Abstract
Long-term recordings of seasonal sleep patterns in captive white-crowned sparrows (Zonotrichia leucophrys gambelii) have shown that these birds markedly reduce sleep time during the migratory period relative to the non-migratory period. It was also found that, despite this sleep reduction, sparrows showed no evidence of neurobehavioral deficits in a standard operant task used to assess the effects of sleep loss. In this study, we performed an extensive microarray analysis of gene expression in the sparrow telencephalon during the migratory season (M), relative to a 78-hour period of enforced sleep restriction during the non-migratory season (SR), and a 6-hour period of normal wakefulness during the non-migratory season (W). Of the estimated 17,100 transcripts that were reliably detected, only 0.17% changed expression as a function of M (relative to both SR and W), and 0.11% as a function of SR (relative to both M and W). Brain transcripts whose expression increased during M include the facilitated glucose transporter GLUT1, the presenilin associated rhomboid-like protein PARL, and several members of the heat shock protein family, such as HSP70, HSP90, GRP78 and BiP. These data suggest that migration is associated with brain cellular stress and enhanced energetic demands.
Introduction
Although sleep is necessary for survival, its expression often interferes with adaptive waking functions. If wakefulness is prolonged, however, neurobehavioral function is inevitably compromised and adaptive waking function impaired (Van Dongen et al. 2003). The struggle between the need for sleep and the need for vigilant wakefulness may be particularly pronounced in the millions of songbirds who, during spring and fall, engage in long-distance migrations between their wintering and breeding grounds. To accomplish this journey, birds must undergo a marked shift in the typical expression of the sleep-wake cycle. Normally diurnal birds commence long-distance nocturnal flight accompanied by intense diurnal foraging, a pattern of activity which leaves little opportunity for sleep (Berthold 1996; Ramenofsky et al. 2003). Somehow, despite this marked sleep loss, migrants continue to engage in adaptive waking behaviors including complex navigation as well as foraging and predator evasion in novel environments.
In the captive white-crowned sparrow (Zonotrichia leucophrys gambelii), long-term recordings of seasonal sleep patterns demonstrated that sparrows chronically reduced total sleep time by an average of 63% in the migratory period relative to the non-migratory period (Rattenborg et al. 2004). Remarkably, despite this sleep reduction, migrating sparrows showed no obvious evidence of deficits on either the accuracy or quantity of responses in a standard operant task used to assess the effects of sleep loss. In contrast, enforced sleep deprivation of a similar duration during the non-migratory period resulted in significant deficits in operant responding, indicating the clear presence of a neurobehavioral susceptibility to sleep loss in the non-migratory period (Rattenborg et al. 2004).
The preservation of neurobehavioral performance of migratory birds, both in the wild and in captivity, is particularly surprising given that cognitive and performance deficits are some of the most consistent consequences of both sleep deprivation and sleep restriction in mammals (Wimmer et al. 1992; Dinges et al. 1997; Harrison and Horne 1999; Doran et al. 2001; Belenky et al. 2003; Van Dongen et al. 2003). It is possible that, in addition to having evolved a number of behavioral, physiological and morphological adaptations to the demands of migration, (Dawson et al. 1983; Andrews 1995; McWilliams and Karasov 2001; Kullberg et al. 2003; Mettke-Hofmann and Gwinner 2003; Wikelski et al. 2003; Mouritsen and Ritz 2005; Pravosudov et al. 2006) migratory birds have also evolved adaptations to combat the negative consequences of sleep loss. To date, however, evidence of potential mechanisms for sleep loss compensation in migratory birds is absent.
In the current study, we examine the brain molecular correlates of migratory sleeplessness in the white-crowned sparrow in an attempt to identify potential cellular mechanisms that might aid these birds in maintaining prolonged wakefulness without impairment. To accomplish this goal, we performed an extensive microarray analysis of gene expression in the sparrow telencephalon during the migratory period, relative to a 78-hour period of enforced sleep restriction during the non-migratory period, and a 6-hour period of normal wakefulness during the non-migratory period.
Methods
Animals and experimental conditions
White-crowned sparrows were captured on either their wintering grounds in the Sacramento valley in California (lat 39°00′ N, long 122°00′ E) or on their summering grounds in Alaska (lat 64°49′ N, long 147°52′ E), between May 2003 and March 2005. All birds were collected using mist nets under the authority of a United States Fish and Wildlife Service permit. Birds were transported to the University of Wisconsin-Madison where they were individually housed in galvanized wire cages (L: 35 cm × W: 25 cm × H: 32 cm) in environmentally controlled rooms (40% relative humidity), and each bird was in visual and auditory contact with other birds in the room. Birds were fed mixed-seed and provided water ad libitum, and their diet was supplemented daily with lettuce, dried insects, live mealworms and grit. The colony was exposed to photoperiodic conditions designed to simulate seasonal lighting changes. Diurnal illuminance level was maintained at 540–640 lux measured at the level of the cage floor. Illuminance during the dark phase was less than 0.5 lux. Birds collected for the migration condition were exposed to 12.5L/11.5D (fall photoperiod) for the week prior to, and including, the day of sacrifice. Birds collected for the sleep restriction (SR) and waking (W) conditions were exposed to 10:5L/13:5D (winter photoperiod) for the week prior to, and including, the day of sacrifice. All birds were sacrificed 6 hours after lights-on.
The sleep restriction period began at lights-on and continued for 78 hours. In an effort to mimic the pattern often seen during migratory sleeplessness, sleep in the SR group was permitted for the first 3 h of the dark phase on the three consecutive nights of the restriction protocol. Birds collected for the W condition were sacrificed in the middle of the day after a period of 6 hours of normal wakefulness.
Since male and female white-crowned sparrows are not morphologically distinct, gender matching of experimental groups was not possible. Gender determination for each group was made after death as follows: (M=10; 5 male, 5 female; SR=10, 7 male, 3 female; W=10; W 6 male, 4 female). Animal protocols followed the National Institutes of Health Guide for the Care and Use of Laboratory Animals and were in accordance with institutional guidelines.
Activity monitoring
Bird activity and behavior was recorded using infrared-sensitive motion detection cameras (one per bird) connected to a digital video storage system (Salient Systems Corp., www.salientsys.com). Infrared illuminators provided lighting for the cameras during the dark phase. Camera sensitivity was individually set to detect any motion beyond that of normal respiration, as described in (Jones et al. 2007).
Sleep restriction procedure
Birds were never handled to induce wakefulness; walking past the cages was sufficient to maintain vigilant wakefulness for the entire 78-hour sleep restriction period, although later in the experiment cage tapping was sometimes required. To diminish the effects of stress, birds were acclimated to this procedure for 30 minutes a day during the week preceding the experimental day. This sleep restriction paradigm has been previously shown to lead to a reduction in the accuracy (percentage correct responses) on both the acquisition and performance components of an operant task used to assess the effects of sleep loss. These effects of sleep restriction were evident following the first night of sleep loss and persisted following subsequent nights of sleep restriction (Rattenborg et al. 2004).
RNA extraction
Birds were deeply anesthetized with isoflurane and decapitated within 1 min from the onset of anesthesia. The head was cooled in liquid nitrogen and the whole brain was removed and dissected. Samples were immediately frozen on dry ice and stored at -80°C. Total RNA was isolated from right telencephalon using Trizol (GIBCO-BRL, Gaithersburg, MD) according to the manufacturer’s instructions. RNA concentration and purity were measured by spectrophotometry. RNA concentration was assessed by measuring the absorbance at 260 nm (A260). RNA purity was determined by measuring the ratio of absorbance at 260 nm and 280 nm (A260/A280). To ensure accurate values, and to increase sensitivity to protein contamination, RNA was buffered in 10 mM Tris·Cl (pH 7.5).”
Affymetrix microarray: labeling, hybridization and data analysis
Microarray labeling, hybridization, and expression analysis were performed according to the Affymetrix GeneChip Expression Analysis Technical Manual (Affymetrix Inc., Santa Clara, CA) and essentially as previously described (Cirelli et al. 2004). Briefly, an equal mass amount of total RNA from the right telencephalon of 6 sparrows within each experimental group (M, SR, W; 20 μg/pool) was pooled, converted into first-strand cDNA using Superscript II Rnase H- reverse transcriptase (Invitrogen Life Technologies, Carlsbad, CA), and the second strand was synthesized, all according to the Affymetrix manual. The telencephalon was chosen because it contains structures functionally similar to the mammalian cerebral cortex, and cortical regions are thought to be responsible for many of the behavioral and cognitive deficits accompanying sleep loss in mammals (Horne 1985; Durmer and Dinges 2005).
The cDNA was then converted to biotinylated cRNA using the ENZO BioArray High Yield In Vitro Transcription kit (Enzo Life Sciences, Farmingdale, NY) according to the manufacturer’s instructions. The cRNA was fragmented at 0.5 μg/μl final concentration in 1X fragmentation buffer (40 mM Tris-acetate, pH 8.1, 100 mM potassium acetate, 30 mM magnesium acetate). The size range of cRNA before (0.5 kb and longer) and after (35-200 base fragments) fragmentation was checked by denaturing agarose electrophoresis.
The hybridization reaction and the automated hybridization procedure were performed according to the Affymetrix manual. Each sample was hybridized to an Affymetrix Chicken GeneChip (38535 probe sets) using 4 arrays per experimental condition. Quality of the cDNA and cRNA syntheses was determined by the 3’/5’ ratio of housekeeping genes within the array. Expression measures for each probe set were calculated using the robust multi-array average gcRMA measure advocated by (Katz et al. 2006) and input into GeneSpring 7.2 software (Silicon Genetics, Redwood City, CA, USA) for further analysis. The original data set, which contained 38,535 probe pairs, was scrubbed to retain only those (6,585) whose absolute, non-normalized values were higher than 15 in half of chips. Statistical significance was determined by condition-to-condition comparisons requiring a significance level of 95% or greater (parametric test, variances not assumed equal, Welch t-test). A migration-related transcript was required to increase or decrease its expression in M birds relative to both SR and W birds. Similarly, a sleep-restriction-related transcript was required to change its expression in SR birds relative to both W and M birds. Genes exhibiting 1.2-fold or greater expression and a probability t-test P value of less than 0.05 are reported.
Songbird microarray: production, labeling, hybridization and data analysis
Songbird cDNA microarrays were produced at the Keck Center for Comparative and Functional Genomics at the University of Illinois as part of a multi-institutional collaboration to develop genomic tools for songbird research. A cDNA library containing >34,000 independent cDNA clones (single reads, from the 5’ end of the insert) was constructed from polyadenylated RNA pooled from the telencephalon of zebra finches (Taeniopygia guttatta) of both sexes, ranging in age from embryonic day 30 up to adulthood. These ESTs, which appear to represent the products of ~16,000 non-redundant genes, were amplified by PCR, spotted on Corning GAPS II amine coated slides using a GeneMachines Oninigrid 100 printer with Telechem Chipmaker 2 pins (Genomic Solutions, Inc.). Each array was arranged in 48 blocks with features spotted in a 20 × 21 pattern for a total of 20,160 features. Each block within the array contained several positive-control genes, including those coding for GAPDH, ß-actin and Histone H3. Additional control DNAs on each array include 3 negative-control soybean genes not present in the songbird genome. Complete information on the songbird initiative and array production can be found at (http://titan.biotec.uiuc.edu/songbird/resources/).
Microarray labeling and hybridization was performed using the Pronto Plus Indirect Systems by Promega. Briefly, an equal mass amount of total RNA from the right telecephalon of 6 sparrows within each experimental group (SR, W, M; 5 μg/pool; same animals used for the Affymetrix microarrays) was pooled, and reverse transcribed to cDNA in the presence of Aminoallyl-dNTP mix by ChipShot Reverse Transcriptase. Labeled aminoallyl cDNA samples were purified using a ChipShot Membrane Column. The purified aminoallyl cDNA was added to one dried aliquot of CyDye (either Cy3 or Cy5; Amersham) for conjugation. The CyDye labeled cDNA was purified using the ChipShot Membrane Column and eluted using manufacturers Elution Buffer. To determine quality and concentration of CyDye labeled cDNA quantification at absorbances of 260, 550, 650 nm were taken and frequency of incorporation was determined. Probes were then combined for each desired comparison, dried by Speed-Vac (Thermo Savant) and resuspended in Pronto Hybridization solution and denatured at 95°C for 5 minutes. The resulting samples were hybridized onto microarray slides using a dye-swap reference design as described in (Churchill 2002) for 14-20 hours at 42°C.
Twelve microarrays were used; four replicates in each experimental condition. Two samples in each condition were labeled with Cy3 and two labeled with Cy5. To control for specific dye bias, each sample was co-hybridized with a reference sample labeled in the opposite dye orientation. The reference sample was generated from a pool of equal mass volumes of RNA from all experimental animals. Hybridization reactions were carried out in Corning hybridization chambers at 42°C overnight. Slides were then washed using solutions provided by the Pronto Plus Indirect System. Slides were scanned immediately after drying using an Axon GenePix 4200A scanner (Axon Instruments). PhotoMultiplier Tube (PMT) settings were adjusted manually to achieve good signal intensities for the majority of spots and to minimize the number of spots on the array with saturated signal values. Prior to normalization, quality confidence measurements (spot diameter, spot area, array footprint, spot circularity, signal-to-noise ratio, spot uniformity, background uniformity, and replicate uniformity) were calculated using GenePix Pro Version 3.0 software (Axon Instruments).
The data obtained from the scanned array images were analyzed with Genespring 7.3 software (Agilent Technologies). Background-subtracted, log-transformed median values were subjected to intensity-dependent LOWESS (locally weighted scatterplot smoother) normalization, in which 20% of the data were used for smoothing, as advocated by Yang (Yang and Speed 2002). The Genespring cross-gene error model was used to obtain a more accurate estimate of the measurement precision of a given gene/EST by combining measurement variation and between-sample variation. Statistical significance was determined by condition-to-condition comparisons requiring a significance level of 95% or greater (parametric test, variances not assumed equal, Welch t-test). As for the analysis of the Affymetrix microarrays, a migration-related transcript was required to change its expression in M birds relative to both SR and W birds. Similarly, a sleep restriction-related transcript was required to change its expression in SR birds relative to both M and W birds. Genes exhibiting 1.2-fold or greater expression and a probability t-test P value of less than 0.05 are reported. Microarray data generated in this study meet Minimum Information about Microarray Experiment (MIAME) standards. All data are available upon request.
Real time quantitative PCR (qPCR)
Attempts were made to confirm with qPCR all known transcripts identified as differentially expressed using microarrays. Attempts were not made to confirm the unknown sequences or ESTs. Primers for qPCR were designed based on sequences obtained from either the chicken or the zebra-finch microarray. Five identified genes of interest were not confirmed due to primer design (amplification) problems. For the qPCR, a 20% change indicates a 1.20 fold increase or decrease in gene expression between experimental conditions. All of the qPCR reactions performed are listed in the tables, although a fold change of less than 20% is not considered a confirmation of gene expression change.
Biological verification of microarray results was obtained by real-time qPCR using pooled RNA from 4 additional birds (not previously used for microarray analysis) for each of the three experimental groups (M, SR, W), PCR was performed essentially as described in (Cirelli et al. 2004) with small modifications. Briefly, reverse transcription reactions were carried out in parallel on DNase I digested pooled total RNA from SR, W and M sparrows and cleaned using the Qiagen RNeasy Kit RNA Cleanup protocol. Reverse transcription (RT) reactions were as follows: 50 ng total RNA, 6.25 μl oligo dT16 (100 μM), and H2O to 28.75 μl total. Samples were incubated at 70°C for 10 min, put briefly on ice, and then incubated at 42°C for 2-5 min. Mix #2 (10 μl of 5X Superscript III First Strand Buffer, 5 μl of 0.1 M DTT, 5 μl dNTPmix (10mM each dNTP), and 1.25 μl Superscript III RNAse H- Reverse Transcriptase) was added (21.25 μl for each RT reaction), mixed, and samples were immediately returned to incubate at 42°C for 1 hour. Reactions were stopped by incubation at 70°C for 15 min. PCR reactions to measure levels of sparrow GAPDH cDNA were done to confirm uniformity of reverse transcription within and between sample groups and concentration differences adjusted. Each PCR reaction contained specific forward and reverse primers (200-750nM final concentration), 2X SYBR Green Master Mix, reverse transcription product in a concentration previously determined, and H2O up to a total volume of 25 μl per well. A two step PCR profile was used: 10 min at 95°C denaturation, followed by 40 cycles alternating between 95°C for 15 sec and 60°C for 60 sec. Dilution series (1:10, 1:50, 1:250, 1:1250, 1:6250) standard curves were performed in quadruplicate for each primer pair using reverse transcription products pooled from all conditions. PCR was done in quintuplicate for each sample condition and relative quantities determined based on the equation of the line of best fit derived from the standard curve and normalized to sparrow GAPDH concentrations. GAPDH was selected as the housekeeping gene based on the consistency of the results obtained with both array platforms across all experimental conditions (mRNA levels did not change across conditions by > 6%).
Results
Activity analysis
Activity and behavior recorded using infrared-sensitive motion detection cameras demonstrate that the typical patterns of seasonal changes in migratory restlessness were reliably reproduced in the laboratory setting. Figure 1 shows one representative day and night of baseline activity during the non-migratory period (winter photoperiod) in 6 birds, as well as a representative period of 78 hours of migratory activity (fall photoperiod). A dramatic increase in nocturnal wakefulness is evident during the migratory season, when these birds would normally be migrating between Alaska and California. Based on motion detection analysis, the percentage of time spent in active wakefulness for the M birds during the entire 78-hour period shown in Figure 1 is 40.2 ± 1.5% (mean±SEM, percentage of total recording time, N=6). When considering activity during the dark and light periods separately, M birds were active an average (mean±SEM) of 45.0 ± 1.7% of the dark period on the 3 recording nights, and an average of 35.8 ± 2.2% of the light period (excluding the 6 hour period preceding sacrifice). Figure 1 also shows one baseline day and night of activity for SR birds followed by the 78-hour sleep restriction period. SR birds were active an average (mean±SEM) of 59.0 ± 3.1% of the entire 78 h period (59.4 ± 3.5% of the dark period and 62.3 ± 2.9% of the light period). Sparrows in the spontaneously wakeful condition (Figure 1) were sacrificed at CT6 after 6 hours of normal diurnal activity. The percentage of time spent active during the last 6 hours before sacrifice for all experimental groups was as follows (mean±SEM, percentage of total recording time, N=6 per condition): SR birds=52.6 ± 6.1; M birds=30.2 ± 3.7; W birds=68.0 ± 2.3. The values above refer to the 6 birds per group used for the array analysis. The additional 4 birds per group used for subsequent PCR confirmation showed similar percentages of activity (data not shown).
Figure 1.
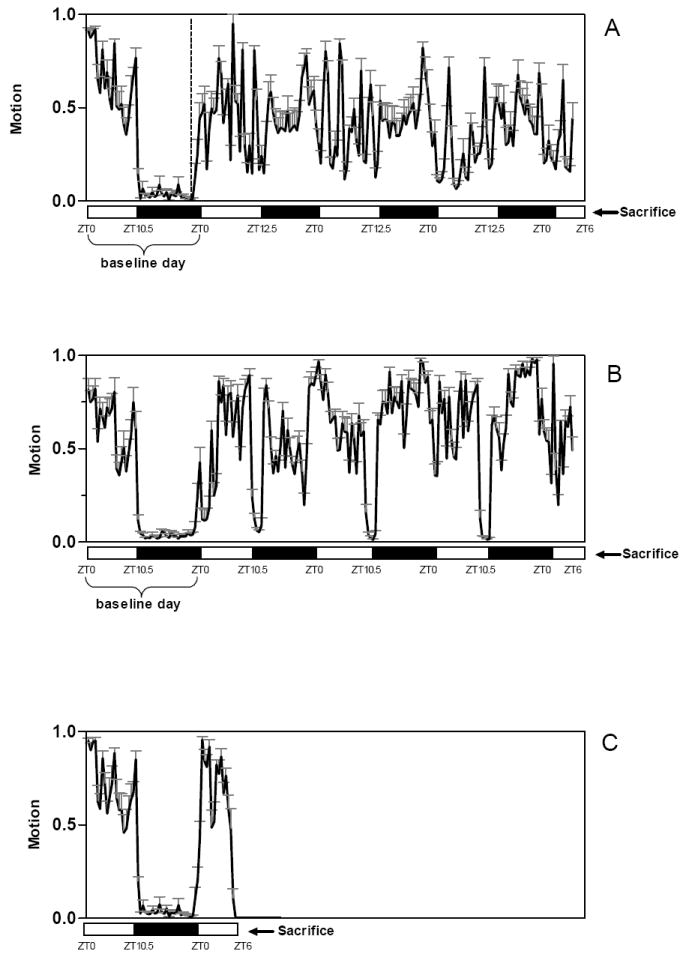
The 3 experimental groups selected to identify gene expression changes associated with migration or sleep restriction. Percent of activity in 30-min bins measured by infrared-sensitive motion detection cameras is shown for all birds. (M) A 24-hour baseline period during the winter photoperiod for the set of migratory birds, and a 78-hour period of migration during the subsequent fall photoperiod. The dashed vertical line indicates that baseline period and migration period represent non-continuous recordings. Note that only 78 hours of migratory activity are shown; however, birds sacrificed for the migratory condition had all been in the migratory state for at least one week before sacrifice. (SR) A 24-hour baseline day, immediately followed by the 78-hour sleep restriction period. (W) A 24-hour baseline day, immediately followed by 6 hours of spontaneous wakefulness (activity data previously published in (Jones et al. 2007)). All birds were sacrificed at ZT6. Values are shown for the 6 birds used for microarray analysis. The additional 4 birds per group used for subsequent qPCR confirmation showed similar percentages of activity (data not shown). White and black bars indicate the light and dark phase, respectively.
It should be pointed out, however, that as demonstrated in a previous study, this activity monitoring fails to adequately quantify wakefulness that is inactive, and thus tends to overestimate sleep and underestimate wakefulness (Jones et al. 2007). Indeed, SR birds, which were kept awake by the experimenter for the entire 6-hour period before death, and therefore awake ~100% of the time, were recorded as actively moving only ~52% of the time, and would thus have been incorrectly scored as asleep for ~48% of the time.
Thus, all birds were sacrificed in the same behavioral state (awake), and at the same circadian time. The inclusion of the spontaneously wakeful group allows us to restrict our search of differentially expressed transcripts to those that are related to migration or sleep restriction rather than to the stress of handling, since spontaneously wakeful birds were not subjected to enforced sleep restriction. It should be noted, however, that this protocol cannot identify genes whose expression is modulated by multiple factors (e.g light + circadian time) or by interactions between factors.
Identification of differentially expressed transcripts and validation of results through qPCR
A microarray platform containing transcripts of the genome of the white-crowned sparrow is unavailable. As a result, we chose to perform two cross-species hybridizations (CSH) in which RNA of the white-crowned sparrow brain was hybridized to two distinct microarray platforms—one containing transcripts from the domestic chicken (Gallus gallus) and the other containing transcripts from the zebra finch (Taeniopygia guttatta). Biologically meaningful information has been obtained using CSH in previous studies (Adjaye et al. 2004; Shah et al. 2004; Nowrousian et al. 2005). In our first attempt at CSH we used the Affymetrix chicken microarrays. However, recent data suggest that cDNA microarrays containing longer oligonucleotides are more suitable for CSH since the long cDNA probes appear to effectively negate small inter-species differences in nucleotide sequences (Chalmers et al. 2005; Bar-Or et al. 2006; Bar-Or et al. 2007). Therefore, in our second CSH, we used custom cDNA microarrays containing transcripts of genes from the zebra finch telencephalon.
Using two microarray platforms containing the genomes of two avian species closely related to the white-crowned sparrow, a total of 29 transcripts were identified as differentially expressed during M (relative to both SR and W), including 21 transcripts with higher expression in M and 8 transcripts with lower expression in M (see Table 1). Of the migration-related transcripts, 22 were identified with the zebra finch array and 7 with the chicken microarray (1 was identified in both arrays). A total of 19 transcripts were identified as differentially expressed during SR (relative to both M and W), including 10 with higher expression in SR and 9 with lower expression in SR (see Table 2). Of these transcripts, 15 were identified using the zebra finch microarray and 6 using the chicken microarray (2 were identified in both arrays). The criteria used to identify migration-related transcripts were strict. First, for both microarrays, transcripts with very low expression level were removed from the analysis. Specifically, on the chicken array, those probe sets whose absolute signal value was below 15 in half of the chips were excluded (6585 out of 38535). In the zebra finch arrays, signal hybridization was defined as fluorescence at least one standard deviation above the background level calculated by GenePix from the regions immediately surrounding each cDNA. Second, the expression difference level between M and SR + W birds (for migration-dependent transcripts) and between SR and W + M birds (for sleep-restriction-dependent transcripts) was required to change by >1.2 fold, because 20% was our detection limit to confirm mRNA changes using qPCR. Third, the expression difference level between transcripts was required to be statistically significant at (p < 0.05).
Table 1. Transcripts differentially expressed in migration relative to sleep restriction and spontaneous wakefulness.
White-crowned sparrow telencephalon genes whose mRNA levels are changed during migration (M) relative to 78 hours of sleep restriction (SR) and 6 hours spontaneous wakefulness (W). (a) transcripts increased (>1.2 fold change, Welch t-test, p<0.05) (b) transcripts decreased (<1.2 fold change, Welch t-test, p<0.05) “% change (array)”, percentage change in M relative to SR and W as indicated by array analysis. “% change (PCR)” indicates transcripts confirmed using qPCR from pooled RNA of 4 additional birds not previously used for microarray analysis
| A. Transcripts with higher expression in migration relative to sleep restriction and spontaneous wakefulness | ||||
|---|---|---|---|---|
| GenBank | Gene description | Gene symbol | % change (array) | % change (PCR) |
| HEAT SHOCK PROTEINS/MOLECULAR CHAPERONES | ||||
| NM_204289 | heat shock protein 90kDa beta (Grp94), member 1 | GRP94 | 1.39;1.27 | 23.2;27.1 |
| NM_204289 | heat shock protein 90kDa beta (Grp94), member 1 | GRP94 | 1.38;1.30 | |
| X07265 | 90kDa heat shock protein class A member 1 (cytosolic) | HSP90AA1 | 1.38;1.39 | 115;62.2 |
| X07265 | 90kDa heat shock protein class A member 1 (cytosolic) | HSP90AA1 | 1.48;1.45 | |
| NM_205003 | heat shock 70kDa protein 8 | BiP | 1.29;1.24 | 28.3;26.1 |
| NM_205003 | heat shock 70kDa protein 8 (HSPA8) | BiP | 1.26;1.47 | |
| EF175165 | heat shock 70kDa (glucose-regulated protein, 78kDa) | HSPA5 | 1.34;1.33 | |
| ENERGY METABOLISM | ||||
| NM_205209 | solute carrier family 2 (facilitated glucose transporter), member 1 | GLUT1 | 1.42;1.46 | 25.8;32.6 |
| MISCELLANEOUS | ||||
| DQ213979 | presenilin associated rhomboid-like (Taeniopygia guttata) | PARL | 1.39;1.25 | 36.9; 105.1 |
| B. Transcripts with lower expression in migration relative to sleep restriction and spontaneous wakefulness | ||||
| GenBank | Gene description | Gene Symbol | % change array | % change PCR |
| MISCELLANEOUS | ||||
| EF191661 | transthyretin (Taeniopygia guttata) | TTR | 1.74;1.71 | 61.3;52.2 |
| EF191661 | transthyretin (Taeniopygia guttata) | TTR | 1.47;1.57 | |
| EF191661 | transthyretin (Taeniopygia guttata) | TTR | 1.47;1.45 | |
| XM_001234688 | heat-responsive protein 12 | HRSP12 | 1.31;1.36 | 33.5;50.7 |
| NM_001093343 | yippee-like 1 | YPEL1 | 1.62;1.52 | 48.8;9.56 |
| XM_418786 | copine IV | CPNE4 | 1.59;1.53 | |
| XM_425011 | solute carrier family 1 glial high affinity glutamate transporter member 3 | SLC1A3 | 1.40;1.20 | 46.6;24.9 |
Table 2. Transcripts differentially expressed in sleep restriction relative to migration and spontaneous wakefulness.
White-crowned sparrow telencephalon genes whose mRNA levels are changed in 78 hours of sleep restriction (SR) relative to migration (M) and 6 hours of spontaneous wakefulness (W). (a) transcripts increased (>1.2 fold change, Welch t-test, p<0.05) (b) transcripts decreased (<1.2 fold change, Welch t-test, p<0.05) “% change (array)”, percentage change in SR relative to M and W as indicated by array analysis. “% change (PCR)” indicates transcripts confirmed using qPCR from pooled RNA of 4 additional birds not previously used for microarray analysis.
| A. Transcripts with higher expression in sleep restriction relative to migration and spontaneous wakefulness | ||||
|---|---|---|---|---|
| Genbank | Gene description | Gene symbol | % Change (array) | % Change (PCR) |
| IMMUNE FUNCTION | ||||
| DQ214165 | immunoglobulin lambda light chain | IGLL | 1.49;1.25 | 54.1;40.6 |
| DQ214165 | immunoglobulin lambda light chain | IGLL | 1.36;1.61 | |
| MISCELLANEOUS | ||||
| NM_001031050 | NIMA (never in mitosis gene a)-related kinase 2 | 1.21;1.34 | ||
| NM_204232 | HIR histone cell cycle regulation defective homolog A | HIRA | 1.78;1.76 | 67.9;23.1 |
| NM_001012824 | mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A | MGAT4A | 1.33;1.24 | |
| XM_420329 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1 | SMARCA1 | 1.46;1.57 | |
| XM_001231767 | ATPase, Ca++ transporting, plasma membrane 2 | ATP2B2 | 1.40;1.31 | 9.3;0.12 |
| B. Transcripts with lower expression in sleep restriction relative to migration and spontaneous wakefulness | ||||
| GenBank | Gene description | Gene symbol | % Change (array) | % Change (PCR) |
| ENZYMES | ||||
| DQ214020 | Glutathione peroxidase (Taeniopygia guttata) | 1.47;1.54 | 53.9;12.02 | |
| NM_204259 | prostaglandin D2 synthase 21kDa | PTGDS | 1.30;1.21 | 29.06;31.5 |
| TRANSCRIPTION FACTORS | ||||
| XM_417039 | general transcription factor IIF | GTF2F2 | 1.38;1.40 | |
| MISCELLANEOUS | ||||
| NM_001004376 | hemoglobin, alpha 1 | HBA1 | 1.45;1.34 | |
| NM_001004375 | hemoglobin, alpha 2 | HBA2 | 1.38;1.29 | 22.8;40.4 |
| NM_001004375 | hemoglobin, alpha 2 | HBA2 | 1.22;1.32 | |
| NM_001004376 | hemoglobin, alpha 1 | HBA1 | 1.52;1.67 | |
| NM_001004376 | hemoglobin, alpha 1 | HBA1 | 1.32;1.25 | |
| XM_419213 | proenkephalin | PENK | 1.21;1.25 | |
We used two distinct microarray platforms—an Affymetrix array containing short oligomer (~25-mer) probes and a custom-constructed cDNA array containing probes up to several hundred nucleotides in length. Although the percent of hybridization of the white-crowned sparrow mRNA to the zebra finch array was greater than the hybridization of the white-crowned sparrow mRNA to the chicken array, (mean±SEM, percent hybridization, chicken array=17.1 ± 0.19; zebra finch array=85 ± .58), there were some consistencies between arrays. Specifically, of the 11 known transcripts identified as migration-related on the zebra finch array, 1 was also identified as migration-related on the chicken array. Similarly, of the 7 known transcripts identified as sleep restriction-related on the zebra finch array, 2 were also identified as sleep restriction-related on the chicken finch array.
The differential expression of a large subset of transcripts was confirmed with real-time qPCR using mRNA from independent groups of M, SR and W birds (biological validation). Initial attempts were made to confirm with qPCR all known transcripts identified as differentially expressed by the microarray analysis. For several sequences, however, our repeated attempts to design effective primers were unsuccessful. The transcripts that remained unconfirmed for technical reasons appear therefore in the Tables without PCR data. Confirmed migration-related transcripts included those coding for the “heat shock protein 90kDa beta (Grp94), member 1”, “90kDa heat shock protein class A member 1 (cytosolic)”, “heat shock 70kDa protein 8 (BiP)”, “heat-responsive protein 12”, “solute carrier family 2 (facilitated glucose transporter), member 1 (GLUT1)”, “presenilin associated rhomboid-like (PARL)”, and “transthyretin (TTR)”. Sleep restriction-related changes were confirmed in transcripts coding for “prostaglandin D2 synthase”, “hemoglobin, alpha 2”, “immunoglobulin lambda light chain”, and “HIR histone cell cycle regulation defective homolog A”. Attempts were also made to confirm the sleep restriction-related transcripts glutathione peroxidase (down in SR) and ATPase, Ca++ transporting, plasma membrane 2 (up in SR). Although the qPCR results were in the predicted direction, they did not meet our criteria of change >20% (“glutathione peroxidase”, 54% M > SR, 12% W > SR; “ATPase, Ca++ transporting, plasma membrane 2” 9.3% M > SR, 0.12% W > SR).
Discussion
In the present experiment we sought to identify brain cellular responses associated specifically with migratory sleeplessness by analyzing gene expression differences between three experimental groups: sleep restricted birds (winter photoperiod), spontaneously wakeful birds (winter photoperiod) and migratory birds (fall photoperiod). Since migration is a complex phenotype involving not only sleeplessness but a host of other physiological adaptations, comparisons between these three conditions cannot fully capture all of the changes that must occur during this state. However, our experimental design did lend itself to the identification of gene expression changes associated with migratory versus enforced sleeplessness. Perhaps the most surprising outcome of this study is that the marked loss of sleep in migrating birds is accompanied by relatively modest reprogramming of telencephalic gene expression. Specifically, of the estimated 17,100 transcripts for which we achieved consistent hybridization, only 0.17% changed expression as a function of migration and 0.11% as a function of sleep restriction.
Transcripts with higher expression during the migratory state include four functionally related members of the heat shock protein family, two cytosolic heat shock proteins, HSP70 and HSP90, as well as two molecular chaperones of the endoplasmic reticulum BiP (GRP78) and GRP94. The upregulation of heat shock proteins and molecular chaperones is part of the unfolded protein response (UPR), which occurs following any perturbations of endoplasmic reticulum function (ER), such as alterations in Ca++ homeostasis, hypoxia, glucose deprivation or heat shock. The induction of these genes, therefore, represents an important pro-survival component of the UPR (Schroder and Kaufman 2005).
GRP78 (Bip) is one of the best-characterized ER chaperone proteins, and has served as a classical marker for UPR activation. The induction of GRP78 is required to alleviate ER stress, maintain ER function and facilitate protein folding. Several studies suggest that BiP may also protect the cell against cell death by suppressing the accumulation of reactive oxygen species and by stabilizing mitochondrial function (Liu et al. 1998; Lee et al. 1999; Yu et al. 1999). BiP has been demonstrated to increase following a few hours of sleep loss in the cerebral cortex of mice (Terao et al. 2003; Naidoo et al. 2005; Mackiewicz et al. 2007) rats (Cirelli et al. 2004; Terao et al. 2006), in the head of fruit flies (Shaw et al. 2000; Naidoo et al. 2007), as well as in the telencephalon of the white-crowned sparrow (Jones et al. 2007). However, its expression is not further increased in the rat cerebral cortex after long-term sleep deprivation relative to short-term sleep loss (Cirelli et al. 2006). A specific migration-associated increase in the expression of BiP and other chaperones provides presumptive evidence that the telencephalon may undergo ER stress during migration. Thus, given that various lines of evidence indicate that BiP has a neuroprotective role against excitotoxicity and apoptosis during the UPR, it is possible that its induction represents a mechanism by which the sparrow brain protects itself from the consequences of stress associated with migration.
Another stress-related transcript upregulated during migration codes for a rhomboid protease of the inner mitochondrial membrane, presenilin-associated rhomboid protease (PARL), originally named for its interaction with presenilin and its role in the processing of amyloid beta precursor protein (Pellegrini et al. 2001). Recent data, however, indicates that PARL plays an important regulatory role in apoptosis. PARL knockout mice undergo progressive multi-tissue atrophy, including atrophy in the thalamus and striatum, mediated by increased apoptosis (Cipolat et al. 2006). The primary regulatory step for mitochondrial-mediated apoptosis (the intrinsic pathway) requires disruption of the mitochondrial membrane followed by the release of cytochrome c from the mitochondrial intermembrane space to the cytoplasm (Danial and Korsmeyer 2004). PARL has been shown to protect cells from apoptosis through its role in preventing the mitochondrial remodeling that leads to cytochrome c release from the intermembrane space of the mitochondria (Jeyaraju et al. 2006; Pellegrini and Scorrano 2007). Interestingly, ER stress associated with the UPR is one of the signals that can lead to early release of Ca2+ from the ER, uptake of Ca2+ into mitochondria; and induction of cytochrome c release, caspase activation and apoptosis (Rao et al. 2004).
We also found in migration an increased expression of the main glucose transporter GLUT1. Increases or decreases in GLUT1 expression have been shown to correlate with increases or decreases in cerebral glucose utilization, respectively, suggesting that glucose transport is a self-limiting step for glucose use in the brain (Barros et al. 2005). The brain relies almost exclusively on glucose as an energy substrate (Magistretti 2006), and the bulk of ATP generated in brain cells depends on the oxidative phosphorylation pathway, with facilitative glucose uptake into cells occurring via glucose transporters such as GLUT1. GLUT 1 has been previously demonstrated to increase in both the rodent cortex and the sparrow telencephalon during waking and short-term sleep deprivation relative to sleep (Cirelli et al. 2004; Jones et al. 2007). Thus, GLUT1 increase during migration suggests that brain metabolism may be still higher in migration relative to waking and enforced sleep restriction.
A total of 5 known transcripts were expressed at lower levels during migration relative to sleep-restriction and spontaneous wakefulness. They do not seem to cluster, however, in any major functional category, and the significance of several other identified transcripts is unknown. Three separate transcripts coding for transthyretin (TTR), the cerebrospinal carrier of the thyroid hormone thyroxine and retinol, were expressed at lower levels in migration. TTR is abundantly synthesized and secreted by the choroid plexus of reptiles, birds and mammals. It is the major carrier of retinol bound to retinol binding protein from liver stores through plasma, and across the choroid plexus to target tissues in the brain (Richardson 2007). Retinoic signaling has been implicated in the regulation of the slow wave activity during non rapid-eye movement sleep (Maret et al. 2005; Kitaoka et al. 2007), but a possible role for TTR remains unclear.
We also identified 12 known sleep restriction-related transcripts. Some of them have been previously identified as changing during mammalian sleep deprivation, and others have been implicated in the regulation of the mammalian sleep-wake cycle. The transcript for prostaglandin D2 synthase (PGDS), for example, is expressed at lower levels during sleep restriction. PGDS is an enzyme necessary for the production of prostaglandin D2 (PGD2), a substance with somnogenic activity in mammals (Huang et al. 2007). The infusion of selective inhibitors of PGDS reduces sleep amounts in rats, concomitant with a reduction of PGD2 content in the brain (Qu et al. 2006), and serum PGDS concentrations are reduced during both enforced sleep deprivation as well as during excessive daytime sleepiness in humans (Jordan et al. 2004; Bassetti et al. 2006). Thus, the differential expression of PGDS during sleep restriction in the sparrow suggests that prostaglandin D2 may play a similar role in avian sleep regulation.
The transcripts for hemoglobin alpha 1 and alpha 2 were also expressed at lower levels during sleep restriction. Hemoglobin reversibly binds oxygen, and is involved in the metabolism and transport of nitric oxide. In the mouse suprachiasmatic nucleus, mRNA levels of both α and ß chains demonstrate robust circadian expression (Ben-Shlomo et al. 2005). In rat cerebral cortex, the mRNA levels of the same genes increase after both spontaneous wakefulness (at night) and short-term sleep deprivation (during the day), suggesting that behavioral state per se, independent of time of day, can induce hemoglobin expression (Cirelli et al. 2004). Hemoglobin expression increases, rather than decreases, in rat cerebral cortex following long-term sleep deprivation (Cirelli et al. 2006). The reason for the difference between rodents and birds is unclear, although an increased expression during long-term deprivation in the rodent cortex could be related to the role of heme proteins as extracellular scavengers of free radicals, and may thus play a role in the cellular stress response.
Finally, we identified a sleep restriction-related increase of immunoglobulin lambda light chain, a transcript also expressed at high levels in the rat cortex following long-term sleep deprivation (Cirelli et al. 2006). Increased expression of this transcript during sleep restriction may indicate an alteration in immune function. Indeed, in addition to the upregulation of several transcripts coding for immunoglobulin molecules during long-term deprivation in the rodent cortex, rats deprived of sleep for a period of 2 to 3 weeks show induction of pro-inflammatory cytokines and increased production of serum immunoglobulins. Ultimately these long-term deprived rats develop septicemia, suggesting that host defenses are compromised by long-term sleep loss (Everson 2005). However, whether the upregulation of immune-related genes following sleep deprivation is related to sleep loss per se, or to the stress associated with the mechanical stimulation used to enforce sleep loss, is still unclear. In the fruit fly brain, the expression of many genes involved in the innate immune response increases following stimulation during a period when the animals are predominantly awake. This increased expression is equal to or greater than the response to stimulation during a period when the animals would be normally sleeping, suggesting a role for the immune system in the brain’s response to mechanical stress (Zimmerman et al. 2006).
Study limitations
This study has several limitations. First, the sleep and activity patterns of birds in the migratory state were recorded in captivity, where migratory restlessness manifests itself in behaviors such as hopping and wing flapping. During migratory flight in the wild, overall motor activity is certainly increased relative to what we observe in caged birds. Although it is unclear to what extent motor activity per se would influence brain gene expression, sleep patterns in wild migrating birds may be quite different from those of captive birds. For example, it is conceivable that SWS could occur either unihemispherically or bihemispherically during migratory flight. Such differences in sleep patterns may result in marked differences in brain gene expression between captive and wild birds, and naturalistic studies are necessary to assess this possibility.
A second important limitation of this study relates to the fact that our experimental groups are not perfectly matched in terms of the duration of sleep loss. Specifically, while the birds in the migratory state had been migrating for an average of 5 days (± 1d), the sleep-restriction protocol lasted for ~3 days (78 hours), and this difference may have influenced our results. However, although migration can represent a highly consolidated state, birds can also migrate for one or two nights and then take a nocturnal period of rest. This pattern made the precise matching of the duration of sleep loss between the two experimental groups difficult. At the end, we selected birds with comparable amount of sleep loss during the last 78 hours, because we assumed that mRNA levels would not be significantly influenced by behavioral state changes that occurred more than 3 days before sacrifice.
Third, we used microarrays from two avian species related to the one under study. Since hybridization is suboptimal because of inter-species gene sequences differences, such an approach necessarily suffers from a loss of sensitivity and coverage of the transcriptome. The difference between our target species and the species used to construct the array cannot, however, account for the relatively low number of transcripts identified as either migration or sleep restriction-related. Using the same two microarray platforms, and the same number of experimental animals, we recently found that a significant number of transcripts (255, 1.4% of 17,100) change their expression as a function of sleep or wakefulness in the telencephalon of the white-crowned sparrow (Jones et al. 2007). The percent of transcripts altered in the white-crowned sparrow telencephalon was similar to that identified as changing in relation to vigilance state in the rat cortex (Cirelli et al. 2004), indicating that, despite the differences between the target and array species, these microarrays are sufficiently sensitive to detect large differences between experimental groups if such differences exist. There was also a lack of concordance in the results of the two arrays. The observed differences may reflect genuine differences in gene expression, as well as methodological artifacts related to gene sequence differences between the species. In support of the latter argument, the hybridization rate of our white-crowned sparrow sample to the zebra finch array was considerably higher (~85%) than the sample hybridization to the chicken microarray (~16%). Importantly, there is a closer phylogenetic relationship between the white-crowned sparrow and the zebra finch than between the white-crowned sparrow and the chicken. Moreover, while the chicken array was based on the whole chicken genome, the zebra finch array was constructed from the telencephalon, the same brain region that we used for our study. These factors may have contributed to the difference in hybridization rate, as well as to the differences in concordance between the results of the two arrays.
Some more general study limitations also warrant mention. First, gene expression changes between groups were assessed in 4 technical replicates of pooled samples from each experimental condition, a strategy employed to limit technical variability. Although such an experimental design does not lend itself to rigorous statistical testing, because it was based on technical, rather than biological replicates, statistical analysis on these replicas is a necessary first step in identifying differentially expressed transcripts. We then used an independent set of animals (biological replicates) and performed extensive qPCR confirmations. Specifically, of the 15 known transcripts identified by microarray analysis as differentially expressed all but two were confirmed when tested with PCR. A second limitation is that we did not use procedures for controlling false discovery rates. However, in our long experience with array analysis we have found that these corrections generate an unacceptable number of false negatives (>50%). As in our previous studies, therefore, our strategy was to avoid correction, but to use criteria stringent enough that the number of false positive was <20%, as determined by extensive PCR confirmation using technical and biological replicas. Lastly, the choice of a 20% change in gene expression as a cutoff for inclusion in this study was based on the detection limit of qPCR used in our previous experiments (Cirelli et al. 2004; Cirelli et al. 2006; Jones et al. 2007). It may still be possible that some changes, although statistically significant, have no functional or biological relevance. On the other hand, it also possible that changes of <20%, although not statistically significant, may be functionally important, perhaps if occurring in abundant transcripts or in transcripts whose levels may represent the limiting step in some cellular processes.
Conclusion
The ease with which migrating birds negotiate the seasonal cycles of sleep restriction suggest that molecular, cellular, as well as higher-level mechanisms have evolved to effectively orchestrate these events. In this study, we sought to identify specific cellular strategies that might aid migratory songbirds in the accomplishment of this feat. Given the extent of sleep reduction during migration, we were somewhat surprised by the qualitatively and, since these changes are consistently less than twofold, quantitatively modest transcriptional changes associated with the migratory phenotype. The transcripts we did identify suggest that the migratory telencephalon is faced with enhanced energetic demands as well as increased cellular stress. The increase in these transcripts during migration relative to other waking states may be part of a set of adaptations that help alleviate the consequences of cellular stress and increased energy demands. Given that the sleeplessness phenotype is more pronounced in the white-crowned sparrow during the spring migratory period, future studies will need to profile animals sampled within the spring season to thoroughly identify the transcriptional changes associated with the migrating phenotype. It will also be important to consider changes in other brain regions, particularly the hypothalamus, since this structure is implicated both in the generation of circadian and seasonal rhythmicity as well as in the regulation of vigilance state.
Supplementary Material
White-crowned sparrow telencephalon genes whose mRNA levels are differentially expressed (>1.2 fold change, Welch t-test, p<0.05) in migration (M) relative to 78 hours of sleep restriction (SR) and 6 hours of spontaneous wakefulness (W). (a) transcripts increased (>1.2 fold change, Welch t-test, p<0.05) (b) transcripts decreased (<1.2 fold change, Welch t-test, p<0.05) “% change (array)”, percentage change in M relative to SR and W as indicated by array analysis. “% change (PCR)” indicates transcripts confirmed using qPCR from pooled RNA of 4 additional birds not previously used for microarray analysis. Transcripts in bold-face have been previously identified as “waking-related” in rat cerebral cortex or the avian telencephalon. Molecular function and biological process annotations are based on the gene ontology (GO) hierarchy and on an extensive analysis of the literature. Unless otherwise stated, all named transcripts in the Tables are based on homology between the nucleotide sequence of the differentially expressed transcript and the nucleotide sequence from the chicken genome. Since the function of many chicken genes is unknown, function categories were constructed based on documented function in other vertebrate species.
White-crowned sparrow telencephalon genes whose mRNA levels are differentially expressed (>1.2 fold change, Welch t-test, p<0.05) in 78 hours of sleep restriction (SR) relative to 78 hours of migration (M) and 6 hours of spontaneous wakefulness (W). a) transcripts increased (>1.2 fold change, Welch t-test, p<0.05) (b) transcripts decreased (<1.2 fold change, Welch t-test, p<0.05) “% change (array)”, percentage change in SR relative to M and W as indicated by array analysis. “% change (PCR)” indicates transcripts confirmed using qPCR from pooled RNA of 4 additional birds not previously used for microarray analysis. Molecular function and biological process annotations are based on the gene ontology (GO) hierarchy and on an extensive analysis of the literature. Unless otherwise stated, all named transcripts in the Tables are based on homology between the nucleotide sequence of the differentially expressed transcript and the nucleotide sequence from the chicken genome. Since the function of many chicken genes is unknown, function categories were constructed based on documented function in other vertebrate species.
Figure 2.
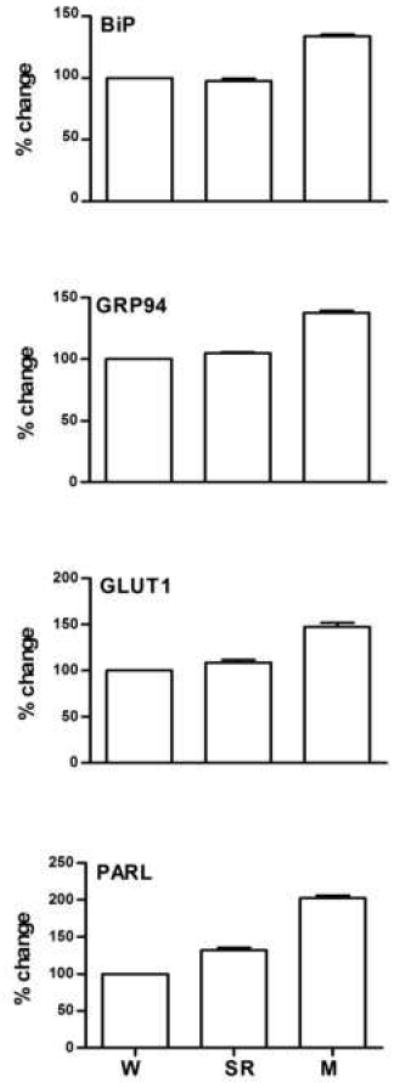
Validation of microarray analysis using quantitative PCR. Expression of four genes in the brain of W, SR and M birds. All values (mean±SEM) are expressed as percentage relative to W=100.
Acknowledgments
This work was supported by a grant from the United States Defense Advanced Research Projects Agency and by NIMH (NIH5/R01/MH071874 to RMB). We thank Dr. Niels C. Rattenborg and for his thoughts and comments as well technical assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adjaye J, Herwig R, Herrmann D, Wruck W, Benkahla A, Brink TC, Nowak M, Carnwath JW, Hultschig C, Niemann H, Lehrach H. Cross-species hybridisation of human and bovine orthologous genes on high density cDNA microarrays. BMC Genomics. 2004;5:83. doi: 10.1186/1471-2164-5-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews JF. Comparative studies on programmes for management of energy supply: torpor, pre-winter fattening and migration. Proc Nutr Soc. 1995;54:301–315. doi: 10.1079/pns19950056. [DOI] [PubMed] [Google Scholar]
- Bar-Or C, Czosnek H, Koltai H. Cross-species microarray hybridizations: a developing tool for studying species diversity. Trends Genet. 2007;23:200–207. doi: 10.1016/j.tig.2007.02.003. [DOI] [PubMed] [Google Scholar]
- Bar-Or C, Bar-Eyal M, Gal TZ, Kapulnik Y, Czosnek H, Koltai H. Derivation of species-specific hybridization-like knowledge out of cross-species hybridization results. BMC Genomics. 2006;7:110. doi: 10.1186/1471-2164-7-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barros LF, Porras OH, Bittner CX. Why glucose transport in the brain matters for PET. Trends Neurosci. 2005;28:117–119. doi: 10.1016/j.tins.2005.01.002. [DOI] [PubMed] [Google Scholar]
- Bassetti CL, Hersberger M, Baumann CR. CSF prostaglandin D synthase is reduced in excessive daytime sleepiness. J Neurol. 2006;253:1030–1033. doi: 10.1007/s00415-006-0153-8. [DOI] [PubMed] [Google Scholar]
- Belenky G, Wesensten NJ, Thorne DR, Thomas ML, Sing HC, Redmond DP, Russo MB, Balkin TJ. Patterns of performance degradation and restoration during sleep restriction and subsequent recovery: a sleep dose-response study. J Sleep Res. 2003;12:1–12. doi: 10.1046/j.1365-2869.2003.00337.x. [DOI] [PubMed] [Google Scholar]
- Ben-Shlomo R, Akhtar RA, Collins BH, Judah DJ, Davies R, Kyriacou CP. Light pulse-induced heme and iron-associated transcripts in mouse brain: a microarray analysis. Chronobiol Int. 2005;22:455–471. doi: 10.1081/CBI-200062353. [DOI] [PubMed] [Google Scholar]
- Berthold P. Control of bird migration. Chapman & Hall; London: 1996. [Google Scholar]
- Chalmers AD, Goldstone K, Smith JC, Gilchrist M, Amaya E, Papalopulu N. A Xenopus tropicalis oligonucleotide microarray works across species using RNA from Xenopus laevis. Mech Dev. 2005;122:355–363. doi: 10.1016/j.mod.2004.09.004. [DOI] [PubMed] [Google Scholar]
- Churchill GA. Fundamentals of experimental design for cDNA microarrays. Nat Genet. 2002;32(Suppl):490–495. doi: 10.1038/ng1031. [DOI] [PubMed] [Google Scholar]
- Cipolat S, Rudka T, Hartmann D, Costa V, Serneels L, Craessaerts K, Metzger K, Frezza C, Annaert W, D’Adamio L, Derks C, Dejaegere T, Pellegrini L, D’Hooge R, Scorrano L, De Strooper B. Mitochondrial rhomboid PARL regulates cytochrome c release during apoptosis via OPA1-dependent cristae remodeling. Cell. 2006;126:163–175. doi: 10.1016/j.cell.2006.06.021. [DOI] [PubMed] [Google Scholar]
- Cirelli C, Gutierrez CM, Tononi G. Extensive and divergent effects of sleep and wakefulness on brain gene expression. Neuron. 2004;41:35–43. doi: 10.1016/s0896-6273(03)00814-6. [DOI] [PubMed] [Google Scholar]
- Cirelli C, Faraguna U, Tononi G. Changes in brain gene expression after long-term sleep deprivation. J Neurochem. 2006;98:1632–1645. doi: 10.1111/j.1471-4159.2006.04058.x. [DOI] [PubMed] [Google Scholar]
- Danial NN, Korsmeyer SJ. Cell death: critical control points. Cell. 2004;116:205–219. doi: 10.1016/s0092-8674(04)00046-7. [DOI] [PubMed] [Google Scholar]
- Dawson WR, Marsh RL, Yacoe ME. Metabolic adjustments of small passerine birds for migration and cold. Am J Physiol. 1983;245:R755–767. doi: 10.1152/ajpregu.1983.245.6.R755. [DOI] [PubMed] [Google Scholar]
- Dinges DF, Pack F, Williams K, Gillen KA, Powell JW, Ott GE, Aptowicz C, Pack AI. Cumulative sleepiness, mood disturbance, and psychomotor vigilance performance decrements during a week of sleep restricted to 4-5 hours per night. Sleep. 1997;20:267–267. [PubMed] [Google Scholar]
- Doran SM, Van Dongen HP, Dinges DF. Sustained attention performance during sleep deprivation: evidence of state instability. Arch Ital Biol. 2001;139:253–267. [PubMed] [Google Scholar]
- Durmer JS, Dinges DF. Neurocognitive consequences of sleep deprivation. Semin Neurol. 2005;25:117–129. doi: 10.1055/s-2005-867080. [DOI] [PubMed] [Google Scholar]
- Everson CA. Clinical assessment of blood leukocytes, serum cytokines, and serum immunoglobulins as responses to sleep deprivation in laboratory rats. Am J Physiol Regul Integr Comp Physiol. 2005;289:R1054–1063. doi: 10.1152/ajpregu.00021.2005. [DOI] [PubMed] [Google Scholar]
- Harrison Y, Horne JA. One Night of Sleep Loss Impairs Innovative Thinking and Flexible Decision Making. Organ Behav Hum Decis Process. 1999;78:128–145. doi: 10.1006/obhd.1999.2827. [DOI] [PubMed] [Google Scholar]
- Horne JA. Sleep function, with particular reference to sleep deprivation. Annals of Clinical Research. 1985;17:199–208. [PubMed] [Google Scholar]
- Huang ZL, Urade Y, Hayaishi O. Prostaglandins and adenosine in the regulation of sleep and wakefulness. Curr Opin Pharmacol. 2007;7:33–38. doi: 10.1016/j.coph.2006.09.004. [DOI] [PubMed] [Google Scholar]
- Jeyaraju DV, Xu L, Letellier MC, Bandaru S, Zunino R, Berg EA, McBride HM, Pellegrini L. Phosphorylation and cleavage of presenilin-associated rhomboid-like protein (PARL) promotes changes in mitochondrial morphology. Proc Natl Acad Sci U S A. 2006;103:18562–18567. doi: 10.1073/pnas.0604983103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S, Pfister-Genskow M, Benca RM, Cirelli C. Molecular correlates of sleep and wakefulness in the brain of the white-crowned sparrow. J Neurochem. 2007 doi: 10.1111/j.1471-4159.2007.05089.x. epub ahead of print. [DOI] [PubMed] [Google Scholar]
- Jordan W, Tumani H, Cohrs S, Eggert S, Rodenbeck A, Brunner E, Ruther E, Hajak G. Prostaglandin D synthase (beta-trace) in healthy human sleep. Sleep. 2004;27:867–874. doi: 10.1093/sleep/27.5.867. [DOI] [PubMed] [Google Scholar]
- Katz S, Irizarry RA, Lin X, Tripputi M, Porter MW. A summarization approach for Affymetrix GeneChip data using a reference training set from a large, biologically diverse database. BMC Bioinformatics. 2006;7:464. doi: 10.1186/1471-2105-7-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitaoka K, Hattori A, Chikahisa S, Miyamoto K, Nakaya Y, Sei H. Vitamin A deficiency induces a decrease in EEG delta power during sleep in mice. Brain Res. 2007;1150:121–130. doi: 10.1016/j.brainres.2007.02.077. [DOI] [PubMed] [Google Scholar]
- Kullberg C, Lind J, Fransson T, Jakobsson S, Vallin A. Magnetic cues and time of season affect fuel deposition in migratory thrush nightingales (Luscinia luscinia) Proc Biol Sci. 2003;270:373–378. doi: 10.1098/rspb.2002.2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Bruce-Keller AJ, Kruman Y, Chan SL, Mattson MP. 2-Deoxy-D-glucose protects hippocampal neurons against excitotoxic and oxidative injury: evidence for the involvement of stress proteins. J Neurosci Res. 1999;57:48–61. doi: 10.1002/(SICI)1097-4547(19990701)57:1<48::AID-JNR6>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Liu H, Miller E, van de Water B, Stevens JL. Endoplasmic reticulum stress proteins block oxidant-induced Ca2+ increases and cell death. J Biol Chem. 1998;273:12858–12862. doi: 10.1074/jbc.273.21.12858. [DOI] [PubMed] [Google Scholar]
- Mackiewicz M, Shockley KR, Romer MA, Galante RJ, Zimmerman JE, Naidoo N, Baldwin DA, Jensen ST, Churchill GA, Pack AI. Macromolecule biosynthesis - a key function of sleep. Physiol Genomics. 2007 doi: 10.1152/physiolgenomics.00275.2006. [DOI] [PubMed] [Google Scholar]
- Magistretti PJ. Neuron-glia metabolic coupling and plasticity. J Exp Biol. 2006;209:2304–2311. doi: 10.1242/jeb.02208. [DOI] [PubMed] [Google Scholar]
- Maret S, Franken P, Dauvilliers Y, Ghyselinck NB, Chambon P, Tafti M. Retinoic acid signaling affects cortical synchrony during sleep. Science. 2005;310:111–113. doi: 10.1126/science.1117623. [DOI] [PubMed] [Google Scholar]
- McWilliams SR, Karasov WH. Phenotypic flexibility in digestive system structure and function in migratory birds and its ecological significance. Comp Biochem Physiol A Mol Integr Physiol. 2001;128:579–593. doi: 10.1016/s1095-6433(00)00336-6. [DOI] [PubMed] [Google Scholar]
- Mettke-Hofmann C, Gwinner E. Long-term memory for a life on the move. Proc Natl Acad Sci U S A. 2003;100:5863–5866. doi: 10.1073/pnas.1037505100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouritsen H, Ritz T. Magnetoreception and its use in bird navigation. Curr Opin Neurobiol. 2005;15:406–414. doi: 10.1016/j.conb.2005.06.003. [DOI] [PubMed] [Google Scholar]
- Naidoo N, Giang W, Galante RJ, Pack AI. Sleep deprivation induces the unfolded protein response in mouse cerebral cortex. J Neurochem. 2005;92:1150–1157. doi: 10.1111/j.1471-4159.2004.02952.x. [DOI] [PubMed] [Google Scholar]
- Naidoo N, Casiano V, Cater J, Zimmerman J, Pack AI. A role for the molecular chaperone protein BiP/GRP78 in Drosophila sleep homeostasis. Sleep. 2007;30:557–565. doi: 10.1093/sleep/30.5.557. [DOI] [PubMed] [Google Scholar]
- Nowrousian M, Ringelberg C, Dunlap JC, Loros JJ, Kuck U. Cross-species microarray hybridization to identify developmentally regulated genes in the filamentous fungus Sordaria macrospora. Mol Genet Genomics. 2005;273:137–149. doi: 10.1007/s00438-005-1118-9. [DOI] [PubMed] [Google Scholar]
- Pellegrini L, Scorrano L. A cut short to death: Parl and Opa1 in the regulation of mitochondrial morphology and apoptosis. Cell Death Differ. 2007;14:1275–1284. doi: 10.1038/sj.cdd.4402145. [DOI] [PubMed] [Google Scholar]
- Pellegrini L, Passer BJ, Canelles M, Lefterov I, Ganjei JK, Fowlkes BJ, Koonin EV, D’Adamio L. PAMP and PARL, two novel putative metalloproteases interacting with the COOH-terminus of Presenilin-1 and -2. J Alzheimers Dis. 2001;3:181–190. doi: 10.3233/jad-2001-3203. [DOI] [PubMed] [Google Scholar]
- Pravosudov VV, Kitaysky AS, Omanska A. The relationship between migratory behaviour, memory and the hippocampus: an intraspecific comparison. Proc Biol Sci. 2006;273:2641–2649. doi: 10.1098/rspb.2006.3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu WM, Huang ZL, Xu XH, Aritake K, Eguchi N, Nambu F, Narumiya S, Urade Y, Hayaishi O. Lipocalin-type prostaglandin D synthase produces prostaglandin D2 involved in regulation of physiological sleep. Proc Natl Acad Sci U S A. 2006;103:17949–17954. doi: 10.1073/pnas.0608581103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramenofsky M, Agatsuma R, Barga M, Cameron R, Harm J, Landys MM, Ramfar T. Migratory Behavior: New Insights from Captive Studies. In: Berthold P, Gwinner E, Sonnenschein E, editors. Avian Migration. Springer; Berlin: 2003. pp. 97–111. [Google Scholar]
- Rao RV, Ellerby HM, Bredesen DE. Coupling endoplasmic reticulum stress to the cell death program. Cell Death Differ. 2004;11:372–380. doi: 10.1038/sj.cdd.4401378. [DOI] [PubMed] [Google Scholar]
- Rattenborg NC, Mandt BH, Obermeyer WH, Winsauer PJ, Huber R, Wikelski M, Benca RM. Migratory sleeplessness in the white-crowned sparrow (Zonotrichia leucophrys gambelii) PLoS Biol. 2004;2:E212. doi: 10.1371/journal.pbio.0020212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson SJ. Cell and molecular biology of transthyretin and thyroid hormones. Int Rev Cytol. 2007;258:137–193. doi: 10.1016/S0074-7696(07)58003-4. [DOI] [PubMed] [Google Scholar]
- Schroder M, Kaufman RJ. The mammalian unfolded protein response. Annu Rev Biochem. 2005;74:739–789. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- Shah G, Azizian M, Bruch D, Mehta R, Kittur D. Cross-species comparison of gene expression between human and porcine tissue, using single microarray platform--preliminary results. Clin Transplant. 2004;18(Suppl 12):76–80. doi: 10.1111/j.1399-0012.2004.00223.x. [DOI] [PubMed] [Google Scholar]
- Shaw PJ, Cirelli C, Greenspan RJ, Tononi G. Correlates of sleep and waking in Drosophila melanogaster. Science. 2000;287:1834–1837. doi: 10.1126/science.287.5459.1834. [DOI] [PubMed] [Google Scholar]
- Terao A, Wisor JP, Peyron C, Apte-Deshpande A, Wurts SW, Edgar DM, Kilduff TS. Gene expression in the rat brain during sleep deprivation and recovery sleep: an Affymetrix GeneChip study. Neuroscience. 2006;137:593–605. doi: 10.1016/j.neuroscience.2005.08.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terao A, Steininger TL, Hyder K, Apte-Deshpande A, Ding J, Rishipathak D, Davis RW, Heller HC, Kilduff TS. Differential increase in the expression of heat shock protein family members during sleep deprivation and during sleep. Neuroscience. 2003;116:187–200. doi: 10.1016/s0306-4522(02)00695-4. [DOI] [PubMed] [Google Scholar]
- Van Dongen HP, Maislin G, Mullington JM, Dinges DF. The cumulative cost of additional wakefulness: dose-response effects on neurobehavioral functions and sleep physiology from chronic sleep restriction and total sleep deprivation. Sleep. 2003;26:117–126. doi: 10.1093/sleep/26.2.117. [DOI] [PubMed] [Google Scholar]
- Wikelski M, Tarlow EM, Raim A, Diehl RH, Larkin RP, Visser GH. Avian metabolism: Costs of migration in free-flying songbirds. Nature. 2003;423:704. doi: 10.1038/423704a. [DOI] [PubMed] [Google Scholar]
- Wimmer F, Hoffmann RF, Bonato RA, Moffitt AR. The effects of sleep deprivation on divergent thinking and attention processes. J Sleep Res. 1992;1:223–230. doi: 10.1111/j.1365-2869.1992.tb00043.x. [DOI] [PubMed] [Google Scholar]
- Yang YH, Speed T. Design issues for cDNA microarray experiments. Nat Rev Genet. 2002;3:579–588. doi: 10.1038/nrg863. [DOI] [PubMed] [Google Scholar]
- Yu Z, Luo H, Fu W, Mattson MP. The endoplasmic reticulum stress-responsive protein GRP78 protects neurons against excitotoxicity and apoptosis: suppression of oxidative stress and stabilization of calcium homeostasis. Exp Neurol. 1999;155:302–314. doi: 10.1006/exnr.1998.7002. [DOI] [PubMed] [Google Scholar]
- Zimmerman JE, Rizzo W, Shockley KR, Raizen DM, Naidoo N, Mackiewicz M, Churchill GA, Pack AI. Multiple mechanisms limit the duration of wakefulness in Drosophila brain. Physiol Genomics. 2006;27:337–350. doi: 10.1152/physiolgenomics.00030.2006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
White-crowned sparrow telencephalon genes whose mRNA levels are differentially expressed (>1.2 fold change, Welch t-test, p<0.05) in migration (M) relative to 78 hours of sleep restriction (SR) and 6 hours of spontaneous wakefulness (W). (a) transcripts increased (>1.2 fold change, Welch t-test, p<0.05) (b) transcripts decreased (<1.2 fold change, Welch t-test, p<0.05) “% change (array)”, percentage change in M relative to SR and W as indicated by array analysis. “% change (PCR)” indicates transcripts confirmed using qPCR from pooled RNA of 4 additional birds not previously used for microarray analysis. Transcripts in bold-face have been previously identified as “waking-related” in rat cerebral cortex or the avian telencephalon. Molecular function and biological process annotations are based on the gene ontology (GO) hierarchy and on an extensive analysis of the literature. Unless otherwise stated, all named transcripts in the Tables are based on homology between the nucleotide sequence of the differentially expressed transcript and the nucleotide sequence from the chicken genome. Since the function of many chicken genes is unknown, function categories were constructed based on documented function in other vertebrate species.
White-crowned sparrow telencephalon genes whose mRNA levels are differentially expressed (>1.2 fold change, Welch t-test, p<0.05) in 78 hours of sleep restriction (SR) relative to 78 hours of migration (M) and 6 hours of spontaneous wakefulness (W). a) transcripts increased (>1.2 fold change, Welch t-test, p<0.05) (b) transcripts decreased (<1.2 fold change, Welch t-test, p<0.05) “% change (array)”, percentage change in SR relative to M and W as indicated by array analysis. “% change (PCR)” indicates transcripts confirmed using qPCR from pooled RNA of 4 additional birds not previously used for microarray analysis. Molecular function and biological process annotations are based on the gene ontology (GO) hierarchy and on an extensive analysis of the literature. Unless otherwise stated, all named transcripts in the Tables are based on homology between the nucleotide sequence of the differentially expressed transcript and the nucleotide sequence from the chicken genome. Since the function of many chicken genes is unknown, function categories were constructed based on documented function in other vertebrate species.


