1 was chosen as the most predictive of outcome (hazard ratio for death, 6.99; 95% CI 0.93 to 52.4; P = 0.06). The coordinate expression of the four genes in a subset of LMS cases associated with poor outcome indicates a functional relationship. In fact, each of the four markers shows significant correlation in expression in pairwise analyses, all with P < 0.019 as shown in Supplementary Table 6 (see http://ajp.amjpathol.org).
These results show that the presence of CSF1 mRNA in the tumor cells and coordinate expression of the CSF1-associated genes (CTSL1, FCGR3a, CD163) in the tumor stroma is an indicator of poor prognosis in LMS. In our previous study the correlation between outcome and the number of histiocytes did not reach statistical significance in gynecological LMS.17 Using CSF1 and three TGCT related markers we now find statistically significant differences in survival in both the gynecological and nongynecological LMS.
Discussion
Recently we reported that macrophage infiltrates, as detected by CD163 and CD68 expression, are associated with poor outcome in LMS.17 In that report we focused primarily on genes known to be expressed by macrophages. In an effort to further evaluate the role of tumor-associated macrophages in LMS we focused in the current study on a major biological chemoattractant for macrophages, CSF1. CSF1 (M-CSF) is a cytokine that binds to the tyrosine kinase receptor CSF1R and contributes to macrophage recruitment. Here we show that CSF1 is expressed by the LMS tumor cells, and CSF1R is expressed in both macrophages and cancer cells.
In addition, we sought to evaluate the expression of other genes whose expression might be driven by CSF1 in LMS. We previously defined a CSF1 response gene set by identifying genes highly and specifically expressed in TGCT, a lesion driven by a chromosomal translocation involving CSF1,18 and showed that a core subset of these genes show coordinate expression in the breast carcinoma microenvironment.20 We have labeled this core set of 112 genes, the “CSF1 response gene set.” In the current study, we show that this CSF1 core response gene set is variably expressed only in LMS but not in several other sarcomas. We have evaluated the expression of CSF1 and three genes from the core response gene set (FCGR3a, CD163, and CTSL1) on LMS TMAs. The use of multiple prognostic markers has been proven to be superior to the use of a single marker in several cancers.29,35 Univariate analysis of disease-specific survival for CSF1, FCGR3a, CD163, and CTSL1 showed that with the exception of CTSL1, none of these markers alone showed significant correlation with survival for both types of LMS. In contrast, the coordinate expression of CSF1 and the three CSF1 response genes was significantly associated with poor survival for both gynecological and nongynecological LMS, and the coordinate expression of the four markers showed stronger association with survival than CTSL1 alone.
The majority of the CSF1 response genes are related to inflammation and immune response.20 In fact, through ontological analysis we found that most of the genes were linked to groups related to inflammation including response to biotic stimulus group, defense response group, immune response group, response to pest, pathogen, or parasite group, and response to other organism group (all groups with a P < 9.92E-16). Additional detailed information can be downloaded from the supplemental information from Beck and colleagues.20
Filtering the TGCT-related genes through breast carcinoma before applying the reduced core CSF1 response signature to the LMS, may result in the identification of a core gene signature that is present in breast cancer but not necessarily present in LMS. To evaluate this possibility, we performed unsupervised hierarchical clustering of the 59 soft tissue sarcomas including 16 LMS with the full list of 459 TGCT-related genes that passed filtering criteria. This analysis demonstrates a distinct cluster of LMS cases that is defined by increased expression of the core CSF1 response genes identified previously in breast cancer with significantly increased expression of the core CSF1 response genes compared with the noncore TGCT-related genes. These findings support our hypothesis that the CSF1 response signature observed in a subset of breast carcinoma is also seen in a subset of leiomyosarcomas.
Based on our findings, we propose a model of macrophage-tumor interaction in LMS where CSF1 plays a central role. CSF1 is produced by the leiomyosarcoma tumor cells and plays an important role in attraction of CD163+FCGR3a+CTSL1+ macrophages into the tumor (paracrine effect), although other cytokines can also be involved in this function, such as CCL236 (which is also present in the CSF1 core response gene set). Subsequently these macrophages may produce cytokines that promote tumor growth, angiogenesis, extracellular matrix breakdown, invasion, and metastasis.1,2 CSF1 may also promote tumor growth through the interaction with CSF1R on the tumor cells (autocrine effect). Both the macrophage cytokines and CSF1 may be necessary to produce an adverse effect in the LMS patients. This model can also be implicated not only in breast carcinoma and leiomyosarcoma but also in other tumors. However, the presence of the same host response in different tumors does not necessarily imply same clinical behavior. In fact, macrophage infiltration can be associated with different outcome depending on the tumor type3,4,5,6,7,8,9 Alternatively, there is data suggesting that different macrophages may mediate opposite effects such as activity against tumor cells versus tumor progression (angiogenesis).37
The potential role of the CSF1 pathway in LMS pathogenesis has important clinical implications, because CSF1 can now be targeted through anti-CSF1 antibodies or CSF1R inhibitors.38 Recently, imatinib was shown to inhibit CSF1R and was successfully used to treat a patient with relapsing pigmented villonodular synovitis/TGCT.39 These observations suggest the potential therapeutic utility of measuring expression of CSF1 and the three TGCT-related proteins in LMS tumor samples to identify patients most likely to respond to CSF1-targeted therapies.
The most frequently used grading systems for soft tissue sarcomas are those defined by the National Cancer Institute and the FNCLCC.40 Little is known about the performance of these grading systems within soft tissue leiomyosarcomas. Coindre and colleagues41 showed that the FNCLCC grading system was the strongest predictor for metastasis in pleomorphic sarcomas, unclassified sarcomas and synovial sarcomas. However in 148 cases of leiomyosarcoma studied, they found that the FNCLCC grading system performed less well, with bone or neurovascular involvement forming a more powerful predictor while tumor size had a similar prognostic effect on outcome as grade.41 In contrast, Gustafson and colleagues42,43 found that vascular invasion and necrosis were independent predictors for poor prognosis in soft tissue LMS, but grade did not correlate with outcome. In the series of 73 cases of soft tissue leiomyosarcomas that are the subject of this study, the FNCLCC grading system did not correlate with outcome.17 Therefore, the ability to predict outcome in nongynecological LMS using the CSF1 core genes described in this study may aid clinicians in the management of these tumors. The FNCLCC grading system was not designed for gynecological LMS. The most important prognostic factor for uterine LMS is tumor stage.44,45 Other factors such as mitotic index remain controversial. Pautier and colleagues44 found that mitotic index was the second best prognostic marker, after stage, in a multivariate analysis with 78 uterine LMS. Mayerhofer and colleagues45 studied 71 uterine LMS and the mitotic index did not correlate with survival in a multivariate analysis with stage (FIGO), age, and mitotic count. In the 76 gynecological LMS cases studied in the current report, the mitotic count correlated with survival (P = 0.003, Table 3) in a univariate analysis. However, it was not independently associated with survival in a multivariate analysis. The coordinated expression of CSF1 and the three CSF1 related proteins was the only factor associated with clinical outcome in multivariate analysis.
In this study, we have shown that the co-expression of CSF1 and CSF1 response proteins (CD163, FCGR3a, and CTSL1) is correlated with poor prognosis in both nongynecological and gynecological LMS. This finding offers insight into the pathogenesis of LMS and suggests that the measurement of CSF1 and CSF1 response proteins, if confirmed on an independent sample set in LMS, could be useful in clinical practice to identify those LMS patients with the highest risk of poor outcome. Identification of these patients may aid clinicians in determining which patients require more aggressive treatment. Furthermore, this is the first study to demonstrate that CSF1 is produced by LMS tumor cells in a subset of cases suggesting that CSF1 production by LMS tumor cells plays an important role in tumor-associated macrophage recruitment. Conceivably, measurement of the expression of CSF1 and CSF1 response genes in LMS could assist in not only predicting prognosis in LMS but also identifying patients most likely to respond to therapies targeted at CSF1 and the CSF1 response pathway.
Supplementary Material
Footnotes
Address reprint requests to Matt van de Rijn, Dept. of Pathology, L-235, Stanford University Medical Center, 300 Pasteur Dr., Stanford, CA 94305. E-mail: mrijn@stanford.edu.
Supported by the National Institutes of Health (grant CA112270), the National Leiomyosarcoma Foundation, the LMSarcoma Direct Research Foundation, and the British Columbia Cancer Agency’s Musculoskeletal Tumor Group (to the University of British Columbia).
I.E. and A.H.B. contributed equally to this study.
Supplemental material for this article can be found on http://ajp.amjpathol.org.
References
- Lewis CE, Pollard JW. Distinct role of macrophages in different tumor microenvironments. Cancer Res. 2006;66:605–612. doi: 10.1158/0008-5472.CAN-05-4005. [DOI] [PubMed] [Google Scholar]
- Condeelis J, Pollard JW. Macrophages: obligate partners for tumor cell migration, invasion, and metastasis. Cell. 2006;124:263–266. doi: 10.1016/j.cell.2006.01.007. [DOI] [PubMed] [Google Scholar]
- Tsutsui S, Yasuda K, Suzuki K, Tahara K, Higashi H, Era S. Macrophage infiltration and its prognostic implications in breast cancer: the relationship with VEGF expression and microvessel density. Oncol Rep. 2005;14:425–431. [PubMed] [Google Scholar]
- Lissbrant IF, Stattin P, Wikstrom P, Damber JE, Egevad L, Bergh A. Tumor associated macrophages in human prostate cancer: relation to clinicopathological variables and survival. Int J Oncol. 2000;17:445–451. doi: 10.3892/ijo.17.3.445. [DOI] [PubMed] [Google Scholar]
- Ohno S, Ohno Y, Suzuki N, Kamei T, Koike K, Inagawa H, Kohchi C, Soma G, Inoue M. Correlation of histological localization of tumor-associated macrophages with clinicopathological features in endometrial cancer. Anticancer Res. 2004;24:3335–3342. [PubMed] [Google Scholar]
- Hanada T, Nakagawa M, Emoto A, Nomura T, Nasu N, Nomura Y. Prognostic value of tumor-associated macrophage count in human bladder cancer. Int J Urol. 2000;7:263–269. doi: 10.1046/j.1442-2042.2000.00190.x. [DOI] [PubMed] [Google Scholar]
- Hamada I, Kato M, Yamasaki T, Iwabuchi K, Watanabe T, Yamada T, Itoyama S, Ito H, Okada K. Clinical effects of tumor-associated macrophages and dendritic cells on renal cell carcinoma. Anticancer Res. 2002;22:4281–4284. [PubMed] [Google Scholar]
- Koide N, Nishio A, Sato T, Sugiyama A, Miyagawa S. Significance of macrophage chemoattractant protein-1 expression and macrophage infiltration in squamous cell carcinoma of the esophagus. Am J Gastroenterol. 2004;99:1667–1674. doi: 10.1111/j.1572-0241.2004.30733.x. [DOI] [PubMed] [Google Scholar]
- Farinha P, Masoudi H, Skinnider BF, Shumansky K, Spinelli JJ, Gill K, Klasa R, Voss N, Connors JM, Gascoyne RD. Analysis of multiple biomarkers shows that lymphoma-associated macrophage (LAM) content is an independent predictor of survival in follicular lymphoma (FL). Blood. 2005;106:2169–2174. doi: 10.1182/blood-2005-04-1565. [DOI] [PubMed] [Google Scholar]
- Forssell J, Oberg A, Henriksson ML, Stenling R, Jung A, Palmqvist R. High macrophage infiltration along the tumor front correlates with improved survival in colon cancer. Clin Cancer Res. 2007;13:1472–1479. doi: 10.1158/1078-0432.CCR-06-2073. [DOI] [PubMed] [Google Scholar]
- Ohno S, Inagawa H, Dhar DK, Fujii T, Ueda S, Tachibana M, Suzuki N, Inoue M, Soma G, Nagasue N. The degree of macrophage infiltration into the cancer cell nest is a significant predictor of survival in gastric cancer patients. Anticancer Res. 2003;23:5015–5022. [PubMed] [Google Scholar]
- Piras F, Colombari R, Minerba L, Murtas D, Floris C, Maxia C, Corbu A, Perra MT, Sirigu P. The predictive value of CD8, CD4, CD68, and human leukocyte antigen-D-related cells in the prognosis of cutaneous malignant melanoma with vertical growth phase. Cancer. 2005;104:1246–1254. doi: 10.1002/cncr.21283. [DOI] [PubMed] [Google Scholar]
- Murdoch C, Giannoudis A, Lewis CE. Mechanisms regulating the recruitment of macrophages into hypoxic areas of tumors and other ischemic tissues. Blood. 2004;104:2224–2234. doi: 10.1182/blood-2004-03-1109. [DOI] [PubMed] [Google Scholar]
- Lin EY, Nguyen AV, Russell RG, Pollard JW. Colony-stimulating factor 1 promotes progression of mammary tumors to malignancy. J Exp Med. 2001;193:727–740. doi: 10.1084/jem.193.6.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kacinski BM. CSF-1 and its receptor in breast carcinomas and neoplasms of the female reproductive tract. Mol Reprod Dev. 1997;46:71–74. doi: 10.1002/(SICI)1098-2795(199701)46:1<71::AID-MRD11>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Chambers SK, Kacinski BM, Ivins CM, Carcangiu ML. Overexpression of epithelial macrophage colony-stimulating factor (CSF-1) and CSF-1 receptor: a poor prognostic factor in epithelial ovarian cancer, contrasted with a protective effect of stromal CSF-1. Clin Cancer Res. 1997;3:999–1007. [PubMed] [Google Scholar]
- Lee CH, Espinosa I, Vrijaldenhoven S, Subramanian S, Montgomery KD, Zhu S, Marinelli RJ, Peterse JL, Poulin N, Nielsen TO, West RB, Gilks CB, van de Rijn M. Prognostic significance of macrophage infiltration in leiomyosarcomas. Clin Cancer Res. 2008;14:1423–1430. doi: 10.1158/1078-0432.CCR-07-1712. [DOI] [PubMed] [Google Scholar]
- West RB, Rubin BP, Miller MA, Subramanian S, Kaygusuz G, Montgomery K, Zhu S, Marinelli RJ, De Luca A, Downs-Kelly E, Goldblum JR, Corless CL, Brown PO, Gilks CB, Nielsen TO, Huntsman D, van de Rijn M. A landscape effect in tenosynovial giant-cell tumor from activation of CSF1 expression by a translocation in a minority of tumor cells. Proc Natl Acad Sci USA. 2006;103:690–695. doi: 10.1073/pnas.0507321103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cupp JS, Miller MA, Montgomery KD, Nielsen TO, O'Connell JX, Huntsman D, van de Rijn M, Gilks CB, West RB. Translocation and expression of CSF1 in pigmented villonodular synovitis, tenosynovial giant cell tumor, rheumatoid arthritis and other reactive synovitides. Am J Surg Pathol. 2007;31:970–976. doi: 10.1097/PAS.0b013e31802b86f8. [DOI] [PubMed] [Google Scholar]
- Beck AH, Espinosa I, Edris B, Li R, Montgomery K, Zhu S, Varma S, Marinelli RJ, van de Rijn M, West RB. The macrophage colony-stimulating factor 1 response signature in breast carcinoma. Clin Cancer Res. 2009;15:778–787. doi: 10.1158/1078-0432.CCR-08-1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillou L, Coindre JM, Bonichon F, Nguyen BB, Terrier P, Collin F, Vilain MO, Mandard AM, Le Doussal V, Leroux A, Jacquemier J, Duplay H, Sastre-Garau X, Costa J. Comparative study of the National Cancer Institute and French Federation of Cancer Centers Sarcoma Group grading systems in a population of 410 adult patients with soft tissue sarcoma. J Clin Oncol. 1997;15:350–362. doi: 10.1200/JCO.1997.15.1.350. [DOI] [PubMed] [Google Scholar]
- Nielsen TO, West RB, Linn SC, Alter O, Knowling MA, O'Connell JX, Zhu S, Fero M, Sherlock G, Pollack JR, Brown PO, Botstein D, van de Rijn M. Molecular characterisation of soft tissue tumours: a gene expression study. Lancet. 2002;359:1301–1307. doi: 10.1016/S0140-6736(02)08270-3. [DOI] [PubMed] [Google Scholar]
- St Croix B, Rago C, Velculescu V, Traverso G, Romans KE, Montgomery E, Lal A, Riggins GJ, Lengauer C, Vogelstein B, Kinzler KW. Genes expressed in human tumor endothelium. Science. 2000;289:1197–1202. doi: 10.1126/science.289.5482.1197. [DOI] [PubMed] [Google Scholar]
- Iacobuzio-Donahue CA, Ryu B, Hruban RH, Kern SE. Exploring the host desmoplastic response to pancreatic carcinoma: gene expression of stromal and neoplastic cells at the site of primary invasion. Am J Pathol. 2002;160:91–99. doi: 10.1016/S0002-9440(10)64353-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West RB, Corless CL, Chen X, Rubin BP, Subramanian S, Montgomery K, Zhu S, Ball CA, Nielsen TO, Patel R, Goldblum JR, Brown PO, Heinrich MC, van de Rijn M. The novel marker, DOG1, is expressed ubiquitously in gastrointestinal stromal tumors irrespective of KIT or PDGFRA mutation status. Am J Pathol. 2004;165:107–113. doi: 10.1016/S0002-9440(10)63279-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinelli RJ, Montgomery K, Liu CL, Shah NH, Prapong W, Nitzberg M, Zachariah ZK, Sherlock GJ, Natkunam Y, West RB, van de Rijn M, Brown PO, Ball CA. The Stanford Tissue Microarray Database. Nucleic Acids Res. 2008;6:D871–D877. doi: 10.1093/nar/gkm861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CL, Montgomery KD, Natkunam Y, West RB, Nielsen TO, Cheang MC, Turbin DA, Marinelli RJ, van de Rijn M, Higgins JP. TMA-Combiner, a simple software tool to permit analysis of replicate cores on tissue microarrays. Mod Pathol. 2005;18:1641–1648. doi: 10.1038/modpathol.3800491. [DOI] [PubMed] [Google Scholar]
- Liu CL, Prapong W, Natkunam Y, Alizadeh A, Montgomery K, Gilks CB, van de Rijn M. Software tools for high-throughput analysis and archiving of immunohistochemistry staining data obtained with tissue microarrays. Am J Pathol. 2002;161:1557–1565. doi: 10.1016/S0002-9440(10)64434-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makretsov NA, Huntsman DG, Nielsen TO, Yorida E, Peacock M, Cheang MC, Dunn SE, Hayes M, van de Rijn M, Bajdik C, Gilks CB. Hierarchical clustering analysis of tissue microarray immunostaining data identifies prognostically significant groups of breast carcinoma. Clin Cancer Res. 2004;10:6143–6151. doi: 10.1158/1078-0432.CCR-04-0429. [DOI] [PubMed] [Google Scholar]
- Iannello A, Ahmad A. Role of antibody-dependent cell-mediated cytotoxicity in the efficacy of therapeutic anti-cancer monoclonal antibodies. Cancer Metastasis Rev. 2005;24:487–499. doi: 10.1007/s10555-005-6192-2. [DOI] [PubMed] [Google Scholar]
- Menzel K, Hausmann M, Obermeier F, Schreiter K, Dunger N, Bataille F, Falk W, Scholmerich J, Herfarth H, Rogler G. Cathepsins B, L and D in inflammatory bowel disease macrophages and potential therapeutic effects of cathepsin inhibition in vivo. Clin Exp Immunol. 2006;146:169–180. doi: 10.1111/j.1365-2249.2006.03188.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strojnik T, Zidanik B, Kos J, Lah TT. Cathepsins B and L are markers for clinically invasive types of meningiomas. Neurosurgery. 2001;48:598–605. doi: 10.1097/00006123-200103000-00029. [DOI] [PubMed] [Google Scholar]
- Chauhan SS, Goldstein LJ, Gottesman MM. Expression of cathepsin L in human tumors. Cancer Res. 1991;51:1478–1481. [PubMed] [Google Scholar]
- Dohchin A, Suzuki JI, Seki H, Masutani M, Shiroto H, Kawakami Y. Immunostained cathepsins B and L correlate with depth of invasion and different metastatic pathways in early stage gastric carcinoma. Cancer. 2000;89:482–487. [PubMed] [Google Scholar]
- Alkushi A, Clarke BA, Akbari M, Makretsov N, Lim P, Miller D, Magliocco A, Coldman A, van de Rijn M, Huntsman D, Parker R, Gilks CB. Identification of prognostically relevant and reproducible subsets of endometrial adenocarcinoma based on clustering analysis of immunostaining data. Mod Pathol. 2007;20:1156–1165. doi: 10.1038/modpathol.3800950. [DOI] [PubMed] [Google Scholar]
- Colombo MP, Mantovani A. Targeting myelomonocytic cells to revert inflammation-dependent cancer promotion. Cancer Res. 2005;65:9113–9116. doi: 10.1158/0008-5472.CAN-05-2714. [DOI] [PubMed] [Google Scholar]
- Allavena P, Sica A, Garlanda C, Mantovani A. The Yin-Yang of tumor-associated macrophages in neoplastic progression and immune surveillance. Immunol Rev. 2008;222:155–161. doi: 10.1111/j.1600-065X.2008.00607.x. [DOI] [PubMed] [Google Scholar]
- Conway JG, McDonald B, Parham J, Keith B, Rusnak DW, Shaw E, Jansen M, Lin P, Payne A, Crosby RM, Johnson JH, Frick L, Lin MH, Depee S, Tadepalli S, Votta B, James I, Fuller K, Chambers TJ, Kull FC, Chamberlain SD, Hutchins JT. Inhibition of colony-stimulating-factor-1 signaling in vivo with the orally bioavailable cFMS kinase inhibitor GW2580. Proc Natl Acad Sci USA. 2005;102:16078–16083. doi: 10.1073/pnas.0502000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blay JY, El Sayadi H, Thiesse P, Garret J, Ray-Coquard I. Complete response to imatinib in relapsing pigmented villonodular synovitis/tenosynovial giant cell tumor (PVNS/TGCT). Ann Oncol. 2008;19:821–822. doi: 10.1093/annonc/mdn033. [DOI] [PubMed] [Google Scholar]
- Fletcher CDM, Unni KK, Mertens F. Pathology and genetics of tumours of soft tissue and bone. Lyon: IARC Press,; World Health Organisation Classification of Tumors. 2002:pp 131–134. [Google Scholar]
- Coindre JM, Terrier P, Guillou L, Le Doussal V, Collin F, Ranchere D, Sastre X, Vilain MO, Bonichon F, N′Guyen Bui B. Predictive value of grade for metastasis development in the main histologic types of adult soft tissue sarcomas: a study of 1240 patients from the French Federation of Cancer Centers Sarcoma Group. Cancer. 2001;91:1914–1926. doi: 10.1002/1097-0142(20010515)91:10<1914::aid-cncr1214>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- Gustafson P, Willen H, Baldetorp B, Ferno M, Akerman M, Rydholm A. Soft tissue leiomyosarcoma. A population-based epidemiologic and prognostic study of 48 patients, including cellular DNA content. Cancer. 1992;70:114–119. doi: 10.1002/1097-0142(19920701)70:1<114::aid-cncr2820700119>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- Gustafson P. Soft tissue sarcoma. Epidemiology and prognosis in 508 patients. Acta Orthop Scand Suppl. 1994;259:1–31. [PubMed] [Google Scholar]
- Pautier P, Genestie C, Rey A, Morice P, Roche B, Lhomme C, Haie-Meder C, Duvillard P. Analysis of clinicopathologic prognostic factors for 157 uterine sarcomas and evaluation of a grading score validated for soft tissue sarcoma. Cancer. 2000;88:1425–1431. [PubMed] [Google Scholar]
- Mayerhofer K, Obermair A, Windbichler G, Petru E, Kaider A, Hefler L, Czerwenka K, Leodolter S, Kainz C. Leiomyosarcoma of the uterus: a clinicopathologic multicenter study of 71 cases. Gynecol Oncol. 1999;74:196–201. doi: 10.1006/gyno.1999.5436. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.