Abstract
It is now becoming generally accepted that a significant amount of human genetic variation is due to structural changes of the genome rather than to base-pair changes in the DNA. As for base-pair changes, knowledge of gene and genome function has been informed by structural alterations that convey clinical phenotypes. Genomic disorders are a class of human conditions that result from structural changes of the human genome that convey traits or susceptibility to traits. The path to the delineation of genomic disorders is intertwined with the evolving technologies that have enabled the resolution of human genome analyses to continue increasing. Similarly, the ability to perform high-resolution human genome analysis has fueled the current and future clinical implementation of such discoveries in the evolving field of genome medicine.
Genomic disorders are diseases that result from rearrangements of the human genome rather than from DNA sequence base changes. Moreover, such rearrangements occur because of architectural features of the genome that incite genome instability. The idea of genomic disorders emanated from locus-specific studies of the common autosomal dominant peripheral neuropathies: Charcot-Marie-Tooth disease type 1A (CMT1A; Mendelian Inheritance in Man (MIM) database ID 118220 [1]) and hereditary neuropathy with liability to pressure palsies (HNPP; MIM 162500). A careful re-read of the early reports on these conditions reveals nearly all the key concepts of genomic disorders, including genomic duplication [2,3] and deletion [4], gene dosage (PMP22) [5-8] and specific gene copy number variation (CNV) [6-8]. The concepts of genome architecture and low-copy repeats (LCRs) or segmental duplications (SDs) were well described before there was either a draft or a finished reference genome sequence [9,10] (Figure 1). The term LCR was first introduced by Bernice Morrow following her studies of DiGeorge syndrome (MIM 188400) rearrangement breakpoints [11] whereas the term SD was introduced by Evan Eichler [12,13] to explain his observations from genome-wide studies. The concepts of non-allelic homologous recombination (NAHR [9], although the specific term NAHR was not introduced until later [14]), reciprocal recombination resulting in duplication/deletion of the same genomic interval [9,10], recombination hotspots [15,16] and the effects of CNV (such as duplication) on the interpretation of the segregation of marker genotypes [2,17] also began to emerge at this early stage.
Figure 1.
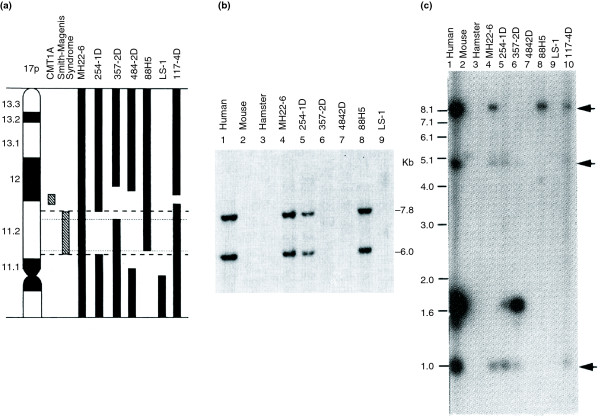
Low-copy repeats (LCRs) flanking the Charcot-Marie-Tooth disease type 1A duplication (CMT1A-REP) and the Smith-Magenis deletion (SMS-REP). (a) A somatic cell hybrid panel with a chromosome 17p ideogram (left) and vertical bars representing the regions retained in the individual human hybrid cell lines listed at the top. (b) Southern hybridization with a CMT1A-REP probe. There are two cross-hybridizing signals in human genomic DNA (lane 1), none in the mouse and hamster genomic DNA (lanes 2 and 3), and the same two in a monochromosomal hybrid (MH22-6, lane 4) retaining human chromosome 17. Both copies map to the CMT1A duplication region at 17p12. This is interpreted as showing that there are two copies of CMT1A-REP, both mapping to the CMT1A duplication locus, and both of which evolved late in the mammalian radiation as they are not present in mouse or hamster [9]. (c) Three copies of SMS-REP (arrows) on chromosome 17 [21]. We used the term REP because at the time my laboratory was working with prokaryotic repeated sequences (REP) and had developed a technique we referred to as rep-PCR [157,158].
Nevertheless, progress was blocked by both technological and conceptual limitations. Technically, we had no way to view the entire human genome simultaneously at a level of resolution that would enable insights into molecular mechanisms. Conceptually, locus-specific thinking had permeated genetics for over a century, with genocentric (gene-specific) views and base-pair changes as the one form of mutation predominating during the latter half of the 20th century and often blindly biasing genetic thinking to this day. The significant heritability and uncertain molecular basis of common disorders has been approached with such geno-centric and 'point mutation' genetic thinking. Even now, we witness this as a recurrent theme with an excessive focus on genome-wide association studies (GWASs) evaluating ancient SNPs, as contrasted with the potential involvement of recent or new mutations and/or CNV.
At the time of the early studies leading to the concept of genomic disorders, the one way to visualize the entire human genome was through chromosome studies and usually by the G-banded karyotype provided from clinical cytogenetics. We were thus fascinated and excited to find that our studies of a microdeletion syndrome, the Smith-Magenis syndrome (SMS; MIM 182290), which results from a 3.7 Mb genomic deletion rearrangement large enough to be visualized by microscopy, revealed similar observations to those found for CMT1A/HNPP, including recurrent breakpoints [18-20], a surrounding genomic architecture consisting of LCRs (repeat gene clusters in this case) [21], reciprocal recombination [22,23] and occurrence by NAHR [21] (Figures 1 and 2).
Figure 2.
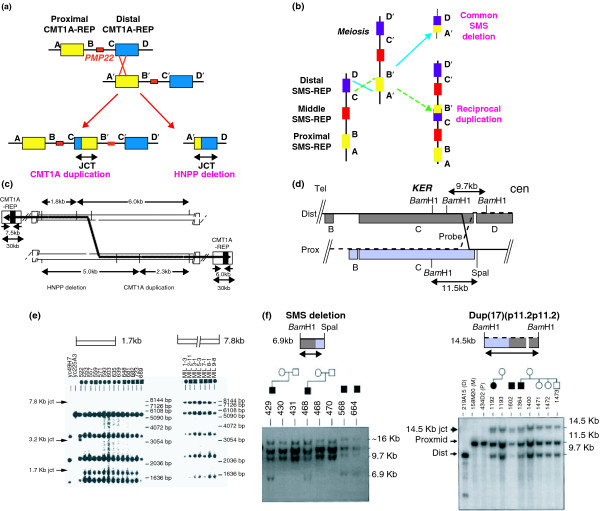
Reciprocal recombinations at the Charcot-Marie-Tooth disease type 1A (CMT1A) duplication locus in 17p12 and the Smith-Magenis syndrome (SMS) locus in 17p11.2. (a) The non-allelic homologous recombination (NAHR) in which the low-copy repeat (CMT1A-REP) substrates lead to reciprocal CMT1A duplication and HNPP deletion [15]. (b, d, f) Analogous data for the SMS deletion and its predicted reciprocal duplication [23]. (c) The model for the crossover and the predicted junction fragments; (e) the Southern analysis supporting this model. Note that these are the same molecular mechanism (NAHR), but it is shown horizontally (as usually depicted by molecular biologists) in (a) and vertically (as usually depicted by cytogeneticists) in (b). Abbreviations: cen, centromeric; dist, distal; mid, middle; prox, proximal; tel, telomeric.
These findings crystallized and solidified the concept of genomic disorders [24]. The concept of genomic disorders is predicated on two general ideas: firstly, that genomic disorders occur by rearrangements of our genome (the human genome is disordered) and not by DNA-sequence-based changes (that is, not by base-pair changes or by SNPs that cause disease); and secondly, that genome architecture incites genome instability. This article stated that structural characteristics of the human genome predispose it to rearrangements that result in human disease traits, and that genome alterations can occur through many mechanisms, including homologous recombination between region-specific LCRs [24]. This first mechanism was later termed NAHR [14]. The term NAHR stresses the mechanism by which these particular rearrangements of the human genome occur, including the requirement for homologous substrates and the observations of gene conversion and recombination hotspots. Furthermore, NAHR can cause duplication, deletion and inversion. In contrast, unequal crossing-over usually refers to the segregation of marker genotypes and can lead to duplication or deletion chromosomes [25-27]. Admittedly, almost all of the cases used to bolster the argument for genomic disorders in the original article on the topic [24] occurred mechanistically by NAHR. However, both Pelizaeus-Merzbacher disease (MIM 312080), caused by genomic duplications, and spinal muscular atrophy (MIM 25330), associated with genomic deletion, were mentioned as other diseases commonly caused by DNA rearrangements that might reflect genomic instability due to unique genome structural features [24].
The same article [24] also suggested that for disorders caused by genomic deletion rearrangements, the reciprocal duplications might be under-recognized. Examples were provided of contiguous-gene-deletion syndromes, such as Williams-Beuren (WBS; MIM 194050), Prader-Willi (MIM 176270), Angelman (MIM 105830) and DiGeorge/velocardio-facial syndromes (DG/VCFS; MIM 188400), that might result from a molecular mechanism similar to that of SMS and suggested the reciprocal duplication, as seen for SMS, may occur [24]. It was also pointed out that such patients with duplications might have different clinical findings and milder phenotypic features than those with deletions, because excess information is usually less detrimental to the organism than deficiency. Therefore, these cases could escape identification through under-ascertainment or be missed by routine cytogenetic analysis because of the further technical challenges required to recognize duplications compared with deletions [24].
The first predicted reciprocal microduplication syndrome was identified shortly thereafter, the duplication of the genomic interval deleted in SMS [28] (Figure 2), but it would take another 7 years to systematically study and describe the phenotypic variability of what has come to be known as the Potocki-Lupski syndrome [29] (PTLS; MIM 610883). Interestingly, these clinical studies showed that autism, as defined by objective psychological testing, was one feature of PTLS [29], thus linking the autism trait to a specific CNV. The apparent predicted reciprocal duplications for both the DG/VCFS [30-32] (MIM 608363) and WBS regions [33-35] (MIM 609757) followed rapidly. Reciprocal duplication syndromes are now being defined for almost all microdeletion syndromes in which the deletion is flanked by LCRs/SDs and that occur by NAHR (for example, dup(17)q21.31q21.31 [36] and duplication of the Sotos syndrome (MIM 117550) region [37]); these are often described within the same year [38,39] or even the same paper [40-44] as the microdeletion syndromes themselves.
After several years of study, the rules for NAHR were elucidated [14,24]. A hallmark experimental approach based on an understanding and implementation of the new knowledge of the NAHR mechanism was executed by Evan Eichler and colleagues. With a reference human genome sequence in hand [45-47] and the technology of genome-wide array comparative genomic hybridization (aCGH) [48], they designed a research array to interrogate genomic intervals flanked by LCRs greater than 10 kb in length, over 95% sequence identical, in direct orientation, and mapping within 50 kb to 5 Mb of each other [49,50]. These arrays were then used to assay patient cohorts with idiopathic mental retardation and other birth defects. In this manner, they defined five new microdeletion syndromes (deletions of 17q21.31 [50-54], 17q12, 15q24, 15q13.3 and 1q21.1) within less than 2 years [44,50,55-57]. Interestingly, the 17q12 deletion was found to be associated with maturity-onset diabetes of the young [56,58] a common, albeit genetically heterogeneous, disorder. The latter two deletions, 15q13.3 and 1q21.1, have also been associated with schizophrenia [59-61], whereas 15q13.3 has also been associated with idiopathic seizures [57,62], mental retardation [57], autism [63,64] and behavioral abnormalities with antisocial behavior [64].
Many other common and complex disorders are being shown to be due to CNV in some fraction of patients. Thus, genomic disorders encompass not only rare multiple congenital anomaly and mental retardation syndromes, but also common and complex traits, such as autism and schizophrenia, as well as other neurobehavioral phenotypes. For instance, deletion and duplication 16p11.2 can also cause autism [40,65]. Both duplications and/or deletion CNVs of the human genome have been associated with HIV susceptibility [66], Crohn's disease [67-69], glomerulonephritis [70], psoriasis [71], systemic lupus erythematosus [72,73], pancreatitis [74] and many other human diseases. Furthermore, animal models for SMS and PTLS show that obesity and several of the objectively assayed behavioral traits can result from a specific gene CNV (i.e. the mouse Rai1 gene [75]).
In the past decade, many important basic science questions have also been addressed through studies of genomic disorders. NAHR hotspots [15,16] had been identified long before allelic homologous recombination (AHR) hotspots [76] were generally appreciated through studies that emerged from the HapMap Project [77,78]. NAHR and AHR hotspots were found to coincide at the two loci where they were studied [79]: the CMT1A duplication/HNPP deletion locus [80] and the neurofibromatosis type 1 deletion locus at 17q11.2 [81]. Fundamental insights into human recombination have been gleaned from studies of genomic rearrangements and genomic disorders [82-86]. Importantly, locus-specific mutation rates for de novo genomic rearrangements that result in CNV were shown both theoretically [87] and experimentally [88] to occur at frequencies of 100 to 10,000 times greater than locus-specific mutation rates for de novo SNPs. Interestingly, the deletions can outweigh duplications about 2:1 at selected autosomal loci and about 4:1 on the Y chromosome at a given locus for rearrangements generated by NAHR [88]. Studies of genomic disorders have also provided fundamental insights into human gene [89-93] and genome [94-100] evolution. Such studies were among the first to provide examples of exon accretion by segmental duplication in the evolution of novel gene functions [91], gene duplication/triplication by de novo CNV formation [92,93], accumulation of LCRs/SDs during primate genome evolution [98,99], and LCRs/SDs at evolutionary chromosomal breakpoints [95,98] and at breaks in synteny between the mouse and human genome [94,100].
As genome-wide tools became more readily available after the consecutive completion of the draft, reference and finished human haploid genome [45-47], many laboratories shifted their experimental approach from locus-specific and genocentric thinking to genomic studies. And as a result, the field of genomic disorders exploded. First, it became apparent that structural variation including CNV [101] of the normal human genome was much greater than anticipated [102-105]. In fact, any two individuals vary more as a result of CNV in terms of numbers of base-pairs involved than all the SNPs combined [104]. Moreover, the clinical implementation of genomic techniques enables high-resolution human genome analysis and can resolve CNVs 10, 100 and even 1,000 times smaller than the 3-5 Mb resolution afforded by a clinical G-banded karyotype. This has revolutionized medical genetics and bolstered the emerging field of genome medicine [106-121]. Array-based technologies can resolve pathogenic subtelomeric CNV better than can subtelomere fluorescent in situ hybridization [119] and can reveal genomic rearrangements in patients with apparently balanced translocations [120,121]. Moreover, these technologies also enable mosaicism to be detected as a cause of a clinical phenotype [114,115]. This was not visualized previously because of stimulation of selected cell types for karyotype analysis [114,115]. Such techniques have also enabled prenatal detection of submicroscopic abnormalities [122-126] and the detection of de novo genomic rearrangement events causing sporadic birth defects [127]. Submicroscopic duplications as a cause of X-linked mental retardation [128,129] and other mental retardation syndromes [130,131] are now revealed. Many new genomic disorders caused by submicroscopic duplications and deletions continue to be described and are catalogued in the DECIPHER database [132].
Continued systematic investigations of rearrangements associated with genomic disorders have uncovered a new mechanism for rearrangements within our genome. As explained above, research on recurrent rearrangements with breakpoint clustering at LCRs/SDs enabled the elucidation of the NAHR mechanism. Recent studies of genomic disorders caused by non-recurrent rearrangements (rearrangements of different sizes and with different breakpoints in each individual) have uncovered a new replication-based human genomic rearrangement mechanism termed FoSTeS (fork stalling and template switching). First unveiled through studies of PLP1 duplications associated with Pelizaeus Merzbacher disease [133], a genomic disorder by the criteria originally defined [24], the mechanism has now been shown to cause some LIS1 duplications [134], MECP2 duplications [93], PMP22 and RAI1 duplications [135], PMP22 exon deletions [135] and some interstitial 9q34 deletions thought to represent terminal deletions [136]. The FoSTeS mechanism, as described based upon the phenomenology of breakpoint/join point sequence analysis in human genomic disorders, has been generalized and the molecular details refined, including through genetic and genomic observations on chromosomal rearrangements in other model organisms (for example, Escherichia coli and yeast), and resulting in the microhomology mediated break induced replication (MMBIR) model that may be operative in all life forms [137]. MMBIR can explain many complex rearrangements [137], such as duplication-triplication-duplication (Figure 3). It may be a novel repair pathway for one-ended, double-stranded DNA generated from collapsed replication forks [137]. Such collapsed forks can occur as a replication fork proceeds through a nick or single-strand region generated by local genome architecture. Furthermore, MMBIR predicts that complex human genomic rearrangements will often be accompanied by extensive loss of heterozygosity and, in some cases, by loss of imprinting because the chromosome that is copied may be either the sister or the homolog [137]. Such loss of heterozygosity could lead to regional uniparental disomy [138] as a novel mechanism for disease.
Figure 3.
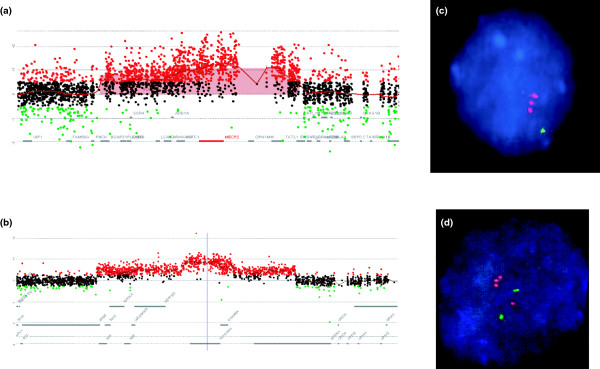
Complex genomic rearrangements. Shown are examples of complex duplication-triplication-duplication rearrangements at MECP2 [93] and LIS1 [134]. (a, b) Array CGH using Agilent custom-designed arrays with interrogating oligonucleotides every few hundred base-pairs from the regions of the genome containing (a) MECP2 and (b) LIS1. Red dots indicate gain of copy number in relation to sex-matched reference DNA; black dots, copy number neutral; green dots, loss of copy number. (c, d) fluorescent in situ hybridization confirmation of the triplication of (c) MECP2 and (d) LIS1 (red, probe interrogating the indicated gene; green, control probe from same chromosome). Note that MECP2 (c) is on the one X chromosome in this male patient, whereas LIS1 (d) is on an autosome and shows both the duplicated (two red signals paired with one green control) with the normal chromosome 17 homologue, with only one copy of LIS1 paired with the green control signal.
In addition to NAHR and FoSTeS/MMBIR, other mechanisms may remain to be uncovered that fulfill the original conception of genomic disorders. Genome architecture may be different for individuals as a result of structural variation within a particular population [50-54,139], so particular individuals may be more susceptible than others to having either a genomic disorder or an offspring with one. Furthermore, other mechanisms, such as nonhomologous end joining and retrotransposition, can lead to structural variation that results in genomic disorders [140], and unique genome architectural features other than LCR/SD, such as AT-rich palindromes [141,142] and non-B DNA conformations [86,143], can incite genome instability. Systematic studies of disorders that occur by such mechanisms may provide insights into local genome architecture that could potentially influence susceptibility to rearrangement; they may thus delineate the 'rules' for FoSTeS/MMBIR as was done for NAHR.
It was initially not known whether human genomic rearrangements reflected random DNA breaks or perhaps selection/survival of genomic regions that could tolerate the gains and losses of CNV. Over the past decade, our thinking has evolved and we can now speak of specific mechanisms (NAHR, MMBIR/FoSTeS, nonhomologous end joining and retrotransposition), and elucidation of the rules for such mechanisms has enabled powerful predictions that have had a direct clinical impact. We have also learnt some of the 'rules' regarding genome architecture. It seems that each rearrangement mechanism can occur anywhere in the human genome, but one mechanism may be preferred over another at a given locus depending on local genome architecture (for example, LCR/SD or non-B DNA). We have realized that CNVs are as important as SNPs to human mutation and perhaps even more important with regard to human sporadic traits [87,127]. Whether CNV or SNP is the more favored mutational event at a given locus may again reflect what the local genome architecture is around that locus [140]. The elucidation of both the mechanisms of CNV formation [144] and how CNVs affect genes to convey phenotypes [145], whether the latter occurs through altered copy number [75,146], gene dysregulation or position effect, has to a large extent come from studies of genomic disorders [147]. The clinical phenotype allows the ascertainment of the genomic rearrangement from the population to enable the molecular studies.
The 'rules' for MMBIR/FoSTeS remain to be further defined with respect to the human genome architecture that might stimulate the events [93,133]. Unquestionably, many more genomic disorders are still to be defined and many Mendelian and complex traits may be shown to be caused by CNV, rather than SNPs of a given gene in selected patients. Thus, a potentially more fruitful and cost-efficient approach to the study of human complex traits may be to examine a few hundred patients for CNV associated with the trait, rather than perform SNP-based GWASs. Such an approach recently yielded insights into Wolf-Parkinson-White syndrome, a common pre-excitation phenomenon resulting in a characteristic electrocardiographic pattern [148]. Certainly all GWASs should look for CNV and not just focus on SNPs [149].
Perhaps the most significant findings regarding the human genome that were not anticipated by the human genome project [45-47,77,78] were the elucidation of genomic disorders and the discovery of the extent to which we vary from each other genetically as a result of CNV. In fact, the establishment of a reference haploid versus diploid genome truly reflects our naiveté with regards to the importance of CNV for human traits. With further widespread clinical implementation of high-resolution human genome analysis, submicroscopic genomic duplications and deletions will probably be identified at an increasing rate. Potentially, the vast majority of the human genome could be involved in CNV, perhaps more of the genome will be subject to, or tolerate, duplication CNV than deletion as observed for chromosomal studies [150,151], and 'reverse genomics' could be used to systematically delineate genomotype-phenotype correlations [134]. The genomic change accompanying a CNV results in a genomotype that may include either more than one, or no genes involved in conveying the specific phenotype and thus is distinct from a genotype.
Such studies will directly address the question: what is the genomic code? This is needed because the genetic code has only addressed the functions of under 2% of the human genome: the coding exons. Systematic analyses of the size, extent and genomic content of CNV and associated phenotypes might lead to a new understanding of 'cis-genetics', the phenotypic consequences of CNV encompassing multiple genes and/or regulatory sequences on one chromosome homolog, as opposed to the 'trans-genetics' focus of Mendelian segregation and transmission of homologous chromosomes. Furthermore, the extents to which human genomic rearrangements occur somatically in mitotic cells are only beginning to be explored [135,152-156]. Thus, genomic disorders will probably continue to be a fruitful area for ongoing and future research.
Abbreviations
AHR: allelic homologous recombination; CMT1A: Charcot-Marie-Tooth disease type 1A; CGH: comparative genomic hybridization; CNV: copy number variation; DECIPHER: database of chromosomal imbalance and phenotype in humans using Ensembl resources; FoSTeS: fork stalling and template switching; GWAS: genome-wide association study; HNPP: hereditary neuropathy with liability to pressure palsies; LCR: low-copy repeat; MMBIR: microhomology mediated break induced replication; NAHR: non-allelic homologous recombination; PTLS: Potocki-Lupski syndrome; SD: segmental duplication; SMS: Smith-Magenis syndrome; SNP: single nucleotide polymorphism.
Competing interests
The author is a consultant for Athena Diagnostics, 23andMe and Ion Torrent Systems, Inc, and holds multiple United States and European patents for DNA diagnostics.
Acknowledgements
I appreciate the critical reviews of Art Beaudet, Weimin Bi, Claudia Car-valho, Evan Eichler, Matt Hurles, Bernice Morrow, Pawel Stankiewicz and Feng Zhang. I apologize, but take full responsibility for, omissions of citations given space limitations. Work in my laboratory has been supported by the Charcot-Marie-Tooth Association, the Muscular Dystrophy Association, the March of Dimes, the Texas Children's Hospital General Clinical Research Center, Baylor College of Medicine Mental Retardation Research Center, Baylor Intellectual and Developmental Disabilities Research Center, and The National Institutes of Health (National Institute of Neurological Disorders and Stroke, R01 NS27042, National Institute of Child Health and Human Development, P01 HD39420, National Eye Institute, R01 EY1325, National Cancer Institute, P01 CA75719, National Institute of Dental and Craniofacial Research, R01 DE015210).
References
- Online Mendelian Inheritance in Man. http://hhttp://www.ncbi.nlm.nih.gov/sites/entrez?db=omim
- Chakravarti A, Patel PI. DNA duplication associated with Charcot-Marie-Tooth disease type 1A. Cell. 1991;66:219–232. doi: 10.1016/0092-8674(91)90613-4. [DOI] [PubMed] [Google Scholar]
- Raeymaekers P, Timmerman V, Nelis E, De Jonghe P, Hoogendijk JE, Baas F, Barker DF, Martin JJ, De Visser M, Bolhuis PA. Duplication in chromosome 17p11.2 in Charcot-Marie-Tooth neuropathy type 1a (CMT 1a). The HMSN Collaborative Research Group. Neuromuscul Disord. 1991;1:93–97. doi: 10.1016/0960-8966(91)90055-W. [DOI] [PubMed] [Google Scholar]
- Chance PF, Alderson MK, Leppig KA, Lensch MW, Matsunami N, Smith B, Swanson PD, Odelberg SJ, Disteche CM, Bird TD. DNA deletion associated with hereditary neuropathy with liability to pressure palsies. Cell. 1993;72:143–151. doi: 10.1016/0092-8674(93)90058-X. [DOI] [PubMed] [Google Scholar]
- Lupski JR, Wise CA, Kuwano A, Pentao L, Parke JT, Glaze DG, Led-better DH, Greenberg F, Patel PI. Gene dosage is a mechanism for Charcot-Marie-Tooth disease type 1A. Nat Genet. 1992;1:29–33. doi: 10.1038/ng0492-29. [DOI] [PubMed] [Google Scholar]
- Patel PI, Roa BB, Welcher AA, Schoener-Scott R, Trask BJ, Pentao L, Snipes GJ, Garcia CA, Francke U, Shooter EM, Lupski JR, Suter U. The gene for the peripheral myelin protein PMP-22 is a candidate for Charcot-Marie-Tooth disease type 1A. Nat Genet. 1992;1:159–165. doi: 10.1038/ng0692-159. [DOI] [PubMed] [Google Scholar]
- Roa BB, Garcia CA, Suter U, Kulpa DA, Wise CA, Mueller J, Welcher AA, Snipes GJ, Shooter EM, Patel PI, Lupski JR. Charcot-Marie-Tooth disease type 1A. Association with a spontaneous point mutation in the PMP22 gene. N Engl J Med. 1993;329:96–101. doi: 10.1056/NEJM199307083290205. [DOI] [PubMed] [Google Scholar]
- Nicholson GA, Valentijn LJ, Cherryson AK, Kennerson ML, Bragg TL, DeKroon RM, Ross DA, Pollard JD, Mcleod JG, Bolhuis PA, Baas F. A frame shift mutation in the PMP22 gene in hereditary neuropathy with liability to pressure palsies. Nat Genet. 1994;6:263–266. doi: 10.1038/ng0394-263. [DOI] [PubMed] [Google Scholar]
- Pentao L, Wise CA, Chinault AC, Patel PI, Lupski JR. Charcot-Marie-Tooth type 1A duplication appears to arise from recombination at repeat sequences flanking the 1.5 Mb monomer unit. Nat Genet. 1992;2:292–300. doi: 10.1038/ng1292-292. [DOI] [PubMed] [Google Scholar]
- Chance PF, Abbas N, Lensch MW, Pentao L, Roa BB, Patel PI, Lupski JR. Two autosomal dominant neuropathies result from reciprocal DNA duplication/deletion of a region on chromosome 17. Hum Mol Genet. 1994;3:223–228. doi: 10.1093/hmg/3.2.223. [DOI] [PubMed] [Google Scholar]
- Edelmann L, Pandita RK, Spiteri E, Funke B, Goldberg R, Palanisamy N, Chaganti RS, Magenis E, Shprintzen RJ, Morrow BE. A common molecular basis for rearrangement disorders on chromosome 22q11. Hum Mol Genet. 1999;8:1157–1167. doi: 10.1093/hmg/8.7.1157. [DOI] [PubMed] [Google Scholar]
- Bailey JA, Yavor AM, Massa HF, Trask BJ, Eichler EE. Segmental duplications: organization and impact within the current human genome project assembly. Genome Res. 2001;11:1005–1017. doi: 10.1101/gr.GR-1871R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey JA, Gu Z, Clark RA, Reinert K, Samonte RV, Schwartz S, Adams MD, Myers EW, Li PW, Eichler EE. Recent segmental duplications in the human genome. Science. 2002;297:1003–1007. doi: 10.1126/science.1072047. [DOI] [PubMed] [Google Scholar]
- Stankiewicz P, Lupski JR. Genome architecture, rearrangements and genomic disorders. Trends Genet. 2002;18:74–82. doi: 10.1016/S0168-9525(02)02592-1. [DOI] [PubMed] [Google Scholar]
- Reiter LT, Murakami T, Koeuth T, Pentao L, Muzny DM, Gibbs RA, Lupski JR. A recombination hotspot responsible for two inherited peripheral neuropathies is located near a mariner transposon-like element. Nat Genet. 1996;12:288–297. doi: 10.1038/ng0396-288. [DOI] [PubMed] [Google Scholar]
- Timmerman V, Rautenstrauss B, Reiter LT, Koeuth T, Löfgren A, Liehr T, Nelis E, Bathke KD, De Jonghe P, Grehl H, Martin JJ, Lupski JR, Van Broeckhoven C. Detection of the CMT1A/HNPP recombination hotspot in unrelated patients of European descent. J Med Genet. 1997;34:43–49. doi: 10.1136/jmg.34.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matise TC, Chakravarti A, Patel PI, Lupski JR, Nelis E, Timmerman V, Van Broeckhoven C, Weeks DE. Detection of tandem duplications and implications for linkage analysis. Am J Hum Genet. 1994;54:1110–1121. [PMC free article] [PubMed] [Google Scholar]
- Greenberg F, Guzzetta V, Montes de Oca-Luna R, Magenis RE, Smith AC, Richter SF, Kondo I, Dobyns WB, Patel PI, Lupski JR. Molecular analysis of the Smith-Magenis syndrome: a possible contiguous-gene syndrome associated with del(p11.2). Am J Hum Genet. 1991;49:1207–1218. [PMC free article] [PubMed] [Google Scholar]
- Guzzetta V, Franco B, Trask BJ, Zhang H, Saucedo-Cardenas O, Montes de Oca-Luna R, Greenberg F, Chinault AC, Lupski JR, Patel PI. Somatic cell hybrids, sequence-tagged sites, simple repeat polymorphisms, and yeast artificial chromosomes for physical and genetic mapping of proximal 17p. Genomics. 1992;13:551–559. doi: 10.1016/0888-7543(92)90124-B. [DOI] [PubMed] [Google Scholar]
- Juyal RC, Figuera LE, Hauge X, Elsea SH, Lupski JR, Greenberg F, Baldini A, Patel PI. Molecular analyses of 17p11.2 deletions in 62 Smith-Magenis syndrome patients. Am J Hum Genet. 1996;58:998–1007. [PMC free article] [PubMed] [Google Scholar]
- Chen KS, Manian P, Koeuth T, Potocki L, Zhao Q, Chinault AC, Lee CC, Lupski JR. Homologous recombination of a flanking repeat gene cluster is a mechanism for a common contiguous gene deletion syndrome. Nat Genet. 1997;17:154–163. doi: 10.1038/ng1097-154. [DOI] [PubMed] [Google Scholar]
- Shaw CJ, Bi W, Lupski JR. Genetic proof of unequal meiotic crossovers in reciprocal deletion and duplication of 17p11.2. Am J Hum Genet. 2002;71:1072–1081. doi: 10.1086/344346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi W, Park SS, Shaw CJ, Withers MA, Patel PI, Lupski JR. Reciprocal crossovers and a positional preference for strand exchange in recombination events resulting in deletion or duplication of chromosome 17p11.2. Am J Hum Genet. 2003;73:1302–1315. doi: 10.1086/379979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupski JR. Genomic disorders: structural features of the genome can lead to DNA rearrangements and human disease traits. Trends Genet. 1998;14:417–422. doi: 10.1016/S0168-9525(98)01555-8. [DOI] [PubMed] [Google Scholar]
- Phillips JA 3rd, Vik TA, Scott AF, Young KE, Kazazian HH Jr, Smith KD, Fairbanks VF, Koenig HM. Unequal crossing-over: a common basis of single alpha-globin genes in Asians and American blacks with hemoglobin-H disease. Blood. 1980;55:1066–1069. [PubMed] [Google Scholar]
- Higgs DR, Vickers MA, Wilkie AO, Pretorius IM, Jarman AP, Weatherall DJ. A review of the molecular genetics of the human alpha-globin gene cluster. Blood. 1989;73:1081–1104. [PubMed] [Google Scholar]
- Nathans J, Piantanida TP, Eddy RL, Shows TB, Hogness DS. Molecular genetics of inherited variation in human color vision. Science. 1986;232:203–210. doi: 10.1126/science.3485310. [DOI] [PubMed] [Google Scholar]
- Potocki L, Chen KS, Park SS, Osterholm DE, Withers MA, Kimonis V, Summers AM, Meschino WS, Anyane-Yeboa K, Kashork CD, Shaffer LG, Lupski JR. Molecular mechanism for duplication 17p11.2-the homologous recombination reciprocal of the Smith-Magenis microdeletion. Nat Genet. 2000;24:84–87. doi: 10.1038/71743. [DOI] [PubMed] [Google Scholar]
- Potocki L, Bi W, Treadwell-Deering D, Carvalho CM, Eifert A, Fried-man EM, Glaze D, Krull K, Lee JA, Lewis RA, Mendoza-Londono R, Robbins-Furman P, Shaw C, Shi X, Weissenberger G, Withers M, Yatsenko SA, Zackai EH, Stankiewicz P, Lupski JR. Characterization of Potocki-Lupski syndrome (dup(p11.2p11.2)) and delineation of a dosage-sensitive critical interval that can convey an autism phenotype. Am J Hum Genet. 2007;80:633–649. doi: 10.1086/512864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ensenauer RE, Adeyinka A, Flynn HC, Michels VV, Lindor NM, Dawson DB, Thorland EC, Lorentz CP, Goldstein JL, McDonald MT, Smith WE, Simon-Fayard E, Alexander AA, Kulharya AS, Ketterling RP, Clark RD, Jalal SM. Microduplication 22q11.2, an emerging syndrome: clinical, cytogenetic, and molecular analysis of thirteen patients. Am J Hum Genet. 2003;73:1027–1040. doi: 10.1086/378818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassed SJ, Hopcus-Niccum D, Zhang L, Li S, Mulvihill JJ. A new genomic duplication syndrome complementary to the velocardiofa-cial (22q11 deletion) syndrome. Clin Genet. 2004;65:400–404. doi: 10.1111/j.0009-9163.2004.0212.x. [DOI] [PubMed] [Google Scholar]
- Yobb TM, Somerville MJ, Willatt L, Firth HV, Harrison K, MacKenzie J, Gallo N, Morrow BE, Shaffer LG, Babcock M, Chernos J, Bernier F, Sprysak K, Christiansen J, Haase S, Elyas B, Lilley M, Bamforth S, McDermid HE. Microduplication and triplication of 22q11.2: a highly variable syndrome. Am J Hum Genet. 2005;76:865–876. doi: 10.1086/429841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville MJ, Mervis CB, Young EJ, Seo EJ, del Campo M, Bamforth S, Peregrine E, Loo W, Lilley M, Pérez-Jurado LA, Morris CA, Scherer SW, Osborne LR. Severe expressive-language delay related to duplication of the Williams-Beuren locus. N Engl J Med. 2005;353:1694–1701. doi: 10.1056/NEJMoa051962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg JS, Brunetti-Pierri N, Peters SU, Kang SH, Fong CT, Salamone J, Freedenberg D, Hannig VL, Prock LA, Miller DT, Raffalli P, Harris DJ, Erickson RP, Cunniff C, Clark GD, Blazo MA, Peiffer DA, Gunderson KL, Sahoo T, Patel A, Lupski JR, Beaudet AL, Cheung SW. Speech delay and autism spectrum behaviors are frequently associated with duplication of the 7q11.23 Williams-Beuren syndrome region. Genet Med. 2007;9:427–441. doi: 10.1097/gim.0b013e3180986192. [DOI] [PubMed] [Google Scholar]
- Depienne C, Heron D, Betancur C, Benyahia B, Trouillard O, Bouteiller D, Verloes A, LeGuern E, Leboyer M, Brice A. Autism, language delay and mental retardation in a patient with 7q11 duplication. J Med Genet. 2007;44:452–458. doi: 10.1136/jmg.2006.047092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchhoff M, Bisgaard AM, Duno M, Hansen FJ, Schwartz M. A 17q21.31 microduplication, reciprocal to the newly described 17q21.31 microdeletion, in a girl with severe psychomotor developmental delay and dysmorphic craniofacial features. Eur J Med Genet. 2007;50:256–263. doi: 10.1016/j.ejmg.2007.05.001. [DOI] [PubMed] [Google Scholar]
- Kirchhoff M, Bisgaard AM, Bryndorf T, Gerdes T. MLPA analysis for a panel of syndromes with mental retardation reveals imbalances in 5.8% of patients with mental retardation and dysmorphic features, including duplications of the Sotos syndrome and Williams-Beuren syndrome regions. Eur J Med Genet. 2007;50:33–42. doi: 10.1016/j.ejmg.2006.10.002. [DOI] [PubMed] [Google Scholar]
- Ben-Shachar S, Ou Z, Shaw CA, Belmont JW, Patel MS, Hummel M, Amato S, Tartaglia N, Berg J, Sutton VR, Lalani SR, Chinault AC, Cheung SW, Lupski JR, Patel A. 22q11.2 distal deletion: a recurrent genomic disorder distinct from DiGeorge syndrome and velocardiofa-cial syndrome. Am J Hum Genet. 2008;82:214–221. doi: 10.1016/j.ajhg.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou Z, Berg JS, Yonath H, Enciso VB, Miller DT, Picker J, Lenzi T, Keegan CE, Sutton VR, Belmont J, Chinault AC, Lupski JR, Cheung SW, Roeder E, Patel A. Microduplications of 22q11.2 are frequently inherited and are associated with variable phenotypes. Genet Med. 2008;10:267–277. doi: 10.1097/GIM.0b013e31816b64c2. [DOI] [PubMed] [Google Scholar]
- Weiss LA, Shen Y, Korn JM, Arking DE, Miller DT, Fossdal R, Saemu-ndsen E, Stefansson H, Ferreira MA, Green T, Platt OS, Ruderfer DM, Walsh CA, Altshuler D, Chakravarti A, Tanzi RE, Stefansson K, Santangelo SL, Gusella JF, Sklar P, Wu BL, Daly MJ. Autism Consortium. Association between microdeletion and microduplication at 16p11.2 and autism. N Engl J Med. 2008;358:667–675. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- Brunetti-Pierri N, Berg JS, Scaglia F, Belmont J, Bacino CA, Sahoo T, Lalani SR, Graham B, Lee B, Shinawi M, Shen J, Kang SH, Pursley A, Lotze T, Kennedy G, Lansky-Shafer S, Weaver C, Roeder ER, Grebe TA, Arnold GL, Hutchison T, Reimschisel T, Amato S, Geragthy MT, Innis JW, Obersztyn E, Nowakowska B, Rosengren SS, Bader PI, Grange DK. Recurrent reciprocal 1q21.1 deletions and duplications associated with microcephaly or macrocephaly and developmental and behavioral abnormalities. Nat Genet. 2008;40:1466–1471. doi: 10.1038/ng.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballif BC, Theisen A, Coppinger J, Gowans GC, Hersh JH, Madan-Khetarpal S, Schmidt KR, Tervo R, Escobar LF, Friedrich CA, McDonald M, Campbell L, Ming JE, Zackai EH, Bejjani BA, Shaffer LG. Expanding the clinical phenotype of the 3q29 microdeletion syndrome and characterization of the reciprocal microduplication. Mol Cytogenet. 2008;1:8. doi: 10.1186/1755-8166-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willatt L, Cox J, Barber J, Cabanas ED, Collins A, Donnai D, Fitz-Patrick DR, Maher E, Martin H, Parnau J, Pindar L, Ramsay J, Shaw-Smith C, Sistermans EA, Tettenborn M, Trump D, de Vries BB, Walker K, Raymond FL. 3q29 microdeletion syndrome: clinical and molecular characterization of a new syndrome. Am J Hum Genet. 2005;77:154–160. doi: 10.1086/431653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mefford HC, Sharp AJ, Baker C, Itsara A, Jiang Z, Buysse K, Huang S, Maloney VK, Crolla JA, Baralle D, Collins A, Mercer C, Norga K, de Ravel T, Devriendt K, Bongers EM, de Leeuw N, Reardon W, Gimelli S, Bena F, Hennekam RC, Male A, Gaunt L, Clayton-Smith J, Simonic I, Park SM, Mehta SG, Nik-Zainal S, Woods CG, Firth HV. Recurrent rearrangements of chromosome 1q21.1 and variable pediatric phenotypes. N Engl J Med. 2008;359:1685–1699. doi: 10.1056/NEJMoa0805384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium. The Sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- Snijders AM, Nowak N, Segraves R, Blackwood S, Brown N, Conroy J, Hamilton G, Hindle AK, Huey B, Kimura K, Law S, Myambo K, Palmer J, Ylstra B, Yue JP, Gray JW, Jain AN, Pinkel D, Albertson DG. Assembly of microarrays for genome-wide measurement of DNA copy number. Nat Genet. 2001;29:263–264. doi: 10.1038/ng754. [DOI] [PubMed] [Google Scholar]
- Sharp AJ, Locke DP, McGrath SD, Cheng Z, Bailey JA, Vallente RU, Pertz LM, Clark RA, Schwartz S, Segraves R, Oseroff VV, Albertson DG, Pinkel D, Eichler EE. Segmental duplications and copy-number variation in the human genome. Am J Hum Genet. 2005;77:78–88. doi: 10.1086/431652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp AJ, Hansen S, Selzer RR, Cheng Z, Regan R, Hurst JA, Stewart H, Price SM, Blair E, Hennekam RC, Fitzpatrick CA, Segraves R, Richmond TA, Guiver C, Albertson DG, Pinkel D, Eis PS, Schwartz S, Knight SJ, Eichler EE. Discovery of previously unidentified genomic disorders from the duplication architecture of the human genome. Nat Genet. 2006;38:1038–1042. doi: 10.1038/ng1862. [DOI] [PubMed] [Google Scholar]
- Koolen DA, Vissers LE, Pfundt R, de Leeuw N, Knight SJ, Regan R, Kooy RF, Reyniers E, Romano C, Fichera M, Schinzel A, Baumer A, Anderlid BM, Schoumans J, Knoers NV, van Kessel AG, Sistermans EA, Veltman JA, Brunner HG, de Vries BB. A new chromosome 17q21.31 microdeletion syndrome associated with a common inversion polymorphism. Nat Genet. 2006;38:999–1001. doi: 10.1038/ng1853. [DOI] [PubMed] [Google Scholar]
- Shaw-Smith C, Pittman AM, Willatt L, Martin H, Rickman L, Gribble S, Curley R, Cumming S, Dunn C, Kalaitzopoulos D, Porter K, Prig-more E, Krepischi-Santos AC, Varela MC, Koiffmann CP, Lees AJ, Rosenberg C, Firth HV, de Silva R, Carter NP. Microdeletion encompassing MAPT at chromosome 17q21.3 is associated with developmental delay and learning disability. Nat Genet. 2006;38:1032–1037. doi: 10.1038/ng1858. [DOI] [PubMed] [Google Scholar]
- Lupski JR. Genome structural variation and sporadic disease traits. Nat Genet. 2006;38:974–976. doi: 10.1038/ng0906-974. [DOI] [PubMed] [Google Scholar]
- Koolen DA, Sharp AJ, Hurst JA, Firth HV, Knight SJ, Goldenberg A, Saugier-Veber P, Pfundt R, Vissers LE, Destrée A, Grisart B, Rooms L, Aa N Van der, Field M, Hackett A, Bell K, Nowaczyk MJ, Mancini GM, Poddighe PJ, Schwartz CE, Rossi E, De Gregori M, Antonacci-Fulton LL, McLellan MD 2nd, Garrett JM, Wiechert MA, Miner TL, Crosby S, Ciccone R, Willatt L. Clinical and molecular delineation of the 17q21.31 microdeletion syndrome. J Med Genet. 2008;45:710–720. doi: 10.1136/jmg.2008.058701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp AJ, Selzer RR, Veltman JA, Gimelli S, Gimelli G, Striano P, Coppola A, Regan R, Price SM, Knoers NV, Eis PS, Brunner HG, Hennekam RC, Knight SJ, de Vries BB, Zuffardi O, Eichler EE. Characterization of a recurrent 15q24 microdeletion syndrome. Hum Mol Genet. 2007;16:567–572. doi: 10.1093/hmg/ddm016. [DOI] [PubMed] [Google Scholar]
- Mefford HC, Clauin S, Sharp AJ, Moller RS, Ullmann R, Kapur R, Pinkel D, Cooper GM, Ventura M, Ropers HH, Tommerup N, Eichler EE, Bellanne-Chantelot C. Recurrent reciprocal genomic rearrangements of 17q12 are associated with renal disease, diabetes, and epilepsy. Am J Hum Genet. 2007;81:1057–1069. doi: 10.1086/522591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp AJ, Mefford HC, Li K, Baker C, Skinner C, Stevenson RE, Schroer RJ, Novara F, De Gregori M, Ciccone R, Broomer A, Casuga I, Wang Y, Xiao C, Barbacioru C, Gimelli G, Bernardina BD, Torniero C, Giorda R, Regan R, Murday V, Mansour S, Fichera M, Castiglia L, Failla P, Ventura M, Jiang Z, Cooper GM, Knight SJ, Romano C. A recurrent 15q13.3 microdeletion syndrome associated with mental retardation and seizures. Nat Genet. 2008;40:322–328. doi: 10.1038/ng.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellanné-Chantelot C, Clauin S, Chauveau D, Collin P, Daumont M, Douillard C, Dubois-Laforgue D, Dusselier L, Gautier JF, Jadoul M, Laloi-Michelin M, Jacquesson L, Larger E, Louis J, Nicolino M, Subra JF, Wilhem JM, Young J, Velho G, Timsit J. Large genomic rearrangements in the hepatocyte nuclear factor-1beta (TCF2) gene are the most frequent cause of maturity-onset diabetes of the young type 5. Diabetes. 2005;54:3126–3132. doi: 10.2337/diabetes.54.11.3126. [DOI] [PubMed] [Google Scholar]
- International Schizophrenia Consortium. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:237–241. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefansson H, Rujescu D, Cichon S, Pietiläinen OP, Ingason A, Steinberg S, Fossdal R, Sigurdsson E, Sigmundsson T, Buizer-Voskamp JE, Hansen T, Jakobsen KD, Muglia P, Francks C, Matthews PM, Gylfason A, Halldorsson BV, Gudbjartsson D, Thorgeirsson TE, Sigurdsson A, Jonasdottir A, Jonasdottir A, Bjornsson A, Mattiasdottir S, Blondal T, Haraldsson M, Magnusdot-tir BB, Giegling I, Möller HJ, Hartmann A. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232–236. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupski JR. Schizophrenia: incriminating genomic evidence. Nature. 2008;455:178–179. doi: 10.1038/455178a. [DOI] [PubMed] [Google Scholar]
- Helbig I, Mefford HC, Sharp AJ, Guipponi M, Fichera M, Franke A, Muhle H, de Kovel C, Baker C, von Spiczak S, Kron KL, Steinich I, Kleefuss-Lie AA, Leu C, Gaus V, Schmitz B, Klein KM, Reif PS, Rosenow F, Weber Y, Lerche H, Zimprich F, Urak L, Fuchs K, Feucht M, Genton P, Thomas P, Visscher F, de Haan GJ, Møller RS. 15q13.3 microdeletions increase risk of idiopathic generalized epilepsy. Nat Genet. 2009;41:160–162. doi: 10.1038/ng.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DT, Shen Y, Weiss LA, Korn J, Anselm I, Bridgemohan C, Cox GF, Dickinson H, Gentile J, Harris DJ, Hegde V, Hundley R, Khwaja O, Kothare S, Luedke C, Nasir R, Poduri A, Prasad K, Raf-falli P, Reinhard A, Smith SE, Sobeih M, Soul J, Stoler J, Takeoka M, Tan WH, Thakuria J, Wolff P, Yusupov R, Gusella JF, Daly MJ, Wu BL. Microdeletion/duplication at 15q13.2q13.3 among individuals with features of autism and other neuropsychiatric disorders. J Med Genet. 2008;46:242–248. doi: 10.1136/jmg.2008.059907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Shachar S Lanpher B German JR Potocki L Nagamani SCS Franco LM El-Hhakam LM Freedenberg D Bottenfield GW Spence JE Shaw CA Patel A Cheung SW Lupski JR Beaudet AL Sahoo T Recurrent 15q13.3 microdeletions are associated with autism and a wide variety of neurodevelopmental phenotypes. J Med Genet in press 19289393
- Kumar RA, KaraMohamed S, Sudi J, Conrad DF, Brune C, Badner JA, Gilliam TC, Nowak NJ, Cook EH Jr, Dobyns WB, Christian SL. Recurrent 16p11.2 microdeletions in autism. Hum Mol Genet. 2008;17:628–638. doi: 10.1093/hmg/ddm376. [DOI] [PubMed] [Google Scholar]
- Gonzalez E, Kulkarni H, Bolivar H, Mangano A, Sanchez R, Catano G, Nibbs RJ, Freedman BI, Quinones MP, Bamshad MJ, Murthy KK, Rovin BH, Bradley W, Clark RA, Anderson SA, O'connell RJ, Agan BK, Ahuja SS, Bologna R, Sen L, Dolan MJ, Ahuja SK. The influence of CCL3L1 gene-containing segmental duplications on HIV-1/AIDS susceptibility. Science. 2005;307:1434–1440. doi: 10.1126/science.1101160. [DOI] [PubMed] [Google Scholar]
- Fellermann K, Wehkamp J, Herrlinger KR, Stange EF. Crohn's disease: a defensin deficiency syndrome? Eur J Gastroenterol Hepatol. 2003;15:627–634. doi: 10.1097/00042737-200306000-00008. [DOI] [PubMed] [Google Scholar]
- Fellermann K, Stange DE, Schaeffeler E, Schmalzl H, Wehkamp J, Bevins CL, Reinisch W, Teml A, Schwab M, Lichter P, Radlwimmer B, Stange EF. A chromosome 8 gene-cluster polymorphism with low human beta-defensin 2 gene copy number predisposes to Crohn disease of the colon. Am J Hum Genet. 2006;79:439–448. doi: 10.1086/505915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarroll SA, Huett A, Kuballa P, Chilewski SD, Landry A, Goyette P, Zody MC, Hall JL, Brant SR, Cho JH, Duerr RH, Silverberg MS, Taylor KD, Rioux JD, Altshuler D, Daly MJ, Xavier RJ. Deletion polymorphism upstream of IRGM associated with altered IRGM expression and Crohn's disease. Nat Genet. 2008;40:1107–1112. doi: 10.1038/ng.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aitman TJ, Dong R, Vyse TJ, Norsworthy PJ, Johnson MD, Smith J, Mangion J, Roberton-Lowe C, Marshall AJ, Petretto E, Hodges MD, Bhangal G, Patel SG, Sheehan-Rooney K, Duda M, Cook PR, Evans DJ, Domin J, Flint J, Boyle JJ, Pusey CD, Cook HT. Copy number polymorphism in Fcgr3 predisposes to glomerulonephritis in rats and humans. Nature. 2006;439:851–855. doi: 10.1038/nature04489. [DOI] [PubMed] [Google Scholar]
- Hollox EJ, Huffmeier U, Zeeuwen PL, Palla R, Lascorz J, Rodijk-Olthuis D, Kerkhof PC van de, Traupe H, de Jongh G, den Heijer M, Reis A, Armour JA, Schalkwijk J. Psoriasis is associated with increased beta-defensin genomic copy number. Nat Genet. 2008;40:23–25. doi: 10.1038/ng.2007.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanciulli M, Norsworthy PJ, Petretto E, Dong R, Harper L, Kamesh L, Heward JM, Gough SC, de Smith A, Blakemore AI, Froguel P, Owen CJ, Pearce SH, Teixeira L, Guillevin L, Graham DS, Pusey CD, Cook HT, Vyse TJ, Aitman TJ. FCGR3B copy number variation is associated with susceptibility to systemic, but not organ-specific, autoimmunity. Nat Genet. 2007;39:721–723. doi: 10.1038/ng2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willcocks LC, Lyons PA, Clatworthy MR, Robinson JI, Yang W, Newland SA, Plagnol V, McGovern NN, Condliffe AM, Chilvers ER, Adu D, Jolly EC, Watts R, Lau YL, Morgan AW, Nash G, Smith KG. Copy number of FCGR3B, which is associated with systemic lupus erythematosus, correlates with protein expression and immune complex uptake. J Exp Med. 2008;205:1573–1582. doi: 10.1084/jem.20072413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Marechal C, Masson E, Chen JM, Morel F, Ruszniewski P, Levy P, Ferec C. Hereditary pancreatitis caused by triplication of the trypsino-gen locus. Nat Genet. 2006;38:1372–1374. doi: 10.1038/ng1904. [DOI] [PubMed] [Google Scholar]
- Walz K, Paylor R, Yan J, Bi W, Lupski JR. Rai1 duplication causes physical and behavioral phenotypes in a mouse model of dup(p11.2p11.2). J Clin Invest. 2006;116:3035–3041. doi: 10.1172/JCI28953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupski JR. Hotspots of homologous recombination in the human genome: not all homologous sequences are equal. Genome Biol. 2004;5:242. doi: 10.1186/gb-2004-5-10-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International HapMap Consortium. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers SR, McCarroll SA. New insights into the biological basis of genomic disorders. Nat Genet. 2006;38:1363–1364. doi: 10.1038/ng1206-1363. [DOI] [PubMed] [Google Scholar]
- Lindsay SJ, Khajavi M, Lupski JR, Hurles ME. A chromosomal rearrangement hotspot can be identified from population genetic variation and is coincident with a hotspot for allelic recombination. Am J Hum Genet. 2006;79:890–902. doi: 10.1086/508709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raedt TD, Stephens M, Heyns I, Brems H, Thijs D, Messiaen L, Stephens K, Lazaro C, Wimmer K, Kehrer-Sawatzki H, Vidaud D, Kluwe L, Marynen P, Legius E. Conservation of hotspots for recombination in low-copy repeats associated with the NF1 microdeletion. Nat Genet. 2006;38:1419–1423. doi: 10.1038/ng1920. [DOI] [PubMed] [Google Scholar]
- Reiter LT, Hastings PJ, Nelis E, De Jonghe P, Van Broeckhoven C, Lupski JR. Human meiotic recombination products revealed by sequencing a hotspot for homologous strand exchange in multiple HNPP deletion patients. Am J Hum Genet. 1998;62:1023–1033. doi: 10.1086/301827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam KW, Jeffreys AJ. Processes of copy-number change in human DNA: the dynamics of {alpha}-globin gene deletion. Proc Natl Acad Sci USA. 2006;103:8921–8927. doi: 10.1073/pnas.0602690103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato T, Inagaki H, Yamada K, Kogo H, Ohye T, Kowa H, Nagaoka K, Taniguchi M, Emanuel BS, Kurahashi H. Genetic variation affects de novo translocation frequency. Science. 2006;311:971. doi: 10.1126/science.1121452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam KW, Jeffreys AJ. Processes of de novo duplication of human alpha-globin genes. Proc Natl Acad Sci USA. 2007;104:10950–10955. doi: 10.1073/pnas.0703856104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki H, Ohye T, Kogo H, Kato T, Bolor H, Taniguchi M, Shaikh TH, Emanuel BS, Kurahashi H. Chromosomal instability mediated by non-B DNA: cruciform conformation and not DNA sequence is responsible for recurrent translocation in humans. Genome Res. 2009;19:191–198. doi: 10.1101/gr.079244.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupski JR. Genomic rearrangements and sporadic disease. Nat Genet. 2007;39:S43–S47. doi: 10.1038/ng2084. [DOI] [PubMed] [Google Scholar]
- Turner DJ, Miretti M, Rajan D, Fiegler H, Carter NP, Blayney ML, Beck S, Hurles ME. Germline rates of de novo meiotic deletions and duplications causing several genomic disorders. Nat Genet. 2008;40:90–95. doi: 10.1038/ng.2007.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami T, Reiter LT, Lupski JR. Genomic structure and expression of the human heme A:farnesyltransferase (COX10) gene. Genomics. 1997;42:161–164. doi: 10.1006/geno.1997.4711. [DOI] [PubMed] [Google Scholar]
- Reiter LT, Murakami T, Koeuth T, Gibbs RA, Lupski JR. The human COX10 gene is disrupted during homologous recombination between the 24 kb proximal and distal CMT1A-REPs. Hum Mol Genet. 1997;6:1595–1603. doi: 10.1093/hmg/6.9.1595. [DOI] [PubMed] [Google Scholar]
- Inoue K, Dewar K, Katsanis N, Reiter LT, Lander ES, Devon KL, Wyman DW, Lupski JR, Birren B. The 1.4-Mb CMT1A duplication/HNPP deletion genomic region reveals unique genome architectural features and provides insights into the recent evolution of new genes. Genome Res. 2001;11:1018–1033. doi: 10.1101/gr.180401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Gaudio D, Fang P, Scaglia F, Ward PA, Craigen WJ, Glaze DG, Neul JL, Patel A, Lee JA, Irons M, Berry SA, Pursley AA, Grebe TA, Freedenberg D, Martin RA, Hsich GE, Khera JR, Friedman NR, Zoghbi HY, Eng CM, Lupski JR, Beaudet AL, Cheung SW, Roa BB. Increased MECP2 gene copy number due to genomic duplication in neurodevel-opmentally delayed males. Genet Med. 2006;8:784–792. doi: 10.1097/01.gim.0000250502.28516.3c. [DOI] [PubMed] [Google Scholar]
- Carvalho CM, Zhang F, Liu P, Patel A, Sahoo T, Bacino C, Peacock S, Shaw C, Pursley A, Tavyev J, Ramocki MB, Nawara M, Obertsztyn E, Vianna-Morgante AM, Stankiewicz P, Zoghbi HY, Cheung SW, Lupski JR. Complex rearrangements in MECP2 duplicated patients suggest Fork Stalling and Template Switching (FoSTeS) as a major mechanism. Hum Mol Genet. in press . [DOI] [PMC free article] [PubMed]
- Probst FJ, Chen KS, Zhao Q, Wang A, Friedman TB, Lupski JR, Camper SA. A physical map of the mouse shaker-2 region contains many of the genes commonly deleted in Smith-Magenis syndrome (del17p11.2p11.2). Genomics. 1999;55:348–352. doi: 10.1006/geno.1998.5669. [DOI] [PubMed] [Google Scholar]
- Stankiewicz P, Park SS, Inoue K, Lupski JR. The evolutionary chromosome translocation 4;19 in Gorilla gorilla is associated with microdupli-cation of the chromosome fragment syntenic to sequences surrounding the human proximal CMT1A-REP. Genome Res. 2001;11:1205–1210. doi: 10.1101/gr.181101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi W, Yan J, Stankiewicz P, Park SS, Walz K, Boerkoel CF, Potocki L, Shaffer LG, Devriendt K, Nowaczyk MJ, Inoue K, Lupski JR. Genes in a refined Smith-Magenis syndrome critical deletion interval on chromosome 17p11.2 and the syntenic region of the mouse. Genome Res. 2002;12:713–728. doi: 10.1101/gr.73702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SS, Stankiewicz P, Bi W, Shaw C, Lehoczky J, Dewar K, Birren B, Lupski JR. Structure and evolution of the Smith-Magenis syndrome repeat gene clusters, SMS-REPs. Genome Res. 2002;12:729–738. doi: 10.1101/gr.82802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stankiewicz P, Shaw CJ, Withers M, Inoue K, Lupski JR. Serial segmental duplications during primate evolution result in complex human genome architecture. Genome Res. 2004;14:2209–2220. doi: 10.1101/gr.2746604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stankiewicz P, Lupski JR. Molecular-evolutionary mechanisms for genomic disorders. Curr Opin Genet Dev. 2002;12:312–319. doi: 10.1016/S0959-437X(02)00304-0. [DOI] [PubMed] [Google Scholar]
- Zody MC, Garber M, Adams DJ, Sharpe T, Harrow J, Lupski JR, Nicholson C, Searle SM, Wilming L, Young SK, Abouelleil A, Allen NR, Bi W, Bloom T, Borowsky ML, Bugalter BE, Butler J, Chang JL, Chen CK, Cook A, Corum B, Cuomo CA, de Jong PJ, DeCaprio D, Dewar K, FitzGerald M, Gilbert J, Gibson R, Gnerre S, Goldstein S. DNA sequence of human chromosome 17 and analysis of rearrangement in the human lineage. Nature. 2006;440:1045–1049. doi: 10.1038/nature04689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen J. Genomics. DNA duplications and deletions help determine health. Science. 2007;317:1315–1317. doi: 10.1126/science.317.5843.1315. [DOI] [PubMed] [Google Scholar]
- Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, Scherer SW, Lee C. Detection of large-scale variation in the human genome. Nat Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- Sebat J, Lakshmi B, Troge J, Alexander J, Young J, Lundin P, Månér S, Massa H, Walker M, Chi M, Navin N, Lucito R, Healy J, Hicks J, Ye K, Reiner A, Gilliam TC, Trask B, Patterson N, Zetterberg A, Wigler M. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, Fiegler H, Shapero MH, Carson AR, Chen W, Cho EK, Dallaire S, Freeman JL, González JR, Gratacòs M, Huang J, Kalaitzopoulos D, Komura D, Mac-Donald JR, Marshall CR, Mei R, Montgomery L, Nishimura K, Okamura K, Shen F, Somerville MJ, Tchinda J, Valsesia A, Woodwark C, Yang F. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Database of Genomic Variants. http://projects.tcag.ca/variation/
- Shaw-Smith C, Redon R, Rickman L, Rio M, Willatt L, Fiegler H, Firth H, Sanlaville D, Winter R, Colleaux L, Bobrow M, Carter NP. Microar-ray based comparative genomic hybridisation (array-CGH) detects submicroscopic chromosomal deletions and duplications in patients with learning disability/mental retardation and dysmorphic features. J Med Genet. 2004;41:241–248. doi: 10.1136/jmg.2003.017731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejjani BA, Saleki R, Ballif BC, Rorem EA, Sundin K, Theisen A, Kashork CD, Shaffer LG. Use of targeted array-based CGH for the clinical diagnosis of chromosomal imbalance: is less more? Am J Med Genet A. 2005;134:259–267. doi: 10.1002/ajmg.a.30621. [DOI] [PubMed] [Google Scholar]
- Cheung SW, Shaw CA, Yu W, Li J, Ou Z, Patel A, Yatsenko SA, Cooper ML, Furman P, Stankiewicz P, Lupski JR, Chinault AC, Beaudet AL. Development and validation of a CGH microarray for clinical cyto-genetic diagnosis. Genet Med. 2005;7:422–432. doi: 10.1097/01.gim.0000170992.63691.32. [DOI] [PubMed] [Google Scholar]
- Schoumans J, Ruivenkamp C, Holmberg E, Kyllerman M, Anderlid BM, Nordenskjold M. Detection of chromosomal imbalances in children with idiopathic mental retardation by array based comparative genomic hybridisation (array-CGH). J Med Genet. 2005;42:699–705. doi: 10.1136/jmg.2004.029637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries BB, Pfundt R, Leisink M, Koolen DA, Vissers LE, Janssen IM, Reijmersdal S, Nillesen WM, Huys EH, Leeuw N, Smeets D, Sister-mans EA, Feuth T, van Ravenswaaij-Arts CM, van Kessel AG, Schoen-makers EF, Brunner HG, Veltman JA. Diagnostic genome profiling in mental retardation. Am J Hum Genet. 2005;77:606–616. doi: 10.1086/491719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer LG, Kashork CD, Saleki R, Rorem E, Sundin K, Ballif BC, Bejjani BA. Targeted genomic microarray analysis for identification of chromosome abnormalities in 1500 consecutive clinical cases. J Pediatr. 2006;149:98–102. doi: 10.1016/j.jpeds.2006.02.006. [DOI] [PubMed] [Google Scholar]
- Menten B, Maas N, Thienpont B, Buysse K, Vandesompele J, Melotte C, de Ravel T, Van Vooren S, Balikova I, Backx L, Janssens S, De Paepe A, De Moor B, Moreau Y, Marynen P, Fryns JP, Mortier G, Devriendt K, Speleman F, Vermeesch JR. Emerging patterns of cryptic chromosomal imbalance in patients with idiopathic mental retardation and multiple congenital anomalies: a new series of 140 patients and review of published reports. J Med Genet. 2006;43:625–633. doi: 10.1136/jmg.2005.039453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg C, Knijnenburg J, Bakker E, Vianna-Morgante AM, Sloos W, Otto PA, Kriek M, Hansson K, Krepischi-Santos AC, Fiegler H, Carter NP, Bijlsma EK, van Haeringen A, Szuhai K, Tanke HJ. Array-CGH detection of micro rearrangements in mentally retarded individuals: clinical significance of imbalances present both in affected children and normal parents. J Med Genet. 2006;43:180–186. doi: 10.1136/jmg.2005.032268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballif BC, Rorem EA, Sundin K, Lincicum M, Gaskin S, Coppinger J, Kashork CD, Shaffer LG, Bejjani BA. Detection of low-level mosaicism by array CGH in routine diagnostic specimens. Am J Med Genet A. 2006;140:2757–2767. doi: 10.1002/ajmg.a.31539. [DOI] [PubMed] [Google Scholar]
- Cheung SW, Shaw CA, Scott DA. Microarray-based CGH detects chromosomal mosaicism not revealed by conventional cytogenetics. Am J Hum Genet. 2007;143:1679–1686. doi: 10.1002/ajmg.a.31740. [DOI] [PubMed] [Google Scholar]
- Lu X, Shaw CA, Patel A, Li J, Cooper ML, Wells WR, Sullivan CM, Sahoo T, Yatsenko SA, Bacino CA, Stankiewicz P, Ou Z, Chinault AC, Beaudet AL, Lupski JR, Cheung SW, Ward PA. Clinical implementation of chromosomal microarray analysis: summary of 2513 postnatal cases. PLoS ONE. 2007;2:e327. doi: 10.1371/journal.pone.0000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou Z, Kang SH, Shaw CA, Carmack CE, White LD, Patel A, Beaudet AL, Cheung SW, Chinault AC. Bacterial artificial chromosome-emulation oligonucleotide arrays for targeted clinical array-comparative genomic hybridization analyses. Genet Med. 2008;10:278–289. doi: 10.1097/GIM.0b013e31816b4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballif BC, Hornor SA, Sulpizio SG, Lloyd RM, Minier SL, Rorem EA, Theisen A, Bejjani BA, Shaffer LG. Development of a high-density peri-centromeric region BAC clone set for the detection and characterization of small supernumerary marker chromosomes by array CGH. Genet Med. 2007;9:150–162. doi: 10.1097/gim.0b013e3180312087. [DOI] [PubMed] [Google Scholar]
- Shao L, Shaw CA, Lu XY, Sahoo T, Bacino CA, Lalani SR, Stankiewicz P, Yatsenko SA, Li Y, Neill S, Pursley AN, Chinault AC, Patel A, Beaudet AL, Lupski JR, Cheung SW. Identification of chromosome abnormalities in subtelomeric regions by microarray analysis: a study of 5,380 cases. Am J Med Genet A. 2008;146A:2242–2251. doi: 10.1002/ajmg.a.32399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gribble SM, Prigmore E, Burford DC, Porter KM, Ng BL, Douglas EJ, Fiegler H, Carr P, Kalaitzopoulos D, Clegg S, Sandstrom R, Temple IK, Youings SA, Thomas NS, Dennis NR, Jacobs PA, Crolla JA, Carter NP. The complex nature of constitutional de novo apparently balanced translocations in patients presenting with abnormal phenotypes. J Med Genet. 2005;42:8–16. doi: 10.1136/jmg.2004.024141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baptista J, Mercer C, Prigmore E, Gribble SM, Carter NP, Maloney V, Thomas NS, Jacobs PA, Crolla JA. Breakpoint mapping and array CGH in translocations: comparison of a phenotypically normal and an abnormal cohort. Am J Hum Genet. 2008;82:927–936. doi: 10.1016/j.ajhg.2008.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickman L, Fiegler H, Shaw-Smith C, Nash R, Cirigliano V, Voglino G, Ng BL, Scott C, Whittaker J, Adinolfi M, Carter NP, Bobrow M. Pre-natal detection of unbalanced chromosomal rearrangements by array CGH. J Med Genet. 2006;43:353–361. doi: 10.1136/jmg.2005.037648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahoo T, Cheung SW, Ward P, Darilek S, Patel A, del Gaudio D, Kang SH, Lalani SR, Li J, McAdoo S, Burke A, Shaw CA, Stankiewicz P, Chinault AC, Veyver IB Van den, Roa BB, Beaudet AL, Eng CM. Prenatal diagnosis of chromosomal abnormalities using array-based comparative genomic hybridization. Genet Med. 2006;8:719–727. doi: 10.1097/01.gim.0000245576.47154.63. [DOI] [PubMed] [Google Scholar]
- Bi W, Breman AM, Venable SF, Eng PA, Sahoo T, Lu XY, Patel A, Beaudet AL, Cheung SW, White LD. Rapid prenatal diagnosis using uncultured amniocytes and oligonucleotide array CGH. Prenat Diagn. 2008;28:943–949. doi: 10.1002/pd.2087. [DOI] [PubMed] [Google Scholar]
- Simovich MJ, Yatsenko SA, Kang SH, Cheung SW, Dudek ME, Pursley A, Ward PA, Patel A, Lupski JR. Prenatal diagnosis of a 9q34.3 microdeletion by array-CGH in a fetus with an apparently balanced translocation. Prenat Diagn. 2007;27:1112–1117. doi: 10.1002/pd.1841. [DOI] [PubMed] [Google Scholar]
- Veyver IB Van den, Patel A, Shaw CA, Pursley AN, Kang SH, Simovich MJ, Ward PA, Darilek S, Johnson A, Neill SE, Bi W, White LD, Eng CM, Lupski JR, Cheung SW, Beaudet AL. Clinical use of array comparative genomic hybridization (aCGH) for prenatal diagnosis in 300 cases. Prenat Diagn. 2009;29:29–39. doi: 10.1002/pd.2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu XY, Phung MT, Shaw CA, Pham K, Neil SE, Patel A, Sahoo T, Bacino CA, Stankiewicz P, Kang SH, Lalani S, Chinault AC, Lupski JR, Cheung SW, Beaudet AL. Genomic imbalances in neonates with birth defects: high detection rates by using chromosomal microarray analysis. Pediatrics. 2008;122:1310–1318. doi: 10.1542/peds.2008-0297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froyen G, Corbett M, Vandewalle J, Jarvela I, Lawrence O, Meldrum C, Bauters M, Govaerts K, Vandeleur L, Van Esch H, Chelly J, Sanlav-ille D, van Bokhoven H, Ropers HH, Laumonnier F, Ranieri E, Schwartz CE, Abidi F, Tarpey PS, Futreal PA, Whibley A, Raymond FL, Stratton MR, Fryns JP, Scott R, Peippo M, Sipponen M, Partington M, Mowat D, Field M. Submicroscopic duplications of the hydroxysteroid dehydrogenase HSD17B10 and the E3 ubiquitin ligase HUWE1 are associated with mental retardation. Am J Hum Genet. 2008;82:432–443. doi: 10.1016/j.ajhg.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugtenberg D, de Brouwer AP, Kleefstra T, Oudakker AR, Frints SG, Schrander-Stumpel CT, Fryns JP, Jensen LR, Chelly J, Moraine C, Turner G, Veltman JA, Hamel BC, de Vries BB, van Bokhoven H, Yntema HG. Chromosomal copy number changes in patients with non-syndromic X linked mental retardation detected by array CGH. J Med Genet. 2006;43:362–370. doi: 10.1136/jmg.2005.036178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Zhang F, Brundage E, Scheuerle A, Lanpher B, Erickson RP, Powis Z, Robinson HB, Trapane PL, Stachiw-Hietpas D, Keppler-Noreuil KM, Lalani SR, Sahoo T, Chinault AC, Patel A, Cheung SW, Lupski JR. Genomic duplication resulting in increased copy number of genes encoding the sister chromatid cohesion complex conveys clinical consequences distinct from Cornelia de Lange. J Med Genet. 2009. in press . [DOI] [PMC free article] [PubMed]
- Stankiewicz P, Beaudet AL. Use of array CGH in the evaluation of dysmorphology, malformations, developmental delay, and idiopathic mental retardation. Curr Opin Genet Dev. 2007;17:182–192. doi: 10.1016/j.gde.2007.04.009. [DOI] [PubMed] [Google Scholar]
- The DECIPHER Database. https://decipher.sanger.ac.uk/
- Lee JA, Carvalho CM, Lupski JR. A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell. 2007;131:1235–1247. doi: 10.1016/j.cell.2007.11.037. [DOI] [PubMed] [Google Scholar]
- Bi W, Sapir T, Shchelochkov OA, Zhang F, Withers MA, Hunter JV, Levy T, Shinder V, Peiffer DA, Gunderson KL, Nezarati MM, Shotts VA, Amato SS, Savage SK, Harris DJ, Day-Salvatore DL, Horner M, Lu XY, Sahoo T, Yanagawa Y, Beaudet AL, Cheung SW, Martinez S, Lupski JR, Reiner O. Duplications in the 17p13.3 Miller-Dieker syndrome region: LIS1 increased expression affects human and mouse brain development. Nat Genet. 2009;41:168–177. doi: 10.1038/ng.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Khajavi M, Connolly A, Towne CF, Batish SV, Lupski JR. The DNA replication FoSTeS/MMBIR mechanism can cause human genomic, genic, and exonic complex rearrangements. Nat Genet. 2009. in press . [DOI] [PMC free article] [PubMed]
- Yatsenko SA, Brundage E, Roney EK, Cheung S-W, Lupski JR. Molecular mechanisms for subtelomeric rearrangements associated with the 9q34.3 microdeletion syndrome. Hum Mol Genet. 2009. in press . [DOI] [PMC free article] [PubMed]
- Hastings PJ, Ira G, Lupski JR. A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS Genet. 2009;5:e1000327. doi: 10.1371/journal.pgen.1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence JE, Perciaccante RG, Greig GM, Willard HF, Ledbetter DH, Hejtmancik JF, Pollack MS, O'Brien WE, Beaudet AL. Uniparental disomy as a mechanism for human genetic disease. Am J Hum Genet. 1988;42:217–226. [PMC free article] [PubMed] [Google Scholar]
- Cusco I, Corominas R, Bayes M, Flores R, Rivera-Brugues N, Cam-puzano V, Perez-Jurado LA. Copy number variation at the 7q11.23 segmental duplications is a susceptibility factor for the Williams-Beuren syndrome deletion. Genome Res. 2008;18:683–694. doi: 10.1101/gr.073197.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Gu W, Hurles M, Lupski JR. Copy number variation in health, disease, and evolution. Annu Rev Genomics Hum Genet. 2009. [DOI] [PMC free article] [PubMed]
- Edelmann L, Spiteri E, Koren K, Pulijaal V, Bialer MG, Shanske A, Goldberg R, Morrow BE. AT-rich palindromes mediate the constitutional t(11;22) translocation. Am J Hum Genet. 2001;68:1–13. doi: 10.1086/316952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurahashi H, Emanuel BS. Long AT-rich palindromes and the constitutional t(11;22) breakpoint. Hum Mol Genet. 2001;10:2605–2617. doi: 10.1093/hmg/10.23.2605. [DOI] [PubMed] [Google Scholar]
- Bacolla A, Jaworski A, Larson JE, Jakupciak JP, Chuzhanova N, Abeysinghe SS, O'Connell CD, Cooper DN, Wells RD. Breakpoints of gross deletions coincide with non-B DNA conformations. Proc Natl Acad Sci USA. 2004;101:14162–14167. doi: 10.1073/pnas.0405974101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Zhang F, Lupski JR. Mechanisms for human genomic rearrangements. Pathogenetics. 2008;1:4. doi: 10.1186/1755-8417-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupski JR, Stankiewicz P. Genomic disorders: molecular mechanisms for rearrangements and conveyed phenotypes. PLoS Genet. 2005;1:e49. doi: 10.1371/journal.pgen.0010049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carelle-Calmels N, Saugier-Veber P, Girard-Lemaire F, Rudolf G, Doray B, Guérin E, Kuhn P, Arrivé M, Gilch C, Schmitt E, Fehren-bach S, Schnebelen A, Frébourg T, Flori E. Genetic compensation in a human genomic disorder. N Engl J Med. 2009;360:1211–1216. doi: 10.1056/NEJMoa0806544. [DOI] [PubMed] [Google Scholar]
- Lupski JR, Stankiewicz P, (Eds) Genomic Disorders - The Genomic Basis of Disease. Totowa: Humana Press; 2006. [DOI] [PubMed] [Google Scholar]
- Lalani SR, Thakuria JV, Cox GF, Wang X, Bi W, Bray MS, Shaw C, Cheung SW, Chinault AC, Boggs BA, Ou Z, Brundage EK, Lupski JR, Gentile J, Waisbren S, Pursley A, Ma L, Khajavi M, Zapata G, Fried-man R, Kim JJ, Towbin JA, Stankiewicz P, Schnittger S, Hansmann I, Ai T, Sood S, Wehrens XH, Martin JF, Belmont JW. 20p12.3 microdeletion predisposes to Wolff-Parkinson-White syndrome with variable neurocognitive benefits. J Med Genet. 2009;46:168–175. doi: 10.1136/jmg.2008.061002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupski JR. Structural variation in the human genome. N Engl J Med. 2007;356:1169–1171. doi: 10.1056/NEJMcibr067658. [DOI] [PubMed] [Google Scholar]
- Brewer C, Holloway S, Zawalnyski P, Schinzel A, FitzPatrick D. A chromosomal duplication map of malformations: regions of suspected haplo- and triplolethality--and tolerance of segmental aneu-ploidy - in humans. Am J Hum Genet. 1999;64:1702–1708. doi: 10.1086/302410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer C, Holloway S, Zawalnyski P, Schinzel A, FitzPatrick D. A chromosomal deletion map of human malformations. Am J Hum Genet. 1998;63:1153–1159. doi: 10.1086/302041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores M, Morales L, Gonzaga-Jauregui C, Domínguez-Vidaña R, Zepeda C, Yañez O, Gutiérrez M, Lemus T, Valle D, Avila MC, Blanco D, Medina-Ruiz S, Meza K, Ayala E, García D, Bustos P, González V, Girard L, Tusie-Luna T, Dávila G, Palacios R. Recurrent DNA inversion rearrangements in the human genome. Proc Natl Acad Sci USA. 2007;104:6099–6106. doi: 10.1073/pnas.0701631104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruder CE, Piotrowski A, Gijsbers AA, Andersson R, Erickson S, de Ståhl TD, Menzel U, Sandgren J, von Tell D, Poplawski A, Crowley M, Crasto C, Partridge EC, Tiwari H, Allison DB, Komorowski J, van Ommen GJ, Boomsma DI, Pedersen NL, den Dunnen JT, Wirdefeldt K, Dumanski JP. Phenotypically concordant and discordant monozy-gotic twins display different DNA copy-number-variation profiles. Am J Hum Genet. 2008;82:763–771. doi: 10.1016/j.ajhg.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbouti A, Stankiewicz P, Nusbaum C, Cuomo C, Cook A, Höglund M, Johansson B, Hagemeijer A, Park SS, Mitelman F, Lupski JR, Fiore-tos T. The breakpoint region of the most common isochromosome, i(17q), in human neoplasia is characterized by a complex genomic architecture with large, palindromic, low-copy repeats. Am J Hum Genet. 2004;74:1–10. doi: 10.1086/380648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho CM, Lupski JR. Copy number variation at the breakpoint region of isochromosome 17q. Genome Res. 2008;18:1724–1732. doi: 10.1101/gr.080697.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell PJ, Stephens PJ, Pleasance ED, O'Meara S, Li H, Santarius T, Stebbings LA, Leroy C, Edkins S, Hardy C, Teague JW, Menzies A, Goodhead I, Turner DJ, Clee CM, Quail MA, Cox A, Brown C, Durbin R, Hurles ME, Edwards PA, Bignell GR, Stratton MR, Futreal PA. Identification of somatically acquired rearrangements in cancer using genome-wide massively parallel paired-end sequencing. Nat Genet. 2008;40:722–729. doi: 10.1038/ng.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupski JR, Weinstock GM. Short, interspersed repetitive DNA sequences in prokaryotic genomes. J Bacteriol. 1992;174:4525–4529. doi: 10.1128/jb.174.14.4525-4529.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versalovic J, Koeuth T, Lupski JR. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 1991;19:6823–6831. doi: 10.1093/nar/19.24.6823. [DOI] [PMC free article] [PubMed] [Google Scholar]