Abstract
We report a simple and robust magnetomechanical system for direct visual observation of the DNA packaging behavior of the bacteriophage phi29 in real time. The system comprises a micron-sized magnetic bead attached to the free end of the viral DNA, a magnet and a bright-field microscope. We show that the phi29 DNA packaging activity can be observed and dynamically analyzed at the single molecular level in bright field with a relatively simple system. With this system we also visually demonstrate the phi29 motor transporting a cargo 10 000 times the viral size.
Bacteriophage phi29 can package its 19.3 kbs (kilobase pairs) double-stranded DNA (dsDNA) into its 42×54 nm2 prohead by means of adenosine triphosphate (ATP) hydrolysis which produces energy to reel in the DNA.1, 2, 3 The phi29 DNA-packaging motor can generate a force up to ∼57 pN and finish the packaging process in 5.5 min, making it one of the most powerful biomolecular motors up to date.4 Understanding the physical and mechanical behavior of this packaging process brings out various potentials of the phi29 packaging motor, some of whose components have been studied as potential gene delivery vehicles for treatment of cancer and viral infections.5, 6, 7 To study the DNA packaging behavior of the phi29 motor at the single-molecular level, a relatively sophisticated and costly technique, optical tweezers, has been used.1, 4, 8, 9 The packaging process has also been observed by using fluorescence microscopy to inspect the Brownian motion of a fluorescent bead (conjugated to the open end of a prepackaged DNA) whereby the translational motion of the bead was interpreted via its Brownian motion perpendicular to the primary translational direction.10, 11 Here we present a simple and robust approach, based on a combination of conventional microscopy and magnetomechanics, for direct bright-field analysis of the phi29 motor packaging activity. This technique allows us to view directly the translation as well as the Brownian motion of a magnetic bead attached to the free end of the viral DNA, in bright field. Using this technique, we show that the phi29 motor can transport a cargo 10 000 times its viral size and describe a quantitative analysis of the DNA packaging dynamics based on the recorded bright-field images and videos.
The operation of this system is illustrated in Fig. 1. The system comprises a bright-field optical microscope and a microfluidic chamber in which previously “stalled” phi29 intermediates, with partially prepackaged DNAs, are immobilized on a vertical glass wall. The open end of each tether DNA is conjugated with a micron-sized magnetic bead. Upon providing a magnetic field, the magnetic bead is pulled away from the glass wall stretching the tether DNA. The phi29 motor is too small to be observed by conventional optical microscopy. Hence, the magnetic bead serves as a motion indicator due to its large size and its ability to stretch the DNA. By monitoring the motion of the magnetic bead bound to the tether DNA, the packaging event is visually observed. The motion of the bead, and hence changes in the DNA length is analyzed using a SPOTTRACKER software.12
Figure 1.
Schematic of the magnetomechanical system for bright-field analysis of phi29 DNA packaging.
To prepare the stalled phi29 motor intermediates containing prepackaged DNA, a nonhydrolyzable ATP analogue, γ-S-ATP, was used. The stalled motors were attached to a glass wall coated with phi29 IgG (antiprohead) which was placed in the chamber. A suspension of magnetic beads (Chemagen, M-PVA Ak11) conjugated with antibodies against phi29 DNA’s end protein gp3 (anti-gp3 beads) was subsequently introduced into the chamber. The anti-gp3 beads recognized and bound to gp3 on the open ends of prepackaged DNAs. By washing with a TMS buffer solution (50 mM tris-HCL, 10 mM MgCl2, 100 mM NaCl), most unbound beads were removed. This resulted in attachment of magnetic beads to the open ends of DNA whose other ends were prepackaged into the phi29 proheads. After providing a magnetic field to remove the remaining unbound magnetic beads, a tethered bead was located using an optical microscope. The restart buffer (containing ATP, DNA packaging enzyme gp16, and TMS buffer) was introduced to reactivate the stalled motor. The preparation of phi29 components, the antibody beads, and the reactivation of the stalled motors, were performed as described in previous studies.11, 13, 14, 15, 16, 17, 18, 19, 20
In the experimental setup, a small glass slide (5×5×0.22 mm3) was settled in a microfluidic chamber (total volume ∼8 μl) which was subjected to investigation by a conventional optical microscope (Nikon Eclipse 80i). The edge of the glass slide served as the “glass wall” for immobilizing the motor intermediates. A charge coupled device camera was mounted on the microscope and connected to a computer for recording the real-time bead activity. A permanent magnet, used to exert force on the magnetic bead, was fixed on a three dimensional translation stage (Thorlabs LT3) and placed beside the microscope. By finely adjusting the position of the magnet, the magnetic force applied to the tethered bead was maintained around 1 pN in order not to overstretch the DNA and∕or stall the motor.4, 21 This was achieved by measuring the magnetic field profile as a function of distance from the magnet and calculating the resulting force according to Eq. 1 which shows the force applied on a sphere by a magnetic field,22
| (1) |
Here, B is magnetic field, μ0 is permeability of free space (4π×10−7 N A−2), μr is relative permeability of the particular superparamagnetic bead (∼8), and d is the bead diameter. The calibration of magnetic force applied on the bead is shown in supplemental material.23
Upon adding the restart buffer into the chamber, the motion of the bead was recorded. Figure 2 shows a series of micrographs of two separate DNA packaging events observed in real time (the corresponding movies are available online).23 In the first case, there was only one bead (diameter ∼1.8 μm, volume ∼3 μm3) bound to the DNA [motor 1, Fig. 2a], while in the second case, a cluster of three beads (diameters ∼1.9, 1.6, and 1 μm, total volume ∼6 μm3) bound to the DNA [motor 2, Fig. 2b]. The real-time visualization of the packaging events revealed that the phi29 motor can transport cargo, attached to the end of tether DNA, which is more than 10 000 times the size of the virus (volume ∼2.7×10−4 μm3). In neither case did the stalled motor immediately start to repackage the DNA upon the addition of restart buffer. Also the restart time varied from one case to the other (motor 1 restarted the packaging ∼2 min after the introduction of restart buffer while motor 2 restarted after ∼12 min).
Figure 2.
Temporal micrographs of beads bound to the tether DNAs of (a) motor 1 and (b) motor 2, showing the phi29 motor packaging activities.
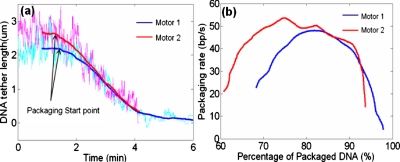
By tracking the motions of the tethered beads we further analyzed the DNA packaging kinetics. The DNA tether lengths were obtained from the beads’ translational motions in the packaging direction (toward the glass wall where the virus is immobilized). Figure 3 shows the plots of DNA tether length against time after restarting the motors, and the packaging rate against the amount of DNA packaged. In both cases, about 2∕3 of the DNAs were prepackaged into the proheads before the motors were restarted (the initial DNA tether length of motor 2 was ∼0.5 μm longer than that of motor 1). The curves shown in Fig. 3a have been shifted on the time axis in order to clearly compare the packaging kinematics of the two motors. Interestingly, the DNA tether length of motor 2 drops suddenly toward the end of the packaging event [time ≈4 min on time axis of Fig. 3a]. This is attributed to the sudden nonspecific binding when the cluster was too close to the glass wall. Noticeably, the packaging rates of both motors first start with some 20 bp∕s and increase as more DNA is packaged [Fig. 3b]. Only when around 20% of DNAs (∼1.3 μm) are left do the packaging rates start to decrease. This phenomenon is different from the consistently decreasing packaging rates reported in the previous studies.1, 4 Diffusion of ATP to the phi29 motor could be a possible reason for the slow packaging rate in the beginning, although we minimized this effect by using a saturating concentration of ATP in the restart buffer1 and by using an overflowing injection volume. It is also possible that the magnetic force applied on the tethered bead (∼1 pN) was in the entropic elasticity range of the dsDNA and thus the tether DNA was not completely stretched to its normal contour length.21 As a result, at beginning of the reactivation, the motor could have packaged more DNA than depicted from the motion of the tethered bead revealing a lower observed packaging rate. When more DNA was packaged, the distance between the bead and the glass approached to the normal contour length of the tether DNA, hence the packaging rate started to decrease.
Figure 3.
(a) DNA tether length against time for two different motors. Bold curves are 500-point moving averages of the light-colored raw data (curves have been shifted on time axis). (b) DNA packaging rate against the amount of DNA packaged relative to the original phi29 DNA length (19.3 kbs). The rates were observed by differentiating the bold curves in (a) and computing 250-point moving averages.
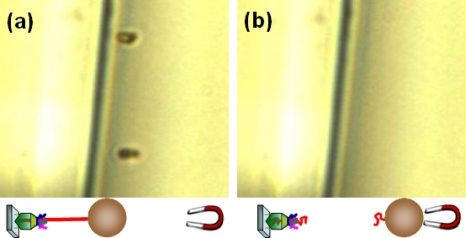
To verify the existence of tether DNA between the magnetic bead and glass wall, a control experiment was performed. The bead-conjugated prepackaged motors were immobilized on the glass wall following the same protocols. After visually confirming the existence of the tethered beads, DNase I (Sigma D4527) was introduced to digest the tether DNA. As expected, the tethered beads were observed to move away upon the application of a magnetic force after the introduction of DNase I, verifying the existence of the tether DNA (Fig. 4).
Figure 4.
Control experiment verifying the existence of tether DNA: the position of tethered beads (a) before and (b) after DNase I introduction.
We demonstrated the bright-field optical observation of the phi29 motor’s DNA packaging event in real time using a magnetomechanical system. Our study shows that the DNA packaging activity of a virus can be visually observed and studied at the single molecular level with a relatively simple system and also visually demonstrates that the phi29 virus can transport a cargo that is four orders of magnitude larger than itself. We anticipate that this system can facilitate the development of robust systems for analyzing mechanical activities of various viruses at the single molecular level.
Acknowledgments
This work was supported by the National Institutes of Health Nanomedicine Development Center: “Phi29 DNA Packaging Motor for Nanomedicine” (Grant PN2EY018230) through the NIH Roadmap for Medical Research.
References
- Chemla Y. R., Aathavan K., Michaelis J., Grimes S., Jardine P. J., Anderson D. L., and Bustamante C., Cell 10.1016/j.cell.2005.06.024 122, 683 (2005). [DOI] [PubMed] [Google Scholar]
- Guo P., Peterson C., and Anderson D., J. Mol. Biol. 10.1016/0022-2836(87)90121-5 197, 229 (1987). [DOI] [PubMed] [Google Scholar]
- Tao Y., Olson N., Xu W., Anderson D., Rossmann M., and Baker T., Cell 10.1016/S0092-8674(00)81773-0 95, 431 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D. E., Tans S. J., Smith S. B., Grimes S., Anderson D. L., and Bustamante C., Nature (London) 10.1038/35099581 413, 748 (2001). [DOI] [PubMed] [Google Scholar]
- Guo P., J. Nanosci. Nanotechnol. 10.1166/jnn.2005.446 5, 1964 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S., Shu D., Simon M. N., and Guo P., Gene 315, 145 (2003). [DOI] [PubMed] [Google Scholar]
- Khaled A., Guo S., Li F., and Guo P., Nano Lett. 10.1021/nl051264s 5, 1797 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickgauer J., Fuller D., Grimes S., Jardine P., Anderson D., and Smith D., Biophys. J. 94, 159 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller D., Rickgauer J., Jardine P., Grimes S., Anderson D., and Smith D., Proc. Natl. Acad. Sci. U.S.A. 10.1073/pnas.0701323104 104, 11245 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugel T., Michaelis J., Hetherington C. L., Jardine P. J., Grimes S., Walter J. M., Falk W., Anderson D. L., and Bustamante C., PLoS Biol. 5, e59 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu D., Zhang H., Jin J., and Guo P., EMBO J. 10.1038/sj.emboj.7601506 26, 527 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sage D., Neumann F., Hediger F., Gasser S., and Unser M., IEEE Trans. Image Process. 10.1109/TIP.2005.852787 14, 1372 (2005). [DOI] [PubMed] [Google Scholar]
- Acharya G., Chang C. L., Doorneweerd D. D., Vlashi E., Henne W. A., Hartmann L. C., Low P. S., and Savran C. A., J. Am. Chem. Soc. 10.1021/ja073094m 129, 15824 (2007). [DOI] [PubMed] [Google Scholar]
- Shu D. and Guo P., Virology 309, 108 (2003). [DOI] [PubMed] [Google Scholar]
- Guo P., Grimes S., and Anderson D., Proc. Natl. Acad. Sci. U.S.A. 10.1073/pnas.83.10.3505 83, 3505 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo P., Rajagopal B., Anderson D., Erickson S., and Lee C., Virology 185, 395 (1991). [DOI] [PubMed] [Google Scholar]
- Guo P., Zhang C., Chen C., Garver K., and Trottier M., Mol. Cell 10.1016/S1097-2765(00)80124-0 2, 149 (1998). [DOI] [PubMed] [Google Scholar]
- Huang L. and Guo P., Virology 312, 449 (2003). [DOI] [PubMed] [Google Scholar]
- Reilly B., Tosi M., and Anderson D., J. Virol. 16, 1010 (1975). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Lee C., and Guo P., Virology 201, 77 (1994). [DOI] [PubMed] [Google Scholar]
- Smith S., Cui Y., and Bustamante C., Science 10.1126/science.274.5288.795 271, 795 (1996). [DOI] [PubMed] [Google Scholar]
- Fisher J., Cummings J., Desai K., Vicci L., Wilde B., Keller K., Weigle C., Bishop G., Taylor R., Davis C., Boucher R., O’Brien E., and Superfine R., Rev. Sci. Instrum. 10.1063/1.1914777 76, 053711 (2005). [DOI] [Google Scholar]
- See EPAPS Document No. E-APPLAB-93-014842 for the calibration of applied magnetic force and two movie files. For more information on EPAPS, see http://www.aip.org/pubservs/epaps.html.