Abstract
The peroxisome proliferator-activated receptors (PPARs) are nuclear hormone receptors that regulate glucose and lipid homeostasis. The PPARγ subtype plays a central role in the regulation of adipogenesis and is the molecular target for the 2,4-thiazolidinedione class of antidiabetic drugs. Structural studies have revealed that agonist ligands activate the PPARs through direct interactions with the C-terminal region of the ligand-binding domain, which includes the activation function 2 helix. GW0072 was identified as a high-affinity PPARγ ligand that was a weak partial agonist of PPARγ transactivation. X-ray crystallography revealed that GW0072 occupied the ligand-binding pocket by using different epitopes than the known PPAR agonists and did not interact with the activation function 2 helix. In cell culture, GW0072 was a potent antagonist of adipocyte differentiation. These results establish an approach to the design of PPAR ligands with modified biological activities.
The nuclear hormone receptors are ligand-activated transcription factors that regulate target genes essential for mammalian physiology and development (1). The peroxisome proliferator-activated receptors (PPARs) are nuclear receptors activated by fatty acids and their eicosanoids metabolites, which regulate genes involved in the biosynthesis, storage, and metabolism of these ligands (2). The pharmacology of synthetic PPAR ligands demonstrated the role of these receptors in regulating glucose and lipid homeostasis and established their utility as molecular targets for the development of drugs for the treatment of diabetes and cardiovascular disease (3).
Biochemical and structural studies with several nuclear receptors revealed that hormone binding induces allosteric changes in the conformation of the ligand-binding domain, which promote recruitment of transcriptional coactivator proteins such as steroid receptor coactivator 1 (SRC1) (4) and CREB binding protein (CBP) (5). We recently reported x-ray crystallographic analysis of the ternary complex of PPARγ with the 2,4-thiazolidinedione (TZD) rosiglitazone (Fig. 1A) and the coactivator SRC1 (6), as well as the complexes of PPARδ with either the fibrate GW2433 or the essential fatty acid eicosapentaenoic acid (7). Despite differences in their gross chemical structure, all of these small molecule PPAR agonists share a common binding mode, in which the acidic head groups form a network of hydrogen bonds with Y473, H449, and H323 within the ligand-binding pocket. These interactions stabilize a charge clamp (6) between the C-terminal activation function 2 (AF-2) helix and a conserved lysine residue on the surface of the receptor, through which coactivator proteins are recruited to the receptor.
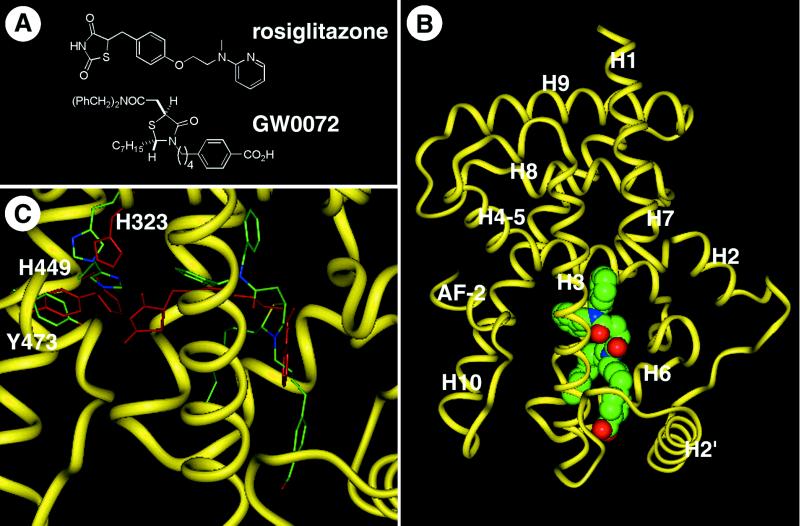
Figure 1.
GW0072 is a PPARγ ligand with a unique binding mode. (A) Chemical structures of the TZD rosiglitazone and the thiazolidine acetamide GW0072. (B) Cocrystal structure of the PPARγ ligand-binding domain with GW0072. The polypeptide backbone is illustrated as a yellow worm schematic, and GW0072 is shown as a Van der Waals space-filling representation with each atom type colored: carbon, green; oxygen, red; nitrogen, blue; and sulfur, yellow. (C) The PPARγ polypeptide backbone atoms from the cocrystal structure with GW0072 superimposed on the cocrystal structure with rosiglitazone (6). The polypeptide backbone of the GW0072 cocrystal structure is shown as a thin yellow ribbon; GW0072 as well as H323, H449, and Y473 of the protein complex are shown as carbon, green; oxygen, red; nitrogen, blue; and sulfur, yellow. Rosiglitazone and the same three amino acids for its respective protein complex are colored red. Differences between the rosiglitazone and GW0072 cocrystal structures exist in the side-chain amino acid positions and not in the overall polypeptide conformation except for the loop between helix 2′ and helix 3, which is the result of crystal packing. GW0072 occupies an epitope of the ligand-binding pocket of PPARγ that precludes it from interactions with Y473, H323, and H449.
Synthetic antagonists of several nuclear receptors have been identified that counter the activity of their cognate hormones. The cocrystal structures of estrogen receptor α with raloxifene and tamoxifen revealed that these antagonists force the AF-2 helix into a conformation that physically blocks the binding of coactivator proteins (8, 9). Antagonist-bound receptors also promote the recruitment of corepressor proteins, such as nuclear receptor corepressor (NCoR) (10) and silencing mediator of retinoid and thyroid hormone receptors (SMRT) (11), which inhibit basal transcriptional activity (10–13). The PPAR crystal structures uncovered a large ligand binding pocket of >1,300 Å3 (6, 7), 2–3 times larger than the corresponding pockets in the steroid hormone, thyroid hormone, or retinoic acid receptors (14). The bulk of this pocket lay distal to the Tyr and His residues involved in allosteric receptor activation (6), but proximal to the proposed ligand entry pocket (7). Given these observations, we postulated that PPAR ligands might be identified that occupied the binding pocket without activating the receptor. In this report we describe the identification of GW0072 (Fig. 1B), a PPARγ ligand that is a partial agonist of receptor transactivation and inhibitor of adipocyte differentiation. Crystallographic and functional analysis suggest that the biological profile of GW0072 is the result of a binding mode that differs from the known agonist ligands.
MATERIALS AND METHODS
Compound Synthesis.
(±)-(2S,5S)-4-(4-(4-Carboxyphenyl)butyl)-2-heptyl-4-oxo-5-thiazolidine N,N-dibenzylacetamide (GW0072, Fig. 1A) was synthesized in the Glaxo Wellcome Medicinal Chemistry Department as a white solid: m.p. 94–97°C; 1H NMR (CDCl3, 400 MHz) 7.96 (d, 2H, J = 8.2 Hz), 7.40–7.20 (m, 10H), 7.14 (d, 2H, J = 7.2 Hz), 4.74 (d, 1H, J = 14.7 Hz), 4.54–4.28 (m, 5H), 3.72–3.62 (m, 1H), 3.47 (dd, 1H, J = 3.0, 13.8 Hz), 3.04–2.95 (m, 1H), 2.78–2.60 (m, 3H), 1.90–1.80 (m, 1H), 1.70–1.50 (m, 8H), 1.44–1.30 m, 8H), 0.88 (t, 3H, J = 6.8 Hz); Anal. (C37H46N2SO4) C, 72.31; H, 7.49; N, 4.56; S, 5.21 Found C, 72.24; H, 7.54; N, 4.56; S, 5.16. Full experimental details will be available elsewhere (J.L.C., unpublished work).
Crystallography.
Crystals of GW0072 complexed to the ligand binding domain of hPPARγ (6) were grown by the vapor phase diffusion method, adding equal volumes of concentrated protein (10 mg/ml, 25 mM Tris, pH 8.0, 5 mM DTT, 200 mM NaCl, 1.5 mM EDTA) and a crystallization buffer (15% polyethylene glycol 4000, 100 mM Hepes, pH 7.5) at a 10:1 molar ratio of GW0072 to protein. Crystals formed in the space group P2 with unit cell dimensions of 92.7 Å, 61.5 Å, 118.8 Å, 90.0°, 102.6°, and 90.0° and were solved by using molecular replacement with the 1PRG coordinates (6). The GW0072 cocrystal structure was refined to 2.7-Å resolution and an R factor of 24.3% with a free R factor (6% omitted) of 29.4% by using x-plor (15). This structure was nearly identical to a cocrystal structure of the di-3-iodobenzyl derivative of GW0072, which was readily interpreted because of the strong iodine electron density peaks (M.V.M. and A.B.M., unpublished results).
Binding Assay.
Binding affinity to the hPPARγ ligand-binding domain was measured by scintillation proximity assay (16) using [3H]rosiglitazone as the competitive radioligand.
Transfections and Cell Culture.
Reporter gene assays were run as described in CV-1 cells by using either the ligand binding domain of hPPARγ as a GAL4 chimera and a (UAS)5-tk-SPAP reporter plasmid (17, 18), or full-length hPPARγ2 and an aP2-tk-CAT reporter plasmid (17). Mammalian two-hybrid transfection mixes included 100 ng (UAS)5-tk-CAT reporter plasmid, 50 ng VP16-hPPARγ2 expression plasmid, 25 ng GAL4 expression plasmid, 200 ng β-galactosidase expression plasmid as internal control, and 25 ng carrier plasmid. Transfections were performed with Lipofectamine (Life Technologies, Grand Island, NY) according to the manufacturer’s instructions. Cell extracts were prepared and assayed for secreted placental alkaline phosphatase, chloramphenicol acetyltransferase, and β-galactosidase activities as described (17, 18).
Plasmids.
VP16-hPPARγ2 expression vector was generated by insertion of the cDNA encoding hPPARγ2 fused to amino acids 410–490 of the VP16 viral activation domain, into the expression vector pSG5 (Stratagene). GAL4-SRC1, GAL4-CBP, GAL4-NCoR, and GAL4-SMRT were generated by insertion of cDNAs encoding amino acids 592–782 (4), 1–115 (5), 2239–2300 (10), and 1281–1344 (11), respectively, fused to amino acids 1–147 of the yeast transcription factor GAL4, including the DNA-binding domain, into the pSG5 expression vector.
Adipocyte Differentiation Assay.
C3H10T1/2 cells were grown on gelatin-coated 24-well plates in DMEM supplemented with 10% FBS as described (17). Medium and compound were changed daily. Cells were stained at day 6 with Oil-red O and photographed. For Northern analyses, cells were harvested on days 3 and 6, and total RNA was prepared by using Trizol Reagent (CLONTECH).
RESULTS
Identification of a Thiazolidine Acetamide PPARγ Ligand.
A weak PPARγ ligand was identified through the synthesis and screening of a combinatorial library of thiazolidine acetamides (19–21). Optimization of this hit by using focused chemical libraries yielded GW0072 (Fig. 1A), which bound to PPARγ with a Ki = 70 nM. Although GW0072 was a high-affinity PPARγ ligand, it showed only 15–20% induction of reporter activity in a standard PPARγ-GAL4 functional assay (Fig. 2A) (17, 18). These data, combined with the chemical structure of GW0072, suggested that it might have a novel biological profile.
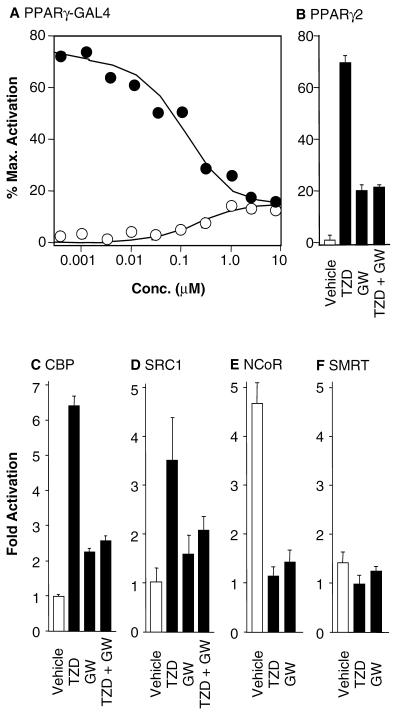
Figure 2.
GW0072 is a PPARγ ligand with a unique functional profile. (A) Dose response on the PPARγ-GAL4 chimera for GW0072 (○) and GW0072 plus 100 nM rosiglitazone (●). Reporter activity was expressed as the % of the maximal activation by 1 μM rosiglitazone. GW0072 demonstrates competitive antagonism of rosiglitazone but retains weak agonist activity at μM concentrations. (B) Activity on full-length PPARγ2 for 100 nM rosiglitazone (TZD), 10 μM GW0072 (GW), and 100 nM rosiglitazone plus 10 μM GW0072 (TZD + GW). Vehicle was 0.1% DMSO. Reporter activity was expressed as the % of the maximal activation by 1 μM rosiglitazone. (C–F) The functional activity of GW0072 is paralleled by its effects on coactivator recruitment to PPARγ2 in a mammalian two-hybrid assay. GW0072 (GW) (10 μM) antagonizes recruitment of the coactivators CBP and SRC1 promoted by 1 μM rosiglitazone (TZD). GW0072 (GW) (10 μM) does not recruit the corepressors NCoR or SMRT.
Structural Analysis of the GW0072-PPARγ Cocrystal Complex.
The initial docking studies of GW0072 in the PPARγ crystal structure (6) identified several potential binding orientations that were different to those of known PPAR agonists. To explore this issue, the cocrystal structure of GW0072 with the PPARγ ligand-binding domain was solved to a resolution of 2.7 Å (Fig. 1B). Remarkably, the structure revealed a binding mode in which GW0072 occupied the region of the receptor pocket bounded by helices 3, 6, and 7 (Fig. 1B). Unlike the known PPAR agonists (3, 7), the carboxylic acid of GW0072 was orientated toward the loop region between H2′ and H3 and did not contact the AF-2 helix or the adjacent His residues (Fig. 1C). The dibenzylamide and heptyl side chains occupied the pocket between H3 and H7, which was lined with hydrophobic residues (Fig. 1B) and accounted for the high affinity binding of this ligand. Notably, the residues Y473, H449, and H323 adopted conformations shifted from their agonist-bound positions, but similar to the apo-PPARγ crystal structure (6) (Fig. 1C). This structural information suggested that the binding mode of GW0072 resulted in a receptor conformation in which the charge clamp was not stabilized through direct interactions with the ligand.
Functional Analysis of GW0072.
Full dose-response analysis in the PPARγ-GAL4 functional assay showed that GW0072 was a weak partial agonist with a relative efficacy = 15–20% of rosiglitazone (Fig. 2A). When assayed in the presence of 100 nM rosiglitazone, GW0072 was a competitive antagonist with an IC50 = 110 nM (Fig. 2A), inhibiting reporter activity to the level of its own partial agonism. Similar results were obtained by using a reporter gene assay using the full-length PPARγ2 (17, 22) (Fig. 2B). Thus, the interaction of GW0072 with the ligand-binding domain of PPARγ was primarily responsible for its functional profile on the full-length receptor.
A mammalian two-hybrid assay was used to examine the ability of GW0072 to recruit the coactivators CBP and SRC1 or the corepressors NCoR and SMRT to PPARγ (Fig. 2 C-F). The assay used full-length PPARγ2 fused to the activation domain of VP16, and the previously identified nuclear receptor interaction domains of CBP, SRC1, NCoR, and SMRT (10, 11, 23, 24) fused to the DNA-binding domain of GAL4. CBP was efficiently recruited to PPARγ2 by 1 μM rosiglitazone, leading to a 6.5-fold increase in reporter activity (Fig. 2C). By contrast, 10 μM GW0072 caused only a 2-fold increase and was able to antagonize the recruitment of CBP induced by rosiglitazone (Fig. 2C). Similar effects were seen with SRC1 (Fig. 2D), where GW0072 antagonized the 3.5-fold increase in reporter activity induced by rosiglitazone. Thus, the low efficacy of GW0072 in PPARγ transactivation assays was paralleled by its limited capacity to recruit the coactivator proteins CBP and SRC1. The corepressor NCoR showed constitutive interaction with PPARγ2 in the absence of ligand (Fig. 2E), which was dissociated by both rosiglitazone and GW0072. SMRT did not interact with PPARγ2 (Fig. 2F) in the presence or absence of either rosiglitazone or GW0072. Thus, GW0072 did not promote the recruitment of the corepressor proteins NCoR or SMRT to the receptor.
GW0072 Is an Adipogenic Antagonist.
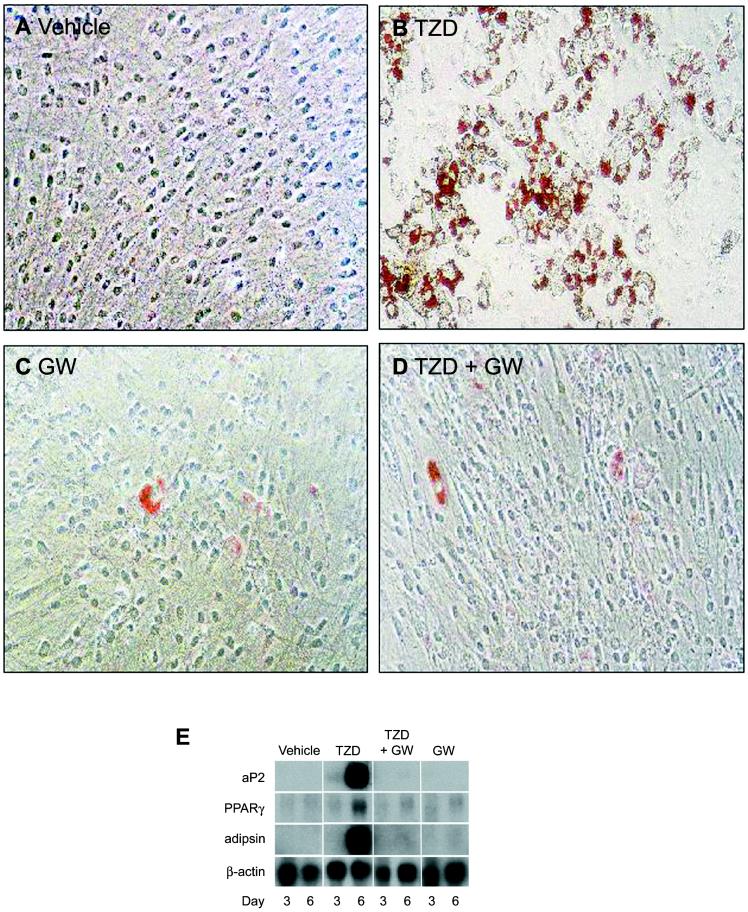
To further characterize the profile of GW0072, we examined its effects on adipocyte differentiation (Fig. 3). PPARγ agonists are known to promote the conversion of a variety of preadipocyte and stem cell lines into mature adipocytes (25). Incubation of 10T1/2 cells with 1 μM rosiglitazone for 6 days resulted in their efficient conversion to adipocytes, as indicated by the increase in Oil-red O staining (Fig. 3B) and the induction of adipocyte fatty acid binding protein, PPARγ, and adipsin gene expression (Fig. 3E). By contrast, when 10T1/2 cells were incubated with 10 μM GW0072 very little conversion to the adipocyte phenotype was observed, as indicated by the virtual absence of Oil-red O staining (Fig. 3C) and the lack of induction of adipocyte-specific genes (Fig. 3E). Notably, GW0072 was able to inhibit the conversion of 10T1/2 cells to adipocytes induced by 1 μM rosiglitazone as measured by Oil-red O staining (Fig. 3D) as well as adipocyte fatty acid binding protein, adipsin, and PPARγ mRNA levels (Fig. 3E). Thus, in cell culture, GW0072 is unable to effect conversion of multipotential stem cells into adipocytes and is a potent antagonist of adipocyte differentiation. The use of GW0072 as a chemical tool in these and other cells promises to be powerful approach for dissecting PPARγ biology.
Figure 3.
GW0072 is a potent antagonist of adipocyte differentiation. (A–D) Oil-red O staining of 10T1/2 cells incubated for 6 days with 0.1% DMSO (vehicle), 1 μM rosiglitazone (TZD), 10 μM GW0072 (GW), or 1 μM rosiglitazone plus 10 μM GW0072 (TZD + GW). (E) Northern blot analysis of 10T1/2 cells for adipocyte-specific genes after treatment of cells for 3 and 6 days. aP2, adipocyte fatty acid binding protein.
DISCUSSION
Our molecular understanding of nuclear receptor signal transduction has been greatly enhanced by the analysis of their cocrystal structures with agonist and antagonist ligands (6–9, 14). These structures, combined with biochemical data, led to important insights into how agonists and antagonists differentially modulate transcriptional activity through the recruitment of coactivator and corepressor proteins (Fig. 4). From a chemical perspective, although agonist and antagonist ligands often are structurally related, the antagonists generally contain an additional chemical appendage that is critical for their inhibitory activity (26). As a result, these agonist and antagonist ligands use the same binding epitopes within the receptor pocket, but the larger total volume of the antagonist (28) leads to repositioning of the AF-2 helix and displacement of coactivator proteins (Fig. 4). Although less well understood, the antagonist-bound receptor complex is also permissive to corepressor binding (10–13).
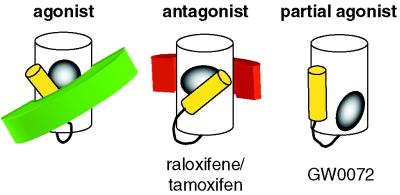
Figure 4.
GW0072 defines an additional class of nuclear receptor ligands. Agonist ligands (gray) shift the AF-2 helix (yellow) into a position that stabilizes recruitment of coactivator (green) to the receptor ligand-binding domain (white). Antagonist ligands (gray) bind to the receptor by using the same epitopes, but their larger size shifts the AF-2 helix into a position that displaces the coactivator. Antagonist ligands also recruit corepressor (red) to the receptor ligand-binding domain. The partial agonist GW0072 (gray) binds to its receptor by using different epitopes, such that it does not directly interact with the AF-2 helix.
With GW0072, we have identified a PPAR ligand that occupies epitopes within the receptor pocket distinct from the known agonist ligands (Fig. 4). GW0072 does not directly affect the positioning of AF-2 or residues involved in coactivator recruitment. Instead, the GW0072-bound receptor adopts a conformation similar to the unliganded apo-receptor. The dissociation of the corepressor NCoR and weak recruitment of coactivator proteins that was effected by GW0072 may be the consequence of either subtle changes in the conformation of the ligand-binding domain or allosteric effects on the heterodimeric partner RXR (28). Thus, GW0072 is an example of a nuclear receptor ligand that does not interact with AF-2 and is mechanistically distinct from conventional agonists and antagonists. The biological profile of this class of ligands is likely to be complex, dependent on both the promoter context of their target genes as well as the cellular concentration of coactivators and corepressors (29). In the case of the GW0072, it is a PPARγ partial agonist that is unable to effect conversion of multipotential stem cells into adipocytes and is a potent antagonist of TZD-induced adipocyte differentiation. Because the PPARs are important molecular targets for the development of diabetes, obesity, and cardiovascular drugs (3), the identification of partial agonists with modified pharmacological profiles may offer benefit for the treatment of human metabolic diseases (30).
Acknowledgments
We thank Donald McDonnell for critical reading of the manuscript.
ABBREVIATIONS
- PPAR
peroxisome proliferator-activated receptor
- TZD
2,4-thiazolidinedione
- SRC1
steroid receptor coactivator 1
- CBP
CREB binding protein
- AF-2
activation function 2
- NCoR
nuclear receptor corepressor
- SMRT
silencing mediator of retinoid and thyroid hormone receptors
Footnotes
Data deposition: The coordinates of the GW0072-PPARγ cocrystal structure have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 4PRG).
References
- 1.Mangelsdorf D J, Thummel C, Beato M, Herrlich P, Schuetz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, et al. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kliewer S A, Sundseth S S, Jones S A, Brown P J, Wisely G B, Koble C, Devchand P, Wahli W, Willson T M, Lenhard J M, et al. Proc Natl Acad Sci USA. 1997;94:4318–4323. doi: 10.1073/pnas.94.9.4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Willson T M, Wahli W. Curr Opin Chem Biol. 1997;1:235–241. doi: 10.1016/s1367-5931(97)80015-4. [DOI] [PubMed] [Google Scholar]
- 4.Onate S A, Tsai S Y, Tsai M-J, O’Malley B W. Science. 1995;270:1354–1357. doi: 10.1126/science.270.5240.1354. [DOI] [PubMed] [Google Scholar]
- 5.Kamei Y, Xu L, Heinzel T, Torchia J, Kurokawa R, Gloss B, Lin S-C, Heyman R A, Rose D W, Glass C K, et al. Cell. 1996;85:403–414. doi: 10.1016/s0092-8674(00)81118-6. [DOI] [PubMed] [Google Scholar]
- 6.Nolte R T, Wisely G B, Westin S, Cobb J E, Lambert M H, Kurokawa R, Rosenfeld M G, Willson T M, Glass C K, Milburn M V. Nature (London) 1998;395:137–143. doi: 10.1038/25931. [DOI] [PubMed] [Google Scholar]
- 7.Xu H E, Lambert M H, Montana V G, Parks D J, Blanchard S G, Brown P J, Sternbach D D, Lehmann J M, Wisely G B, Willson T M, et al. Mol Cell. 1999;3:397–403. doi: 10.1016/s1097-2765(00)80467-0. [DOI] [PubMed] [Google Scholar]
- 8.Brzozowski A M, Pike A C W, Dauter Z, Hubbard R E, Bonn T, Engstrom O, Ohman L, Greene G L, Gustafsson J-A, Carlquist M. Nature (London) 1997;389:753–758. doi: 10.1038/39645. [DOI] [PubMed] [Google Scholar]
- 9.Shiau A K, Barstad D, Loria P M, Cheng L, Kushner P J, Agard D A, Greene G L. Cell. 1998;95:927–937. doi: 10.1016/s0092-8674(00)81717-1. [DOI] [PubMed] [Google Scholar]
- 10.Horlein A J, Naar A M, Heinzel T, Torchia J, Gloss B, Kurokawa R, Ryan A, Kamei Y, Soderstrom M, Glass C K, et al. Nature (London) 1995;377:397–404. doi: 10.1038/377397a0. [DOI] [PubMed] [Google Scholar]
- 11.Chen J D, Evans R M. Nature (London) 1995;377:454–457. doi: 10.1038/377454a0. [DOI] [PubMed] [Google Scholar]
- 12.Smith C L, Nawaz Z, O’Malley B W. Mol Endocrinol. 1997;11:657–666. doi: 10.1210/mend.11.6.0009. [DOI] [PubMed] [Google Scholar]
- 13.Wagner B L, Norris J D, Knotts T A, Weigel N L, McDonnell D P. Mol Cell Biol. 1998;18:1369–1378. doi: 10.1128/mcb.18.3.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moras D, Gronemeyer H. Curr Opin Cell Biol. 1998;10:384–391. doi: 10.1016/s0955-0674(98)80015-x. [DOI] [PubMed] [Google Scholar]
- 15.Brunger A. x-plorVersion 3.0: A System for Crystallography and NMR. New Haven, CT: Yale Univ. Press; 1992. [Google Scholar]
- 16.Nichols J S, Parks D J, Consler T G, Blanchard S G. Anal Biochem. 1998;257:112–119. doi: 10.1006/abio.1997.2557. [DOI] [PubMed] [Google Scholar]
- 17.Lehmann J M, Moore L B, Smith-Oliver T A, Wilkison W O, Willson T M, Kliewer S A. J Biol Chem. 1995;270:12953–12956. doi: 10.1074/jbc.270.22.12953. [DOI] [PubMed] [Google Scholar]
- 18.Willson T M, Cobb J E, Cowan D J, Wiethe R W, Correa I D, Prakash S R, Beck K D, Moore L B, Kliewer S A, Lehmann J M. J Med Chem. 1996;39:665–668. doi: 10.1021/jm950395a. [DOI] [PubMed] [Google Scholar]
- 19.Holmes C P, Jones D G. J Org Chem. 1995;60:2318–2319. [Google Scholar]
- 20.Holmes C P, Chinn J P, Look G C, Gordon E M, Gallop M A. J Org Chem. 1995;60:7328–7333. [Google Scholar]
- 21.Look G C, Schullek J R, Holmes C P, Chinn J P, Gordon E M, Gallop M A. Bioorg Med Chem Lett. 1996;6:707–712. [Google Scholar]
- 22.Elbrecht A, Chen Y, Cullinan C A, Hayes N, Leibowitz M D, Moller D E, Berger J. Biochem Biophys Res Commun. 1996;224:431–437. doi: 10.1006/bbrc.1996.1044. [DOI] [PubMed] [Google Scholar]
- 23.Heery D M, Kalkhoven E, Hoare S, Parker M G. Nature (London) 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- 24.Torchia J, Rose D W, Inostroza J, Kamei Y, Westin S, Glass C K, Rosenfeld M G. Nature (London) 1997;387:677–684. doi: 10.1038/42652. [DOI] [PubMed] [Google Scholar]
- 25.Tontonoz P, Hu E, Spiegelman B M. Cell. 1994;79:1147–1156. doi: 10.1016/0092-8674(94)90006-x. [DOI] [PubMed] [Google Scholar]
- 26.Katzenellenbogen J A, Katzenellenbogen B S. Chem Biol. 1996;3:529–536. doi: 10.1016/s1074-5521(96)90143-x. [DOI] [PubMed] [Google Scholar]
- 27.Bogan A A, Cohen F E, Scanlan T S. Nat Struct Biol. 1998;5:679–681. doi: 10.1038/1372. [DOI] [PubMed] [Google Scholar]
- 28.Westin S, Kurokawa R, Nolte R T, Wisely G B, McInerney E M, Rose D W, Milburn M V, Rosenfeld M G, Glass C K. Nature (London) 1998;395:199–202. doi: 10.1038/26040. [DOI] [PubMed] [Google Scholar]
- 29.Lavinsky R M, Jepsen K, Heinzel T, Torchia J, Mullen T-M, Schiff R, Del-Rio A L, Ricote M, Ngo S, Gemsch J, et al. Proc Natl Acad Sci USA. 1998;95:2920–2925. doi: 10.1073/pnas.95.6.2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kliewer S A, Willson T M. Curr Opin Genet Dev. 1998;8:576–581. doi: 10.1016/s0959-437x(98)80014-2. [DOI] [PubMed] [Google Scholar]