Abstract
The pattern of DNA methylation plays an important role in regulating different genome functions. To test the hypothesis that DNA methylation is a reversible biochemical process, we purified a DNA demethylase from human cells that catalyzes the cleavage of a methyl residue from 5-methyl cytosine and its release as methanol. We show that similar to DNA methyltransferase, DNA demethylase shows CpG dinucleotide specificity, can demethylate mdCpdG sites in different sequence contexts, and demethylates both fully methylated and hemimethylated DNA. Thus, contrary to the commonly accepted model, DNA methylation is a reversible signal, similar to other physiological biochemical modifications.
Many lines of evidence have established that modification of cytosine moieties residing in the dinucleotide sequence CpG in vertebrate genomes plays a role in regulating a number of genome functions such as parental imprinting (1–2), X chromosome inactivation (3–4), suppression of expression of ectopic genes (5–6), and differential gene expression (7–10). DNA methylation can perform its function of differentially marking genes because CpG dinucleotides are differentially methylated, forming a pattern of methylation (11). It is clear that the pattern of methylation is fashioned by a sequence of methylation and demethylation events (12–15) during development and is maintained in the fully differentiated cell (7–10, 16).
Most biological modifications such as protein phosphorylation are reversible, and enzymes exist that can catalyze either the modification or its removal. This reversibility is essential for their functioning as biological signals that can respond to changing physiological cues. DNA methylation has been considered to be an exception because removal of a methyl group from DNA must involve a cleavage of a carbon–carbon bond, which has been considered an unlikely reaction. Although it has been accepted that demethylation must occur during development, indirect mechanisms involving base excision and repair have been previously proposed to be responsible for removal of methyl groups from DNA at different stages of development (17–21). Because it has been believed that removal of methyl groups from DNA is a cumbersome process, the accepted model has been that DNA methylation is a heritable and stable signal (7–10). We have recently cloned a human cDNA that encodes a DNA demethylase (dMTase) activity that can catalyze the cleavage of a methyl residue from 5-methyl cytosine (22). This has provided molecular proof that true demethylation of DNA is biochemically feasible. If biological DNA methylation is a truly a reversible process in living cells, nuclei of mammalian cells should bear an activity that removes the specific products of the DNA methylation reaction. This report describes the testing of this hypothesis by purification of a dMTase from nuclear extracts of human cells and definition of its substrate and sequence specificity.
MATERIALS AND METHODS
Purification of DNA Demethylase Activity.
Nuclear extracts were prepared from A549 (American Type Culture Collection CCL 185) cultures at near confluence as described (24). A freshly prepared nuclear extract (1 ml = 6 mg) was diluted to a conductivity equivalent to 0.2 M NaCl in buffer L (10 mM Tris⋅HCl, pH 7.5/10 mM MgCl2) and applied onto a DEAE–Sephadex A-50 column (Amersham Pharmacia) (2.0 × 1 cm) that was preequilibrated with buffer L at a flow rate of 1 ml/min. After a 15-ml wash with buffer L, proteins were eluted with a 5-ml linear gradient of NaCl (0.2–5.0 M). Fractions of 0.5-ml volume were collected and assayed for dMTase activity. dMTase eluted between 4.9 and 5.0 M NaCl. Active DEAE–Sephadex column fractions were pooled, adjusted to 0.2 M NaCl by dilution, and loaded onto an SP-Sepharose column (Amersham Pharmacia) (2.0 × 1 cm). After washing of the column as described above, the proteins were eluted with 5 ml of a linear NaCl gradient (0.2–5.0 M). Fractions of 0.5-ml volume were collected and assayed for demethylase activity. Demethylase activity eluted at ≈5.0 M NaCl. Active fractions were pooled, adjusted to 0.2 M NaCl by dilution, and applied to a Q-Sepharose (Amersham Pharmacia) column (2.0 × 1 cm), and proteins were eluted as described above. The demethylase activity eluted at ≈4.8–5.0 M NaCl. The pooled fractions of Q-Sepharose column were loaded onto a 2.0 × 2.0 cm DEAE–Sephacel column (Amersham Pharmacia) and eluted with 10 ml of buffer L. The activity was detected at fraction 4, which is very near the void volume. A silver-stained SDS/PAGE analysis of the active fractions revealed three polypeptides of 38–40 kDa that coeluted with dMTase activity in the last three chromatographic steps (data not shown). We have not yet resolved which of these polypeptides is the demethylase enzyme or whether they form a complex. A batch of 40 purified column fractions was pooled and subjected to cold vacuum evaporation followed by dialysis in 1 liter of buffer L (no salt) at 4°C with three changes at every 6-hour interval. The dMTase from 40 preparations was suspended in 0.1 ml of buffer L (no NaCl). Preparations so obtained were assayed for enzyme activity and used for subsequent experiments.
Assay of the Conversion of Methyl-dCMP (mdCMP) to dCMP.
To assay mdCMP conversion to dCMP, we used methods described (24). One nanogram of α-32P-labeled, fully methylated poly[mdC32pdG]n or a control nonmethylated poly[dC32pdG]n substrate were prepared as described (24), reacted with 50 μl of each column fraction for 3 hours at 37°C, purified by phenol/chloroform extraction, and subjected to micrococcal nuclease digestion (100 μg at 10 μl) (Amersham Pharmacia) to 3′ mononucleotides for 15 hours at 37°C and calf spleen phosphodiesterase (2 μg) (Boehringer Mannheim) for 2 hours at 37°C. The digestion products were loaded onto a TLC plate (Kodak, 13255 cellulose), developed in medium containing 132 ml of isobutyric acid, 40 ml of water, and 4 ml of ammonia solution, autoradiographed, and the intensity of the different spots was determined by using a phosphorimager (Fuji, BAS 2000). Poly[mdC32pdA] and poly[mdC32pdT] substrates were prepared as follows. 0.5 μg of 20-mer oligonucleotides 5′-(GG)10-3′, 5′-(GT)10-3′, and 5′(GA)10-3′ were boiled and annealed at room temperature with oligonucleotide 5′-CCCCCC-3′, 5′-CACACA-3′, and 5′-CTCTCT-3′ respectively. The complementary strand was extended with Klenow fragment by using m5dCTP (Boehringer Mannheim) and either [α-32P]dATP (100 μCi, 3,000 Ci/mmol; 1 Ci = 37 GBq) or [α-32P]dTTP (100 μCi, 3,000 Ci/mmol), respectively.
GC/MS Analysis.
The demethylation reactions (volume 50 μl) were run in conical vials having a total internal volume of 350 μl. The vials were closed with a Teflon-lined screw cap and left at room temperature for 18 hours. The vials were cooled in an ice bath and opened, and 10 mg of NaCl and 50 μl of toluene was added. The vials were frequently shaken over a period of 1 hour. The toluene phases were pipetted into clean vials in such a manner as to rigorously exclude water carryover. Anhydrous sodium sulfate (5 mg) was added to the toluene extracts to remove water, and the toluene phases were pipetted into autoinjector vials for GC/MS analysis. Aliquots of 3 μl were analyzed under the following instrumental conditions: instrument, Hewlett-Packard 5988A; column, 30 m × 0.25 mm i.d. fused quartz capillary with 0.25-μm DB-1 liquid phase, programmed after an initial hold for 1 min at 70 deg at 5°C/min to 80°C, then ramped ballistically to 280°C for bake-out for 5 min; injector and interface temperatures, 250°C; helium flow rate, 1.5 ml/min; mass spectrometer: ion source, 200°C; 70 eV electron impact ionization, scanning from m/z 10–50 in full scan mode was begun 6 s after injection and halted at 1.5 min to avoid acquisition of the intense toluene solvent peak.
Bisulfite-Treated DNA Methylation Analysis.
The primers used for the DNA methyltransferase genomic region (GenBank accession no. M84387) were: MET5′1, 5′-ggattttggtttatagtattgt-3′ MET 5′ (nested) 5′-ggaattttaggtttttatatgtt-3′; MET3′, 1 5′-ctcttcataaactaaatattataa-3′ and MET3′ (nested) 5′-tccaaaactcaacataaaaaaat-3′.
RESULTS
Purification of a dMTase Activity from Mammalian Nuclei.
Because it has been previously proposed that the extensive hypomethylation observed in cancer cells might be a consequence of activation of dMTase activity by oncogenic pathways (23–24), we have used the human lung carcinoma cell line A549 as a source for purification of a DNA demethylation activity. To directly measure the conversion of 5-mdCMP in DNA to dCMP, we have used a completely methylated 32P-labeled [mdC32pdG]n double-stranded oligomer that we previously described (24). After incubation with the different fractions collected from chromatographic separation of nuclear extracts of A549 cells, the DNA was purified and subjected to cleavage with micrococcal nuclease to 3′ mononucleotides. The 3′-labeled mdCMP and dCMP were separated by using TLC, and the conversion of mdCMP to dCMP was directly determined (24). This assay provides a stringent test for demethylation and discriminates it from previously described 5 mCpG replacement activities (17–21).
Nuclear extracts prepared from A549 cells were subjected to a sequence of protein purification steps by using DEAE-Sephadex, Sp-Sepharose, Q-Sepharose and DEAE-Sephacel matrix chromatography, and active dMTase fractions were identified (Fig. 1A and Table 1). The purified dMTase activity is a protein because it is abolished after proteinase K treatment and is not inhibited but rather is enhanced after RNase treatment. ddCTP (500 μM), which inhibits DNA polymerase β (25–26), does not inhibit demethylation of the [mdC32pdG]n substrate, nor is it inhibited by high concentrations of methyl-dCTP (500 μM) (Fig. 1A), which is consistent with the hypothesis that demethylation does not involve an excision and replacement mechanism. If a replacement mechanism is involved in demethylation, the presence of mdCTP should result in incorporation of methylated cytosines and essential inhibition of demethylation. Thus, the dMTase identified here is a protein and not an RNA and is unequivocally different from the previously published glycosylase based dMTase activities (17–21).
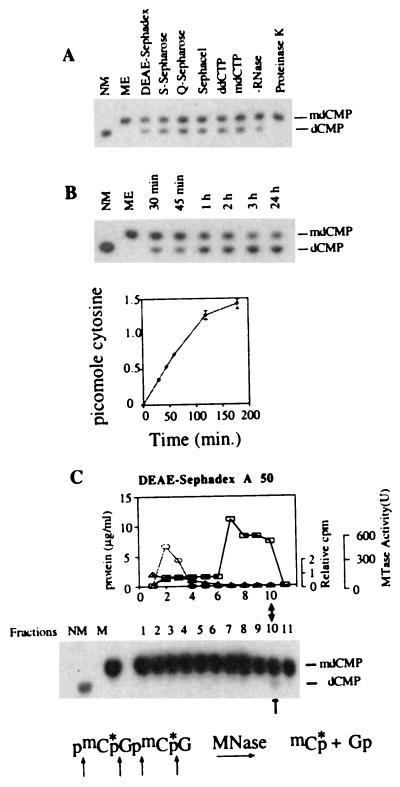
Figure 1.
Purification of a dMTase from human cells. (A) [mdC32pdG]n double-stranded oligomer (1 ng) was incubated for 1 hour at 37°C with either buffer L (ME), 0.3 μg of active dMTase fractions from the different chromatography steps in the presence of 100 μg/ml RNase as indicated, with DEAE-Sephacel dMTase fraction in the presence of RNase, and either ddCTP (500 μM) or mdCTP (500 μM), with DEAE-Sephacel dMTase fraction in the absence of RNase, or with DEAE-Sephacel dMTase fraction after preincubation for 30 min. with proteinase K (200 μg/ml). The reaction products were digested to 3′ mononucleotides and analyzed on TLC (24). Methylated (ME) and nonmethylated (NM) [dC32pdG]n substrates were digested to 3′ mononucleotides and loaded on the TLC plate as controls. (B) Kinetics of dMTase activity. [mdC32pdG]n double-stranded oligomer (1 ng) was incubated with either DEAE-Sephacel dMTase fraction (0.3 μg) for different time points and analyzed for demethylation as above. A representative chromatogram is shown. Chromatograms were quantified by a PhosphorImager, and the rate of transformation of mdCMP to dCMP was calculated and plotted. The results are a summary of three independent determinations ± SD. (C) Nuclear extracts were fractionated on a DEAE-Sephadex column. Fifty-microliter samples from each fraction (total fraction volume was 500 μl) were incubated with 1 ng of [mdC32pdG]n double-stranded oligomer, digested to 3′ mononucleotides, and analyzed on TLC (24). Control methylated (ME) and nonmethylated (NM) [dC32pdG]n substrates were digested to 3′ mononucleotides and loaded on the TLC plate to indicate the expected position of dCMP and mdCMP. The active fraction is indicated by an arrow. DNA methyltransferase activity was determined by using a hemimethylated DNA substrate and [3H]AdoMet33 (○). One unit equals the amount of enzyme that catalyzes transfer of 1 pmol of tritiated methyl group on hemimethylated DNA per minute. The results are an average of three independent determinations. Protein concentration was determined by using the Bio-Rad Bradford kit (□). To test whether dCTP copurifies with dMTase, the elution profile of 20 μM of [α-32P]dCTP (1.1 × 106 dpm) incubated with the protein was determined by scintillation counting of the different DEAE fractions (▵) and presented as fraction of dCTP loaded on the column.
Table 1.
Purification of dMTase from A549 cells
| Purification step | Total protein, μg | Specific activity, pmol/hour/μg | Fold purification |
|---|---|---|---|
| Nuclear extract | 6,000 | 1.83 × 10−5 | — |
| DEAE-Sephadex | 3.75 | 0.156 | 844.5 |
| SP-Sepharose | 0.77 | 0.663 | 35,939.84 |
| Q-Sepharose | 0.46 | 1.13 | 62,860 |
| DEAE-Sephacel | 0.018 | 10.19 | 552,243 |
Nuclear extract from A549 cells (1 ml, 6 mg) was applied sequentially onto a DEAE-Sephadex, SP-Sepharose, Q-Sepharose, and DEAE-Sephacel columns as described in Materials and Methods. dMTase was assayed by using a 50-μl sample of each 0.5-ml fraction using the assay described in Fig. 1 and Ref. 11. The specific activity of the active fractions after each purification step is represented as picomoles of CH3 released per hour per microgram of protein. The ratio of specific activity of dMTase after each step to that of the nuclear extract is shown as fold purification.
The dMTase reaction proceeds without any requirement for additional substrates such as dCTP, redox factors such as NADH and NADPH, or energy sources such as ATP (data not shown). As shown in Fig. 1B, the dMTase reaction maintains its initial velocity up to 90 minutes and continues up to 120 minutes. This time course is inconsistent with dependence on enzyme-bound additional nonreplenishable substrates such as dCTP or ATP or a nonreplenishable redox factor such as NADH or NADPH. Exhausting the nonreplenishable substrate or redox factor would have resulted in rapid deceleration of the initial velocity.
As shown in Fig. 1C, a clear peak of dMTase activity is eluted at the high salt fraction 10. The first chromatography step purified the dMTase activity from the bulk of nuclear protein (Fig. 1C) and separated dMTase away from the DNA methyltransferase, suggesting that they are independent proteins. DEAE-Sephadex chromatography also purifies dCTP away from the dMTase at least 1 × 106-fold (Fig. 1C), thus excluding the possibility that the demethylation activity in fraction 10 is a glycosylase–apyrimidine nuclease-repair activity. If any dCTP is present in the nuclear extract, the remaining concentration after fractionation on DEAE is well below the Km of the known DNA polymerases (25–26).
dMTase Reaction Produces Methanol.
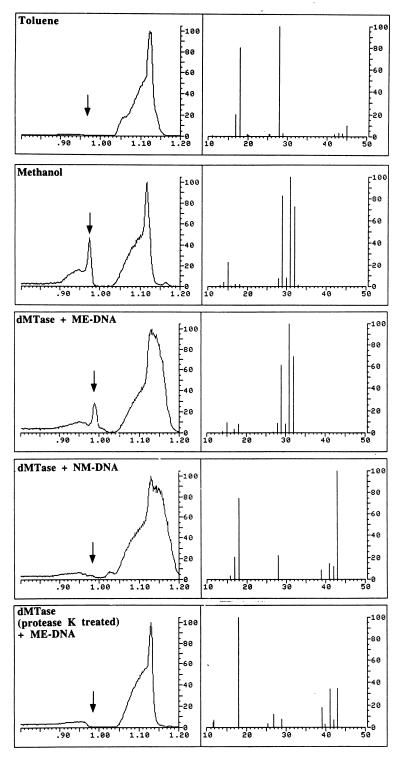
To determine the product of the demethylation reaction, we identified the leaving group. As illustrated in Fig. 2, incubation of methylated DNA with dMTase (dMTase + ME–DNA) results in release of a peak with the retention time and mass spectrum (peaks are identified at 32, 31, and 29 atomic mass, which are the atomic masses of methanol and ionized methanol, respectively), which is consistent with its identification as methanol. Incubation of dMTase with nonmethylated DNA does not release methanol, indicating that methanol is a product of the demethylation reaction. No methanol is released when the samples are incubated with dMTase pretreated with protease K, indicating that the release of methanol from methylated DNA is catalyzed by an enzymatic activity. These results demonstrate that we have purified a dMTase from a human cell.
Figure 2.
Purified dMTase releases the methyl group in the form of methanol. Shown are GC/MS analyses of reference materials and incubated samples for methanol. (Left) A portion of the gas chromatogram produced by the injected sample. The abscissa is the retention time in minutes and the ordinate is relative intensity in arbitrary units. (Right) The mass spectrum measured at the retention time of methanol (marked with an arrow, Left). The abscissa is the mass/charge (m/z) ratio for the ions detected, whereas the ordinate is the relative intensity of the ions detected. The large irregularly shaped peaks in the gas chromatograms are caused by hydrocarbon impurities in the toluene solvent that would normally be of no consequence and not detected except that the instrumental sensitivity required for methanol detection is such that they are also detected. “Toluene” illustrates the results of the GC/MS analysis of the toluene solvent used in this study. The mass spectrum recorded at the retention time for methanol does not present ions that are prominent in the mass spectrum for dilute authentic methanol in toluene solution (1:1,000 by volume, m/z 15, 29, 31, and 32, “Methanol”). “dMTase + ME DNA” illustrates the results obtained for the DNA demethylation reaction [with methylated CpG DNA (400 ng) and purified dMTase (4.8 μg) in a volume of 50 μl at 37°C overnight] and demonstrates that a peak with the retention time and mass spectrum consistent with its identification as methanol is detected. An additional ion at m/z 18 is most likely caused by residual water in the toluene extract. “dMTase + NM-DNA” and “dMTase (protease K treated) + ME-DNA” are the results of the analyses for demethylation reaction with CpG DNA with purified dMTase and protease K-treated dMTase with methylated CpG DNA [these reaction were performed with 4.8 μg of purified dMTase and 400 ng of DNA in a reaction volume of 50 μl with overnight incubation at 37°C, 10 units of protease K was used for treatment of 4.8 μg of dMTase in a reaction volume of 42 μl at 50°C for 3 hours], respectively, and display none of the ions associated with methanol. The ion at m/z 18 is because of residual water in the extract. Prominent ions in the range m/z 39–43 are because of trace hydrocarbons in the toluene.
dMTase Displays mCpG Sequence Specificity.
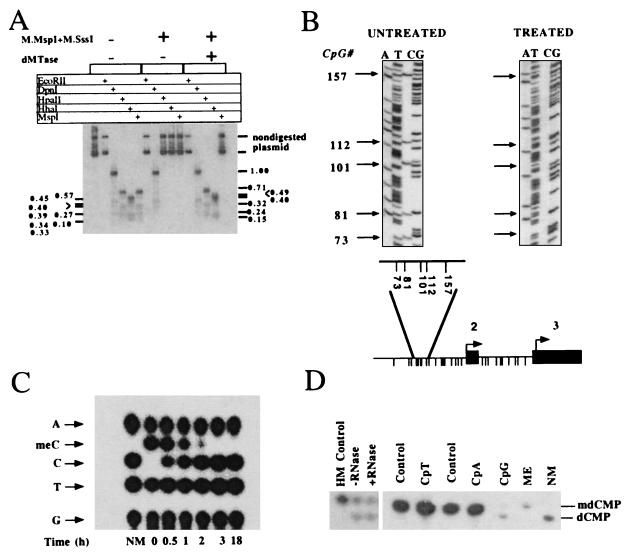
If DNA methylation is a reversible process, the demethylase should be able to remove the specific products of the reaction catalyzed by DNA methyltransferase. DNA methyltransferase catalyzes the transfer of methyl groups to CG dinucleotides residing in many possible sequence contexts generating either hemimethylated or fully methylated DNA (27–28). The results presented in Fig. 3 demonstrate that dMTase is a general dMTase activity that demethylates fully or hemimethylated dCpdG in DNA flanked by a variety of sequences that are distributed at different frequencies, but does not demethylate methylated adenines or methylated cytosines that do not reside in the dinucleotide CG. First, as shown in Fig. 3A, a plasmid DNA methylated in vitro at all dCpdG sites with SssI methylase, all d*CpdCpdGpdG sites with MspI methylase (which methylates the external C in the sequence *CCGG, thus enabling the determination of demethylation at the CC dinucleotide) and in vivo with the Escherichia coli DNA cytosine methylase at dCmdCdA/dTdGdG sites and with the DNA adenine methylase at dGmdAdTdC sites (adenine methylated) was treated with dMTase. The state of methylation of the plasmid was determined by using the indicated methylation-sensitive restriction enzymes. dMTase demethylates CpG methylated sites as indicated by the sensitivity of the dMTase-treated plasmid to HpaII and HhaI but does not demethylate C*C, C*A, or C*T methylated sites as indicated by the resistance to MspI and EcoRII restriction enzymes, or adenine methylation, as indicated by its sensitivity to DpnI. Second, bisulfite mapping analysis of methylation of five methylated C*G sites residing in a SssI methylase in vitro methylated pMetCAT plasmid after dMTase treatment shows that all C*G sites are demethylated irrespective of their flanking sequences thus excluding the possibility that demethylation is limited to CCGG or CGCG sequences (Fig. 3B). Third, dMTase does not demethylate two fully methylated cytosine-bearing oligomers [dmC32pdA]n,[mdC32pdT]n (15), demonstrating that mdCpdA and mdCpdT are not demethylated by dMTase (Fig. 3D). Fourth, dMTase demethylates a hemimethylated synthetic substrate [dCpdG]n*[mdC32pdG]n (Fig. 3D). Demethylation of the plasmid DNA is complete under these conditions (Fig. 3A), whereas demethylation of a methylated [mdCpdG]n substrate is not complete under the same conditions (Fig. 3D). This can reflect differences in the sequence composition of the substrate and the frequency of methylated cytosines. The [mdCpdG]n contains on average of 16-fold more methylated cytosines per molecule than plasmid DNA. Alternatively, these differences might reflect discrepancies in the assays used, restriction enzyme digestion vs. a nearest neighbor analysis. To address this discrepancy, we have labeled a fully methylated plasmid DNA with [α-32P]dGTP, 5-methyl-dCTP, dATP, and dTTP, subjected it to dMTase treatment, and digested it to mononucleotides at different time points after the initiation of the reaction, and subjected the samples to a TLC analysis. As shown in Fig. 3C, the plasmid DNA is fully demethylated by 3 hours, which is consistent with the results obtained with methylation-sensitive restriction enzymes (Fig. 3A).
Figure 3.
Substrate specificity of dMTase. (A) Demethylation of methylated SK plasmid. Plasmid pBluescript SK (5 ng) was methylated with SssI methylase and MspI methylase. dMTase-treated (1.2 μg of concentrated DEAE Sephacel fraction for 3 hours) (methylated SK + dMTase) and nontreated plasmid (methylated SK) as well as nonmethylated SK plasmid were digested with EcoRII, DpnI, HhaI, MspI, and HpaII. The digestion products were analyzed on a 2% agarose gel, blotted onto a Hybond N+ filter, and hybridized with a 32P-labeled SK probe. The expected HpaII/MspI fragments are indicated on the right side of the figure, the expected HhaI fragments are indicated on the left side of the figure. Note that control plasmids are not digested with MspI because the plasmid is methylated with MspI methyltransferase). (B) Bisulfite methylation analysis of dMTase-treated pMetCAT plasmid. pMetCAT plasmid (24), methylated to completion with SssI, was subjected to a dMTase reaction as above. The dMTase-treated plasmid and nontreated control were subjected to bisulfite treatment as described (31), and the genomic region bearing five CpG sites upstream of the second exon of DNA methyltransferase was amplified by using PCR (31) and sequenced. Twenty clones were sequenced per DNA sample. Unmethylated cytosines are converted to thymidines by this protocol, whereas methylated cytosines are protected and are visualized as cytosines. One representative sequencing gel of bisulfite treated DNA is presented per condition. Arrows indicate the specific CpG sites by their position. The numbering is according to GenBank accession no. M84387. (Lower) The physical map of the genomic region residing upstream of exon 2 of DNA methyltransferase (indicated by filled boxes). Intronic sequence is indicated by a line. A blowup of the region amplified after bisulfite treatment is shown above the physical map. The different CpG sites in the fragment are presented as descending lines. Under these conditions, all the clones of the demethylase-treated plasmids were demethylated. (C) Plasmid pBluescript SK (5 ng) was methylated with SssI methylase and MspI methylase. The methylated pBluescript SK was used as template to DNA synthesis by using DNA polymerase and [α32-P]dGTP, mdCTP, dTTP, dATP, and hexanucleotide primers. After two rounds of purification on NAP-5 column, the substrate was subjected to demethylation by using 0.12 μg of purified dMTase for different time intervals as indicated. (D) Demethylation of hemimethylated [mdC32pdG]n·[dCpdG]n (HM), [mdC32pdA]n (CpA), and [mdC32pdT]n (CpT) DNA substrates (15). One nanogram of the indicated substrates, as well as a double methylated substrate [mdC32pdG]n (CpG), were incubated either in the presence (0.3 μg) or absence of dMTase (control for CpT and CpA and ME for CpG) and were then digested to 3′ mononucleotides and analyzed by TLC. Hemimethylated DNA was incubated with dMTase in the presence or absence of RNase (100 μg/ml). Nonmethylated [dC32pdG]n served as a control for the position of dCMP.
The Km of dMTase for hemimethylated and fully methylated DNA was determined by measuring the initial velocity of the reaction at different concentrations of substrate (Table 2). The calculated Km for hemimethylated DNA is 6 nM, which is 2-fold higher than the Km for DNA methylated on both strands, 2.5–3 nM (Table 2). It is unclear yet whether this small difference in affinity for the substrate has any significance in a cellular context. Thus, similar to the DNA methyltransferase, dMTase shows dinucleotide sequence selectivity, but differs from DNA methyltransferase (which has preference for hemimethylated substrates), because it prefers fully methylated DNA, which is consistent with a role for dMTase in altering established methylation patterns.
Table 2.
Kinetic parameters for dMTase
| Substrate | Km (DNA), nM | Vmax, pmol/hour |
|---|---|---|
| Methylated CpG | 2.5 | 340 |
| hemimethylated CpG | 6.0 | 402 |
| methylated SK-DNA | 3.3 | 40.42 |
For determination of kinetic parameters, the dMTase reactions were performed as in Fig. 1 except that varying DNA concentrations, from 0.1 nM to 50 nM, were used in a total volume of 100 μl containing 1.2 μg of DEAE-Sephacel dMTase fraction. Because it has been established that the reaction proceeds for at least 3 hours (Fig. 1B), the initial velocity of reaction was measured at 1-hour intervals. The velocity data was collected at each substrate DNA concentration. The Km and Vmax values for demethylase activity were determined from double-reciprocal plots of velocity vs. substrate concentration.
DISCUSSION
Demethylase performs the reverse reaction to DNA methyltransferase and is an excellent candidate to be one of its important partners in shaping the methylation pattern of genomes. It remains to be seen whether lower organisms that bear DNA methyltransferases also harbor dMTases or whether this activity is unique to organisms where DNA demethylation plays a critical role in development and regulation of genome function.
Direct demethylation of DNA has in the past been considered highly unlikely. The identification of methanol as the leaving group of the demethylation reaction and water as a possible reactant provides an explanation for the thermodynamic feasibility of DNA demethylation. However, the questions remains whether there is a kinetic pathway that can lower the activation barrier enough for the reaction to take place? Cedar and Verdine (31) have recently proposed that one possibility is that dMTase, like methyltransferases, stabilizes the reaction intermediate by a combination of covalent catalysis at C6 and proton shuffling at N3. Once such an intermediate is stabilized by dMTase, a hydroxide ion can then attack the C5 methyl group.
Methylation patterns exhibit site specificity, yet one remaining question is how can a general enzyme demethylate certain sites and not others? One possible hypothesis that has previously been proposed is that an interplay between local factors such as transcription factors and the general availability of dMTase and methyltransferase determines the pattern of methylation (23–24, 29–30). The identification of dMTase enables future experiments to test this hypothesis.
The existence of a general dMTase(s) adds unexpected potential plasticity to DNA methylation patterns. The presence of a general enzyme that can remove well established methylation patterns and the potential for modulation of its activity by general signals (24) points to the possibility that the covalent structure of the genome could be modified postmitotically in response to extracellular signals, pathological as well as physiological.
Acknowledgments
We thank Dr. O. A. Memer for help with the spectrometry. This study was supported by the National Cancer Institute of Canada.
ABBREVIATIONS
- dMTase
DNA demethylase
- mdCMP
methyl-dCMP
Footnotes
A Commentary on this article begins on page 5894.
References
- 1.Lalande M. Annu Rev Genet. 1996;30:173–195. doi: 10.1146/annurev.genet.30.1.173. [DOI] [PubMed] [Google Scholar]
- 2.Efstratiadis A. Curr Opin Genet Dev. 1994;4:265–280. doi: 10.1016/s0959-437x(05)80054-1. [DOI] [PubMed] [Google Scholar]
- 3.Beard C, Li E, Jaenish R. Genes Dev. 1995;9:2325–2334. doi: 10.1101/gad.9.19.2325. [DOI] [PubMed] [Google Scholar]
- 4.Mohandas T, Sparkes R S, Shapiro L J. Science. 1981;211:393–396. doi: 10.1126/science.6164095. [DOI] [PubMed] [Google Scholar]
- 5.Challita P M, Skelton D, el-Khoueiry A, Yu X J, Weinberg K, Kohn D B. J Virol. 1995;69:748–755. doi: 10.1128/jvi.69.2.748-755.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shinar D, Yoffe O, Shani M, Yaffe D. Differentiation. 1989;41:116–126. doi: 10.1111/j.1432-0436.1989.tb00739.x. [DOI] [PubMed] [Google Scholar]
- 7.Razin A, Riggs A D. Science. 1980;210:604–610. doi: 10.1126/science.6254144. [DOI] [PubMed] [Google Scholar]
- 8.Eden S, Cedar H. Curr Opin Genet Dev. 1994;4:255–259. doi: 10.1016/s0959-437x(05)80052-8. [DOI] [PubMed] [Google Scholar]
- 9.Nan X, Campoy F J, Bird A. Cell. 1997;88:471–481. doi: 10.1016/s0092-8674(00)81887-5. [DOI] [PubMed] [Google Scholar]
- 10.Siegfried Z, Cedar H. Curr Biol. 1997;7:305–307. doi: 10.1016/s0960-9822(06)00144-8. [DOI] [PubMed] [Google Scholar]
- 11.Razin A, Szyf M. Biochim Biophys Acta. 1984;782:331–342. doi: 10.1016/0167-4781(84)90043-5. [DOI] [PubMed] [Google Scholar]
- 12.Brandeis M, Ariel M, Cedar H. BioEssays. 1993;15:709–713. doi: 10.1002/bies.950151103. [DOI] [PubMed] [Google Scholar]
- 13.Kafri T, Gao X, Razin A. Proc Natl Acad Sci USA. 1993;90:10558–10562. doi: 10.1073/pnas.90.22.10558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Monk M, Boubelik M, Lehnert S. Development. 1987;99:371–382. doi: 10.1242/dev.99.3.371. [DOI] [PubMed] [Google Scholar]
- 15.Shemer R, Eisenberg S, Breslow J L, Razin A. J Biol Chem. 1991;266:23676–23681. [PubMed] [Google Scholar]
- 16.Stein R, Gruenbaum Y, Pollack Y, Razin A, Cedar H. Proc Natl Acad Sci USA. 1982;79:61–65. doi: 10.1073/pnas.79.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Razin A, Szyf M, Kafri T, Roll M, Giloh H, Scarpa S, Carotti D, Cantoni G L. Proc Natl Acad Sci USA. 1986;83:2827–2831. doi: 10.1073/pnas.83.9.2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vairapandi M, Duker N J. Nucleic Acids Res. 1993;21:5323–5327. doi: 10.1093/nar/21.23.5323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jost J P, Siegmann M, Sun L J, Leung R. J Biol Chem. 1995;270:9734–9739. doi: 10.1074/jbc.270.17.9734. [DOI] [PubMed] [Google Scholar]
- 20.Weiss A, Keshet I, Razin A, Cedar H. Cell. 1996;87:709–718. doi: 10.1016/s0092-8674(00)80146-4. [DOI] [PubMed] [Google Scholar]
- 21.Fremont M, Siegmann M, Gaulis S, Matthies R, Hess D, Jost J P. Nucleic Acids Res. 1997;25:2375–2380. doi: 10.1093/nar/25.12.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bhattacharya S K, Ramchandani S, Cervoni N, Szyf M. Nature (London) 1999;397:579–584. doi: 10.1038/17533. [DOI] [PubMed] [Google Scholar]
- 23.Szyf M. Trends Pharmacol Sci. 1994;7:233–238. doi: 10.1016/0165-6147(94)90317-4. [DOI] [PubMed] [Google Scholar]
- 24.Szyf M, Theberge J, Bozovic V. J Biol Chem. 1995;270:12690–12696. doi: 10.1074/jbc.270.21.12690. [DOI] [PubMed] [Google Scholar]
- 25.Fisher P A, Wang T S, Korn D. J Biol Chem. 1979;254:6128–6137. [PubMed] [Google Scholar]
- 26.Copeland W C, Wang T S. J Biol Chem. 1991;266:22739–22748. [PubMed] [Google Scholar]
- 27.Gruenbaum Y, Cedar H, Razin A. Nature (London) 1982;295:620–622. doi: 10.1038/295620a0. [DOI] [PubMed] [Google Scholar]
- 28.Bestor T, Laudano A, Mattaliano R, Ingram V. J Mol Biol. 1988;203:971–983. doi: 10.1016/0022-2836(88)90122-2. [DOI] [PubMed] [Google Scholar]
- 29.Szyf M. Biochem Cell Biol. 1991;69:764–767. doi: 10.1139/o91-117. [DOI] [PubMed] [Google Scholar]
- 30.Szyf M, Tanigawa G, McCarthy P L. Mol Cell Biol. 1990;10:4396–4340. doi: 10.1128/mcb.10.8.4396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clark S J, Harrison C L, Paul C L, Frommer M. Nucleic Acids Res. 1994;22:2990–2997. doi: 10.1093/nar/22.15.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cedar H, Verdine G L. Nature (London) 1999;397:579–580. doi: 10.1038/17492. [DOI] [PubMed] [Google Scholar]