Abstract
AIM: To investigate the clinicopathological roles of Bmi1 in esophageal squamous cell carcinoma (ESCC).
METHODS: Quantitative real-time polymerase chain reaction and immunohistochemical staining for Bmi1 were performed in cancerous and adjacent non-cancerous paraffin-embedded esophageal specimens.
RESULTS: The Bmi1 expression level was unaffected by gender and age. The level of Bmi1 mRNA in ESCC was significantly higher than that in the adjacent non-cancerous tissues (2.181 ± 2.158 vs 0.931 ± 0.894, P = 0.0152), and its over-expression was aggressively associated with lymph node metastasis (3.580 ± 2.487 vs 1.703 ± 0.758, P = 0.0003), poorer cell differentiation (P = 0.0000) and advanced pathological stage (3.827 ± 2.673 vs 1.590 ± 0.735, P = 0.0001). The patients were divided into high-expression and low-expression groups based on the median expression level of Bmi1 mRNA, and a shorter overall survival time in the former group was observed. Immunohistochemistry for Bmi1 oncoprotein showed diffusely positive, focally positive and negative expression in 44, 16 and 10 of 70 ESCC cases, respectively, compared with three, two and five of 10 adjacent non-cancerous cases (P = 0.027). The positive rate of the oncoprotein in samples of histological grade III was higher than that of grade II (P = 0.031), but its expression had no relation to the lymph node metastasis and pathological staging. In 70 ESCC samples, Bmi1 showed high intense expression in the cytoplasm and less or even no expression in the nucleus.
CONCLUSION: Bmi1 was over-expressed in ESCC. Increased Bmi1 mRNA expression was significantly associated with ESCC progression, and the oncoprotein was largely distributed in the cytoplasm of tumor cells.
Keywords: Esophageal squamous cell carcinoma, Bmi1, Quantitative real-time polymerase chain reaction, Immunohistochemistry, Clinicopathology
INTRODUCTION
Esophageal cancer is one of the most frequently occurring malignancies and the seventh leading cause of cancer-related deaths in the world. It exhibits considerable geographic variation, and 95% of tumors are esophageal squamous cell carcinoma (ESCC)[1]. Besides the impact of the environment, the process of esophageal tumorigenesis at the molecular level is related to disorders of cell amplification, differentiation, senescence and apoptosis. The genetic bases underlying esophageal tumorigenesis have been partly understood in the past few years, including a loss of the anti-oncogene p53 and over-expression of epidermal growth factor receptor or c-Myc[2]. However, other molecular mechanisms involved in esophageal tumorigenesis progress are still largely unknown.
Bmi1, located in 10p11.23, is a member of the polycomb group (PcG) and a component of the polycomb repressive complex 1. It was initially identified as an oncogene cooperating with c-Myc in the generation of lymphomas in double transgenic mice[3–5]. Several lines of evidence imply that Bmi1 plays an important role in the regulation of cell proliferation and senescence and is required for maintenance of adult hematopoietic and neural stem cells[6–9]. Bmi1 gene amplification is observed mainly in mantle cell lymphomas[10], and recent serial studies have shown that Bmi1 is overexpressed in many somatic solid tumors such as colon carcinoma, non-small cell lung cancer, breast cancer, head and neck squamous cell carcinoma and gastric carcinoma[11–15], and it may be of diagnostic and prognostic relevance. However, to date, no report about the role of Bmi1 in ESCC has been made. The up-regulation of c-Myc and the down-regulation of p53 and p16 in ESCC[2] tissues make it plausible that Bmi1 may play an important role in the initiation and development of ESCC. This study was designed to investigate Bmi1 expression in ESCC tissues and its impact on patients with ESCC.
MATERIALS AND METHODS
Ethics
The use of study specimens for analyses was approved by the Research Ethics Committee of Nanjing Medical University. Informed written consent was obtained from all the patients.
Case selection
From June 1997 to February 2000, 80 ESCC and 15 adjacent non-cancerous paraffin-embedded samples were obtained from the tumor center of Nanjing First Hospital affiliated to Nanjing Medical University. There were 52 male and 28 female patients with a mean age of 60 years (range: 41-82). The patients were given preoperative examination including biopsy for diagnosis, barium X-ray, CT and ultrasonic endoscopy for clinical staging, and no treatment was given before operation. Radical resection was performed in each patient, and all the samples underwent postoperative pathological examination. There were 54 cases of stage I-II and 26 cases of stage III-IV cancer according to the American Joint Committee on Cancer staging manual (AJCC, 2002)[16]. With regard to postoperative histological results, 16 were well-differentiated, 40 moderately differentiated and 24 poorly differentiated. Another 70 ESCC and 10 non-cancerous paraffin-embedded samples were enlisted from January 2002 to December 2003 in the same institution. There were 48 male and 22 female patients with a mean age of 61 years (range: 38-89). All the patients were assessed for physiological ability and endoscopy and CT scan were performed for clinical staging prior to routine surgery for ESCC. The postoperative pathological examination found 56 cases of stage I-II and 14 cases of III-IV cancer according to AJCC (2002) pTNM standards[16]. Clinical follow-up after surgery and diagnosis was based on periodic visits (every 3 mo during the first year, every 6 mo the second year, and then yearly until relapse).
RNA extraction and quantitative real-time polymerase chain reaction (PCR)
Real-time quantitative PCR was performed on paraffin-embedded sections from 80 ESCC patients and 15 adjacent non-cancerous samples. Briefly, total RNA was extracted by Recover All Total Nucleic Acid Isolation kit (Ambion), and 10 mg of DNase-treated total RNA was used for reverse transcription with Superscript III (Invitrogen, Carlsbad, CA, USA). An aliquot representing 100 ng input RNA was amplified by quantitative real-time PCR using the TaqMan PCR reagent kit and assay-on-demand gene expression products (FAM/Sybr, Foster City, CA, USA). RNA extracted from a non-cancerous lesion in one patient was used as a standard. After reverse transcription, standard cDNA was serially diluted to obtain five standard solutions for use in PCR to generate the reference curve. Sequences of the Bmi1 bidirectional primers were designed using Primer 5.0 rotor-gene 6.0 (Corbett Research) as follows: Bmi1 sense 5'-GTATTCCCTCCACCTCTTCTTG-3', Bmi1 antisense 5'-TGCTGATGACCCATTTACTGAT-3'. House-keeping gene: β-actin sense 5'-CCTGTACGCCAACACAGTGC-3', antisense 5'-ATACTCCTGCTTGCTGATCC-3'. Quantitative real-time PCR was carried out in a Rotor-Gene 3000 PCR kit (Corbett Research) with 10000 × Syber Green (Molecular Probes). After reverse transcription, standard cDNA was serially diluted to obtain five standard solutions for use in PCR to generate the reference curve. The relative amount of cDNA in each sample was measured by interpolation using the standard curve (Figure 1), and then the relative ratio of Bmi1 to β-actin (housekeeping gene) expression was calculated for each ESCC sample.
Figure 1.

Amplification curve (A) and standard curve (B) of quantitative real-time PCR for Bmi1 mRNA. Total RNA was extracted for subsequent reverse transcription, and standard cDNA was serially diluted to obtain five standard solutions (1 × 10-1, 1 × 10-2, 1 × 10-3, 1 × 10-4, 1 × 10-5) for use in PCR to generate the reference curve, slope rate of the straight line (a) = -3.42089, intercept (b) = 5.97517, correlation coefficient (r) = 0.9996. The strength of Bmi1 and β-actin was directly generated by the machine.
Immunohistochemistry
Histopathological evaluation was performed on 4-μm slides stained with hematoxylin and eosin (HE) (Figure 2). Commercially available rabbit monoclonal antibodies against Bmi-1 (1:100, Santa Cruz Biotechnology) were used as primary antibodies. A paraffin section of the ESCC sample was deparaffinized and rehydrated in graded alcohol to water. Antigenic enhancement was performed by submerging in citrate buffer (pH 6.0) and microwaving. Endogenous peroxide activity was quenched by applying 0.3% hydrogen peroxide for 10 min, followed by incubation with 1% BSA to block the non-specific binding. The primary monoclonal anti-Bmi1 antibody was incubated for 60 min at 37°C. After washing, the tissue section was reacted with the biotinylated anti-rabbit IgG, and visualized using a Dako Envision System horseradish peroxidase for monoclonal antibodies. The slides were immersed in the prepared diaminobenzidine solution, which produces a brown precipitate at the level of the antigen-primary antibody. Slides were then counterstained with hematoxylin, dehydrated through alcohols of increasing concentration, placed in xylene, coverslipped using Permount, and analyzed under light microscopy. Each section was evaluated by at least two independent professional pathologists, the distribution of Bmi-1 was scored on a semi-quantitative scale, the percentage of positive tumor cells was recorded and divided as follows: negative (< 10% of tumor cells positive), locally positive (10%-50% of tumor cells positive), and diffusely positive (> 50% of tumor cells positive).
Figure 2.

Staining of ESCC tissues. The tumor cells of cancerous tissues were stained as violet in the nucleus and pink in the cytoplasm (× 100).
Statistical analysis
Data were expressed as mean ± SD and analyzed using the Stata v9.0-CYGiSO bin (Computer Resource Center, USA). The significance of differences among groups was determined by Student’s t test and χ2 test or Fisher’s exact test. The difference in free survival between groups was analyzed by the Kaplan-Meier method and log-rank test. The starting point for calculating free survival was the date of surgery, and the endpoint was the date of death. Statistical significance was assessed at the two sided 5% level, and P values less than 0.05 were considered statistically significant.
RESULTS
Quantitative real-time PCR analysis
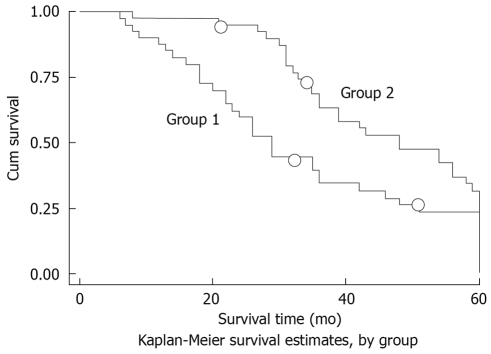
Clinical follow-up was made in 76 patients. The comparative expression levels were determined as a ratio between the Bmi1 and the housekeeping gene (β-actin) to correct for variation in the amounts of mRNA. The 5-year survival rate was 37.46%. The Bmi1 mRNA level was higher in the cancerous tissues than that in the non-cancerous tissues (2.181 ± 2.158 vs 0.931 ± 0.894, P = 0.0152). The Bmi1 expression level was unaffected by gender and age. The expression level of Bmi1 mRNA was much lower at the I-II stage than that at the III-IV stage, which varied inversely with the differentiation grade, and was lower in cases without metastatic lymph nodes than in those with metastatic lymph nodes (Table 1). Based on the detection of Bmi-1 median expression level (1.085), patients were divided into the down-expression group (Bmi1 mRNA level < 1.085) and the up-expression group (Bmi1 mRNA level > 1.085), and the accumulated survival rate was higher in the former than that in the latter (Figure 3).
Table 1.
Associations of Bmi-1 mRNA expression in ESCC tissues with clinicopathological characteristics (mean ± SD)
| Parameter | Bmi-1 mRNA | P |
| Age (yr) | ||
| ≥ 60 | 2.312 ± 2.171 | 0.3816 |
| < 60 | 2.167 ± 2.045 | |
| Gender | ||
| Male | 2.402 ± 2.359 | 0.2133 |
| Female | 1.770 ± 1.690 | |
| Lymph node metastases | ||
| Yes | 3.580 ± 2.487 | 0.0003 |
| No | 1.703 ± 0.758 | |
| Stage | ||
| III/IV | 3.827 ± 2.673 | 0.0001 |
| I/II | 1.590 ± 0.735 | |
| Histological grade | ||
| I | 0.881 ± 0.418 | 0.000 |
| II | 1.858 ± 0.979 | |
| III | 3.580 ± 2.487 |
Quantitative real-time PCR was employed to identify Bmi1 mRNA expression. The comparative expression levels were determined as a ratio between Bmi1 and housekeeping gene (β-actin) to correct variation in the amount of mRNA.
Figure 3.
Effects of Bmi1 mRNA expression on prognosis of ESCC patients. Two patients did not complete the 5-year follow-up in each group (small circles). The survival rate was higher in the down-expression group (group 2) than in the up-expression group (group 1), χ2 = 4.41, P = 0.0356.
Protein analysis
Bmi1 oncoprotein expression was diffusely positive, focally positive and negative in 44, 16 and 10 of 70 ESCC cases, respectively. Compared with three, two and five of 10 adjacent non-cancerous cases, Bmi1 protein was significantly increased in ESCC samples (P = 0.027). Analysis of protein localization in ESCC cells was made, and the tumor cells with Bmi1 staining were divided into three categories, referring to both the nucleus and cytoplasm. In all 70 tested ESCC tissues, Bmi1 presented highly intense expression in both nucleus and cytoplasm with varied degrees, accompanied by less or even no expression in the nucleus, with significant differences (Figure 4). The positivity of the oncoprotein in samples of histological grade III was more frequent than that of grade II, but no significant differences were observed between other differentiated grades (P = 0.031). No relationship was found between the Bmi1 protein expression and lymph node metastases, pathological staging and cell differentiation (Table 2). Clinical follow-up was made in 67 patients. The 5-year survival rate was 40.01%. There was no statistical difference in survival rates between groups according to the Bmi1 expression (P = 0.1704).
Figure 4.
Immunohistochemical staining of ESCC using antibody to Bmi-1. A: Bmi1 protein expression in adjacent non-cancerous tissues (× 400); B: Cytoplasm staining of Bmi1 in ESCC cells (× 400); C: Intense Bmi-1 staining in the nucleus in ESCC cells (× 400). The brown staining under light microscopy indicates positivity.
Table 2.
Relationship between Bmi-1 protein expression in ESCC tissues and clinicopathological status
| Parameter | > 50% | 10%-50% | < 10% | P |
| Age (yr) | ||||
| ≥ 60 | 23 | 9 | 4 | 0.784 |
| < 60 | 21 | 7 | 6 | |
| Histological grade | ||||
| I | 7 | 3 | 3 | 0.079 |
| II | 28 | 5 | 6 | |
| III | 9 | 8 | 1 | |
| Lymph node metastases | ||||
| + | 11 | 9 | 3 | 0.073 |
| - | 33 | 7 | 7 | |
| Stage | ||||
| III/IV | 9 | 4 | 1 | 0.691 |
| I/II | 35 | 12 | 9 | |
| Location | ||||
| Nucleus | 0 | 7 | 8 | 0.000 |
| Cytoplasm | 30 | 18 | 16 | |
| Both nucleus and cytoplasm | 5 | 15 | 20 |
ESCC was subjected to immunohistochemistry using antibodies to Bmi1. χ2 test was used to detect the difference between Bmi1 oncoprotein and clinicopathological status.
DISCUSSION
ESCC is a major cause of morbidity and mortality worldwide[1], and it is significant to identify a biological genetic molecular marker related to its pathophysiological processes.
Epigenetic aberrations, the heritable changes in gene expression that occur in chromatin structure including DNA methylation, histone post-translational modifications and nucleosomal remodeling, rather than the DNA sequence, are involved in cancer development[17–20]. Bmi1, the first PcG protein found, is a chromatin modifier implicated in the tumorigenesis through negatively regulating the gene expression such as the INK4A locus, which is thought to regulate p53 and the Rb signaling pathway in cooperation with c-myc[5,6,19,21]. In this retrospective study, we examined the Bmi1 expression and investigated its impact on ESCC patients.
Bmi1 mRNA expression was significantly higher in the ESCC samples than in the adjacent non-cancerous tissues, and so was Bmi1 protein expression, which indicated that Bmi1 plays an important role in the development of ESCC, and has diagnostic value.
Dirks[22] has reported that Bmi1-deficient tumors may be less aggressive because they have fewer stem cells. Bmi1 expression is also found inversely correlated with the differentiation grade of clear cell carcinoma and is involved in tumor progression[23]. Our data are in agreement with the findings by previous publications that the acquisition of metastatic ability of tumor cells is considered a late event in the evolution of malignant tumors. We found that the Bmi1 mRNA expression was higher in the stage I/II tissues than in stage III/IV, significantly lower in patients without metastatic lymph nodes, and inversely related to cell differentiation. The oncoprotein was more frequently observed in tissues with poorer differentiation. These results suggest that Bmi1 expression may not be required for initiation of ESCC but is required for its progression. It may be a guide for postoperative therapy and a differentiation marker in ESCC with high malignancy. Furthermore, we discovered that the accumulated survival of patients in the up-expression group was much shorter than that of patients in the down-expression group, which may predict survival in ESCC patients.
It was interesting to note that Bmi1 protein expression was negatively correlated with malignant grade, including lymph node metastasis and advanced pathological stage. This may have been because the samples for protein analysis were obtained at different periods than those for mRNA analysis, which resulted in a different selection bias. Also, the number of lymph nodes resected by different operators varied, and the lymph nodes removed during surgery for pathological diagnosis may have been misdiagnosed as metastatic lymph nodes.
The PcG protein Bmi1 showed abundant nuclear expression in prostate cancer, colorectal cancer and gastric carcinoma[11,15,24]. However, in our study, cytoplasmic staining appeared in most of the tumor cells with less or even no expression in the nucleus alone, which suggests that Bmi1 produces a marked effect on the development of ESCC, mainly in the cytoplasm. This is inconsistent with the PcG pathway activation hypothesis that states that increased Bmi1 expression in cancer cells is associated with elevated levels of H2AubiK119 and H3metK27 histones, which suppress the expression of the INK4a/ARF locus in the nucleus[21].
The mechanisms of Bmi1 up-expression that induce adverse pathological and clinical features in ESCC patients are poorly understood. Some previous studies have shown that Bmi1 expression is a potential escape mechanism and associated with markedly increased likelihood of treatment failure and disease relapse after surgery[25,26]. Qin et al[27] have found that down-regulation of Bmi-1 enhances 5-fluorouracil-induced apoptosis in nasopharyngeal carcinoma cells and have suggested that the combination of 5-FU treatment and Bmi-1 depletion might be a potential clinical strategy for cancer chemotherapy.
However, we believe that further investigations on larger series of ESCC patients, including clinical follow-up and novel molecular techniques, are needed to confirm our conclusions. Whether Bmi1 can be used for accurate prediction of ESCC and its potential chemosensitivity to current pharmaceutical treatment needs further study.
COMMENTS
Background
Bmi1, a member of the polycomb group and a component of the polycomb repressive complex1, has been considered as a oncogene involved in many solid and hematological malignant tumors, and it may be of diagnostic and prognostic relevance. However, to date, no report about the role of Bmi1 in esophageal squamous cell carcinoma (ESCC) has been made. However, the up-regulation of c-Myc and the down-regulation of p53 and p16 in ESCC tissues make it plausible that Bmi1 may play an important role in the initiation and progression of ESCC.
Research frontiers
This research, for the first time, investigated the expression of Bmi1 and its clinicopathological role in patients with ESCC.
Innovations and breakthroughs
A significant upregulation of Bmi1 was observed in cancerous tissues in contrast to adjacent non-cancerous paraffin-embedded esophageal specimens at the mRNA and the protein level by quantitative real-time polymerase chain reaction (PCR) and immunohistochemistry. Furthermore, overexpression of Bmi1 positively correlated with lymph node metastases, pathological stage and differentiation grade at the mRNA level.
Applications
Bmi1 may act as a guide for the postoperative therapy and a differentiation marker of ESCC with high malignancy, and for prediction of the survival of ESCC patients.
Peer review
The authors investigated the expression of Bmi1 at the mRNA and protein level in patients with ESCC in comparison with healthy adjacent tissue, by real-time PCR and immunohistochemistry. They observed a significant up-regulation of Bmi1 in tumor tissues in contrast to healthy control tissues. Up-regulation correlated positively with lymph node metastases, stage and histological grading at the mRNA level. However, the histological analysis showed no such correlation. The authors conclude that Bmi1 expression is common in ESCC and may serve as a marker to predict lymph node metastasis and survival in ESCC patients.
Supported by Nanjing First Hospital, Nanjing Medical University and Nanjing Health Bureau, No. ZKX0114
Peer reviewer: Bruno Stieger, Professor, Department of Medicine, Division of Clinical Pharmacology and Toxicology, University Hospital, Zurich 8091, Switzerland
S- Editor Cheng JX L- Editor Ma JY and Kerr C E- Editor Zheng XM
References
- 1.Fisichella PM, Patti MG. Esophageal cancer: eMedicine: oncology, 2009-03-04. Available from: URL: http://emedicine.medscape.com/article/277930-overview. [Google Scholar]
- 2.Kuwano H, Kato H, Miyazaki T, Fukuchi M, Masuda N, Nakajima M, Fukai Y, Sohda M, Kimura H, Faried A. Genetic alterations in esophageal cancer. Surg Today. 2005;35:7–18. doi: 10.1007/s00595-004-2885-3. [DOI] [PubMed] [Google Scholar]
- 3.Haupt Y, Alexander WS, Barri G, Klinken SP, Adams JM. Novel zinc finger gene implicated as myc collaborator by retrovirally accelerated lymphomagenesis in E mu-myc transgenic mice. Cell. 1991;65:753–763. doi: 10.1016/0092-8674(91)90383-a. [DOI] [PubMed] [Google Scholar]
- 4.Haupt Y, Bath ML, Harris AW, Adams JM. bmi-1 transgene induces lymphomas and collaborates with myc in tumorigenesis. Oncogene. 1993;8:3161–3164. [PubMed] [Google Scholar]
- 5.Jacobs JJ, Scheijen B, Voncken JW, Kieboom K, Berns A, van Lohuizen M. Bmi-1 collaborates with c-Myc in tumorigenesis by inhibiting c-Myc-induced apoptosis via INK4a/ARF. Genes Dev. 1999;13:2678–2690. doi: 10.1101/gad.13.20.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jacobs JJ, Kieboom K, Marino S, DePinho RA, van Lohuizen M. The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature. 1999;397:164–168. doi: 10.1038/16476. [DOI] [PubMed] [Google Scholar]
- 7.Park IK, Qian D, Kiel M, Becker MW, Pihalja M, Weissman IL, Morrison SJ, Clarke MF. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature. 2003;423:302–305. doi: 10.1038/nature01587. [DOI] [PubMed] [Google Scholar]
- 8.Molofsky AV, Pardal R, Iwashita T, Park IK, Clarke MF, Morrison SJ. Bmi-1 dependence distinguishes neural stem cell self-renewal from progenitor proliferation. Nature. 2003;425:962–967. doi: 10.1038/nature02060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Molofsky AV, He S, Bydon M, Morrison SJ, Pardal R. Bmi-1 promotes neural stem cell self-renewal and neural development but not mouse growth and survival by repressing the p16Ink4a and p19Arf senescence pathways. Genes Dev. 2005;19:1432–1437. doi: 10.1101/gad.1299505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beà S, Tort F, Pinyol M, Puig X, Hernández L, Hernández S, Fernandez PL, van Lohuizen M, Colomer D, Campo E. BMI-1 gene amplification and overexpression in hematological malignancies occur mainly in mantle cell lymphomas. Cancer Res. 2001;61:2409–2412. [PubMed] [Google Scholar]
- 11.Kim JH, Yoon SY, Kim CN, Joo JH, Moon SK, Choe IS, Choe YK, Kim JW. The Bmi-1 oncoprotein is overexpressed in human colorectal cancer and correlates with the reduced p16INK4a/p14ARF proteins. Cancer Lett. 2004;203:217–224. doi: 10.1016/j.canlet.2003.07.009. [DOI] [PubMed] [Google Scholar]
- 12.Vonlanthen S, Heighway J, Altermatt HJ, Gugger M, Kappeler A, Borner MM, van Lohuizen M, Betticher DC. The bmi-1 oncoprotein is differentially expressed in non-small cell lung cancer and correlates with INK4A-ARF locus expression. Br J Cancer. 2001;84:1372–1376. doi: 10.1054/bjoc.2001.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo WJ, Zeng MS, Yadav A, Song LB, Guo BH, Band V, Dimri GP. Mel-18 acts as a tumor suppressor by repressing Bmi-1 expression and down-regulating Akt activity in breast cancer cells. Cancer Res. 2007;67:5083–5089. doi: 10.1158/0008-5472.CAN-06-4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prince ME, Sivanandan R, Kaczorowski A, Wolf GT, Kaplan MJ, Dalerba P, Weissman IL, Clarke MF, Ailles LE. Identification of a subpopulation of cells with cancer stem cell properties in head and neck squamous cell carcinoma. Proc Natl Acad Sci USA. 2007;104:973–978. doi: 10.1073/pnas.0610117104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu JH, Song LB, Zhang X, Guo BH, Feng Y, Li XX, Liao WT, Zeng MS, Huang KH. Bmi-1 expression predicts prognosis for patients with gastric carcinoma. J Surg Oncol. 2008;97:267–272. doi: 10.1002/jso.20934. [DOI] [PubMed] [Google Scholar]
- 16.Greene FL, American Joint Committee on Cancer, American Cancer Society. AJCC cancer staging manual. 6th ed. New York: Springer; 2002. pp. 91–98. [Google Scholar]
- 17.Fraga MF, Esteller M. DNA methylation: a profile of methods and applications. Biotechniques. 2002;33:632, 634, 636–649. doi: 10.2144/02333rv01. [DOI] [PubMed] [Google Scholar]
- 18.Quina AS, Buschbeck M, Di Croce L. Chromatin structure and epigenetics. Biochem Pharmacol. 2006;72:1563–1569. doi: 10.1016/j.bcp.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 19.Sparmann A, van Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nat Rev Cancer. 2006;6:846–856. doi: 10.1038/nrc1991. [DOI] [PubMed] [Google Scholar]
- 20.Cao R, Tsukada Y, Zhang Y. Role of Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing. Mol Cell. 2005;20:845–854. doi: 10.1016/j.molcel.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 21.Shilatifard A. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu Rev Biochem. 2006;75:243–269. doi: 10.1146/annurev.biochem.75.103004.142422. [DOI] [PubMed] [Google Scholar]
- 22.Dirks P. Bmi1 and cell of origin determinants of brain tumor phenotype. Cancer Cell. 2007;12:295–297. doi: 10.1016/j.ccr.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 23.Kozakowski N, Soleiman A, Pammer J. BMI-1 expression is inversely correlated with the grading of renal clear cell carcinoma. Pathol Oncol Res. 2008;14:9–13. doi: 10.1007/s12253-008-9006-3. [DOI] [PubMed] [Google Scholar]
- 24.van Leenders GJ, Dukers D, Hessels D, van den Kieboom SW, Hulsbergen CA, Witjes JA, Otte AP, Meijer CJ, Raaphorst FM. Polycomb-group oncogenes EZH2, BMI1, and RING1 are overexpressed in prostate cancer with adverse pathologic and clinical features. Eur Urol. 2007;52:455–463. doi: 10.1016/j.eururo.2006.11.020. [DOI] [PubMed] [Google Scholar]
- 25.Liu S, Dontu G, Wicha MS. Mammary stem cells, self-renewal pathways, and carcinogenesis. Breast Cancer Res. 2005;7:86–95. doi: 10.1186/bcr1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berezovska OP, Glinskii AB, Yang Z, Li XM, Hoffman RM, Glinsky GV. Essential role for activation of the Polycomb group (PcG) protein chromatin silencing pathway in metastatic prostate cancer. Cell Cycle. 2006;5:1886–1901. doi: 10.4161/cc.5.16.3222. [DOI] [PubMed] [Google Scholar]
- 27.Qin L, Zhang X, Zhang L, Feng Y, Weng GX, Li MZ, Kong QL, Qian CN, Zeng YX, Zeng MS, et al. Downregulation of BMI-1 enhances 5-fluorouracil-induced apoptosis in nasopharyngeal carcinoma cells. Biochem Biophys Res Commun. 2008;371:531–535. doi: 10.1016/j.bbrc.2008.04.117. [DOI] [PubMed] [Google Scholar]