Abstract
During prolonged stress or chronic treatment with neurotoxins, robust compensatory mechanisms occur which maintain sufficient levels of catecholamine neurotransmitters in terminal regions. One of these mechanisms is the up-regulation of tyrosine hydroxylase (TH), the enzyme that controls catecholamine biosynthesis. In neurons of the periphery and locus coeruleus, this up-regulation is associated with an initial induction of TH mRNA. In contrast, this induction either does not occur or is nominal in mesencephalic dopamine neurons. The reasons for this lack of compensatory TH mRNA induction remain obscure, because so little is known about the regulation of TH expression in these neurons. In this report we test whether activation of the cAMP signaling pathway regulates TH gene expression in two rodent models of midbrain dopamine neurons, ventral midbrain organotypic slice cultures and MN9D cells. Our results demonstrate that elevation of cAMP leads to induction of TH protein and TH activity in both model systems; however, TH mRNA levels are not up-regulated by cAMP. The induction of TH protein is the result of a novel post-transcriptional mechanism that activates TH mRNA translation. This translational activation is mediated by sequences within the 3′UTR of TH mRNA. Our results support a model in which cAMP induces or activates trans-factors that interact with the TH mRNA 3′UTR to increase TH protein synthesis. An understanding of this novel regulatory mechanism may help to explain the control of TH gene expression and consequently dopamine biosynthesis in midbrain neurons under different physiological and pathological conditions.
Tyrosine hydroxylase (TH) is the enzyme that controls the biosynthesis of dopamine, norepinephrine and epinephrine. It is one of the most meticulously regulated enzymes in the nervous system (Kumer and Vrana, 1996; Sabban and Kvetnansky, 2001). Its regulation is mediated by short-term and long-term mechanisms, both of which are adaptive responses to increased release of catecholamines during nerve stimulation, permitting neurons to maintain an appropriate supply of these vital neurotransmitters. Short-term mechanisms control the activity of pre-existing enzyme via phosphorylation of serine sites within its N-terminal domain. Long-term mechanisms involve induction of TH enzyme protein; these mechanisms are activated during chronic stimulation, such as long-term stress or repeated drug treatment. In adrenal medulla and locus coeruleus, long-term regulation is mediated by mechanisms that modulate TH gene transcription rate and TH mRNA stability (Sabban and Kvetnansky, 2001; Wong and Tank, 2007).
Much less is known about regulation of TH gene expression in mesencephalic dopaminergic neurons. Some workers have shown that severe stress, chronic nicotine treatment or reserpine treatment induces TH gene expression in midbrain, with those neurons having their cell bodies in the ventral tegmentum being more responsive than those with their cell bodies in the substantia nigra (Ortiz et al., 1996; Pasinetti et al., 1990; Serova and Sabban, 2002). However, this induction has not been observed in other studies and when observed, the effects are usually small or short-lived compared to those seen in adrenal medulla or locus coeruleus (Biguet et al., 1986; Melia et al., 1992; Smith et al., 1991).
In a previous collaborative study (Bowyer et al., 1998), we used high doses of amphetamine to destroy dopaminergic nerve terminals in striatum without producing significant loss of dopaminergic cell bodies in substantia nigra. Our expectation was that this loss of nerve terminals would lead to an adaptive induction of TH mRNA in midbrain cell bodies. To our surprise, midbrain TH mRNA levels were unaffected by this amphetamine-induced loss of striatal nerve terminals. One explanation for this lack of response in midbrain is that the signaling pathways mediating TH mRNA induction are not activated in this amphetamine neurotoxicity paradigm. However, to test this hypothesis is difficult, because so little is known about the regulation of TH in midbrain neurons.
Cyclic AMP is a powerful inducer of TH gene transcription rate and TH mRNA in many model systems (Kumer and Vrana, 1996; Sabban and Kvetnansky, 2001). Furthermore, it has been reported that forskolin induces TH mRNA, when it is injected into the rat substantia nigra under in vivo conditions (Leviel et al., 1991). In the present study we test the effect of cAMP on TH gene expression in two rodent models of midbrain dopamine neurons under culture conditions. Unexpectedly, our results demonstrate that cAMP does not induce TH mRNA in these two midbrain models. Instead, cAMP induces TH protein by activating a novel post-transcriptional mechanism that enhances the translation of pre-existing TH mRNA.
MATERIALS AND METHODS
Ventral midbrain slice cultures
Brains were removed from Sprague-Dawley rat pups on postnatal day 7-10 and immediately placed in oxygenated (95% O2/5% CO2), ice-cold artificial spinal fluid (ACSF; 125 mM NaCl, 26 mM NaHCO3, 2.5 mM KCl, 1.25 mM NaH2PO4, 2 mM CaCl2, 1 mM MgSO4 (anhydrous) and 25 mM glucose, pH 7.4). The midbrain was dissected with the use of a brain block. The first cut was made midway through the third ventricle and a second cut was made below the pons-midbrain junction. The cortex was removed and the ventral midbrain was isolated and placed in 5% low melting agarose (dissolved in ACSF), which was then placed in an ice bath. Coronal slices (350-400 um) were obtained using an Integraslice 7550 PSDS Vibraslicer (Campden Instruments) and perfused in ice-cold oxygenated ACSF for 1-2 hr. The slices from each rat pup were placed on millicell inserts (Millipore) in a single well of a 6-well plate and cultured at 35° in an atmosphere of 95% air and 5% CO2 in medium containing 50% Earles MEM, 25 mM Hepes, 25% Hanks basal salt solution (with phenol red), 2 mM L-glutamine, 0.01 ug/ml penicillin/streptomycin, 25% heat-inactivated horse serum (GIBCO) and 6.4 g/l glucose. This procedure is a modification of the method of Stoppini et al (1991). Unless otherwise stated, the slices were cultured for 7 days in vitro (DIV) prior to experimentation.
Previous studies using anatomical and electrophysiological analyses indicated that neonatal rat brain slices cultured under these conditions survived well for many weeks in vitro (Stoppini et al., 1991; Thomas et al., 1998). In our initial studies using these midbrain cultures, TH-positive cells appeared morphologically healthy for at least 14 DIV. They had intact cell bodies along with extended fibers; this was apparent at each time point examined. The slices possessed small and large unipolar, bipolar and multipolar TH-immunoreactive cells. Nissl body staining also depicted healthy and viable neurons that remained viable for at least 14 DIV. Biochemical analyses of numerous slice cultures demonstrated initial decreases after 24 hr in vitro in TH mRNA and TH activity that varied in extent compared to that observed in freshly-dissected midbrain tissues. However, from 1 DIV to 7 DIV, basal TH mRNA levels and TH activity in the cultures did not change significantly. These initial results suggested that after an initial variable loss of neurons that occurred rapidly after placing the slices into explant culture, neurons remained healthy and viable in the midbrain slices and basal TH expression remained relatively constant for at least 14 DIV.
Cell cultures
MN9D cells were obtained from Dr. Lisa Opanashuk in the Department of Environmental Medicine at the University of Rochester Medical Center. These cells were created by Heller and coworkers (Choi et al., 1992) and are a hybrid cell line derived from the fusion of mouse neuroblastoma N18TG2 cells with embryonic mouse mesencephalic neurons. The cells were cultured as described by Choi et al (1992) in DMEM supplemented with 10% fetal bovine serum (Invitrogen) in an atmosphere of 95% air and 5% CO2. PC18 and cath.a cells were cultured as previously described (Lewis-Tuffin et al., 2004; Tank et al., 1986).
Construction of TH-luciferase plasmids
The modified GL3 plasmid was constructed by first deleting the SV40 promoter in pGL3-Control (Promega Corporation) using HindIII and BglII and inserting into this site the SV40 early enhancer/promoter from the pRL-SV40 vector (Promega Corporation), which was excised from pRL-SV40 using the same two enzymes. This modified GL3 was used as a control vector in some experiments and was also the starting backbone for the TH-luciferase vectors used in this study. To construct TH(5′3′U)-Luc (see Figure 6A), the 35 bp TH 5′UTR was inserted just upstream of the luciferase gene by cutting the modified GL3 vector with HindIII and NcoI and inserting an oligonucleotide encoding these sequences with HindIII and NcoI sites on the appropriate ends. The SV40 enhancer and SV40 late poly(A) signal sequences (downstream of the luciferase gene) in the modified GL3 vector were excised using XbaI and BamHI. TH cDNA sequences were then inserted into this XbaI and BamHI site; these TH cDNA sequences encoded 160 bp of coding sequence within the 5′ end of exon 13 of the TH gene, 280 bp of TH mRNA 3′UTR (encoding the 3′ end of exon 13) and 80 bp of genomic DNA downstream of the TH mRNA poly(A) addition site. TH(Δ5′U)-Luc was created by excising the TH mRNA 5′ UTR sequences using HindIII and NcoI, filling in the protruding ends with Klenow DNA polymerase I and religation. TH(ΔEx13)-Luc was created by excising the TH mRNA coding sequences in exon 13, using XbaI and KpnI followed by blunt-ending and religation. Both the 5′UTR and exon 13 coding sequences were excised to create TH(3′U)-Luc. TH(3′UΔKS)-Luc was generated by excising the TH mRNA 3′UTR sequences from KpnI to SphI from TH(3′U)-Luc, followed by blunt-ending and religation.
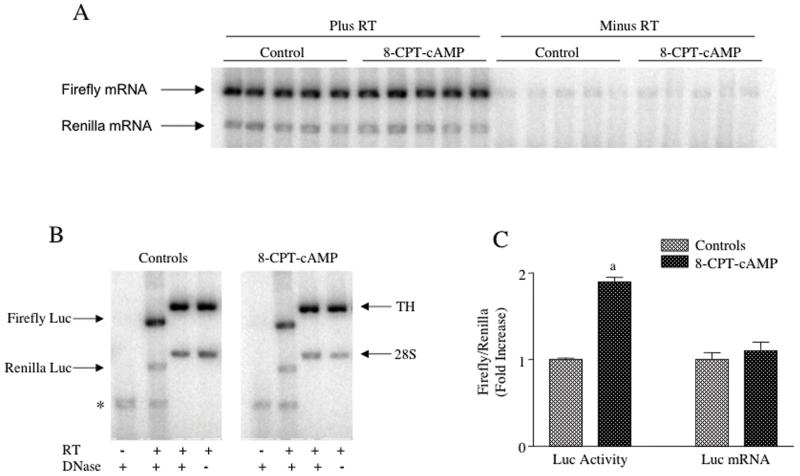
Figure 6. TH-luciferase mRNA levels were not increased by treatment of transiently-transfected MN9D cells with 8-CPT-cAMP.
MN9D cells were transfected with TH(3′U)-luciferase and the Renilla luciferase expression vector. Twenty-four hr after transfection, the cells were treated for another 24 hr with or without 0.5 mM 8-CPT-cAMP. The cells were then harvested and total cellular RNA was isolated. The RNA samples were treated with 2 units of TURBO-DNase (unless otherwise stated) prior to RT-PCR. (A) Autoradiograms showing both firefly and Renilla luciferase mRNA levels detected by semiquantitive RT-PCR with or without RT are shown in the figure. (B) Autoradiograms are shown depicting semiquantitative RT-PCR assays run under different conditions as noted in the figure measuring firefly and Renilla luciferase mRNAs, along with TH mRNA and 28S rRNA in the same samples. (C) Results are shown of 4 experiments in which both luciferase activity and luciferase mRNA levels expressed in MN9D cells transiently-transfected with the TH(3′U)-luciferase construct were measured in the same samples. The results represent the means ± SE from 17-18 dishes.
a: p < .01 compared to controls.
Enzyme assays and TH protein measurement
All procedures were performed on ice or in a cold room unless otherwise stated. For measurement of TH activity, midbrain slices or cells were homogenized in a buffer containing 30 mM potassium phosphate (pH 6.8), 50 mM NaF and 10 mM EDTA. The resulting supernatant was assayed for TH activity at 30°C under Vmax conditions (unless otherwise stated) employing the coupled decarboxylase assay as described previously (Tank et al., 1986), using 6-methyl-5,6,7,8-tetrahydropterin (6MPH4) as cofactor. TH activity was expressed as pmoles of 14CO2 formed per minute divided by mg of protein. TH protein was measured using western analysis as described previously (Yoshimura et al., 2004). The same slice or cell extracts used for assaying TH activity were used for measuring TH protein. Firefly and Renilla luciferase activities were measured using the Dual-Reporter Luciferase assay system from Promega Corporation. Protein determinations were made by the method of Bradford (1976), using bovine serum albumin as standard.
TH mRNA measurements
TH mRNA levels were measured routinely using a semiquantitative RT-PCR assay as described previously (Sun et al., 2004; Yoshimura et al., 2004). Briefly, 0.2-0.8 ug of total cellular RNA isolated from midbrain slices or MN9D cells were subjected to RT using random hexamer primers. Aliquots of the resulting single-stranded cDNA products were used along with the appropriate primers (see below) in the PCR to incorporate 32P-dATP (0.5 uCi per reaction) into double-stranded products encoding 537 bp TH cDNA or 294 bp 28S cDNA. The 5′ TH mRNA sense primer encoded rat or mouse cDNA sequences 390 to 410; the 3′ antisense primers were complementary to rat or mouse TH cDNA sequences 888 to 908. These primer sequences were based on the rat TH cDNA sequence; however, the mouse TH cDNA sequences in these primers diverged by only 1 (5′-primer) or 2 (3′-primer) bases and hence, these primers were successful for assaying TH mRNA in both rat and mouse RNA samples. The sequences for these primers were as follows: 5′ TH primer, 5′-ccc cac ctg gag tat ttt gtg-3′; 3′ TH primer, 5′-atc acg ggc gga cag tag acc-3′. The primer sequences used for PCR of 28S rRNA were identical in both rat and mouse cDNAs. The 5′ 28S rRNA primer encoded sequences 1 to 22 of rat or mouse 28S cDNA (5′-gtg aac agc agt tga aca tggg-3′); the 3′ 28S primer was complementary to rat or mouse 28S cDNA sequences 276 to 295 (5′-aac cgc gac gct ttc caag-3′). For each experiment, the linearity of the RT-PCR was assessed with respect to both ug RNA added to the RT reaction and the number of PCR cycles. Densitometric values of the bands corresponding to the TH mRNA and 28S rRNA PCR products observed on the dried-down electrophoretic gels were quantified using Phosphorimager analysis. TH mRNA values were normalized to 28S rRNA values for each sample. These normalized values (TH mRNA/28S rRNA) were then expressed as fold-increases over control values for each experiment and presented as such in the figures.
In select experiments (see Figure 2), TH mRNA was quantified using a quantitative RT-PCR assay employing an Applied Biosystems Prism 7000 real-time PCR cycler. Initial experiments were performed with RNA extracts from each cell line to determine the linearity of the TH mRNA and GAPDH mRNA signals with respect to total cellular RNA concentrations added to the RT reactions. For MN9D, PC18 and cath.a cells, linearity was achieved using total cellular RNA concentrations between 0.05 to 0.2 ug. We routinely used 0.1 ug total cellular RNA in each assay. The RT reaction was performed as described for the semiquantitative assay (see above). Real-time PCR was performed as recommended by Applied Biosystems using a 2 ul aliquot of the cDNA produced in the 20 ul RT reaction and SYBR green indicator to measure the amount of cDNA product formed in each PCR cycle. Forward and reverse primers were present at 1 uM in the PCR reactions. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) mRNA was measured in each sample for normalization purposes, using sample cDNA transcribed from the same RT reaction as that used for measuring TH mRNA. The TH primers were as follows: rat forward primer encoded sequences 620 to 639 of rat TH cDNA (5′-tcg gaa gct gat tgc aga ga-3′) and rat reverse primer was complementary to sequences 695 to 705 (5′-ttc cgc tgt gta ttc cac atg-3′); mouse forward primer encoded sequences 1016 to 1033 of mouse TH cDNA (5′-tgt tgg ctg acc gca cat-3′) and mouse reverse primer was complementary to sequences 1058 to 1076 (5′-gcc ccc aga gat gca agt c-3′). GAPDH mRNA primers were the same for both rat and mouse samples and were as follows: forward primer encoded sequences 507 to 517 of rodent GAPDH cDNA (5′-gga agg gct cat gac cac agt-3′) and reverse primer was complementary to sequences 570 to 590 (5′-acc ttt cga cac cgc act acc-3′). Standard curves for TH mRNA and GAPDH mRNA were created for each experiment. Cloned TH cDNA and GAPDH cDNA sequences encompassing the amplified regions were transcribed in vitro using an Ambion MAXIScript Transcription kit. Known amounts (pmols) of the in vitro transcribed mRNAs were used to construct these standard curves using the same RT-PCR conditions employed for the sample mRNAs. The CT values derived from the real-time PCRs for different concentrations of standard TH mRNA or GAPDH mRNA were plotted against the known concentrations of the standard mRNAs used in the RT reaction to construct these curves. The CT values for the sample TH mRNAs and GAPDH mRNAs were then compared to the CT values on the standard curves to obtain pmol of TH mRNA and GAPDH mRNA. The TH mRNA values were then normalized to the GAPDH mRNA values for each sample.
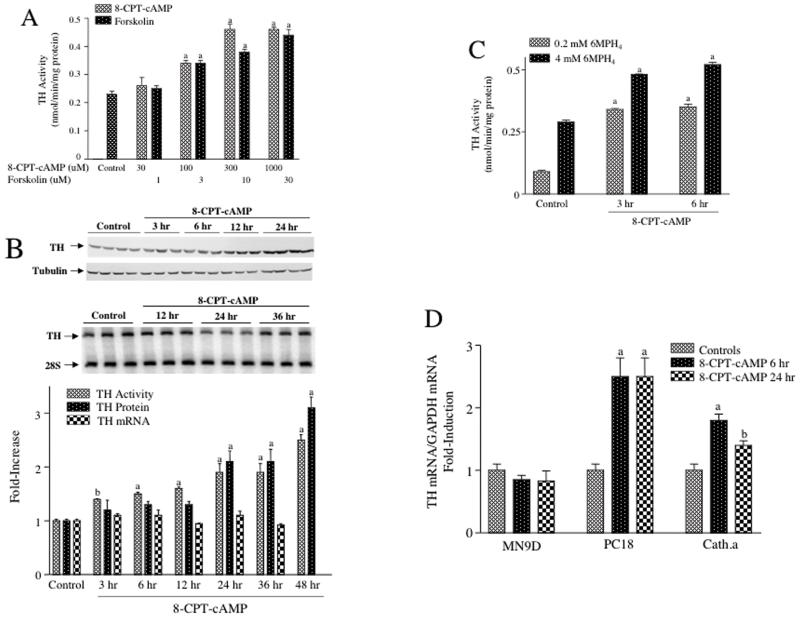
Figure 2. TH activity and TH protein, but not TH mRNA were induced by 8-CPT-cAMP in MN9D cells.
(A) MN9D cells were treated for 48 hr with either 8-CPT-cAMP or forskolin at the concentrations designated in the figure. Control cells for the 8-CPT-cAMP experiments were left untreated, whereas controls for the forskolin experiments were treated with vehicle (dimethylsulfoxide). Cells were harvested, quick-frozen and stored at -80°C until assayed for TH activity. The results represent the means ± SE from 3 dishes. (B) MN9D cells were treated with 1 mM 8-CPT-cAMP for the time periods designated in the figure; control cells were left untreated. The top panel is a representative autoradiogram of a western blot measuring TH protein and tubulin, which was used to control for variability in sample loading or protein transfer to the membrane. The middle panel is a representative autoradiogram of semiquantitative RT-PCR assays measuring TH mRNA and 28S rRNA, which was used for normalization purposes. The bar graph in the lower panel depicts a time course for TH activity (measured under apparent Vmax conditions), TH protein (normalized to tubulin) and TH mRNA (normalized to 28S rRNA), each expressed as fold increase over controls. The results represent the means ± SE from 3-8 dishes. (C) MN9D cells were treated with 1 mM 8-CPT-cAMP for 3 or 6 hr and then harvested and stored at -80°C until assayed. Control cells were untreated. TH activity was assayed using either 0.2 mM or 4 mM 6MPH4 as cofactor. The results represent the means ± SE from 3 dishes. (D) MN9D, PC18 and cath.a cells were treated with 0.5 mM 8-CPT-cAMP for 6 or 24 hr; controls were left untreated. TH mRNA was measured using quantitative RT-PCR and normalized to GAPDH mRNA measured in the same samples. The results represent the means ± SE from 5-6 dishes.
a: p < .01 compared to controls. b: p < .05 compared to controls.
Luciferase mRNA measurements
Firefly and Renilla luciferase mRNA levels were measured using semiquantitative RT-PCR, essentially as described above with minor modifications. Due to significant contamination of the RNA samples with plasmid DNA, each sample was treated with 2 units of TURBO-DNase (Ambion, Inc) prior to the RT step. Furthermore, all samples were assayed with or without the RT step, to check for DNA contamination; only those assays that showed negligible or very low levels of signal in the absence of the RT step (relative to that observed in assays run in the presence of the RT step) were used for quantitation of luciferase mRNA. As for the TH mRNA assays, all RT-PCR reactions were run under conditions of linearity with respect to both RNA input and PCR cycle number. The primers used for firefly luciferase mRNA were as follows: 5′ primer encoded sequences 92 to 112 of the luciferase mRNA coding region (5′-aga gat acg ccc tgg ttc ctg-3′); the 3′ primer was complementary to sequences 469 to 492 of the coding region (5′-cat cga ctg aaa tcc ctg gta atc-3′). The primers used for Renilla luciferase mRNA were: 5′ primer encoded sequences 657 to 676 of the coding region (5′-gcc tcg tga aat ccc gtt ag-3′); 3′ primer was complementary to sequences 865 to 886 of the coding region (5′-cca ttt cat cag gtg cat cttc-3′).
Measurement of TH rate of synthesis
The rate of TH protein synthesis in midbrain slices or MN9D cells was estimated using pulse-labeling procedures essentially as described by Tank et al (1986). Slices or cells were incubated in the presence or absence of 1 mM 8-CPT-cAMP for 24 hr, at which time the medium was changed to leucine-free DMEM (supplemented with 10% fetal bovine serum) containing 25 uCi/ml of [4,5-3H]-leucine (60 Ci/mmol). The cells were incubated for an additional 1.5 hr in the labeling medium in the continued presence of the cAMP analog, when appropriate. The cells were then harvested and washed twice in PBS containing 1 mM leucine. Radiolabeled TH protein was isolated by immunoprecipitation and radioactivity was measured in the immunoprecipitated protein as described previously (Tank et al., 1986). The [3H]TH cpm values denoted in the table in Figure 3 represented the amount of radioactivity incorporated into TH protein normalized to the ug total protein present in the cell or slice extract. This value was calculated by dividing the cpm of [3H]leucine incorporated into 50 units of TH by the ug total protein present in that 50 units of TH activity. Incorporation of radiolabeled leucine into total soluble protein was measured as described previously (Tank et al., 1986). Rates of synthesis were calculated as cpm incorporated into TH protein per ug protein divided by cpm incorporated into total soluble protein per ug protein. In select experiments the gel was incubated in EN3HANCE (Perkin-Elmer) for 1 hr as described by the manufacturer, dried down onto Whatman 3MM paper and exposed to X-ray film for 7 days in a -80°C freezer. The radiolabeled band corresponding to TH was then identified on the autoradiogram.
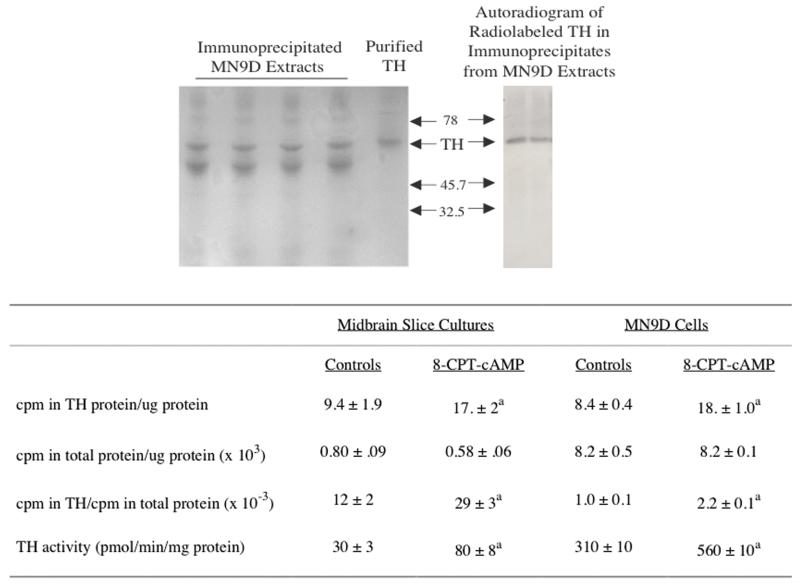
Figure 3. The rate of synthesis of TH protein was increased in cultured rat midbrain slices and MN9D cells treated with 8-CPT-cAMP.
Midbrain slices or MN9D cells were treated with 1 mM 8-CPT-cAMP for 24 hr; control cells were untreated for the same period of time. The cultures were then incubated for an additional 1.5 hr in the presence of leucine-free medium supplemented with serum, 25 uCi/ml 3H-leucine and 1 mM 8-CPT-cAMP, when appropriate. Cultures were harvested and radiolabeled TH protein was isolated using immunoprecipitation procedures. The final immunoprecipitates were subjected SDS-PAGE. After electrophoresis, the gels were stained with Coomassie blue; the left panel in the figure depicts a representative Coomassie blue-stained gel. In a select experiment, a gel was treated with EN3HANCE, dried down onto Whatman 3MM paper and exposed to X-ray film for 7 days. The autoradiographic signal is shown in the right-hand panel in the figure. In most experiments, the TH band was excised from the wet gel, along with a region of the gel that did not contain measurable protein staining (used to determine background radioactivity). These gel pieces were solubilized and radioactivity counted by liquid scintillation spectrometry. The cpm incorporated into TH protein, along with the cpm incorporated into total cell protein are presented in the table. TH activity was assayed in an aliquot of the same supernatants used for immunoprecipitation. The results represent the means ± SE from 3 experiments.
a: p < .01 compared to controls.
Polysome profile analysis of MN9D TH mRNA
Polysome profile analysis was performed as described by Xu et al (2007). Briefly, MN9D cells were homogenized in ice-cold low-salt lysis buffer (20 mM Tris-HCl (pH 7.5), 10 mM NaCl, 3 mM MgCl2, 1 mM RNasin, 1 mM dithiothreitol, 0.3% Triton X-100, and 0.05 M sucrose). Nuclei and the majority of mitochondria were sedimented by centrifugation for 10 min at 10,000g at 4°C. The supernatants (1 ml) were adjusted to 170 mM NaCl and 13 mM MgCl2, layered onto 9 ml linear sucrose gradients (15% to 45% w/w) and centrifuged at 36,000 rpm for 2 hr at 4°C in a SW40 rotor. Fractions (800 ul) were collected after centrifugation, and RNA in each fraction was precipitated and resuspended in 15 ul DEPC-treated water. TH mRNA levels in each fraction was assayed using the semiquantitative RT-PCR assay described above. To control for differences in recovery of RNA from each fraction during the isolation procedure, a known amount (100 ng) of a standard control RNA was added to each fraction before RNA isolation. This standard RNA was obtained by in vitro transcription (using a kit from Ambion, Inc) of TH genomic DNA promoter sequences and detected using specific primers (see Xu et al (2007) for details). The amount of TH mRNA in each fraction was first normalized to the recovery of this standard RNA and then expressed as a percentage of the total mRNA isolated from the gradients. A weighted mean average for each gradient was calculated using the methods of Gore et al (1995).
Transfection of MN9D cells and luciferase assays
MN9D cells were cultured in 6 well dishes (9.6 cm2) and transfected with 2 ug plasmid DNA using Lipofectamine (Invitrogen) as described by the manufacturer. Twenty-four hr after the transfection, the medium was removed and the cells were incubated for another 24 hr in the presence or absence of 8-CPT-cAMP or forskolin in fresh medium. The cells were then washed once with ice-cold PBS, harvested and centrifuged. The cell pellets were stored at -80°C. For stable transfections, the cells were transfected as described above, except that 0.067 ug of a plasmid expressing the neomycin resistance gene (RSV-neo) was cotransfected with 2 ug of the TH(3′U)-luciferase plasmid. Forty-eight hr after the transfection, the cells were treated with 500 ug/ml of G418. The concentration of G418 was decreased to 100 ug/ml after 2-3 days of treatment, at which time most of the MN9D cells were dead. Resistant colonies were observed 2 weeks after the transfection. Twenty of these colonies were isolated and after expansion tested for luciferase activity and luciferase mRNA levels. Two of these colonies, designated U1 and U17 were used for the studies in Figure 7.
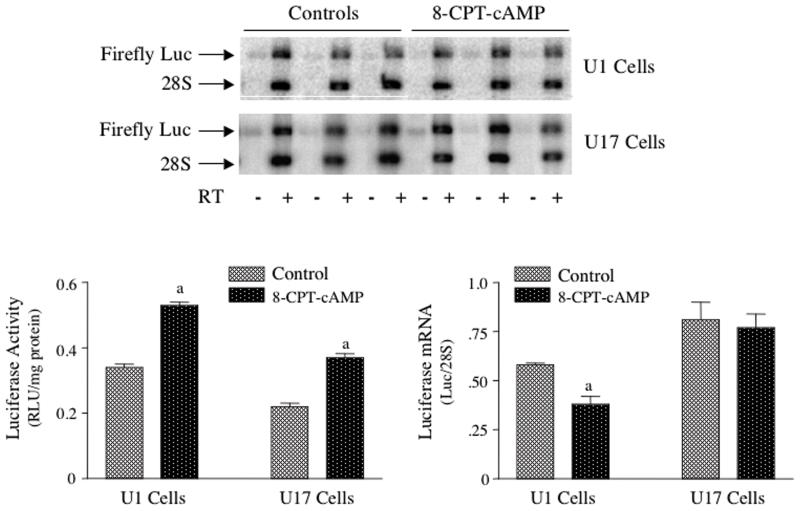
Figure 7. TH-luciferase activity, but not TH-luciferase mRNA levels were increased by treatment of stably-transfected MN9D cells with 8-CPT-cAMP.
MN9D cells were stably transfected with TH(3′U)-luciferase and two clonal colonies expressing luciferase activity and mRNA were isolated; these colonies were designated U1 and U17. These cells were treated with or without 0.5 mM 8-CPT-cAMP for 24 hr and then both luciferase activity and luciferase mRNA levels were measured. Representative autoradiograms depicting the luciferase mRNA levels in each cell line are shown in the top panel. The results represent the means ± SE from 6 dishes.
a: p < .01 compared to controls.
Statistical analyses
The results were analyzed by one-way analysis of variance, using the computer program INSTAT. Comparisons between groups were made using the Dunnett multiple comparisons test or the Student’s t-test, if only two groups were analyzed. A level of p < .05 was considered statistically significant.
RESULTS
TH protein and TH activity were increased in midbrain slice cultures in response to treatment with 8-CPT-cAMP
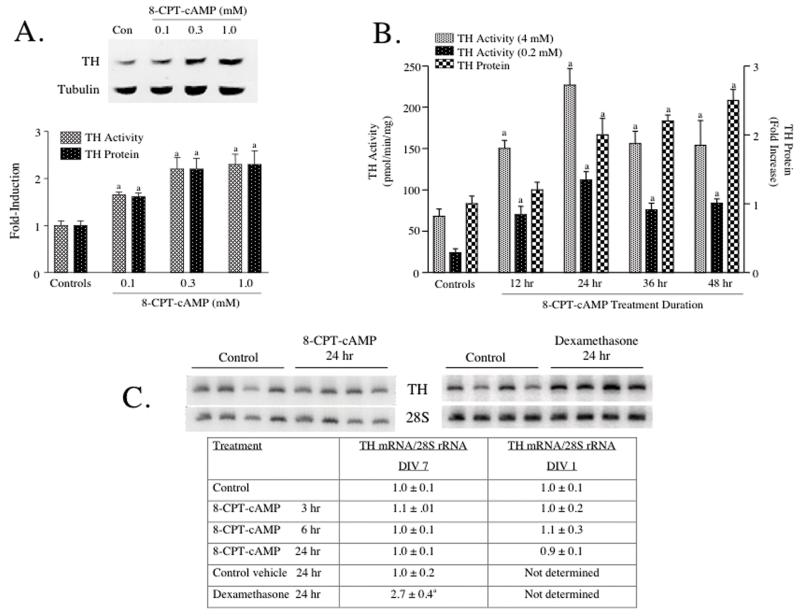
When midbrain slices were treated for 24 hr with the cAMP analog, 8-CPT-cAMP, TH activity measured under saturating cofactor conditions (4 mM 6MPH4) increased in a dose-dependent manner (Figure 1A). A 1.5-fold increase in TH activity was observed using 0.1 mM 8-CPT-cAMP and a maximal increase was observed using 0.3 mM cAMP analog. When assayed under these apparent Vmax conditions, an increase in TH enzyme activity is usually indicative of an increase in TH protein. To test this hypothesis directly, western analysis was performed using the same supernatants assayed for TH activity. A representative western blot is presented above the bar graph in Figure 1A. TH protein levels increased to the same extent as TH activity at each concentration of 8-CPT-cAMP.
Figure 1. TH activity and TH protein, but not TH mRNA were induced in cultured rat midbrain slices by treatment with 8-CPT-cAMP.
(A) Midbrain slices were treated for 24 hr with 8-CPT-cAMP at the concentrations designated in the figure. Control slices were left untreated. (B) Midbrain slices were treated with 1 mM 8-CPT-cAMP for the durations of time designated in the figure. Control slices were left untreated. All slices were harvested at the same time, quick-frozen and stored at -80°C. TH activity was assayed using 4 mM 6MPH4 in panel A and either 0.2 mM or 4 mM 6MPH4 in panel B. Control TH activity in panel A was 27 ± 3 pmol/min/mg protein. TH protein was measured using western analysis. An autoradiogram depicting a representative western blot is shown above the bar graphs in panel A. The results in panels A and B represent the means ± SE from 3-6 slice cultures. (C) Midbrain slices were treated with 1 mM 8-CPT-cAMP or 0.1 uM dexamethasone for the durations of time designated in the figure. Control cultures were left untreated or treated with vehicle (dimethylsulfoxide). All cultures were harvested at the same time, quick-frozen and stored at -80°C. Total cellular RNA was isolated and TH mRNA was assayed using semiquantitative RT-PCR. Autoradiograms depicting representative RT-PCR assays are shown for midbrain slice cultures (DIV 7) treated for 24 hr with either drug. The data in the table represent values for TH mRNA divided by those for 28S rRNA measured in the same samples and expressed as fold-increase over controls. The data represent the means ± SE from 6-12 midbrain slices for the 8-CPT-cAMP experiments and 4 midbrain slices for the dexamethasone experiments.
a: p < .05 compared to controls
A time course of this response to 8-CPT-cAMP is presented in Figure 1B. TH activity assayed using saturating pterin cofactor concentration (4 mM) increased significantly at each time point between 12 hr and 48 hr of treatment with the cAMP analog. Similar increases in TH activity were observed when the enzyme was assayed using a subsaturating pterin cofactor concentration. TH protein levels increased 2-3 fold after 24, 36 or 48 hr of 8-CPT-cAMP treatment. However, in contrast to the observed increase in TH activity, TH protein levels were not elevated at the 12 hr time point. Taken together, these results indicate that TH is induced 2-3 fold by 8-CPT-cAMP in midbrain slice cultures. Similar increases in TH activity and TH protein were observed when midbrain slice cultures were treated with 10 uM forskolin for 24 hr (data not shown).
TH mRNA levels were not affected by treatment of midbrain slices with 8-CPT-cAMP
Midbrain slices were treated with 1 mM 8-CPT-cAMP and total cellular RNA was isolated at different time points. TH mRNA levels were measured using semiquantitative RT-PCR and normalized to the levels of 28S rRNA measured in the same samples (Figure 1C). This assay was utilized in a number of our previous studies to detect induction of TH mRNA in adrenal medulla, locus coeruleus and pheochromocytoma cell lines (Osterhout et al., 2005; Sun et al., 2004; Sun et al., 2003; Yoshimura et al., 2004). An example of an autoradiogram showing results of this assay using midbrain slice RNA samples is presented in Figure 1C; a more extensive time course is shown in the table below the autoradiogram. It is evident from the autoradiogram and the more quantitative analysis in the table that TH mRNA levels did not increase significantly in response to 8-CPT-cAMP at any time point tested. In contrast, as shown in the autoradiogram and table in Figure 1C, dexamethasone (used as a positive control) induced TH mRNA 2-3 fold in midbrain slice cultures.
The induction of TH protein without an induction of TH mRNA in response to cAMP suggested that a post-transcriptional mechanism mediated this response in midbrain dopamine neurons. However, it was possible that this unexpected response was due to changes in the neurons that occurred during culture of the slices for seven days. To minimize this possibility, midbrain slices were prepared as usual, but only cultured for 1 day (DIV 1) prior to treatment with 8-CPT-cAMP (see table in Figure 1C). Similar to the results obtained using the DIV 7 cultures, cAMP treatment did not induce TH mRNA at any time point tested in the DIV 1 cultures. In contrast, TH activity and TH protein increased 2-3-fold after 24 hr of treatment with 8-CPT-cAMP in the DIV 1 cultures (TH activity: controls = 24 ± 3 pmol/min per mg protein, 8-CPT-cAMP-treated = 52 ± 4, N = 10, p < .01; TH protein: controls = 1.0 ± 0.1, 8-CPT-cAMP-treated = 2.4 ± 0.4, N = 8, p < .01).
TH protein and TH activity were increased in MN9D cells in response to treatment with 8-CPT-cAMP
In order to study the mechanisms for this apparent post-transcriptional response in more detail, we tested whether cultured MN9D cells could be used as a model system, in which TH expression responded to cAMP in a similar manner as that observed in the midbrain slice cultures. These results are summarized in Figure 2. MN9D cells were treated for 48 hr with different concentrations of either 8-CPT-cAMP or forskolin (Figure 2A). TH activity was measured under apparent Vmax conditions, using a saturating concentration of pterin cofactor (4 mM 6MPH4). Both drugs produced concentration-dependent increases in TH activity. Maximal or near-maximal effects were observed using 300 um 8-CPT-cAMP or 10 uM forskolin; the concentration-response curve for 8-CPT-cAMP was very similar to that observed in midbrain slice cultures. To test whether these increases in TH activity were due to induction of TH protein, western analysis was performed; a representative immunoblot is presented in the top panel of Figure 2B. As in midbrain slices, it is evident from this blot that TH protein levels increased significantly after 12-24 hr of treatment with 8-CPT-cAMP in MN9D cells. In a more detailed time course study, TH activity and TH protein levels were measured in MN9D cells treated with 1 mM 8-CPT-cAMP for different durations of time (Figure 2B, bar graph). Significant increases in TH activity (measured under apparent Vmax conditions with respect to pterin cofactor) were observed after 3, 6 and 12 hr of treatment with the cAMP analog. These early increases in enzyme activity did not correlate with changes in TH protein levels, which were measured in the same MN9D supernatants. TH activity increased more dramatically (1.9-2.5 fold) after 24, 36 and 48 hr of treatment; these increases correlated closely with changes in TH protein levels. In many experiments performed in MN9D cells over the past 2-3 years, the changes in TH activity and TH protein levels in response to 24 hr of treatment with 8-CPT-cAMP varied from 1.5-2.5-fold; however, in every experiment significant increases in TH activity and TH protein were measured in response to cAMP.
The results of these MN9D time courses, as well as those performed in the midbrain slice cultures suggested that the increases in TH activity observed at the later time points (24, 36 and 48 hr) were due to induction of TH protein, whereas those at the earlier time points (3 and 6 hr) were due to enzyme activation. To test further this latter hypothesis, we assayed TH activity in samples from the early time points using low pterin cofactor concentrations in the assays (Figure 2C). Under these assay conditions, TH activity increased by ∼4-fold at both the 3 and 6 hr time points, when TH protein was not induced. In contrast, TH activity measured in the same samples under saturating cofactor concentrations elicited smaller increases (∼1.7-fold) in TH activity (similar to that seen in the other experiments presented in Figure 2B). These data are suggestive of a cAMP-mediated activation of TH at these early time points, leading to a decrease in the Km for pterin cofactor, as observed in many other studies in other cell types (Kumer and Vrana, 1996); however, a more thorough enzyme kinetic analysis is necessary to confirm this conclusion.
Treatment of MN9D cells with 8-CPT-cAMP was not associated with induction of TH mRNA
We tested whether the cAMP-mediated induction of TH protein in MN9D cells was associated with an initial induction of TH mRNA. A representative autoradiogram depicting TH mRNA and 28S rRNA levels in RNA samples isolated from MN9D cells is shown in Figure 2B; data collected from a number of time course experiments are shown in the bar graph of Figure 2B. TH mRNA levels did not increase in MN9D cells treated with the cAMP analog at any time point tested between 3 and 36 hr; similar results were observed using forskolin as a stimulus (data not shown).
The lack of cAMP-mediated induction of TH mRNA in both midbrain slice cultures and MN9D cells was very unexpected. To further verify that this result was not an artifact of the method used to assay TH mRNA, we developed a quantitative RT-PCR assay for TH mRNA using a real-time PCR cycler. The results of experiments using this quantitative assay (Figure 2D) agreed closely with those obtained using the semiquantitative assay, in that treatment of MN9D cells with 8-CPT-cAMP for 6 or 24 hr did not induce TH mRNA. In contrast, as expected, cAMP treatment induced TH mRNA significantly in both adrenal medulla-derived rat pheochromocytoma PC18 cells and locus coeruleus-derived cath.a cells (Figure 2D).
Treatment with 8-CPT-cAMP was associated with an increased rate of synthesis of TH protein in midbrain slices and MN9D cells
To test whether the cAMP-mediated induction of TH protein was due to an increase in the rate of synthesis of the enzyme, pulse-labeling experiments were performed in both model systems (Figure 3). Midbrain slices or MN9D cells were incubated in the presence or absence of 1 mM 8-CPT-cAMP for 24 hr, at which time the slices or cells were treated for an additional 1.5 hr with 3H-leucine in the continued presence of the cAMP analog, when appropriate. In preliminary experiments using pulse-chase procedures, we found that the apparent half-life of TH protein in MN9D cells was ∼40 hr (data not shown). Hence, the degradation of TH protein during the 1.5 hr pulse period would be predicted to be negligible. The slices or cells were harvested after the 1.5 hr pulse and the incorporation of 3H-leucine into TH protein was determined using immunoprecipitation procedures. Cell extracts were incubated overnight with TH antiserum; the immunoprecipitates were collected by centrifugation and subjected to SDS-PAGE. The gels were stained for protein and the ∼60 kDa TH protein band was visualized. A representative gel stained for protein is shown in Figure 3. A major band that comigrated with purified TH was easily identified, excised from the gel and the radioactivity counted by liquid scintillation spectrometry. In an initial experiment, the specificity of the antiserum was tested by exposing a dried-down gel to X-ray film for 7 days. A single major radioactive band that comigrated with purified TH was observed on the autoradiogram (Figure 3). Using this assay, we measured an ∼2-fold increase in the incorporation of radiolabel into TH protein in both model systems (see table in Figure 3). There was no change in the incorporation of 3H-leucine into total cellular protein in either model. The relative rate of synthesis of TH protein, measured as the incorporation of radiolabeled leucine into TH divided by that into total protein increased by 2-3-fold in either midbrain slices or MN9D cells treated for 24 hr with 8-CPT-cAMP. These increases in rate of synthesis correlated reasonably well with the increases in TH activity measured in the same cell extracts. Even though we have not yet ruled out an effect on TH protein degradation, these results suggest that the induction of TH elicited by the cAMP analog was primarily due to an increase in TH protein synthesis.
Treatment with 8-CPT-cAMP increased the association of TH mRNA with polysomes in MN9D cells
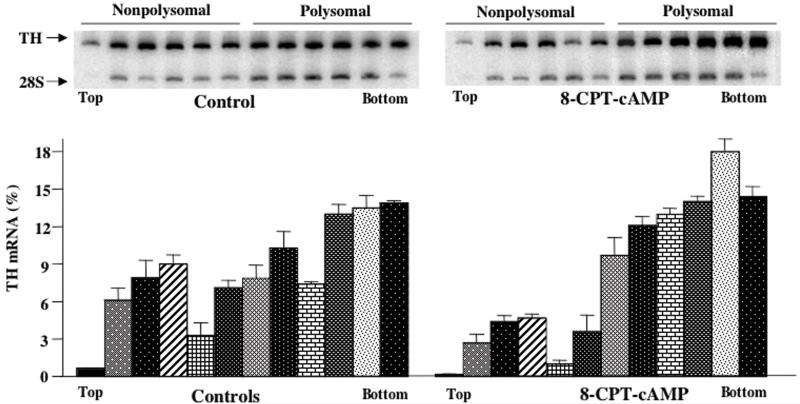
If cAMP treatment was associated with increased translation of pre-existing TH mRNA, then there should be an increase in the association of TH mRNA with polyribosomes in cAMP-treated cells. To test this hypothesis, MN9D cell extracts were subjected to sucrose gradient centrifugation to separate polysomal and non-polysomal RNAs. After centrifugation, fractions were collected from the gradients and TH mRNA levels in each fraction were measured using the semiquantitative RT-PCR assay. Autoradiograms from a representative sucrose gradient fractionation are shown at the top of Figure 4; TH mRNA levels in each fraction collected from three separate experiments are presented in the bar graphs in Figure 4. TH mRNA levels were isolated in fractions throughout the gradient, representing both polysomal-bound RNA (heavier fractions) and non-polysomal-bound RNA (lighter fractions). In cells treated with 8-CPT-cAMP for 24 hr, there was a significant shift in TH mRNA levels to the heavier fractions (Figure 4). No such shift in 28S rRNA was observed. The percentage of TH mRNA isolated in the heaviest six fractions (which are those enriched in 28S rRNA, as might be expected if they represent polysomes) is 66 ± 3% of the total TH mRNA isolated from the gradient in control samples and 84 ± 1% in 8-CPT-cAMP-treated samples (p < .01). A second method to analyze these data is to use a weighted mean average approach as described by Gore et al (1995). This is a measure of the fraction number in the gradient that contains the weighted average amount of TH mRNA relative to that measured throughout the entire gradient and is a method to quantitate a shift in the polysome profile. An increase in the weighted mean average indicates a shift of TH mRNA to heavier fractions containing polysomal-bound TH mRNA. The weighted mean averages for controls and 8-CPT-cAMP-treated cells were 7.8 ± .13 and 8.6 ± .15 (p < .02), respectively. Both of these analyses indicate a significant shift of TH mRNA levels to polysomes in cAMP-treated MN9D cells.
Figure 4. Polysome profile analysis of MN9D cell TH mRNA.
MN9D cells were either untreated or treated with 1 mM 8-CPT-cAMP for 24 hr. Cytoplasmic extracts were prepared and fractionated by sucrose gradient centrifugation. RNA was isolated from each fraction and TH mRNA and 28S rRNA levels were measured using semiquantitative RT-PCR assays. Autoradiograms of representative RT-PCR assays are shown in the top panels. The densities of the images in the autoradiograms were quantitated using Phosphorimager analysis and TH mRNA levels in each fraction were expressed as a percentage of total levels summed over all 12 fractions. The results represent the means ± SE from 3 experiments.
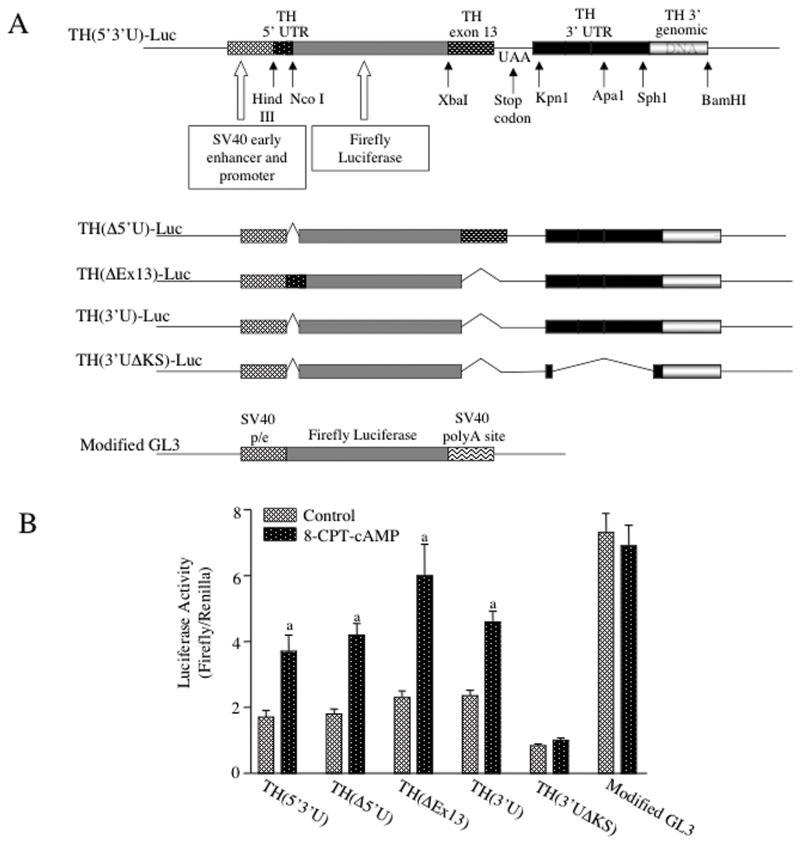
TH mRNA 3′UTR sequences conferred activation of reporter gene expression by 8-CPT-cAMP
Regulation of mRNA translation is often determined by sequences within the 5′ or 3′ UTR of the specific mRNA (Kwon and Hecht, 1991; Ostareck-Lederer et al., 1994). To test which sequences participated in the cAMP-mediated translational activation of TH mRNA, we constructed a reporter gene (designated TH(5′3′U)-luciferase) which encoded the 35 nt 5′UTR and 280 nt 3′UTR of TH mRNA, along with 160 nt of TH mRNA coding sequences that are present in exon 13 of the TH gene and 80 bp of TH 3′ genomic sequence downstream of the polyA+ addition site. These TH mRNA and genomic sequences were fused to the coding region of the firefly luciferase (Luc) gene as shown in Figure 5. This hybrid gene was driven by the SV40 promoter/enhancer. Deletion mutations of this transgene were also constructed as shown in Figure 5. A modified version of the GL3-control construct (Promega Corporation), which was used as the vector backbone for these TH-luciferase constructs, was utilized as a control for these experiments.
Figure 5. TH mRNA 3′UTR conferred cAMP-mediated inducibility on the luciferase gene in MN9D cells.
(A) Diagrams of the TH-luciferase constructs used in these experiments are shown in the figure. (B) MN9D cells were transfected with one of the TH-luciferase constructs along with the Renilla luciferase expression vector used for normalization purposes. Twenty-four hr after transfection, the cells were treated with or without 0.5 mM 8-CPT-cAMP for another 24 hr. The cells were then harvested and firefly and Renilla luciferase activities were measured in cell extracts. The results represent the means ± SE from 11-12 dishes, except for the modified GL3 data which were from 4 dishes. a: p < .01 compared to controls. (C) PC12 or cath.a cells were transfected with the TH(3′U)-luciferase and the Renilla expression vector constructs. Twenty-four hr after transfection, the cells were treated for another 24 hr with or without either 1 mM 8-CPT-cAMP (cath.a cells) or 10 uM forskolin (PC12 cells). The results represent the means ± SE from 4-5 dishes.
a: p < .01 compared to controls
MN9D cells were transiently transfected with one of these vectors and cells were incubated in the presence or absence of 0.5 mM 8-CPT-cAMP for 24 hr. The cells were cotransfected with a vector expressing Renilla luciferase and the expression of this gene was used to control for transfection efficiency. The expression of firefly luciferase activity in cells transfected with the wild type vector, TH(5′3′U)-luciferase, was increased by ∼2-fold in response to cAMP analog treatment (Figure 5). Deletion of either the 5′UTR (TH(Δ5′U)) or the TH mRNA coding sequences at the 5′ end of exon 13 (TH(ΔEx13)) did not diminish the response to 8-CPT-cAMP. Similarly, deletion of both the 5′UTR and exon 13 coding sequences (TH(3′U)) had no effect on the cAMP response. Furthermore, deletion of the 5′UTR and/or exon 13 coding sequences had no significant effect on the basal expression of luciferase in the context of this hybrid gene. In contrast, deletion of the 3′UTR sequences between the Kpn1 and Sph1 sites (TH(3′UΔKS) decreased the basal expression of luciferase by ∼50% (Figure 5). More importantly, the loss of these 3′UTR sequences totally ablated the response of the hybrid gene to cAMP. As expected, cAMP treatment did not elicit an effect on luciferase expression in cells transfected with the control modified GL3-control vector; however, basal luciferase expression in these cells was 3-4 fold greater than that observed in cells transfected with the wild type TH-luciferase construct. This disparity may be due to differences in expression determined by the TH mRNA 3′UTR versus the SV40 3′UTR present in the modified GL3 vector. Alternatively, TH mRNA 3′UTR sequences may confer regulatory effects on luciferase mRNA translation or stability, as observed in analogous studies using 3′UTR sequences from other mRNAs (Kwon and Hecht, 1991; Ostareck-Lederer et al., 1994). To test whether this response to cAMP was specific for MN9D cells, mouse locus coeruleus-derived cath.a cells and rat pheochromocytoma PC12 cells were transfected with TH(3′U)-luciferase and treated for 24 hr with either 8-CPT-cAMP or forskolin. Expression from TH(3′U)-luciferase was not significantly affected by either treatment in the cath.a cells or PC12 cells (data not shown).
Luciferase mRNA expression from TH(3′U)-luciferase was not affected by 8-CPT-cAMP in MN9D cells
The observed increase in TH(3′U)-luciferase activity elicited by cAMP in MN9D cells might be due to an increase in translation and/or stability of TH-luciferase mRNA molecules. Two approaches were used to differentiate between these different possibilities. In the first approach, MN9D cells were transiently transfected with TH(3′U)-luciferase and the Renilla luciferase expression vector. These transfected cells were then treated for 24 hr with 0.5 mM 8-CPT-cAMP or vehicle and total cellular RNA was isolated. Initial studies indicated that the levels of both firefly and Renilla luciferase mRNAs were very low in these transiently-transfected cells. Furthermore, measurement of these mRNAs was confounded by contamination of the RNA samples with transfected plasmid DNAs. To overcome this latter problem, the RNA samples were treated extensively with DNase (TURBO-DNase from Ambion, Inc) prior to the RT-PCR assay. Furthermore, controls were run for each sample in which the RT step was omitted, to detect any residual plasmid DNA. Examples of this type of analysis are shown in Figure 6A. These autoradiograms depict firefly and Renilla luciferase mRNAs measured using the radioactive semiquantitative RT-PCR assay. Both mRNAs were detected in RNA samples treated first with RT to produce corresponding cDNAs. In contrast, no significant signals were detected for Renilla luciferase mRNA and only minor signals were detected for firefly luciferase mRNA in samples that were not treated first with RT. As is evident from the autoradiogram, treatment of the MN9D cells with 8-CPT-cAMP for 24 hr did not significantly increase firefly luciferase mRNA levels (Figure 6A). To verify that the DNase treatment did not alter mRNA levels, TH mRNA and 28S rRNA levels were measured in MN9D RNA samples that were treated with or without TURBO-DNase (Figure 6B). As seen in the first two lanes of the autoradiograms in panel B of Figure 6, DNase treatment totally abolished the signals representing firefly and Renilla mRNAs, when the samples were not first incubated with RT; these results indicated that the contaminating plasmid DNAs were degraded by this DNase treatment. In contrast, both TH mRNA and 28S rRNA signals were not affected by the DNase treatment, suggesting that TURBO-DNase did not significantly degrade RNA in the samples. Quantitative measurements of firefly and Renilla mRNAs using this assay are presented in the bar graph in Figure 6C. Treatment with 8-CPT-cAMP for 24 hr did not affect the levels of firefly luciferase mRNA expressed from the TH(3′U)-luciferase construct. In contrast, firefly luciferase enzyme activity increased ∼1.9-fold in these experiments. These results argued against a stabilization of firefly luciferase mRNA, but were consistent with a translational activation of firefly luciferase mRNA conferred by the TH 3′UTR sequences.
In a second approach, MN9D cells were stably transfected with TH(3′U)-luciferase and the effects of treatment with 8-CPT-cAMP on luciferase mRNA and enzymatic activity were analyzed. This approach was used to avoid the plasmid DNA contamination problems that we encountered when trying to measure luciferase mRNA levels, as well as the very low expression of the luciferase gene using the transient transfection approach. Two stably-transfected MN9D cell lines were isolated that expressed reasonable concentrations of firefly luciferase mRNA; these cell lines were designated U1 and U17. As shown in the autoradiogram in Figure 7, the semiquantitative RT-PCR assay easily detected firefly luciferase mRNA in RNA samples from these cells. Only minor luciferase mRNA bands were observed in samples that were not first treated with RT; these minor bands were presumably due to contaminating genomic DNA. When the stably-transfected cells were treated for 24 hr with 8-CPT-cAMP, luciferase mRNA levels were the same or slightly less than that observed in control cells. In contrast, luciferase activity increased significantly (1.5-2 fold) in response to the cAMP analog in both cell lines. These results also support the hypothesis that cAMP activates translation, but does not stabilize TH(3′U)-luciferase mRNA in MN9D cells.
DISCUSSION
Very little information is available concerning the molecular mechanisms that control TH gene expression in mesencephalic dopamine neurons. This lack of information is especially important when trying to understand the compensatory mechanisms that regulate dopamine biosynthesis in nigrostriatal neurons in neurodegenerative paradigms. When mesencephalic dopamine neurons are completely destroyed in animal models with the use of neurotoxins, a number of cognitive, motor and sensory deficits are observed, from which the animals never recover (Robinson et al., 1990; Zigmond et al., 1990). However, when these neurons are only partially destroyed (<80%), there is considerable recovery of function from these deficits over time. This recovery is due to compensatory mechanisms that occur in the surviving nerve terminals, as well as neuronal sprouting in the nerve terminal regions (Jakowec et al., 2004; Robinson et al., 1990; Rothblat et al., 2001; Sedelis et al., 2000; Zigmond et al., 1990). These mechanisms maintain appropriate concentrations of dopamine at striatal synapses, even when large numbers of nerve terminals are destroyed.
In contrast to the vigorous compensatory mechanisms that occur at nerve terminals, only modest or insignificant increases and even decreases in TH mRNA levels are observed in surviving midbrain dopamine cell bodies after lesioning with neurotoxins (Blanchard et al., 1995; Bowyer et al., 1998; Kastner et al., 1994; Pasinetti et al., 1992; Rothblat et al., 2001). Furthermore, TH gene transcription rate does not increase in surviving midbrain cell bodies after lesioning with 6-hydroxydopamine (Sherman and Moody, 1995). In human Parkinsonian patients, decreases in midbrain TH mRNA levels have been observed (Javoy-Agid et al., 1990; Kastner et al., 1993). This lack of compensatory induction of TH gene expression in the midbrain is puzzling. One would anticipate the existence of robust homeostatic mechanisms that elevate TH gene expression in dopamine cell bodies, in order to increase dopamine biosynthesis in spared striatal nerve terminals and to maintain high levels of TH protein in newly-sprouted striatal nerve terminals. This idea is supported by studies in the peripheral nervous system; it is well-documented that when norepinephrine is depleted from sympathetic nerve terminals by reserpine or 6-hydroxydopamine, TH is induced in ganglionic cell bodies (Black et al., 1985; Mueller et al., 1969; Trocme et al., 2001).
An explanation for this lack of compensatory response is that receptors and/or signaling pathways that mediate the induction of the TH gene may be down-regulated, may not be activated or their signals may be overcome by increased inhibitory input during Parkinson’s disease or after chemical lesioning of striatal terminals. These stimulatory receptors and signaling pathways could be those activated by neurotransmitters released from afferent nerve terminals, those recognizing humoral factors like glucocorticoids or retinoids, and/or those recognizing neurotrophic factors released from glial cells or target tissues. However, there is almost no information concerning which receptors and signaling pathways are responsible for regulating TH gene expression in adult dopaminergic neurons in response to extracellular physiological signals. Leviel et al (1991) have reported that intranigral microinjection of forskolin elicits the induction of TH mRNA, but not TH protein in substantia nigra. This early report suggests that activation of the cAMP signaling pathway may induce midbrain TH gene expression, presumably as observed in most other model systems. However, this finding has never been confirmed or expanded upon.
In the present report we test directly in two different cultured model systems whether the signaling pathways activated by cAMP induce TH expression in midbrain dopamine neurons. We have chosen to study the cAMP signaling pathway, because cAMP is a powerful inducer of TH gene transcription rate, TH mRNA and TH protein in most other model systems (Kumer and Vrana, 1996) and because cAMP has been implicated in the induction of midbrain TH in the previous report (Leviel et al., 1991). As expected, our results demonstrate that TH protein is induced after treatment of either midbrain slices or MN9D cells for 24-48 hr with either forskolin or the cAMP analog, 8-CPT-cAMP. A cAMP-mediated activation of pre-existing TH enzyme molecules is also suggested at earlier time points in the MN9D cells and possibly in midbrain slice cultures. However, in the latter model, the increase in TH activity observed at 12 hr (Figure 1) without a concomitant increase in TH protein appears to be due to an increased apparent Vmax. This result is unexpected, because activation of TH by cAMP is usually associated with a decreased Km for pterin cofactor (Kumer and Vrana, 1996). However, Daubner et al (1992) have shown that PKA-mediated phosphorylation of TH in the presence of inhibitory concentrations of dopamine produces an activation of the enzyme that is characterized by an apparent increase in Vmax. More studies are needed to confirm the precise mechanism(s) mediating this enzyme activation process in this model system.
Surprisingly, our studies show that even though treatment with 8-CPT-cAMP induces TH protein, it does not induce TH mRNA in either DIV 1 or DIV 7 midbrain slice cultures or in MN9D cells. These results are in direct contrast to those reported by Leviel et al (1991), who observed induction of TH mRNA, but not TH protein after microinjection of forskolin into the substantia nigra under in vivo conditions. The reason for this discrepancy is not clear; however, it is likely due to inherent differences using in vivo versus cell culture approaches. For instance, microinjected forskolin may be raising cAMP levels in nondopaminergic afferent nerves or neighboring glial cells, leading to stimulation of TH gene transcription in dopaminergic neurons via indirect mechanisms not related to cAMP generation in the dopamine neuron itself. The use of anesthetic in the microinjection studies may be having an unexpected effect. Alternatively, neuronal damage produced by the microinjection may be causing the release of chemicals that stimulate TH mRNA expression. These confounding issues inherent to the in vivo approach are less likely to be observed in the cultured model systems used in the present study. However, it is important to note that the TH gene promoter has a perfect consensus CRE just 35 bp upstream of the transcriptional start site and many studies have shown that this CRE is responsive to cAMP in pheochromocytoma, neuroblastoma and cath.a cells (Kumer and Vrana, 1996; Lewis-Tuffin et al., 2004). Hence, the lack of an apparent transcriptional response in the cultured dopamine cell model systems is very surprising and how these results relate to the response observed under in vivo conditions requires further investigation. Nevertheless, a significant finding of the present study is that the TH gene in cultured midbrain dopamine neurons apparently responds to cAMP in a different manner than in cultured cells derived from other lineages.
A second major finding in this report is that TH protein is induced by a cAMP-mediated mechanism that regulates TH mRNA translation in midbrain neurons. This conclusion is based on our findings that cAMP increases the rate of synthesis of TH protein without a concomitant increase in TH mRNA levels in both midbrain slices and MN9D cells and increases the percentage of TH mRNA localized with polysomes in MN9D cells. We also show that this translational response is dependent on sequences within the 3′UTR of TH mRNA. These sequences reside in a region that encodes the polypyrimidine-rich binding site described by Czyzyk-Krzeska and coworkers (Paulding and Czyzyk-Krzeska, 1999). These workers have shown that this site plays an important role in mediating the stabilization of TH mRNA that occurs in response to hypoxia in PC12 cells. They have also shown that hypoxia induces the levels of a trans-acting protein, designated PCBP (polyC-binding protein) that apparently binds to this site, leading presumably to a decreased degradation of TH mRNA. It is conceivable that proteins binding to this site (or an adjacent site) in MN9D cells may regulate the translation of TH mRNA in addition to its degradation. According to this model, cAMP may induce a protein (or set of proteins) that binds to the 3′UTR of TH mRNA, leading to its increased association with polysomes and consequently, enhanced translation and synthesis of TH protein. More work is needed to identify the specific sequences within the TH mRNA 3′UTR and the trans-acting factors that mediate this translational response.
This cAMP-mediated translational activation of TH mRNA may be unique to midbrain dopamine neurons. As discussed above, in all other model systems studied so far, cAMP-stimulated increases in TH protein are associated with increases in TH mRNA and TH gene transcription rate. In these models, cAMP is thought to induce TH via signaling pathways that phosphorylate CREB or other CRE-binding proteins, which bind to the TH CRE and transactivate the TH gene promoter, leading to the observed transcriptional stimulation. Even though it has not been directly assessed in these other systems, there is no evidence that cAMP regulates TH mRNA translation, since the increases in TH mRNA and TH protein are usually very similar in size. The evidence in this paper supports that conclusion, since we were unable to detect any cAMP-mediated increases in luciferase reporter expression directed by the TH mRNA 3′UTR in either PC12 cells or cath.a cells. However, in this regard, we have recently shown under in vivo conditions that TH mRNA translation is regulated by stress in rat adrenal medulla (Xu et al., 2007), but there is no evidence yet that this regulation is cAMP-mediated.
The reason for this possibly unique and novel cAMP response in midbrain dopaminergic neurons remains obscure. This mechanism may have developed in these cells, because the more common transcriptional response of the TH gene to cAMP is not functional. Alternatively, this mechanism may be necessary to induce TH in response to nerve stimulation in a particular cellular locale in midbrain neurons. A number of synaptic proteins are known to be regulated by translational mechanisms at local dendritic synapses (Krichevsky and Kosik, 2001). Perhaps, TH mRNA is also localized in dendrites and local elevation of cAMP leads to local synthesis of TH protein and increased dopamine biosynthesis in these dendrites. From a pharmacological standpoint, drugs that block receptors linked to inhibition of adenylyl cyclase, such as antagonists of D2 dopamine receptors, may raise cAMP in dopamine neurons for long periods of time, activating this mechanism. More work is needed to test these hypotheses. Nevertheless, our results clearly demonstrate a novel post-transcriptional mechanism that regulates TH mRNA translation. This mechanism may play an important role in maintaining dopamine levels in midbrain dopamine neurons. Further elucidation of the molecular and cellular mechanisms that mediate this translational response may lead to new therapeutic strategies to up-regulate dopamine biosynthesis in nigrostriatal neurons during Parkinson’s disease. It may also provide new avenues for engineering viral vectors designed to over-express TH and replace dopamine in different disease states.
Acknowledgments
The authors would like to acknowledge the excellent technical assistance of Carol R. Sterling for performing some of the experiments in this paper.
This work was supported by NIH grants DA05014 and NS39415 to AWT. P.R. was supported by training grants ES07026 and DA07232.
Abbreviations
- TH
tyrosine hydroxylase
- 3′ UTR
3′ untranslated region
- ACSF
artificial spinal fluid
- 6MPH4
6-methyl-5,6,7,8-tetrahydropterin
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- 8-CPT-cAMP
8-chlorothio-cyclic AMP
- DIV
days in vitro
REFERENCES
- Biguet NF, Buda M, Lamouroux A, Samolyk D, Mallet J. Time course of the changes of TH mRNA in rat brain and adrenal medulla after a single injection of reserpine. EMBO J. 1986;5:287–291. doi: 10.1002/j.1460-2075.1986.tb04211.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black IB, Chikaraishi DM, Lewis EJ. Trans-synaptic increase in RNA coding for tyrosine hydroxylase in a rat sympathetic ganglion. Brain Res. 1985;339:151–153. doi: 10.1016/0006-8993(85)90635-3. [DOI] [PubMed] [Google Scholar]
- Blanchard V, Chritin M, Vyas S, Savasta M, Feuerstein C, Agid Y, Javoy-Agid F, Raisman-Vozari R. Long-term induction of tyrosine hydroxylase expression: compensatory response to partial degeneration of the dopaminergic nigrostriatal system in the rat brain. J Neurochem. 1995;64:1669–1679. doi: 10.1046/j.1471-4159.1995.64041669.x. [DOI] [PubMed] [Google Scholar]
- Bowyer JF, Frame LT, Clausing P, Nagamoto-Combs K, Osterhout CA, Sterling CR, Tank AW. The long-term effects of amphetamine neurotoxicity on tyrosine hydroxylase mRNA and protein in aged rats. J Pharmacol Exp Ther. 1998;286:1074–1085. [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Choi HK, Won L, Roback JD, Wainer BH, Heller A. Specific modulation of dopamine expression in neuronal hybrid cells by primary cells from different brain regions. Proc Natl Acad Sci U S A. 1992;89:8943–8947. doi: 10.1073/pnas.89.19.8943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daubner SC, Lauriano C, Haycock JW, Fitzpatrick PF. Site-directed mutagenesis of serine 40 of rat tyrosine hydroxylase. J Biol Chem. 1992;267:12639–12646. [PubMed] [Google Scholar]
- Gore AC, Roberts JL. Translational efficiency of gonadotropin-releasing hormone mRNA is negatively regulated by phorbol ester in GT1-7 cells. Endocrinol. 1995;136:1620–1625. doi: 10.1210/endo.136.4.7895672. A. H. [DOI] [PubMed] [Google Scholar]
- Jakowec MW, Nixon K, Hogg E, McNeill T, Petzinger GM. Tyrosine hydroxylase and dopamine transporter expression following 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine-induced neurodegeneration of the mouse nigrostriatal pathway. J Neurosci Res. 2004;76:539–550. doi: 10.1002/jnr.20114. [DOI] [PubMed] [Google Scholar]
- Javoy-Agid F, Hirsch EC, Dumas S, Duyckaerts C, Mallet J, Agid Y. Decreased tyrosine hydroxylase messenger RNA in the surviving dopamine neurons of the substantia nigra in Parkinson’s disease: an in situ hybridization study. Neuroscience. 1990;38:245–253. doi: 10.1016/0306-4522(90)90389-l. [DOI] [PubMed] [Google Scholar]
- Kastner A, Herrero MT, Hirsch EC, Guillen J, Luquin MR, Javoy-Agid F, Obeso JA, Agid Y. Decreased tyrosine hydroxylase content in the dopaminergic neurons of MPTP-intoxicated monkeys: effect of levodopa and GM1 ganglioside therapy. Ann Neurol. 1994;36:206–214. doi: 10.1002/ana.410360213. [DOI] [PubMed] [Google Scholar]
- Kastner A, Hirsch EC, Herrero MT, Javoy-Agid F, Agid Y. Immunocytochemical quantification of tyrosine hydroxylase at a cellular level in the mesencephalon of control subjects and patients with Parkinson’s and Alzheimer’s disease. J Neurochem. 1993;61:1024–1034. doi: 10.1111/j.1471-4159.1993.tb03616.x. [DOI] [PubMed] [Google Scholar]
- Krichevsky AM, Kosik KS. Neuronal RNA granules: a link between RNA localization and stimulation-dependent translation. Neuron. 2001;32:683–696. doi: 10.1016/s0896-6273(01)00508-6. [DOI] [PubMed] [Google Scholar]
- Kumer SC, Vrana KE. Intricate regulation of tyrosine hydroxylase activity and gene expression. J Neurochem. 1996;67:443–462. doi: 10.1046/j.1471-4159.1996.67020443.x. [DOI] [PubMed] [Google Scholar]
- Kwon YK, Hecht NB. Cytoplasmic protein binding to highly-conserved sequences in the 3′ untranslated region of protamine 2 mRNA, a translationally regulated transcript of male germ cells. Proc Natl Acad Sci USA. 1991;88:3584–3588. doi: 10.1073/pnas.88.9.3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leviel V, Guibert B, Mallet J, Faucon-Biguet N. Induction of tyrosine hydroxylase in the rat substantia nigra by local injection of forskolin. J Neurosci Res. 1991;30:427–432. doi: 10.1002/jnr.490300219. [DOI] [PubMed] [Google Scholar]
- Lewis-Tuffin LJ, Quinn PG, Chikaraishi DM. Tyrosine hydroxylase transcription depends primarily on cAMP response element activity, regardless of the type of inducing stimulus. Mol Cell Neurosci. 2004;25:536–547. doi: 10.1016/j.mcn.2003.10.010. [DOI] [PubMed] [Google Scholar]
- Melia KR, Rasmussen K, Terwilliger RZ, Haycock JW, Nestler EJ, Duman RS. Coordinate regulation of the cyclic AMP system with firing rate and expression of tyrosine hydroxylase in the rat locus coeruleus: effects of chronic stress and drug treatments. J Neurochem. 1992;58:494–502. doi: 10.1111/j.1471-4159.1992.tb09748.x. [DOI] [PubMed] [Google Scholar]
- Mueller RA, Thoenen H, Axelrod J. Increase in tyrosine hydroxylase activity after reserpine administration. J Pharmacol Exp Ther. 1969;169:74–79. [PubMed] [Google Scholar]
- Ortiz J, Fitzgerald LW, Lane S, Terwilliger R, Nestler EJ. Biochemical adaptations in the mesolimbic dopamine system in response to repeated stress. Neuropsychopharmacology. 1996;14:443–452. doi: 10.1016/0893-133X(95)00152-4. [DOI] [PubMed] [Google Scholar]
- Ostareck-Lederer A, Ostareck DH, Standart N, Thiele BJ. Translation of 15-lipoxygenase mRNA is inhibited by a protein that binds to a repeated sequence in the 3′ untranslated region. EMBO J. 1994;13:1476–1481. doi: 10.1002/j.1460-2075.1994.tb06402.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterhout CA, Sterling CR, Chikaraishi DM, Tank AW. Induction of tyrosine hydroxylase in the locus coeruleus of transgenic mice in response to stress or nicotine treatment: lack of activation of tyrosine hydroxylase promoter activity. J Neurochem. 2005;94:731–741. doi: 10.1111/j.1471-4159.2005.03222.x. [DOI] [PubMed] [Google Scholar]
- Pasinetti GM, Morgan DG, Johnson SA, Millar SL, Finch CE. Tyrosine hydroxylase mRNA concentration in midbrain dopaminergic neurons is differentially regulated by reserpine. J Neurochem. 1990;55:1793–1799. doi: 10.1111/j.1471-4159.1990.tb04970.x. [DOI] [PubMed] [Google Scholar]
- Pasinetti GM, Osterburg HH, Kelly AB, Kohama S, Morgan DG, Reinhard JF, Stellwagon RH, Finch CE. Slow changes of tyrosine hydroxylase gene expression in dopaminergic brain neurons after neurotoxin lesioning: a model for neuron aging. Mol Brain Res. 1992;13:63–73. doi: 10.1016/0169-328x(92)90045-d. [DOI] [PubMed] [Google Scholar]
- Paulding WR, Czyzyk-Krzeska MF. Regulation of tyrosine hydroxylase mRNA stability by protein-binding, pyrimidine-rich sequence in the 3′-untranslated region. J Biol Chem. 1999;274:2532–2538. doi: 10.1074/jbc.274.4.2532. [DOI] [PubMed] [Google Scholar]
- Robinson TE, Castaneda E, Whishaw IQ. Compensatory changes in striatal dopamine neurons following recovery from injury induced by 6-OHDA or methamphetamine: a review of the evidence from microdialysis studies. Can J Psychol. 1990;44:253–275. doi: 10.1037/h0084241. [DOI] [PubMed] [Google Scholar]
- Rothblat DS, Schroeder JA, Schneider JS. Tyrosine hydroxylase and dopamine transporter expression in residual dopaminergic neurons: Potential contributors to spontaneous recovery from experimental Parkinsonism. J Neurosci Res. 2001;65:254–266. doi: 10.1002/jnr.1149. [DOI] [PubMed] [Google Scholar]
- Sabban EL, Kvetnansky R. Stress-triggered activation of gene expression in catecholaminergic systems: dynamics of transcriptional events. Trends Neurosci. 2001;24:91–98. doi: 10.1016/s0166-2236(00)01687-8. [DOI] [PubMed] [Google Scholar]
- Sedelis M, Hofele K, Auburger GW, Morgan S, Boraud T, Gross CE. Spontaneous long-term compensatory dopaminergic sprouting in MPTP-treated mice. Synapse. 2000;38:363–368. doi: 10.1002/1098-2396(20001201)38:3<363::AID-SYN16>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- Serova L, Sabban EL. Involvement of alpha 7 nicotinic acetylcholine receptors in gene expression of dopamine biosynthetic enzymes in rat brain. J Pharmacol Exp Ther. 2002;303:896–903. doi: 10.1124/jpet.102.039198. [DOI] [PubMed] [Google Scholar]
- Sherman TG, Moody CA. Alterations in tyrosine hydroxylase expression following partial lesions of the nigrostriatal bundle. Mol Brain Res. 1995;29:285–296. doi: 10.1016/0169-328x(94)00259-h. [DOI] [PubMed] [Google Scholar]
- Smith KM, Mitchell SN, Joseph MH. Effects of chronic and subchronic nicotine on tyrosine hydroxylase activity in noradrenergic and dopaminergic neurones in the rat brain. J Neurochem. 1991;57:1750–1756. doi: 10.1111/j.1471-4159.1991.tb06377.x. [DOI] [PubMed] [Google Scholar]
- Stoppini L, Buchs PA, Muller D. A simple method for organotypic cultures of nervous tissue. J Neurosci Methods. 1991;37:173–182. doi: 10.1016/0165-0270(91)90128-m. [DOI] [PubMed] [Google Scholar]
- Sun B, Chen X, Xu L, Sterling CR, Tank AW. Chronic nicotine treatment leads to induction of tyrosine hydroxylase in locus coeruleus neurons: The role of transcriptional activation. Mol Pharmacol. 2004;66:1011–1021. doi: 10.1124/mol.104.001974. [DOI] [PubMed] [Google Scholar]
- Sun B, Sterling CR, Tank AW. Chronic nicotine treatment leads to sustained stimulation of tyrosine hydroxylase gene transcription rate in rat adrenal medulla. J Pharmacol Exp Ther. 2003;304:575–588. doi: 10.1124/jpet.102.043596. [DOI] [PubMed] [Google Scholar]
- Tank AW, Ham L, Curella P. Induction of tyrosine hydroxylase by cyclic AMP and glucocorticoids in a rat pheochromocytoma cell line: effect of the inducing agents alone or in combination on the enzyme levels and rate of synthesis of tyrosine hydroxylase. Mol Pharmacol. 1986;30:486–496. [PubMed] [Google Scholar]
- Thomas MP, Webster WW, Norgren RB, Monaghan DT, Morrisett RA. Survival and functional demonstration of interregional pathways in fore/midbrain slice explant cultures. Neuroscience. 1998;85:615–626. doi: 10.1016/s0306-4522(97)00646-5. [DOI] [PubMed] [Google Scholar]
- Trocme C, Ravassard P, Sassone-Corsi P, Mallet J, Biguet NF. CREM and ICER are differentially implicated in trans-synaptic induction of tyrosine hydroxylase gene expression in adrenal medulla and sympathetic ganglia of rat. J Neurosci Res. 2001;65:91–99. doi: 10.1002/jnr.1132. [DOI] [PubMed] [Google Scholar]
- Wong DL, Tank AW. Stress-induced catecholaminergic function: transcriptional and post-transcriptional control. Stress. 2007;10:121–130. doi: 10.1080/10253890701393529. [DOI] [PubMed] [Google Scholar]
- Xu L, Chen X, Sun B, Sterling C, Tank AW. Evidence for regulation of tyrosine hydroxylase mRNA translation by stress in rat adrenal medulla. Brain Res. 2007;1158:1–10. doi: 10.1016/j.brainres.2007.04.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura R, Xu L, Sun B, Tank AW. Nicotinic and muscarinic acetylcholine receptors are essential for the long-term response of tyrosine hydroxylase gene expression to chronic nicotine treatment in rat adrenal medulla. Mol Brain Res. 2004;126:188–197. doi: 10.1016/j.molbrainres.2004.04.007. [DOI] [PubMed] [Google Scholar]
- Zigmond MJ, Abercrombie ED, Berger TW, Grace AA, Stricker EM. Compensations after lesions of central dopaminergic neurons: some clinical and basic implications. Trends Neurosci. 1990;13:290–296. doi: 10.1016/0166-2236(90)90112-n. see comments. [DOI] [PubMed] [Google Scholar]