Abstract
Hydroxyl radical footprinting is a technique for studying protein structure and binding that entails oxidizing a protein system of interest with diffusing hydroxyl radicals, and then measuring the amount of oxidation of each amino acid. One important issue in hydroxyl radical footprinting is limiting amino acid oxidation by secondary oxidants to prevent uncontrolled oxidation which can cause amino acids to appear more solvent accessible than they really are. Previous work suggested that hydrogen peroxide was the major secondary oxidant of concern in hydroxyl radical footprinting experiments; however, even after elimination of all hydrogen peroxide, some secondary oxidation was still detected. Evidence is presented for the formation of peptidyl hydroperoxides as the most abundant product upon oxidation of aliphatic amino acids. Both reverse phase liquid chromatography and catalase treatment were shown to be ineffective at eliminating peptidyl hydroperoxides. The ability of these peptidyl hydroperoxides to directly oxidize methionine is demonstrated, suggesting the value of methionine amide as an in situ protectant. Hydroxyl radical footprinting protocols require the use of an organic sulfide or similar peroxide scavenger in addition to removal of hydrogen peroxide in order to successfully eradicate all secondary oxidizing species and prevent uncontrolled oxidation of sulfur-containing residues.
Introduction
In the last decade, hydroxyl radical footprinting has proven to be a useful method for analyzing macromolecular structure, ligand binding, and conformational changes [1, 2]. Hydroxyl radical footprinting takes advantage of the fact that the rate of oxidation of each amino acid correlates directly with the solvent accessibility of that amino acid; as the solvent accessibility increases or decreases, the rate of oxidation of that amino acid will increase or decrease in a corresponding manner [3, 4]. Hydroxyl radicals have proven to be a relatively high-resolution footprinting reagent, and have been used to study the configuration of DNA, RNA, and proteins [1, 2]. The sites of oxidation and rates of oxidation at each site are determined by tandem mass spectrometry [5].
Hydroxyl radicals can be efficiently generated in situ by many methods, all of which use hydrogen peroxide as a precursor oxidant (e.g. Fenton chemistry, UV photolysis of peroxide) and/or produce hydrogen peroxide as a product (e.g. X-ray radiolysis, electrochemistry, gamma radiolysis) [1]. After formation, hydroxyl radicals self-quench in solution by recombination to form H2O2, and oxidation of amino acid side chains may also produce H2O2 as a side product [6]. Buffer exchange [7] and protein precipitation [8] to remove the hydrogen peroxide have been reported as strategies to eliminate uncontrolled secondary oxidation. Addition of catalase or methionine amide was found to eliminate much, but not all, of the uncontrolled oxidation in model systems [9].
Previous work has shown that peptidyl hydroperoxides (ROOH) are major products of oxidation of aliphatic amino acids [10]. Aliphatic amino acids are useful targets for radiolytic footprinting, but while previous studies using peptides found significant amounts of peptidyl hydroperoxides (detected as a mass shift of 32 Da in the mass spectrum) [11, 12], they have not been identified in any of the hydroxyl radical protein footprinting results reported [13]. It was previously suggested that peptidyl hydroperoxides were inherently unstable, and thus did not survive the post-oxidation processing and mass spectrometry [9]; however, studies of leucine oxidation indicated that the peptidyl hydroperoxide was sufficiently stable to analyze by mass spectrometry [11]. This is of significant concern, as peptidyl hydroperoxides are capable of two-electron oxidation of sulfur-containing amino acids, which could cause such amino acids to be oxidized in an uncontrolled manner during processing. Additionally, catalase is relatively ineffective at eliminating mixed hydroperoxides [14], and strategies based on removing diffusing secondary oxidants would be ineffective at removing protein-bound secondary oxidants like peptidyl hydroperoxides. Therefore, if standard hydroxyl radical footprinting methodologies do generate substantial amounts of peptidyl hydroperoxides, catalase treatment and buffer exchange by itself is insufficient. In this report, we study the major oxidation products of various aliphatic amino acids to determine if peptidyl hydroperoxides are a major oxidation product, and the ability of peptidyl hydroperoxides to directly oxidize methionine is measured.
Experimental
Materials
Fmoc-Val-OH, Fmoc-Leu-OH, Fmoc-Pro-OH, Fmoc-Ile-OH, and Fmoc-Gly-NovaSyn TGT were purchased from Novabiochem/EMD Biosciences (Gibbstown, NJ). Fmoc-L-Phe-OH and Fmoc-Gly-OH were purchased from Applied Biosystems (Foster City, CA). Coupling reagent 1-H-benzotriazolium-1-[bis(dimethyl-amino)methylene]-5-chloro-hexafluorophosphate (1-),3-oxide (HCTU) and additive 1-hydroxybenzotriazole anhydrous (HOBt) were purchased from Peptides International (Louisville, Kentucky). N,N′-diisopropylethylamine (DIEA), N,N′-dimethylformamide (DMF), trifluoroacetic acid (TFA), dichloromethane (CH2Cl2), methanol, and bovine catalase were purchased from Sigma-Aldrich (St Louis, MO). Toluene was purchased from VWR International (West Chester, PA). Hydrogen peroxide was purchased from J.T. Baker (Phillipsburg, NJ). Methionine amide was purchased from Bachem California Inc. (Torrance, CA). Ultrapure water (18 MΩ) was prepared in-house with a Millipore Mill-Q water purification system (Millipore Bedford, MA).
Tripeptide Synthesis
Tripeptides Gly-Ile-Gly (GIG), Gly-Leu-Gly (GLG), Gly-Pro-Gly (GPG), Gly-Val-Gly (GVG), and Gly-Phe-Gly (GFG) were synthesized following a protocol similar to earlier work [15, 16]. Then 100mg TGT resin was placed in a glass vessel (8.5 ml) containing a porous glass frit for manual synthesis. Fmoc removal was achieved with piperidine:DMF (1:4) for 20 min, followed by 2-h couplings of corresponding amino acid derivatives (4 eq), which were mediated by HCTU (4 eq)/HOBt (4 eq)/DIEA (5 eq) in DMF (2 ml) at 25 °C. Washings between reactions were carried out with DMF and CH2Cl2, then DMF again, and no intermediate capping steps were done. After chain assembly was completed and the N-terminal Fmoc group removed, the resin was washed with DMF and CH2Cl2 and dried in a desiccator overnight. The tripeptide resin was then treated with TFA/H2O (50/50) for 2 h and washed to recover peptides, which were used directly following lyophilization.
Sample Preparation, Irradiation, and Analysis of Peptides
Peptide solutions were prepared at 90μM in a solution of 147mM hydrogen peroxide (H2O2). The peptides were oxidized by hydroxyl radicals, using fast photooxidation of proteins (FPOP) to generate these radicals [19-21]. A one-syringe flow system was used with a capillary inner diameter of 97μm and flow rate of 17.7μL/min. The flowing mixture was illuminated with UV light from one pulse of a KrF excimer laser (GAM Laser, Inc., Orlando FL) at an average laser power of 60mJ/pulse as previously described [17, 18]. Under these conditions, each fraction of sample was irradiated only once, with ∼15% of the total sample volume remaining unirradiated to prevent multiple irradiations. All samples were purified by HPLC (Agilent Technologies 1200 series) using a C18 analytical column to remove all diffusing secondary oxidants. The post-LC samples were examined via direct infusion by nanoelectrospray using a Q-Tof 2 mass spectrometer (Waters, Milford, MA) with a capillary voltage of 3.8kV, cone voltage of 20V, and flow rate of 0.4μL/min. Catalase was added to 20μL 90μM of each oxidized peptide after purification by HPLC to a final concentration of 50nM catalase tetramer. The mixture was incubated for 24 h at room temperature and analyzed by MS. Methionine amide (Me-NH2) was added to 20μL 90μM oxidized tripeptide after purification by HPLC to a final concentration of 1.45 mM and allowed to incubate 24 h at room temperature.
Results and Discussion
A series of tripeptides were generated for these studies, four aliphatic and one aromatic. These tripeptides were made as Gly-X-Gly, as glycine is the most unreactive amino acid.
MS analysis of oxidation products
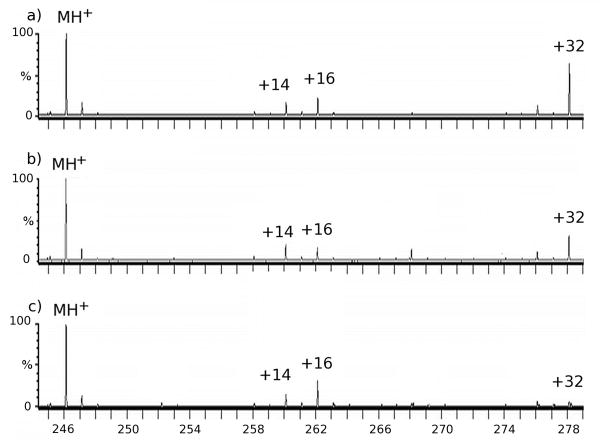
The hydroxyl and carbonyl products are prominent in the mass spectra of the oxidized aliphatic GXG solutions, as exemplified by GIG in Figure 1a. The most abundant oxidation product is M+32, which could be indicative of either a peptidyl hydroperoxide or a dialcohol. This is in sharp contrast to previous reports indicating that the M+16 product is the most abundant [12], but agrees with previous results of leucine oxidation [11]. HPLC separation had no apparent effect on the relative abundance of the oxidized products, indicating that the oxidation is not a result of electrospray-induced activation of hydrogen peroxide. The results of catalase and methionine amide treatment on the M+32 oxidation products of the various GXG peptides are shown in Table 1. After incubating the HPLC-purified GXG with 50nM catalase tetramer for 24 h, very little conversion of M+32 to M+16 was observed (Figure 1b). Upon an addition of 1.45mM methionine amide to the oxidized solution, the MS exhibited a marked but incomplete conversion of M+32 to M+16 (Figure 1c). When methionine amide and catalase were added together to the oxidized solution, the conversion of M+32 to M+16 was very similar to that of methionine amide alone, with no additive effect observed (data not shown).
Figure 1.
Mass spectra of GIG (a) oxidized by FPOP and purified by HPLC (b) oxidized by FPOP, purified by HPLC, and incubated with 50nM catalase for 24 h (c) oxidized by FPOP, purified by HPLC, and incubated with 1.45mM methionine amide 24 h.
Table 1.
Average ratio of oxidation products
| Peptide Sequence | Avg +32/+16 |
|---|---|
| GIG ox | 1.6988 +/- 0.07 |
| GIG ox+cat | 1.5348 +/- 0.1 |
| GIG ox+met | 0.5115 +/- 0.04 |
| GLG ox | 1.0743 +/- 0.1 |
| GLG ox+cat | 0.9734 +/- 0.4 |
| GLG ox+met | 0.4255 +/- 0.05 |
| GPG ox | 1.2801 +/- 0.1 |
| GPG ox+cat | 1.4359 +/- 0.05 |
| GPG ox+met | 0.3023 +/- 0.01 |
| GVG ox | 0.8468 +/- 0.008 |
| GVG ox+cat | 0.5792 +/- 0.01 |
| GVG ox+met | 0.0948 +/- 0.01 |
| GFG ox | 0.5743 +/- 0.02 |
| GFG ox+cat | 0.5402 +/- 0.05 |
| GFG ox+met | 0.2652 +/- 0.008 |
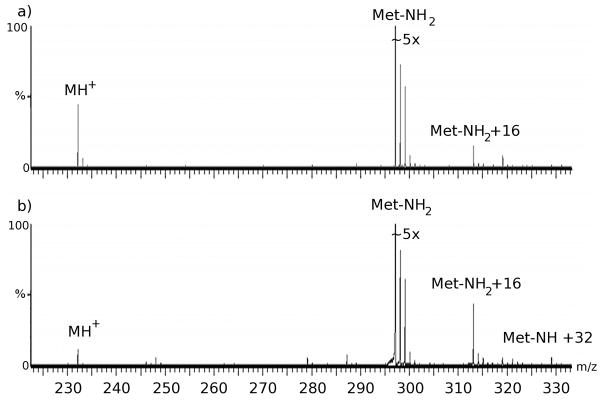
The main area of concern regarding peptidyl hydroperoxides in hydroxyl radical footprinting is the potential that the peptidyl hydroperoxides can directly oxidize the sulfur-containing residues. In order to determine if this is a issue, we measured the amount of oxidation of methionine amide with an unoxidized peptide and with an oxidized peptide. The results show that peptidyl peroxides do oxidize methionine to the methionine sulfoxide (Figure 2). This suggests that the observed M+32 product is indicative of a mixture of peptidyl hydroperoxides, which act as oxidants to modify the methionine amide to the sulfoxide, and peptidyl dialcohols (which are unreactive to the methionine amide). The slight conversion of the M+32 product to M+16 by catalase is probably due to oxidation of sulfur-containing residues in the catalase tetramer by the peptidyl hydroperoxide, and/or the peptidyl hydroperoxide acting as a suicide substrate, inactivating the catalase in the process [19].
Figure 2.
(a) Unoxidized GVG incubated with 1.45mM methionine amide for 24 h. (b) Oxidized GVG incubated with 1.45mM methionine amide for 24 h.
The initiating reaction in aliphatic residues is the abstraction of a hydrogen from the side chain [20, 21], resulting in a peptidyl radical. This radical rapidly scavenges oxygen, forming a peptidyl peroxyl radical that can undergo competing reactions to form carbonyls, alcohols, and peptidyl peroxides [6, 10, 24-26]. In all four aliphatic peptides, the hydroxyl and carbonyl products were present, however the M+32 oxidation peak was consistently found in much greater abundance than the other oxidation products, and the incomplete reactivities of the M+32 products with methionine amide support assignment as peptidyl hydroperoxides with small amounts of isomeric dialcohols. MS/MS data of the M+32 product of oxidized GVG purified by HPLC show a loss of 18 Da (water) and a loss of 34 Da (hydrogen peroxide) [11] (see supplemental data), while MS/MS of the M+16 product show only a loss of 18 Da (data not shown). MS/MS data of the M+32 product of oxidized GVG after incubation with methionine amide show a loss of 18 Da but no loss of 34 Da (see supplemental data), indicating that the M+32 product after incubation consists of dialcohols and not peptidyl hydroperoxides. These data confirm that the M+32 products of the aliphatic residues prior to incubation with methionine amide are mainly comprised of peptidyl hydroperoxides which serve to oxidize the organic sulfide of methionine amide, converting the peptidyl hydroperoxide to the corresponding alcohols.
The oxidation products of the aromatic peptide GFG were also investigated. The M+32 product was less abundant than the M+16 product in the oxidized sample, unlike in the aliphatic peptides. Addition of catalase or methionine amide did result in some conversion of the M+32 product to the M+16 product (Table 1), although the results were not as drastic. Details of the investigation of the GFG oxidation products are given in online supplemental data.
Conclusion
Our studies have found that peptidyl hydroperoxides are major oxidation products of aliphatic amino acids and are capable of oxidizing sulfur-containing amino acids. Removal of these secondary oxidants is necessary to prevent secondary oxidation of the other amino acid residues, especially during the enzymatic digestion process that follows hydroxyl radical labeling in most experiments. Our results suggest that peptidyl hydroperoxide-mediated oxidation can be suppressed in hydroxyl radical footprinting experiments of proteins and peptides with sulfur-containing residues via competitive scavenging by methionine amide. Alternative approaches using catalase and HPLC separation are shown to be ineffective at removing these peptide-based secondary oxidants, allowing secondary oxidation of methionine. Future hydroxyl radical footprinting protocols should include the use of methionine amide or a related peroxide scavenger in addition to other quenching strategies in order to eliminate the contribution of peptidyl hydroperoxides and related secondary oxidants from uncontrolled oxidation of sulfur-containing residues.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health (RR05357).
References
- 1.Xu G, Chance M. Hydroxyl Radical-Mediated Modification of Proteins as Probes for Structural Proteomics. Chem Rev. 2007:3514–3543. doi: 10.1021/cr0682047. [DOI] [PubMed] [Google Scholar]
- 2.Konermann L, Tong X, Pan Y. Protein Structure and Dynamics Studied by Mass Spectrometry: H/D Exchange, Hydroxyl Radical Labeling, and Related Approaches. J Mass Spectrom. 2008;43:1021–36. doi: 10.1002/jms.1435. [DOI] [PubMed] [Google Scholar]
- 3.Chance MR. Unfolding of Apomyoglobin Examined by Synchrotron Footprinting. Biochem Biophys Res Commun. 2001;287:614–621. doi: 10.1006/bbrc.2001.5628. [DOI] [PubMed] [Google Scholar]
- 4.Charvátová O, Foley BL, Bern MW, Sharp JS, Orlando R, Woods RJ. Quantifying Protein Interface Footprinting by Hydroxyl Radical Oxidation and Molecular Dynamics Simulation: Application to Galectin-1. J Am Soc Mass Spectr. 2008;19:1692–1705. doi: 10.1016/j.jasms.2008.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kiselar JG, Maleknia SD, Sullivan M, Downard KM, Chance MR. Hydroxyl Radical Probe of Protein Surfaces Using Synchrotron X-Ray Radiolysis and Mass Spectrometry. Int J Radiat Biol. 2002;78:101–114. doi: 10.1080/09553000110094805. [DOI] [PubMed] [Google Scholar]
- 6.Garrison WM. Reaction Mechanisms in the Radiolysis of Peptides, Polypeptides, and Proteins. Chem Rev. 1987;87:381–398. [Google Scholar]
- 7.Sharp JS, Becker JM, Hettich RL. Protein Surface Mapping by Chemical Oxidation: Structural Analysis by Mass Spectrometry. Anal Biochem. 2003;313:216–225. doi: 10.1016/s0003-2697(02)00612-7. [DOI] [PubMed] [Google Scholar]
- 8.Lim J, Vachet RW. Development of a Methodology Based on Metal-Catalyzed Oxidation Reactions and Mass Spectrometry to Determine the Metal Binding Sites in Copper Metalloproteins. Anal Chem. 2003;75:1164–1172. doi: 10.1021/ac026206v. [DOI] [PubMed] [Google Scholar]
- 9.Xu G, Kiselar J, He Q, Chance MR. Secondary Reactions and Strategies to Improve Quantitative Protein Footprinting. Anal Chem. 2005;77:3029–3037. doi: 10.1021/ac048282z. [DOI] [PubMed] [Google Scholar]
- 10.Denisov ET, Khudyakov IV. Mechanisms of Action and Reactivities of the Free Radicals of Inhibitors. Chem Rev. 1987;87:1313–1357. [Google Scholar]
- 11.Sharp JS, Tomer KB. The Effects of Anion Proximity in Peptide Primary Sequence on the Rate and Mechanism of Leucine Oxidation. Anal Chem. 2006;78:4885–4893. doi: 10.1021/ac060329o. [DOI] [PubMed] [Google Scholar]
- 12.Xu G, Chance MR. Radiolytic Modification and Reactivity of Amino Acid Residues Serving as Structural Probes for Protein Footprinting. Anal Chem. 2005;77:4549–4555. doi: 10.1021/ac050299+. [DOI] [PubMed] [Google Scholar]
- 13.Takamoto K, Chance MR. Radiolytic Protein Footprinting with Mass Spectrometry to Probe the Structure of Macromolecular Complexes. Annu Rev Biophys Biomol Struct. 2006;35:251. doi: 10.1146/annurev.biophys.35.040405.102050. [DOI] [PubMed] [Google Scholar]
- 14.Palcic MM, Dunford HB. The Reaction of Human Erythrocyte Catalase with Hydroperoxides to Form Compound I. J Biol Chem. 1980;255:6128–6132. [PubMed] [Google Scholar]
- 15.Raman J, Fritz TA, Gerken TA, Jamison O, Live D, Liu M, Tabak LA. The Catalytic and Lectin Domains of Udp-Galnac: Polypeptide {Alpha}-N-Acetylgalactosaminyltransferase Function in Concert to Direct Glycosylation Site Selection. J Biol Chem. 2008;283:22942–22951. doi: 10.1074/jbc.M803387200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu M, Borgert A, Barany G, Live D. Conformational Consequences of Protein Glycosylation: Preparation of O-Mannosyl Serine and Threonine Building Blocks, and Their Incorporation into Glycopeptide Sequences Derived from Alpha-Dystroglycan. Biopolymers. 2008;90:358–368. doi: 10.1002/bip.20847. [DOI] [PubMed] [Google Scholar]
- 17.Hambly D, Gross M. Laser Flash Photochemical Oxidation to Locate Heme Binding and Conformational Changes in Myoglobin. Int J Mass Spectrom. 2007;259:124–129. [Google Scholar]
- 18.Hambly DM, Gross ML. Laser Flash Photolysis of Hydrogen Peroxide to Oxidize Protein Solvent-Accessible Residues on the Microsecond Timescale. J Amer Soc Mass Spectrom. 2005;16:2057–2063. doi: 10.1016/j.jasms.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 19.Pichorner H, Jessner G, Ebermann R. Tbooh Acts as a Suicide Substrate for Catalase. Arch Biochem Biophys. 1993;300:258–64. doi: 10.1006/abbi.1993.1036. [DOI] [PubMed] [Google Scholar]
- 20.Brodskaya GA, Sharpatyi VA. Radiolysis of Aqueous Tyrosine Solutions: Dependence on the Medium Ph. Russ J Phys Chem. 1967;41:583. [Google Scholar]
- 21.Dizdaroglu M, Simic MG. In: Oxygen and Oxy-Radicals in Chemistry and Biology. Rodgers MAJ, Powers EL, editors. Academic; New York: 1981. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.