Abstract
This work presents an assay for total thiols and total disulfides in biological samples via HPLC quantification of 5-thio-2-nitrobenzoic acid (TNB) derived from the reaction of thiols with 5,5′-dithiobis(2-nitrobenzoic acid) (DTNB, Ellman’s reagent). This method also provides simultaneous quantification of glutathione (GSH) via the measurement of the GSH DTNB adduct (GSH-TNB). By using 326 nm as the detecting wavelength, the HPLC detection limit for TNB and the GSH-TNB adduct was determined to be 15 pmol and 7.5 pmol respectively. A recovery study with OVCAR-3 cells revealed that the recovery yields for TNB in the procedures for determining non-protein thiols, protein thiols, non-protein disulfides, and protein disulfides were 99.4±1.2% (n = 3), 98.1±5.0% (n = 3), 95.6±0.9% (n = 3), and 96.6±2.3% (n = 3) respectively. The recovery yield for GSH-TNB in the procedures for determining non-protein thiols, protein thiols, non-protein disulfides, and protein disulfides were 99.0±0.3% (n = 3), 95.1±4.9% (n = 3), 96.8±0.6% (n = 3), and 95.1±2.9% (n = 3) respectively. The reproducibility, expressed as the relative standard deviation for the analyte, for TNB was determined to be 2.8% (n = 6) for non-protein thiols, 3.9% (n = 6) for protein thiols, 3.6% (n = 6) for non-protein disulfides and 4.6% (n = 6) for protein disulfides. The reproducibility for GSH-TNB was determined to be 1.6% (n= 6) for non-protein thiols and 2.6% (n = 6) for non-protein disulfides. By comparing the amount of GSH determined in a biological sample before NaBH4 reduction with that after the reduction, this method can provide information associated with thiol glutathionylation which would be useful for protein glutathionylation study. This method should be applicable to cellular, subcellular, protein, or other biomatrix samples for thiol and disulfide quantification and will be a useful analytical method in the study of thiol redox state and thiol glutathionylation.
1. Introduction
Thiol redox state (TRS), reflected by the ratio of thiols (-SH)/disulfides (-S-S-), is a critical parameter associated with various essential biochemical processes, such as regulation of protein function, stabilization of protein structures, protection of proteins against irreversible oxidation of critical cysteine residues, and regulation of enzyme functions and transcription [1–3]. There is also a growing body of evidence demonstrating that an abnormal thiol redox state is involved in the pathogenesis of a variety of diseases, such as chronic heart disease [4–5], rheumatoid arthritis [6], acquired immunodeficiency syndrome (AIDS) [7], Parkinson’s disease, Alzheimer’s disease, Friedreich’s ataxia (FRDA), multiple sclerosis and amyotrophic lateral sclerosis [8,9], cancer [10], diabetes [11], and liver disorder [12]. Therefore, determination of thiol redox states at the cellular, subcellular, protein, or biomatrix level can provide valuable information on various normal or abnormal biochemical processes.
In general, TRS is characterized by the levels of non-protein thiols (NP-SH) and protein thiols (P-SH), as well as various protein and non-protein disulfides (NP-S-S-NP, P-S-S-P, NP-S-S-P). NP-SH mainly include glutathione (GSH), cysteine, γ-glutamylcysteine, homocysteine, and coenzyme A [13]. Among these, GSH is the most abundant thiol in cells and is present in mM concentration [13]. NP-S-S-NP is made up of the oxidized forms of NP-SH and their mixed disulfides. Traditionally, GSH and its oxidized form GSSG are usually the thiol components emphasized in the literature as indicators of TRS whereas the contribution of other NP-SH, P-SH and their disulfides has been largely ignored, despite the fact that GSSG constitutes only a minor portion of the TRS component of NP-S-S-NP [13]. Consequently, numerous analytical methods have been developed for the characterization and quantification of GSH and GSSG while methods for total thiols and total disulfides are relatively limited.
Current analytical methods for total thiols generally involve a thiol selective reagent which can be either a fluorescent or non-fluorescent compound. In the case of a thiol selective fluorescent reagent, total thiols are determined based on fluorescence generated from a product derived from the reaction of thiols with the reagent [14–19]. With a non-fluorescent reagent such as 5,5′-dithiobis(2-nitrobenzoic acid) (DTNB, Ellman’s reagent) [13, 17, 20–22], 4,4′-dithiodipyridine (4-DTDP) [22–24], and n-octyldithionitrobenzoic acid (OTNB) [25], the quantification is generally based on the reaction of the reagent with a thiol to form a thiol-reagent adduct with a concomitant release of one equivalent of another product. Spectrophotometric quantification of the released product indirectly provides the quantity of the thiols. Total disulfides can be determined by reduction of disulfides to thiols with various reducing reagents, such as sodium borohydride (NaBH4), dithiothreitol (DTT), and dithioerythritol [26], followed by thiol determination. Subtraction of the thiols obtained before the reduction from that derived after the reduction provides the quantity of the total disulfides. In addition to these photometric methods, electrochemical assays have also been reported for thiol determination [27, 28]. Among all these methods, spectrophotometric quantification using Ellman’s reagent is by far the most commonly used assay for total thiol and total disulfide quantification [13, 17, 20–22]. Ellman’s reagent reacts with a thiol leading to the formation of a thiol-TNB adduct and a concomitant release of one equivalent of 5-thio-2-nitrobenzoic acid (TNB) (Figure 1). Quantification of the thiols is based on the released TNB which can be measured spectrophotometrically at 412 nm with a molar absorptivity of 14,150 M−1 cm−1 at a neutral pH [29]. One of the major problems of quantification by spectrophotometry is the interference by compounds which exhibit absorption at the measured wavelength, a phenomenon often encountered with a biological sample and already noticed for the Ellman’s spectrophotometric method [30]. In our study of cellular thiol redox states, we failed to accurately determine cellular total thiols and total disulfides with the Ellman’s spectrophotometric method for the same reason. One of the solutions to overcome this problem is to use HPLC to quantify analytes.
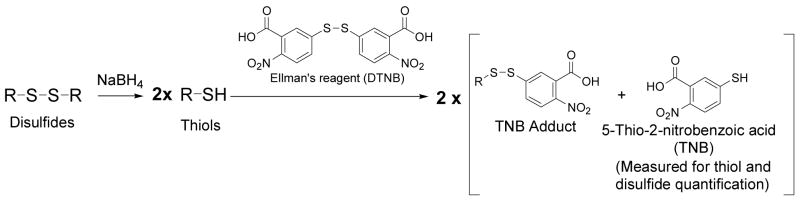
Figure 1.
Principle of thiol or disulfide quantification via measurement of TNB. R-SH can be protein or non-protein thiols. R-S-S-R can be protein or non-protein disulfides.
Recently, Hansen and co-workers reported an HPLC assay for protein thiols and disulfides based on the reaction of thiols with 4-DTDP [31]. 4-DTDP reacts with a thiol to form a 4-DTDP-thiol adduct with a concomitant release of one equivalent 4-thiopyridone (4-TP). HPLC quantification of 4-TP provides the quantities of total thiols or total disulfides after disulfides being reduced to thiols. Interestingly, despite the extensive use of Ellman’s reagent in total thiol quantification by spectrophotometric measurement of TNB, no report has been found in the literature for total thiol assays based on the HPLC quantification of TNB. A lack of an HPLC assay of TNB has been attributed to the low absorptivity of TNB at 412 nm under an acidic condition which is typical for most HPLC assays [31]. Indeed, when we conducted an HPLC analysis, TNB showed minimal absorption at 412 nm under the HPLC condition employed. The maximum absorption was found at 326 nm which was also noticed recently by Landino and co-workers [32]. Based on this observation, we have developed a total thiol and total disulfide assay via HPLC quantification of TNB. Total thiols were obtained based on TNB formed from the reaction of thiols with DTNB. Total disulfides were derived by subtracting total thiols obtained before NaBH4 reduction from that after the reduction. The principle of this assay is illustrated in Figure 1. This method was sensitive enough to detect total thiols and disulfides from samples derived from OVCAR-3 cells, a human ovarian cancer cell line. In addition, this method can also simultaneously quantify GSH through detection of the GSH-TNB adduct. An additional advantage of this method is that by knowing the quantities of GSH before NaBH4 reduction and GSSG in a biological sample, the quantity of GSH obtained after NaBH4 reduction provides information related to thiol glutathionylation, which would be useful in the study of thiol glutathionylation. An example of protein glutathionylation determination is also presented demonstrating the usefulness of this method in the study of protein glutathionylation. The method should be applicable to cellular, subcellular, protein, or other biomatrix samples for total thiol and total disulfide quantification, as well as thiol glutathionylation.
2. Materials and Methods
2.1 Materials
GSH, GSSG, p-aminobenzoic acid, ethylenediaminetetraacetic acid (EDTA), DTNB and trypsin were obtained from Sigma-Aldrich Chemical Co. (Milwaukee, WI). HPLC grade acetonitrile, NaBH4, and formic acid were obtained from Fisher Scientific (Pittsburgh, PA). Sulfosalicylic acid was purchased from J.T. Baker Chemical Co. (Phillipsburg, NJ). RPMI 1640 growth medium, fetal bovine serum (FBS), and penicillin/streptomycin were purchased from Mediatech, Inc (Herndon, VA). OVCAR-3 cells were obtained from the National Cancer Institute. Ultrapure water was prepared using a Milli-Q purification system (Millipore, Bedford, MA). 2-Acetylamino-3-[4-(2-acetylamino-2-carboxy-ethylsulfanylthiocarbonylamino)phenylthiocarbamoylsulfanyl]propionic acid 2-AAPA, a glutathione reductase inhibitor, was synthesized in this laboratory [33].
2.2 Solutions
Solutions of GSH (1.0 mg/ml), GSSG (1.0 mg/ml), sulfosalicylic acid (3%, 10%, w/w), and BSA (1.43 mg/ml) were dissolved in ultrapure water under argon. DTNB (19.8 mg/ml), p-aminobenzoic acid (10.0 mg/ml), and NaBH4 (283.7 mg/ml) were prepared in sodium phosphate buffer (0.15 M, pH 7.5). The stock solutions of GSH, GSSG, and BSA were stored at −80°C. The solution of NaBH4 was prepared fresh and stored over ice for one-day assay. 2-AAPA (1.0 mg/ml) was prepared in RPMI 1640 growth medium and stored at −80°C.
2.3 Preparation of OVCAR-3 cell lysate
Exponentially growing OVCAR-3 cells were cultured in a flask at 37°C in RPMI 1640 growth medium containing 10% FBS and 1% penicillin/streptomycin in a 5% CO2 incubator. Medium was discarded. Cells (5 million) were detached by trypsinization and washed with ice-cold PBS containing 1 mM EDTA (2 ml). One ml of ice-cold sulfosalicylic acid (3%, w/w) was added to the cell mixture. The mixture was sonicated over ice using a Misonix XL2020 sonicator with a cup horn probe (Farmingdale, NY) for 4 min. The lysed cells were centrifuged at 6800 × g for 1 min. The supernatant and lysate pellets were separated. The supernatant was used for the determination of non-protein thiols and non-protein disulfides. The lysate pellets were washed thoroughly with sulfosalicylic acid (3%, w/w, 1 ml × 3) to ensure that no residual non-protein thiols were left before being re-suspended in 1 ml of sulfosalicylic acid (3%, w/w) for the quantification of protein thiols and protein disulfides.
2.4 Preparation of OVCAR-3 cell lysate supernatant for non-protein thiol assays
OVCAR-3 cell lysate supernatant (100 μL) was vortex-mixed with a solution containing sodium phosphate buffer (0.15 M, pH 7.5, 1070 μl), DTNB (19.8 mg/ml, 50 μl), and internal standard p-aminobenzoic acid (10 mg/ml, 20 μl). The sample was allowed to stand at room temperature for 10 min followed by addition of HCl (36.5%, 50 μl) and centrifugation at 6800 × g for 1 min. The supernatant was subjected to HPLC analysis.
2.5 Preparation of OVCAR-3 cell lysate supernatant for non-protein disulfide assays
OVCAR-3 cell lysate supernatant (100 μl) was vortex-mixed with a solution of sodium phosphate buffer (0.5 M, pH 7.5, 200 μL) and NaBH4 (283.7 mg/ml, 120 μl). The mixture was incubated at 60°C for 30 min followed by slow addition of HCl (18.3%, 160 μL) over ice to adjust the pH to 1. The mixture was added to a solution of p-aminobenzoic acid (10 mg/ml, 20 μl) and DTNB (19.8 mg/ml, 70 μl) followed by neutralization with sodium phosphate buffer (0.5 M, pH 10, 590 μl). The mixture was allowed to stand at room temperature for 20 min followed by addition of HCl (36.5%, 50 μl) and centrifugation at 6800 × g for 1 min. The supernatant was subjected to HPLC analysis.
2.6 Preparation of OVCAR-3 cell lysate pellet suspension for protein thiol assays
The cell lysate pellet suspension (100 μl) was treated the same way as described in the assay for non-protein thiols except 1120 μl of sodium phosphate buffer (0.15 M, pH 7.5) was used.
2.7 Preparation of OVCAR-3 cell lysate pellet suspension for protein disulfide assays
The cell lysate pellet suspension (100 μl) was treated the same way as described in the assay for non-protein disulfides except ethanol (50 μl) was added during NaBH4 reduction to reduce foaming.
2.8 Protein glutathionylation determination
2-AAPA, which was found to produce protein glutathionylation in a monkey kidney cell line (unpublished results from this laboratory), was used to produce protein glutathionylation in OVCAR-3 cells. Exponentially growing OVCAR-3 cells (20 million) were suspended in 100 ml of RPMI 1640 growth medium containing 10% FBS, 1% penicillin/streptomycin and 2-AAPA (0.1 mM). The mixture was incubated at 37°C for 20 min in a 5% CO2 incubator. Cell lysate pellets were prepared as described in section 2.3 except the pellets was washed 10 times with 1 ml of sulfosalicylic acid solution (3%, w/w) to ensure that no residual non-protein thiols were left before being re-suspended in 1 ml of sulfosalicylic acid (3%, w/w). GSH released from proteins after NaBH4 reduction was determined as described in section 2.7.
2.9 Preparation of BSA samples for thiol determination (total thiols + total disulfides)
BSA was dissolved in water and purified on a Microcon YM-10 centrifugal filter unit with a molecular weight cut off of 10,000 (Millipore, Bedford, MA) to remove any trace of low-molecular weight thiols and disulfides. Sulfosalicylic acid (10%, 30 μL) was added to the BSA solution (1.43 mg/ml, 70 μL) and the resulting BSA solution was subjected to the procedure of the total protein disulfide assay except additional 100 μl ethanol was added to reduce foaming.
2.10 HPLC analysis
HPLC analysis was carried out on a Beckman Coulter HPLC (System Gold) system, controlled by a system controller (32Karat workstation with PC), equipped with a 125 gradient pumping module, a 508 autosampler with a sample cooling system, and a 168 photodiode array detector. The autosampler was set at 4°C. The HPLC conditions employed an Apollo C18 column (250 mm × 4.6 mm i.d., 5 μm) (Alltech, Deerfield, IL), solvent A [aqueous solution with 0.9% (v/v) formic acid] and solvent B (acetonitrile)]. Solvent B was first increased from 0% to 30% in 9 min, then to 80% in 7 min, and held at 80% for an additional 5 min. All flow-rates were 0.8 ml/min. The injection volume was 50 μl. 326 nm was employed as the detection wavelength.
3. Results
3.1 HPLC analysis
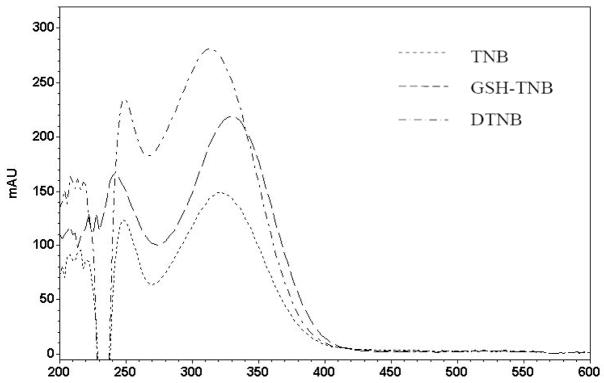
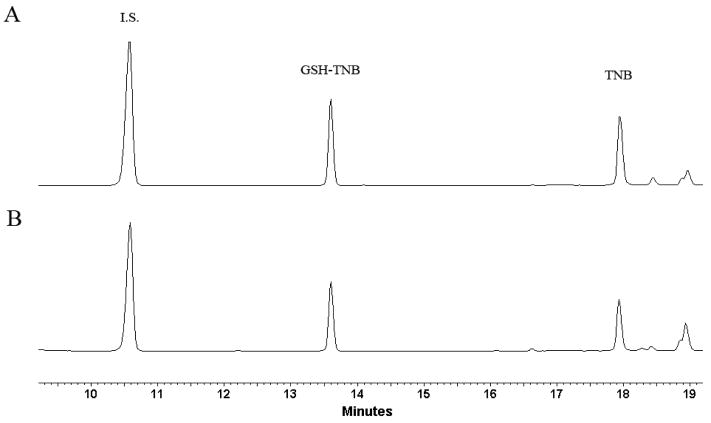
The initial HPLC assay employed 412 nm as the detection wavelength, the wavelength used in the spectrophotometric assay of TNB [29]. The separation of the analytes was readily achieved through a gradient mobile phase of acetonitrile and water containing 0.9% formic acid. However, the sensitivity of TNB at 412 nm was very low. To find an optimal detecting wavelength, an HPLC chromatogram of the analytes with a full scan detection (200 nm to 600 nm) was conducted. After examining the spectrum, TNB was found to exhibit a maximum absorption at 326 nm and minimal absorption at 412 nm under the HPLC condition (Figure 2). The wavelength of 326 nm was also found to be appropriate for the detection of DTNB and the GSH-TNB adduct (Figure 2). Therefore, 326 nm was chosen as the detection wavelength. Figure 3 provides representative chromatograms derived from a mixture of standards (Figure 3A) and a cell lysate sample (Figure 3B). DTNB is not included in the chromatogram due to its off scale. The HPLC detection limits (a signal to noise ratio of 3) for TNB and GSH-TNB were determined to be 15 pmol and 7.5 pmol, respectively. The retention time for p-aminobenzoic acid, GSH-TNB, and TNB were 10.6, 13.7, and 18.1 minutes, respectively.
Figure 2.
UV spectrum of TNB, GSH-TNB and DTNB obtained in a HPLC run of a standard sample containing GSH (15 μg/ml) and DTNB (39.6 μg/ml). The detector was set to monitor the wavelengths from 200 to 600 nm. The composition of the mobile phase when GSH-TNB, TNB, and DTNB were eluated is estimated to be acetonitrile: 0.9% formic acid in water = 3:2, 4:1, and 4:1.
Figure 3.
Representative HPLC chromatograms derived from A: GSH (27 μg/ml) derivatized by DTNB (1.1 mg/ml) in the presence of p-aminobenzoic acid (I.S.) (0.15 μg/ml) with 50 μl loaded to HPLC; B: OVCAR-3 cell lysate supernatant prepared for thiol analysis as described under Materials and Methods.
3.2 Reproducibility
Reproducibility studies were carried out in six replicates for TNB and GSH-TNB in OVCAR-3 cell lysate supernatant and cell lysate pellet suspension. The reproducibility, expressed as the relative standard deviation for the analyte, for TNB was determined to be 2.8% (n = 6) for non-protein thiols, 3.9% (n = 6) for protein thiols, 3.6% (n = 6) for non-protein disulfides and 4.6% (n = 6) for protein disulfides. The reproducibility for GSH-TNB was determined to be 1.6% (n = 6) for non-protein thiols and 2.6% (n = 6) for non-protein disulfides.
3.3 Recovery study
GSH and GSSG were employed to determine the recovery yields of total thiols, total disulfides and GSH. The recovery study was conducted by spiking OVCAR-3 cell lysate supernatant (for non-protein part) or cell lysate pellet suspension (for protein part) with GSH (95.7 nmol/million cells) and GSSG (48.0 nmol/million cells). After subtraction of TNB, GSH-TNB derived from the endogenous thiols and disulfides, the recovery yields for TNB with the procedures for determining non-protein thiols, protein thiols, non-protein disulfides, and protein disulfides were 99.4±1.2% (n = 3), 98.1±5.0% (n = 3), 95.6±0.9% (n = 3), and 96.6±2.3% (n = 3) respectively. The recovery yield for GSH-TNB with the procedures for determining non-protein thiols, protein thiols, non-protein disulfides, and protein disulfides were 99.0±0.3% (n = 3), 95.1±4.9% (n = 3), 96.8±0.6% (n = 3), and 95.1±2.9% (n = 3) respectively.
3.4 Determination of GSH-TNB and TNB stability in cell homogenate
The stability of GSH-TNB and TNB in cell lysate samples for HPLC assays was studied. GSH-TNB was stable at room temperature for one day and no decomposition was observed when stored at −80 °C for one month. TNB did not appear to be stable in the samples. There were 7%, 2%, 26%, and 15% decomposition of TNB observed in the samples for the determination of non-protein thiols, protein thiols, non-protein disulfides, and protein disulfides respectively in 1 h at 4 °C. However, samples after DTNB derivatization but before HCl acidification (please refer to section 2.4 or 2.5 for sample preparation) were stable at −80°C for one month. To avoid TNB decomposition, samples after DTNB derivatization but before HCl acidification were stored at −80 °C until HPLC analysis.
3.5 Quantification of thiols, disulfides, and GSH in cell lysate supernatant and cell lysate pellet suspension of OVCAR-3 cells
Quantification of thiols by HPLC was based on the peak area ratio of TNB to the internal standard (I.S.) and determined by reference to a TNB standard curve expressed as GSH equivalents. The TNB standard curves were constructed based on TNB formed in cell lysate supernatant (for non-protein thiols) or cell lysate pellet suspension (for protein thiols) spiked with various known amounts of GSH, and subsequent subtraction of the TNB detected in the supernatant or suspension with no added GSH. Results of non-protein thiols and protein thiols determined in OVCAR-3 cells are presented in Tables 1 and 2 respectively.
Table 1.
Non-protein thiols, disulfides, and GSH determined in OVCAR-3 cells. The data are presented as the means ± S.D. of three independent determinations of the same sample.
| Thiols | ||
|---|---|---|
| Cell lysate supernatant | Non-protein thiols (NP-SH)a | GSH (nmol/million cells) |
| Before NaBH4 reduction | 12.81±0.25 | 12.16±0.05 |
| After NaBH4 reduction | 15.77±0.41 | 13.52±0.04 |
| Disulfides | ||
| Non-protein disulfidesb | Non-protein disulfides derived from mixed disulfides with GSHc | |
| 1.48±0.20 | 0.68±0.03 | |
expressed as GSH equivalents, nmol/million cells
; (expressed as GSSG equivalents, nmol/million cells)
; (expressed as GSSG equivalents, nmol/million cells)
Table 2.
Protein thiols, protein disulfides and GSH determined in OVCAR-3 cells. The data are presented as the means ± S.D. of three independent determinations of the same sample.
| Thiols | ||
|---|---|---|
| Cell lysate pellet suspension | Protein thiols (P-SH)a | GSH (nmol/million cells) |
| Before NaBH4 reduction | 30.96±0.23 | Not detected |
| After NaBH4 reduction | 33.69±1.43 | Not detected |
| Disulfides | ||
| Protein disulfidesb | Protein disulfides derived from mixed disulfides with GSHc | |
| 1.37±0.83 | Not detected | |
expressed as GSH equivalents, nmol/million cells
; (expressed as GSSG equivalents, nmol/million cells)
; (expressed as GSSG equivalents, nmol/million cells)
Quantification of disulfides was achieved based on the formula presented below. Thiols in the formula was quantified similarly as discussed above except the TNB standard curves for the disulfides were constructed with GSSG instead of GSH. The samples for TNB standard curve construction were subjected to the same procedures for the determination of non-protein disulfides or protein disulfides.
| 1) |
Thiols are expressed in GSH equivalents.
The quantification results of non-protein and protein disulfides are presented in Tables 1 and 2 respectively.
Quantification of GSH was achieved via the measurement of GSH-TNB from the same HPLC chromatogram (Figure 3) and based on the peak area ratio of GSH-TNB to the internal standard and determined by reference to a GSH-TNB standard curve. The GSH-TNB standard curve can be conveniently obtained from the corresponding HPLC chromatograms for the TNB standard curve. GSH was detected in cell lysate supernatant samples both before and after NaNH4 reduction (Table 1). However, no GSH was detected in cell lysate pellet samples (Table 2). In order to observe protein glutathionylation, OVCAR-3 cells were treated with 2-AAPA (0.1 mM). No GSH was detected in 2-AAPA-treated OVCAR-3 cell lysate pellet samples before NaBH4 reduction. However, 2.15±0.12 nmol GSH/million cells were detected upon reduction of the 2-AAPA-treated OVCAR-3 cell lysate pellet samples with NaBH4 revealing that significant protein glutathionylation occurred.
Although no reports are available on the quantities of non-protein thiols, protein thiols, non-protein disulfides, and protein disulfides in OVCAR-3 cells, the quantity of GSH determined to be 12.16±0.05 nmol/million cells before reduction in non-protein thiol samples by this method (Table 1) is in agreement with the literature reported values for the cells, which are between 7.9±1.3 nmol/million cells [34] and 29.5 nmol/million cells [35].
3.6 Determination of the total thiols + total disulfides of BSA
To validate this methodology, the total thiols + total disulfides of BSA were determined. A TNB standard curve was constructed by spiking a solution of BSA in water (1.43 mg/ml, 70 μl) and sulfosalicylic acid (10%, 30 μl) with various amount of GSSG. BSA was determined to contain 35.02±0.83 thiols/BSA molecule (n = 3) after NaBH4 reduction consistent with the reported value (35 thiols/BSA molecule) [31].
4. Discussion
One of the challenges in thiol assay is the oxidation of thiol during sample treatment. Thiol oxidation can lead to the formation of disulfide which changes the ratio of thiol/disulfide. An acidic condition minimizes thiol oxidation by suppressing the ionization of thiols to thiol anions which are much more easily oxidized than unionized thiol. Further, some peptide thiols, such as GSH, can be degraded by enzymes resulting in analytical errors of the analyte. This phenomenon is especially serious when kidney samples are involved [36]. Therefore, an acidic solution was employed for the preparation of cell lysate. An acidic solution has been frequently used in the preparation of biological samples in thiol assays [36]. An additional advantage of using an acidic solution for the preparation of cell lysate is that it makes determination of protein thiols and disulfides easier since proteins are denatured under an acidic condition. Denaturing proteins is essential in determining thiols and disulfides buried inside proteins [31]. However, derivatization of thiols with DTNB occurs much more rapidly under neutral conditions than acidic conditions [37]. To prepare the acidic sample for DTNB derivatization, a high concentration phosphate buffer (pH 7.5) was used to quickly increase the pH of the sample to 7.5. Derivatization was found to be complete within 20 min. It needs to be noted that significant DTNB hydrolysis occurs when pH is >8 [38]. Riener and co-workers reported that 5% DTNB hydrolysis was observed in 48 h when the pH was 8 and the hydrolysis became much more significant as the pH increased [38]. Since the hydrolysis product of DTNB is TNB, over estimation of thiols will occur when the pH of DTNB derivatization is over 8. Therefore, caution needs to be taken to avoid high pH in DTNB derivatization. Under the condition employed in this method, no DTNB hydrolysis was observed.
In disulfide determination, NaBH4 was chosen to reduce disulfides for its convenience, clean reduction, and easy removal by acid [26, 31, 39]. Acid destroys NaBH4 through converting the hydride to H2 which is released in the form of bubble from the sample. A high concentration (0.5 M) of phosphate buffer (pH 7.5) was used to bring the pH of the acidic cell lysate sample to 7.5 for NaBH4 reduction. The time required for reduction of disulfides in samples was 30 min. Ethanol was added to reduce foaming during the reduction, and the residual NaBH4 was removed by HCl. It is essential that enough HCl is added to remove NaBH4 completely since NaBH4 can reduce DTNB to TNB leading to an over estimation of total thiols. The final pH of the solution should be less than 1 in order to completely destroy NaBH4. The acidic solution was then neutralized to pH 7.5 with a sodium phosphate buffer (0.5 M, pH 10) for DTNB derivatization. A strong base, such as NaOH solution, was not used here to avoid DTNB hydrolysis.
The application of this methodology was demonstrated with OVCAR-3 cells. With five millions cells, this method easily detected non-protein thiols, non-protein disulfides, GSH, protein thiols and protein disulfides (Tables 1 and 2). A comparison of the GSH quantities before and after NaBH4 reduction in cell lysate supernatant provides a piece of valuable information related to thiol glutathionylation (Table 1). Under a normal condition, the ratio of GSH:GSSG is about 100:1 [40]. Consistent with this ratio, GSSG in the cell lysate supernatant of OVCAR-3 cells was determined by liquid chromatography/mass spectrometry (LC/MS) to be only 0.060 ± 0.003 nmol/million cells (unpublished data from this laboratory), which indicates that the reduction of GSSG in the supernatant should only lead to an ~0.12 nmol/million cells increase in GSH. However, subtraction of GSH before the reduction (12.16±0.05 nmol/million cells) from that after the reduction (13.52±0.04 nmol/million cells) shows that the NaBH4 reduction led to a 0.68±0.03 nmol/million cells increase in GSH. This amount was much higher than the contribution from GSSG, indicating that the extra increase in GSH was derived from the release of GSH from glutathione-mixed disulfides (NP-S-S-G), a piece of valuable information related to thiol glutathionylation. The ratio of GSH released from NP-S-S-G vs GSH in OVCAR-3 was about 0.1:1 (Table 1) consistent with the ratios reported in most other biological samples [36]. No GSH release from proteins was detected in OVCAR-3 cell lysate pellets (Table 2). To further demonstrate that this method could be employed for the determination of protein glutathionylation, OVCAR-3 cells were treated with 2-AAPA (0.1 mM) for 20 min. Significant protein glutathionylation (2.15±0.12 nmol GSH/million cells) was detected demonstrating that this method can be used for the determination of protein glutathionylation.
To further validate this methodology, we determined the total thiols + total disulfides of BSA. Our data are consistent with the literature reported value which further confirms the validity of the method.
5. Conclusion
In summary, we have developed an HPLC method for the determination of thiols and disulfides. The method overcomes the interference problem associated with spectrophotometric thiol determination by Ellman’s reagent. This method will be useful in the study of thiol redox state and thiol glutathionylation.
Acknowledgments
This work was supported by grants from the National Institutes of Health (CA098810-01, CA120062-01) and 2005 Governor Rounds’ Individual Research Seed Grant Awards.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ghezzi P. Biochem Soc Trans. 2005;33:1378–1381. doi: 10.1042/BST0331378. [DOI] [PubMed] [Google Scholar]
- 2.Ghezzi P, Bonetto V, Fratelli M. Antioxid Redox Signal. 2005;7:964–972. doi: 10.1089/ars.2005.7.964. [DOI] [PubMed] [Google Scholar]
- 3.Biswas S, Chida AS, Rahman I. Biochem Pharmacol. 2006;71:551–564. doi: 10.1016/j.bcp.2005.10.044. [DOI] [PubMed] [Google Scholar]
- 4.Ferrari R, Alfieri O, Curello S, Ceconi C, Cargnoni A, Marzollo P, Pardini A, Caradonna E, Visioli O. Circulation. 1990;81:201–211. doi: 10.1161/01.cir.81.1.201. [DOI] [PubMed] [Google Scholar]
- 5.Burrell CJ, Blake DR. Br Heart J. 1989;61:4–8. doi: 10.1136/hrt.61.1.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reglinski J, Smith WE, Brzeski M, Marabani M, Sturrock RD. J Med Chem. 1992;35:2134–2137. doi: 10.1021/jm00089a026. [DOI] [PubMed] [Google Scholar]
- 7.Kalebic T, Kinter A, Poli G, Anderson ME, Meister A, Fauci AS. Proc Natl Acad Sci U S A. 1991;88:986–990. doi: 10.1073/pnas.88.3.986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calabrese V, Lodi R, Tonon C, D’Agata V, Sapienza M, Scapagnini G, Mangiameli A, Pennisi G, Stella AM, Butterfield DA. J Neurol Sci. 2005;233:145–162. doi: 10.1016/j.jns.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 9.Patsoukis N, Papapostolou I, Zervoudakis G, Georgiou CD, Matsokis NA, Panagopoulos NT. Neurosci Lett. 2005;376:24–28. doi: 10.1016/j.neulet.2004.11.019. [DOI] [PubMed] [Google Scholar]
- 10.Valko M, Rhodes CJ, Moncol J, Izakovic M, Mazur M. Chem Biol Interact. 2006;160:1–40. doi: 10.1016/j.cbi.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 11.Dursun E, Timur M, Dursun B, Suleymanlar G, Ozben TJ. Diabetes Complications. 2005;19:142–146. doi: 10.1016/j.jdiacomp.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 12.Cesaratto L, Vascotto C, D’Ambrosio C, Scaloni A, Baccarani U, Paron I, Damante G, Calligaris S, Quadrifoglio F, Tiribelli C, Tell G. Free Radic Res. 2005;39:255–268. doi: 10.1080/10715760400029603. [DOI] [PubMed] [Google Scholar]
- 13.Patsoukis N, Georgiou CD. Anal Bioanal Chem. 2004;378:1783–1792. doi: 10.1007/s00216-004-2525-1. [DOI] [PubMed] [Google Scholar]
- 14.Toyo’oka T, Imai K. Anal Chem. 1984;56:2461–2484. [Google Scholar]
- 15.Ayers FC, Warner GL, Smith KL, Lawrence DA. Anal Biochem. 1986;154:186–193. doi: 10.1016/0003-2697(86)90513-0. [DOI] [PubMed] [Google Scholar]
- 16.Winters RA, Zukowski J, Ercal N, Matthews RH, Spitz DR. Anal Biochem. 1995;227:14–21. doi: 10.1006/abio.1995.1246. [DOI] [PubMed] [Google Scholar]
- 17.Wright SK, Viola RE. Anal Biochem. 1998;265:8–14. doi: 10.1006/abio.1998.2858. [DOI] [PubMed] [Google Scholar]
- 18.Patsoukis N, Georgiou CD. Anal Bioanal Chem. 2005;383:923–929. doi: 10.1007/s00216-005-0095-5. [DOI] [PubMed] [Google Scholar]
- 19.Pullela PK, Chiku T, Carvan MJ, 3rd, Sem DS. Anal Biochem. 2006;352:265–273. doi: 10.1016/j.ab.2006.01.047. [DOI] [PubMed] [Google Scholar]
- 20.Ellman GL. Arch Biochem Biophys. 1959;82:70–77. doi: 10.1016/0003-9861(59)90090-6. [DOI] [PubMed] [Google Scholar]
- 21.Riddles PW, Blakeley RL, Zerner B. Anal Biochem. 1979;94:75–81. doi: 10.1016/0003-2697(79)90792-9. [DOI] [PubMed] [Google Scholar]
- 22.Riener CK, Kada G, Gruber HJ. Anal Bioanal Chem. 2002;373:266–276. doi: 10.1007/s00216-002-1347-2. [DOI] [PubMed] [Google Scholar]
- 23.Grassetti DR, Murray JF., Jr Arch Biochem Biophys. 1967;119:41–49. doi: 10.1016/0003-9861(67)90426-2. [DOI] [PubMed] [Google Scholar]
- 24.Egwim IOC, Gruber HJ. Anal Biochem. 2001;288:188–194. doi: 10.1006/abio.2000.4891. [DOI] [PubMed] [Google Scholar]
- 25.Faulstich H, Tews P, Heintz D. Anal Biochem. 1993;208:357–362. doi: 10.1006/abio.1993.1061. [DOI] [PubMed] [Google Scholar]
- 26.Bramanti E, Lomonte C, Onor M, Zamboni R, Raspi G, D’Ulivo A. Anal Bioanal Chem. 2004;380:310–318. doi: 10.1007/s00216-004-2746-3. [DOI] [PubMed] [Google Scholar]
- 27.Gevondyan NM, Gevondyan VS, Gavrilyeva EE, Modyanov NN. FEBS Lett. 1989;255:265–268. doi: 10.1016/0014-5793(89)81103-2. [DOI] [PubMed] [Google Scholar]
- 28.Nishiyama J, Kuninori T. Anal Biochem. 1992;200:230–234. doi: 10.1016/0003-2697(92)90457-i. [DOI] [PubMed] [Google Scholar]
- 29.Eyer P, Worek F, Kiderlen D, Sinko G, Stuglin A, Simeon-Rudof V, Reiner E. Anal Biochem. 2003;312:224–227. doi: 10.1016/s0003-2697(02)00506-7. [DOI] [PubMed] [Google Scholar]
- 30.Willig S, Hunter DL, Dass PD, Padilla S. Vet Hum Toxicol. 1996;38:249–253. [PubMed] [Google Scholar]
- 31.Hansen RE, Ostergaard H, Norgaard P, Winther JR. Anal Biochem. 2007;363:77–82. doi: 10.1016/j.ab.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 32.Landino LM, Mall CB, Nicklay JJ, Dutcher SK, Moynihan KL. Nitric Oxide. 2008;18:11–18. doi: 10.1016/j.niox.2007.09.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seefeldt T, Zhao Y, Chen W, Raza A, Carlson L, Herman J, Stoebner A, Hanson S, Foll R, Guan XX. J Biol Chem. 2008 doi: 10.1074/jbc.M802683200. in revision. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ozols RF, Louie KG, Plowman J, Behrens BC, Fine RL, Dykes D, Hamilton TC. Biochem Pharmacol. 1987;36:147–153. doi: 10.1016/0006-2952(87)90392-3. [DOI] [PubMed] [Google Scholar]
- 35.Ferretti A, D’Ascenzo S, Knijn A, Iorio E, Dolo V, Pavan A, Podo F. Br J Cancer. 2002;86:1180–1187. doi: 10.1038/sj.bjc.6600189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cotgreave IA, Moldéus P. J Biochem Biophys Methods. 1986;13:231–249. doi: 10.1016/0165-022x(86)90102-8. [DOI] [PubMed] [Google Scholar]
- 37.Guan X, Hoffman B, Dwivedi C, Matthees DP. J Pharm Biomed Anal. 2003;31:251–261. doi: 10.1016/s0731-7085(02)00594-0. [DOI] [PubMed] [Google Scholar]
- 38.Riener CK, Kada G, Gruber HJ. Anal Bioanal Chem. 2002;373:266–276. doi: 10.1007/s00216-002-1347-2. [DOI] [PubMed] [Google Scholar]
- 39.Brown WD. Biochim Biophys Acta. 1960;44:365–367. [Google Scholar]
- 40.Schirmer RH, Krauth-Siegel RL. In: Glutathione, Chemical, Biochemical, and Medical Aspects. Part A. Dolphin D, Poulson R, editors. III. Wiley; New York: 1999. pp. 553–596. [Google Scholar]