Abstract
Background
PspA is recognized as a major pneumococcal virulence factor and a possible vaccine candidate. The aim of this study was to analyze the PspA family and clade distribution among 112 Spanish pneumococci representatives of dominant clones among patients with invasive disease (n = 66) and nasopharyngeal healthy carriage in children (n = 46).
Results
PspA family 2 was predominant among invasive (63.6%) and carriage (54.3%) pneumococcal isolates. No PspA family 3 isolates were detected and only one strain was PspA negative. Although four clonal complexes contained strains of different clades, a clear association between clade and multi locus sequence typing results was found. Clades 1, 3 and 4 were associated with a wide variety of sequence types (ST) related to multiresistant and antibiotic-susceptible worldwide-disseminated clones. Clade 1 was associated with Spain6B-ST90, Spain14-ST18, Colombia5-ST289, Sweden1-ST306, Denmark14-ST230 and Sweden1-ST304 clones. Clade 3 was associated with Spain23F-ST81, Spain9V-ST156, Tennessee14-ST67, Netherlands3-ST180 and Netherlands7F-ST191 clones. Clade 4 was related to Sweden15A-ST63, Netherlands18C-ST113 and Greece21-ST193 clones. In contrast, PspA clade was not related to serotype, age or clinical origin of the isolates.
Conclusion
PspA clades were associated with genotypes. PspA family 2 and family 1 were dominant among major Spanish pneumococcal clones isolated from patients with invasive disease and nasopharyngeal carriage in children.
Background
Streptococcus pneumoniae is a major cause of serious community-acquired diseases (such as pneumonia, bacteremia or meningitis), especially in children, the elderly, and among patients with immunological disorders [1]. Nasopharyngeal colonization by S. pneumoniae is highly common, particularly among children attending day-care centers and in adults in long-term institutions [2].
Pneumococci are presently divided into 91 serotypes, which are defined by differences in their polysaccharide capsule [3,4]. Two serotype-based vaccines are currently available: the 23-valent polysaccharide vaccine (23V-PSV) which has been shown to be effective in the elderly [5-7], and the heptavalent pneumococcal conjugate vaccine (PCV7) which is used in children below the age of two [5]. In the USA the introduction of PCV7 in children was associated with a decrease in the incidence of invasive pneumococcal diseases (IPD) among children and adults [8]. Moreover, a decrease in IPD caused by resistant strains has also been observed [9]. Although current IPD rates are lower than those observed in the pre-vaccine period, recent reports have shown an increase in IPD caused by non-vaccine serotypes in the USA [10]. In Spain, since the introduction of PCV7, IPD rates due to PCV7 serotypes have decreased in both children and adults, but this improvement has been counterbalanced by an increase in IPD due to non-PCV7 serotypes [11,12].
Currently, two new conjugated vaccines are under development – 10-valent and the 13-valent vaccines, which both contain some emerging serotypes [13]. Alternative vaccines are also being evaluated, such as those based on pneumococcal virulence proteins. Many pneumococcal proteins have been investigated as vaccine candidates, for instance, pneumolysin, PsaA, PspC, and PspA [13,14]. The pneumococcal surface protein A (PspA) is an important virulence factor which interferes with complement deposition on the pneumococcal surface [15] and is detected in almost all pneumococci [16-18]. It is highly immunogenic and protective and has proved to be highly cross-reactive both in various animal models [15,19,20] and in humans [21]. It is hypothesized that a PspA-based vaccine could protect against invasive disease and also eliminate the carrier state [15-22]. PspA is constituted by five domains: a signal peptide, a α-helical charged domain which includes a clade-defining region, a proline-rich region, a choline-binding domain and a C-terminal domain [16]. Although the PspA encoding gene (pspA) is highly genetically variable, the classification by families is based on nucleotide and amino acid identity. Each of the three PspA families is subdivided into different clades: family 1 is composed by two clades (clade 1 and 2), family 2 comprises three clades (clades 3, 4 and 5), and PspA family 3 has only one divergent clade (clade 6) [16].
The aim of this study was to analyze the distribution of the PspA clades among a pneumococcal collection representative of major clones found in two previous studies among healthy children carriers [23] and patients with invasive disease [11].
Methods
Bacterial strains
One hundred and twelve pneumococcal strains previously characterized by pulsed field gel electrophoresis (PFGE) with SmaI restriction enzyme, as described elsewhere [24] and serotyped by Quellung reaction [25], were selected as follows:
a) Forty-nine pneumococci isolated from adults with IPD in Barcelona (NorthEast of Spain) between 1997 and 2007 (Additional file 1). These 49 strains were representative of the 32 major genotypes found among 968 pneumococci causing IPD in adult patients in Barcelona [11]. The pneumococcal strains selected were isolated from blood (n = 36), CSF (n = 11) and other sterile fluids (n = 2);
b) Seventeen pneumococci isolated from patients with IPD in Oviedo (Northern Spain) in 2004–2005 (Additional file 1) selected as representative of the nine major PFGE patterns among 101 consecutive invasive strains (unpublished data). The selected strains were isolated from blood (n = 11), CSF (n = 3) and other sterile fluids (n = 3);
c) Forty-six pneumococci were selected from nasopharyngeal carriers aged from 1 to 4 years old, in Oviedo (Northern Spain) in 2004–2005 [23] (Additional file 1). These strains were representative of 29 dominant PFGE patterns found among 365 pneumococci isolated from children attending 23 day-care centers.
Antimicrobial susceptibility testing
The minimal inhibitory concentration (MIC) was determined by microdilution following CLSI guidelines [26] using a panel of antimicrobials which included penicillin, erythromycin, clindamycin, tetracycline, chloramphenicol and cotrimoxazol. Resistant strains were defined according to CLSI criteria [27]. S. pneumoniae ATCC 49619 was used as control.
Multilocus sequence typing (MLST) and eBURST
MLST was performed as described previously [28]. The allele's number and sequence types (ST) were assigned using the pneumococcal MLST website [29]. Lineage assignment was achieved by eBURST analysis [30,31].
PspA detection
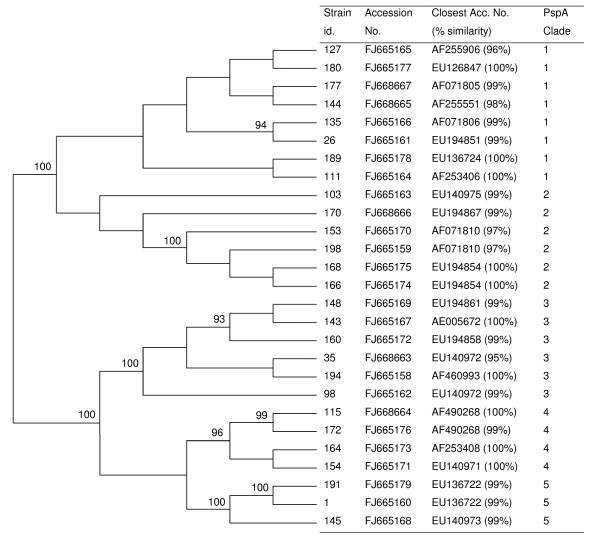
The PCRs were carried out in a standard PCR mixture of 50 μl containing 2.5 mM of MgCl2, 240 μM (each) of deoxynucleoside triphosphates (dNTPs), 0.3 μM of each primer, and 2 U of Taq DNA polymerase (AmpliTaq Gold®, Roche). The cycle conditions consisted of: an initial 94°C (10 min), 30 cycles of 94°C (1 min), 55°C (1 min) and 72°C (3 min), followed by 72°C (10 min). A multiplex PCR reaction was tested [32], but some samples did not amplify with LSM12/SKH63 [32,33] or LSM12/SKH52 [22] primer combinations. The combination of LSM12/SKH2 primers [16] was successfully used for all samples except one. The isolate that did not amplify was retested with the same cycle pattern at an annealing temperature of 52°C and with different primer combinations (LSM12/SKH63, LSM12/SKH52 and LSM12/SKH2). Controls for PspA family 1 (Spain14-ST18) and PspA family 2 (Spain23F-ST81) were run in each reaction set. PCR products were purified and sequenced using SKH2 primer, as described elsewhere [34]. Sequence edition was performed using the SeqScape version 2.1.1 (Applied Biosystems) software, while DNA sequences were assigned using BLAST [35]. Clade type was established when the closest match presented identity higher than 95% (Figure 1). The phylogenetic and molecular evolutionary analyses were conducted using MEGA4 version 4.1 software [36]. The evolutionary history was inferred using the Neighbor-Joining method and the bootstrap consensus tree inferred from 1000 replicates. The evolutionary distances were computed using the Kimura 2-parameter method [36].
Figure 1.
Phylogenetic tree of a 373-bp region that includes pspA clade-defining region. Phylogenetic and molecular evolutionary analyses were conducted with the MEGA4 program (version 4.1) [36] by the Neighbor-Joining method. Only bootstrap confidence intervals exceeding 90% are shown.
Nucleotide sequence accession numbers
The sequences of 27 isolates representing all the different sequences from this collection were deposited in GenBank with accession numbers: [GenBank:FJ665158, GenBank:FJ665159, GenBank:FJ665160, GenBank:FJ665161, GenBank:FJ665162, GenBank:FJ665163, GenBank:FJ665164, GenBank:FJ665165, GenBank:FJ665166, GenBank:FJ665167, GenBank:FJ665168, GenBank:FJ665169, GenBank:FJ665170, GenBank:FJ665171, GenBank:FJ665172, GenBank:FJ665173, GenBank:FJ665174, GenBank:FJ665175, GenBank:FJ665176, GenBank:FJ665177, GenBank:FJ665178, GenBank:FJ665179, GenBank:FJ668663, GenBank:FJ668664, GenBank:FJ668665, GenBank:FJ668666, GenBank:FJ668667].
Results and discussion
PspA families and clade distribution
Among the 112 pneumococci studied, the majority (59.8%, 67/112) were identified as belonging to PspA family 2 (31 isolates of clade 3, 27 of clade 4 and nine of clade 5), while the remaining 39.3% (44/112) belonged to family 1 (29 isolates of clade 1 and 15 of clade 2). One strain was negative. No PspA family 3 isolates were detected. Figure 1 shows the phylogenetic tree of the 27 new PspA sequences found as well as the accession numbers and the percentage of identity to previously published sequences. Sequences of strains of PspA families 1 and 2 were precisely grouped, and all were joined into their respective clades. The similarity of isolates of the same family ranged from 84% to 100%. The percentage of similarity within isolates of the same clade ranged as follows: clade 1 (84 to 95), clade 2 (84 to 100), clade 3 (93 to 99), clade 4 (91 to 98) and clade 5 (96 to 100).
Among the 66 pneumococci isolated from patients with IPD, 63,6% (42/66) were found to be of PspA family 2 (24 isolates of clade 3, 12 of clade 4 and six of clade 5), 34.8% (23/66) of family 1 (20 isolates of clade 1 and three of clade 2) and one isolate was negative. The high prevalence of PspA family 2 among pneumococci isolated from adults with IPD has already been reported in Spain, Canada, Sweden, the USA and France [37,38], although in Australia, the UK and Japan PspA family 1 was the most prevalent [38,39]. The dominance of family 2, clade 3 observed in our study has also been reported in other studies of pneumococci causing IPD in adults in France [37] and in children from Germany [40].
PspA family 2 was also dominant (54.3%, 25/46) among pneumococci isolated from the nasopharynx of healthy children (seven of clade 3, 15 of clade 4 and three of clade 5), while family 1 accounted for 45.7% (21/46) of the strains (nine of clade 1 and 12 of clade 2). These data are in agreement with two PspA studies [32,34] which found PspA family 2 to be dominant among pneumococci isolated from Brazilian children carriers. Moreover, the clade distribution also showed a prevalence of clade 4, followed by clade 1 and clade 3 [34]. A recent publication with data collected from pneumococci isolated from nasopharyngeal carriage in Finnish children showed similar prevalences of PspA family 1 and family 2 [41].
Additional file 1 shows the results for PspA family and clade of the 112 pneumococcal isolates studied with respect to sequence type (ST), serotype, resistance pattern and source. The pneumococci isolated from children carriers or from patients with IPD invasive disease seem to be indistinct, suggesting that PspA type is independent of age or clinical origin, as has been shown elsewhere [32,34].
Relationship between PspA and serotypes
In agreement with previous studies [16,32,42] our results showed that PspA clades are independent of serotypes. Pneumococci of the same serotype were associated with different PspA clades from the same or a different family (Additional file 1). For instance, pneumococci of serotype 6A could have PspA clade 2 (family 1), whereas pneumococci of serotype 6B could express PspA clades 1, 2, 4 or 5 (families 1 and 2). Since PspA is independent of serotype, PspA-based vaccines could improve upon the results obtained with serotype-based vaccines and might avoid a possible serotype replacement, as previously observed [10]. Since a PspA-based vaccine potentially has high coverage due to the fact that it is cross protective and immunogenic among children and adults [21], similar data should be investigated in other geographical areas in order to study the potential coverage of a PspA-based vaccine, and to adapt it to different formulations if necessary.
Relationship between PspA and clones
PspA clade classification was related to genotypes, and all strains with the same ST always presented the same PspA clade (see Additional file 1), regardless of origin or capsular type. In spite of the high genetical variability of pspA gene, all isolates of the same ST showed 100% of identity between their sequences. For instance, among nine pneumococci with ST63 obtained from invasive and carriage samples, four capsular types were found (15A, 19A, 19F and 23F) but all of them had PspA of clade 4 (see Additional file 1). However, other authors have found different PspA families among isolates that shared a common ST [41]. In our study, among 65 STs found, only 7 accounted for more than three isolates (ST63 n = 9, ST156 n = 5, and ST42, ST260, ST180, ST62 and ST81 with four isolates each). This fact may be a limitation of the present study and may affect its capacity to assess the relationship between ST and PspA.
The eBURST analysis reveals the presence of 15 clonal complexes (CC) and 22 singletons (S) (Additional file 1). The association of CC and S with clade was as follows: clade 1 (23 STs: 7 CC and 7 S), clade 2 (11 STs: 4 CC and 2 S), clade 3 (14 STs: 3 CC and 6 S), clade 4 (13 STs: 4 CC and 4 S), and clade 5 (4 STs: 1 CC and 3 S). Four CCs contained only clade 1-associated STs, three CCs contained clade 4-related STs, two CCs contained only clade 2-related STs, and two CC contained clade 3-related STs. Four CCs contained STs related to two different clades of the same or a different PspA family.
The relationship of PspA clade with multiresistant worldwide-disseminated clones described by the Pneumococcal Molecular Epidemiology Network (PMEN) [24] has been reported previously [32,42-44]. Our study provides further information since the majority of CCs found are related to PMEN clones. For instance, the Spain9V-ST156 (CC156) clone, which is one of the most important clones causing IPD worldwide [11,32,42,43], included six STs in the present study. All six STs of this CC had PspA clade 3, suggesting that PspA is highly conserved in this clone, even in SLV or DLV or when expressing capsular type 9 V or 14. Similar results were found among other CCs related to other multiresistant PMEN clones: Spain6B-ST90 (clade 1), Spain14-ST18 (clade 1), Denmark14-ST230 (clade 1), Spain23F-ST81 (clade 3), Greece21-ST193 (clade 4) and Sweden15A-ST63 (clade 4). The CC439 related to PMEN clone Tennessee23F-ST37, which included six STs in our study, had two PspA clades (1 and 4). This finding was in agreement with a study from Finland, which found PspA from families 1 and 2 among isolates within the same or different ST of this CC439 [41].
There is still little information about the relationship between PspA clade and antibiotic-susceptible PMEN clones, since the available data only refer to family level [42]. Our study provides new information about the antibiotic-susceptible clones, which are associated with the increase of IPD observed in recent years in some European countries [11,45] and in the USA [10]. For instance, the Sweden1-ST306 clone had clade 1. This clone has been described as the cause of IPD outbreaks in Europe and its frequency is currently increasing in Spain as cause of IPD and, especially, parapneumonic empyema in children [45]. CCs which were also related to antibiotic-susceptible PMEN clones included clade 1 (Colombia5-ST289 and Sweden1-ST304) and clade 3 (Netherlands7F-ST191, Netherlands3-ST180 and Tennessee14-ST67). Other associations of PspA clade with emerging clones were also observed such as clade 1 for serotype 22-ST433 and serotype 10A-CC97, and clade 5 for serotype 12-ST989. The CC53 (Netherlands8-ST53) included strains of two clades: clade 1 for those isolated with ST53 that were serotype 8, and clade 3 for isolates with ST62 (DLV) that were serotype 11A or non-typeable.
Since PspA type is associated with genotype, and with our knowledge of the clonal distribution of pneumococci causing IPD in Southern Barcelona area [11] we estimate that at least 45.1% would be of PspA family 2, and 23.4% of family 1. The most prevalent clades among invasive pneumococci would be clade 3 (48.2%) and clade 1 (33.7%). Similarly, we estimate that among the pneumococci isolated from children carriage [23] at least 31.6% appear to be PspA family 2 and 29.8% PspA family 1, with clade 3 (26.0%) and clade 1 (22.5%) being the most frequent.
Conclusion
Our study supports previous data [34,38,39,43] demonstrating that PspA family and clade distribution are independent of serotype, age and clinical origin of the isolates, but are highly associated with genotype. This study suggests that PspA family 1 and 2 molecules should be included in future PspA-based vaccine formulations. Further studies are needed to determine the genetic diversity of PspA in each geographical area.
Authors' contributions
CA and JL conceived the study and participated in its design. AF, RM and JL participated in field and clinical aspects of the study. DR and CA carried out the molecular genetic studies and sequence alignment. DR and CA wrote the manuscript, which was coordinated and critically reviewed by JL. All authors read and approved the final manuscript.
Supplementary Material
Table 1. Characteristics of 112 representative pneumococcal strains selected for this study.
Contributor Information
Dora Rolo, Email: dbispo@idibell.cat.
Carmen Ardanuy, Email: c.ardanuy@bellvitgehospital.cat.
Ana Fleites, Email: anamaria.fleites@sespa.princast.es.
Rogelio Martín, Email: r.martin@bellvitgehospital.cat.
Josefina Liñares, Email: fina.linares@bellvitgehospital.cat.
Acknowledgements
DR was supported by a grant from IDIBELL (Institut d'Investigació Biomèdica de Bellvitge). This work was supported by a grant from the Fondo de Investigaciones Sanitarias de la Seguridad Social (PI060647), and by CIBER de Enfermedades Respiratorias (CIBERES – CB06/06/0037), which is an initiative of the ISCIII – Instituto de Salud Carlos III, Madrid, Spain. We thank Dr. Adela G. de la Campa who offered critical review and helpful discussions. We are also grateful to our colleagues L. Calatayud, M. Alegre, E. Pérez and all staff of the Microbiology Laboratory of the Hospital Universitari de Bellvitge for their assistance with this project. We acknowledge the use of the Streptococcus pneumoniae MLST website [29], which is located at Imperial College London and is funded by the Wellcome Trust.
References
- Musher DM. Infections caused by Streptococcus pneumoniae: clinical spectrum, pathogenesis, immunity and treatment. Clin Infect Dis. 1992;14:801–807. doi: 10.1093/clinids/14.4.801. [DOI] [PubMed] [Google Scholar]
- Mato R, Sanches IS, Simas C, Nunes S, Carriço JA, Souza NG, Frazão N, Saldanha J, Brito-Avô A, Almeida JS, Lencastre HD. Natural history of drug-resistant clones of Streptococcus pneumoniae colonizing healthy children in Portugal. Microb Drug Resist. 2005;11:309–322. doi: 10.1089/mdr.2005.11.309. [DOI] [PubMed] [Google Scholar]
- Austrian R. The enduring pneumococcus: unfinished business and opportunities for the future. Microb Drug Resist. 1997;3:111–115. doi: 10.1089/mdr.1997.3.111. [DOI] [PubMed] [Google Scholar]
- Park IH, Pritchard G, Cartee R, Brandao A, Brandileone MCC, Nahm MH. Discovery of a new capsular serotyp (6C) within serogroup 6 of Streptococcus pneumoniae. J Clin Microbiol. 2007;45:1225–1233. doi: 10.1128/JCM.02199-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogaert D, Hermans PWM, Adrian PV, Rümke HC, Groot R. Pneumococcal vaccines: an update on current strategies. Vaccine. 2004;22:2209–2220. doi: 10.1016/j.vaccine.2003.11.038. [DOI] [PubMed] [Google Scholar]
- Mangtani P, Cutts F, Hall AJ. Efficacy of polysaccharide pneumococcal vaccine in adults in more developed countries: the state of the evidence. Lancet Infect Dis. 2003;3:71–78. doi: 10.1016/S1473-3099(03)00514-0. [DOI] [PubMed] [Google Scholar]
- Vila-Córcoles A, Ochoa-Gondar O, Hospital I, Ansa X, Vilanova A, Rodriguez T, Llor C. EVAN Study Group. Protective effects of the 23-valent pneumococcal polysaccharide vaccine in the elderly population: the EVAN-65 study. Clin Infect Dis. 2006;43:860–868. doi: 10.1086/507340. [DOI] [PubMed] [Google Scholar]
- Whitney CG, Farley MM, Hadler J, Harrison L, Bennett NM, Lynfield R, Reingold A, Cieslak PR, Pilishvili T, Jackson D, Facklam RR, Jorgensen H, Schuchat A. Active Bacterial Core Surveillance of the Emerging Infections Program Network. Decline in invasive pneumococcal disease after the introduction of protein-polysaccharide conjugate vaccine. N Engl J Med. 2003;348:1737–1746. doi: 10.1056/NEJMoa022823. [DOI] [PubMed] [Google Scholar]
- Kyaw MH, Lynfield R, Schaffner W, Craig AS, Hadler J, Reingold A, Thomas AR, Harrison LH, Bennett NM, Farley MM, Facklam RR, Jorgensen H, Besser J, Zell ER, Schuchat A, Whitney CG. Active Bacterial Core Surveillance of the Emerging Infections Program Network. Effect of introduction of the pneumococcal conjugate vaccine on drug-resistant Streptococcus pneumoniae. N Engl J Med. 2006;354:1455–1463. doi: 10.1056/NEJMoa051642. [DOI] [PubMed] [Google Scholar]
- Hicks LA, Harrison LH, Flannery B, Hadler JL, Schaffner W, Craig AS, Jackson D, Thomas A, Beall B, Pynfield R, Reingold A, Farley MM, Whitney CG. Active Bacterial Core Surveillance of the Emerging Infections Program Network. Incidence of pneumococcal disease due to non-pneumococcal conjugate vaccine (PCV7) serotypes in the United States during the era of widespread PCV7 vaccination, 1998–2004. J Infect Dis. 2007;196:1346–1354. doi: 10.1086/521626. [DOI] [PubMed] [Google Scholar]
- Ardanuy C, Tubau F, Pallares R, Calatayud L, Ángeles-Domínguez M, Rolo D, Grau I, Martín R, Liñares J. Epidemiology of invasive pneumococcal disease among adult patients in Barcelona before and after pediatric 7-valent pneumococcal conjugate vaccine introduction, 1997–2007. Clin Infect Dis. 2009;48:57–64. doi: 10.1086/594125. [DOI] [PubMed] [Google Scholar]
- Muñoz-Almagro C, Jordan I, Gene A, Latorre C, Garcia-Garcia JJ, Pallares R. Emergence of invasive pneumococcal disease caused by nonvaccine serotypes in the era of 7-valent conjugate vaccine. Clin Infect Dis. 2008;46:174–182. doi: 10.1086/524660. [DOI] [PubMed] [Google Scholar]
- Paton J, Boslego JW. In: Pneumococcal Vaccines: the Impact of Conjugate Vaccine. Siber GR, Klugman K, Mäkelä PH, editor. Washington DC:ASM Press; 2008. Protein Vaccines; pp. 421–35. [Google Scholar]
- Ogunniyi AD, Grabowicz M, Briles DE, Cook J, Paton C. Development of a vaccine against invasive pneumococcal disease based on combinations of virulence proteins of Streptococcus pneumoniae. Infect Immun. 2007;75:350–357. doi: 10.1128/IAI.01103-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren B, Szalai AJ, Thomas O, Hollingshead SK, Briles DE. Both family 1 and family 2 PspA proteins can inhibit complement deposition and confer virulence to a capsular serotype 3 strain of Streptococcus pneumoniae. Infect Immun. 2003;71:75–85. doi: 10.1128/IAI.71.1.75-85.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingshead SK, Becker R, Briles DE. Diversity of PspA: Mosaic genes and evidence for past recombination in Streptococus pneumoniae. Infect Immun. 2000;68:5889–5900. doi: 10.1128/IAI.68.10.5889-5900.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jedrzejas MJ. Pneumococcal virulence factors: structure and function. Microbiol Mol Biol Rev. 2001;65:187–207. doi: 10.1128/MMBR.65.2.187-207.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDaniel LS, Sheffield JS, Delucchi P, Briles DE. PspA, a surface protein of Streptococcus pneumoniae, is capable of eliciting protection against pneumococci of more than one capsular type. Infect Immun. 1991;59:222–228. doi: 10.1128/iai.59.1.222-228.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briles DE, Tart RC, Swiatlo E, Dillard JP, Smith P, Benton KA, Ralph BA, Brooks-Walter A, Crain MJ, Hollingshead SK, McDaniel LS. Pneumococcal diversity: considerations for new vaccine strategies with emphasis on pneumococcal surface protein A (PspA) Clin Microbiol Rev. 1998;11:645–657. doi: 10.1128/cmr.11.4.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darrieux M, Moreno AT, Ferreira DM, Pimenta FC, Andrade AL, Lopes AP, Leite LC, Miyaji EN. Recognition of pneumococcal isolates by antisera raised against PspA fragments from different clades. J Med Microbiol. 2008;57:273–278. doi: 10.1099/jmm.0.47661-0. [DOI] [PubMed] [Google Scholar]
- Nabors GS, Braun PA, Herrmann DJ, Heise ML, Pyle DJ, Gravenstein S, Schilling M, Ferguson LM, Hollingshead SK, Briles DE, Becker RS. Immunization of healthy adults with a single recombinant pneumococcal surface protein A (PspA) variant stimulates broadly cross-reactive antibodies to heterologous PspA molecules. Vaccine. 2000;18:1743–1754. doi: 10.1016/S0264-410X(99)00530-7. [DOI] [PubMed] [Google Scholar]
- Vela-Coral MC, Fonseca N, Castañeda E, Di Fabio JL, Hollingshead SK, Briles DE. Pneumococcal surface protein A of invasive Streptococcus pneumoniae isolates from Colombian children. Emerg Infect Dis. 2001;7:832–836. doi: 10.3201/eid0705.017510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleites A, Valdés E, Trabazo R, Ardanuy C, Fenoll A, Liñares J, The Spanish Pneumococcal Infection Study Network. Streptococcus pneumoniae colonizing healthy children attending Day Care Centers (DCCs): two years of annual surveillance. 46th Intersci Conf Antimicrob Agents Chemother. 2006. p. G-151.
- McGee L, McDougal L, Zhou J, Spratt BG, Tenover FC, George R, Hakenbeck R, Hryniewicz W, Lefévre JC, Tomasz A, Klugman KP. Nomenclature of major antimicrobial-resistant clones of Streptococcus pneumoniae defined by the pneumococcal molecular epidemiology network. J Clin Microbiol. 2001;39:2565–2571. doi: 10.1128/JCM.39.7.2565-2571.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenoll A, Jado I, Vicioso D, Pérez A, Casal J. Evolution of Streptococcus pneumoniae: serotypes and antibiotic resistance in Spain. Update 1990–1996. J Clin Microbiol. 1998;36:3447–3454. doi: 10.1128/jcm.36.12.3447-3454.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute. Methods for dilution antimicrobial susceptibility test for bacteria that growth aerobically. 7. Clinical and Laboratory Standards Institute, USA; 2006. p. M7-A6. [Google Scholar]
- Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing; Eighteenth Informational Supplement. Clinical and Laboratory Standards Institute, USA; 2008. p. M100-S18. [Google Scholar]
- Enright MC, Spratt BG. A multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology. 1998;144:3049–3060. doi: 10.1099/00221287-144-11-3049. [DOI] [PubMed] [Google Scholar]
- Streptococcus pneumoniae MLST database. http://spneumoniae.mlst.net/
- Feil EJ, Li BC, Aanensen DM, Hanage WP, Spratt BG. eBURST: Inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J Bacteriol. 2004;186:1518–1530. doi: 10.1128/JB.186.5.1518-1530.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- eBURST Website. http://eburst.mlst.net
- Brandileone MCC, Andrade ALSS, Teles EM, Zanella RC, Yara TI, Fabio JLD, Hollingshead SK. Typing of pneumococcal surface protein A (PspA) in Streptococcus pneumoniae isolated during epidemiological surveillance in Brazil: towards novel pneumococcal protein vaccines. Vaccine. 2004;22:3890–3896. doi: 10.1016/j.vaccine.2004.04.009. [DOI] [PubMed] [Google Scholar]
- Swiatlo E, Brooks-Walter A, Briles DE, McDaniel LS. Oligonucleotides identify conserved and variable regions of psp A and pspA-like sequences of Streptococcus pneumoniae. Gene. 1997;188:279–284. doi: 10.1016/S0378-1119(96)00823-2. [DOI] [PubMed] [Google Scholar]
- Pimenta FC, Ribeiro-Dias F, Brandileone MCC, Miyaji EN, Leite LCC, Andrade ALSS. Genetic diversity of PspA types among nasopharyngeal isolates collected during an ongoing surveillance study of children in Brazil. J Clin Microbiol. 2006;44:2838–2843. doi: 10.1128/JCM.00156-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basic Local Alignment Search Tool Website. http://blast.ncbi.nlm.nih.gov/Blast.cgi [DOI] [PubMed]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Baril L, Briles DE, Crozier P, King J, Punar M, Hollingshead SK, McCormick JB. Characterization of antibodies to PspA and PsaA in adults over 50 years of age with invasive pneumococcal disease. Vaccine. 2004;23:789–793. doi: 10.1016/j.vaccine.2004.07.033. [DOI] [PubMed] [Google Scholar]
- Hollingshead SK, Baril L, Ferro S, King J, Coan P, Briles DE. Pneumococcal Proteins Epi Study Group. Pneumococcal surface protein A (PspA) family distribution among clinical isolates from adults over 50 years of age collected in seven countries. J Med Microbiol. 2006;55:215–221. doi: 10.1099/jmm.0.46268-0. [DOI] [PubMed] [Google Scholar]
- Ito Y, Osawa M, Isozumi R, Imai S, Ito I, Hirai T. Kansai Community Acquired Pneumococcal Pneumonia Study Group. Pneumococcal surface protein A family types of Streptococcus pneumoniae from community-acquired pneumonia patients in Japan. Eur J Clin Microbiol Infect Dis. 2007;26:739–742. doi: 10.1007/s10096-007-0364-7. [DOI] [PubMed] [Google Scholar]
- Heeg C, Franken C, Linden MVD, Al-Lahham A, Reinert RR. Genetic diversity of pneumococcal surface protein A of Streptococcus pneumoniae meningitis in German children. Vaccine. 2007;25:1030–1035. doi: 10.1016/j.vaccine.2006.09.061. [DOI] [PubMed] [Google Scholar]
- Melin MM, Hollingshead SK, Briles DE, Hanage WP, Lahdenkari M, Kaijalainen T, Kilpi TM, Käyhty HM. Distribution of pneumococcal surface protein A families 1 and 2 among Streptococcus pneumoniae isolates from children in Finland who had acute otitis media or were nasopharyngeal carriers. Clin Vaccine Immunol. 2008;15:1555–1563. doi: 10.1128/CVI.00177-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadowy E, Skoczyñska A, Fiett J, Gniadkowski M, Hryniewicz W. Multilocus sequence types, serotypes, and variants of the surface antigen PspA in Streptococcus pneumoniae isolates from meningitis patients in Poland. Clin Vaccine Immunol. 2006;13:139–144. doi: 10.1128/CVI.13.1.139-144.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beall B, Gheraldi G, Facklam RR, Hollingshead SK. Pneumococcal PspA sequence types of prevalent pneumococcal strains in the United States and of internationally disseminated clones. J Clin Microbiol. 2000;38:3663–3669. doi: 10.1128/jcm.38.10.3663-3669.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dicuonzo G, Gheraldi G, Gertz RE, D'Ambrosio F, Goglio A, Lorino G, Recchia S, Pantosti A, Beall B. Genotypes of invasive pneumococcal isolates recently recovered from Italian patients. J Clin Microbiol. 2002;40:3660–3665. doi: 10.1128/JCM.40.10.3660-3665.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barricarte A, Castilla J, Gil-Setas A, Torroba L, Navarro-Alonso JA, Irisarri F, Arriazu M. Effectiveness of the 7-valent pneumococcal conjugate vaccine: a population-based case-control study. Clin Infect Dis. 2007;44:1436–1441. doi: 10.1086/516779. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table 1. Characteristics of 112 representative pneumococcal strains selected for this study.