Abstract
Using comparative sequencing approaches, we investigated the evolutionary history of the European-enriched 17q21.31 MAPT inversion polymorphism. We present a detailed, BAC-based sequence assembly of the inverted human H2 haplotype and contrast it with the sequence structure and genetic variation of the corresponding 1.5 Mb region for the non-inverted H1 human haplotype and that of chimpanzee and orangutan. We find that inversion of the MAPT region is similarly polymorphic in other great ape species and present evidence that the inversions have occurred independently in both chimpanzee and humans. In humans, the inversion breakpoints correspond to core duplications encoding the LRRC37 gene family. Our analysis favors the H2 configuration and sequence haplotype as the likely great ape/human ancestral state with inversion recurrences during primate evolution. We demonstrate that the H2 architecture has evolved more extensive sequence homology, perhaps explaining its preference to undergo microdeletion associated with mental retardation in European populations.
INTRODUCTION
It has become clear that a large proportion of genetic variability among humans and between humans and chimpanzees involves large-scale genomic structural changes such as deletions, insertions and inversions 1–5. In this regard, the ~970 kb inversion of the MAPT locus on human chromosome 17 represents one of the most structurally complex and evolutionarily dynamic regions of the genome 6–8. This locus occurs in humans as two haplotypes, H1 (direct orientation) and H2 (inverted orientation) 6,9, which show no recombination between them over a region of ~1.5 Mb 10. The two haplotypes have different functional impact. Consistent differences in cortical gene expression has been observed between the two 11. Specific H1 haplotypes are associated with Alzheimer’s disease, amyotrophic lateral sclerosis/PDC of Guam, corticobasal degeneration and progressive supranuclear palsy 9,10,12–15. The H2 haplotype is predisposed to recurrent microdeletions associated with the 17q21.31 microdeletion syndrome 16–18. The H1 locus occurs in all populations and shows a normal pattern of genetic variability and recombination. In contrast, the H2 locus occurs predominantly in European descent populations 19 where it shows limited H2 diversity but extensive diversity (0.3%) when compared to H1, suggesting an ancient coalescent of ~3 million years ago 6,8,20. Both the ancient inversion and the microdeletion event are thought to have arisen as the result of non-allelic homologous recombination between large blocks of segmental duplications (200–500 kbp in length). The goal of this study was to reconstruct the evolutionary history of this region by conducting detailed analysis of its sequence organization and assessing variation in its structure within and between human and non-human primate populations.
RESULTS
Duplication Analysis
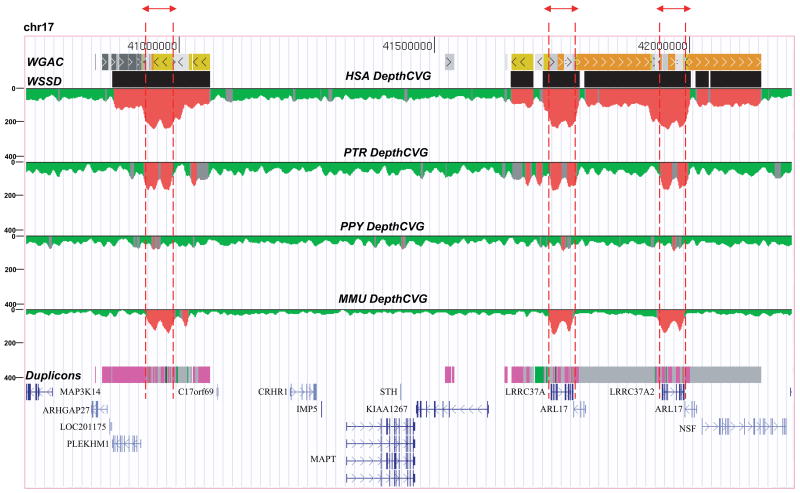
Due to the central role of the duplications in both the microdeletion and the evolution of the inversion, we began our analysis by comparing the duplication architecture among primate species. According to the H1 haplotype organization within the genome assembly, the inversion is flanked by two duplication blocks, 203 kbp (proximal) and 484 kbp (distal) in length. We estimated the evolutionary timing of various segmental duplications by comparing the duplication architecture in human, chimpanzee, orangutan and macaque (Fig. 1). Using whole-genome shotgun sequence data from each species (Methods), we mapped regions of excess read depth and sequence divergence against the human reference genome assembly (build36) (Fig. 1). This approach may be used to accurately predict large (≥10 kbp), high-identity segmental duplications within 21 and between species 5. We find that 71% (486/687 kbp) of the duplication architecture is specific to the human species (i.e. not detected as duplicated in the chimpanzee, orangutan or macaque genome). The analysis predicts that most (≥87%) of the segmental duplications emerged after the divergence of the chimpanzee and human lineage from the orangutan <12 million years ago (this is subsequently confirmed by a more detailed examination of the chimpanzee and orangutan sequence assemblies which show limited evidence of duplications within the orangutan sequence assembly for this locus, Supplementary Note). Interestingly, a core segmental duplication of ~40 kbp, corresponding to the LRRC37 gene family, is distributed throughout chromosome 17 and predicted to be one of the few duplications common to chimpanzee, human and macaque.
Figure 1. Comparative segmental duplication analysis of the 17q21.31 region.
Regions of excess (≥ mean +3 standard deviation, colored red) WGS depth-of-coverage are shown for human (HSA), chimpanzee (PTR), orangutan (PPY) and macaque (MMU) mapped against the human reference genome (build36). This approach detects ≥90% of all segmental duplications which are larger than 10 kbp in length and greater than 94% sequence identity 5. The analysis suggests that the majority (~71%) of the duplication architecture is human-specific except for a core duplicated segment corresponding to the LRRC37A gene family (highlighted by red dashed lines) 32.
Inversion Analysis
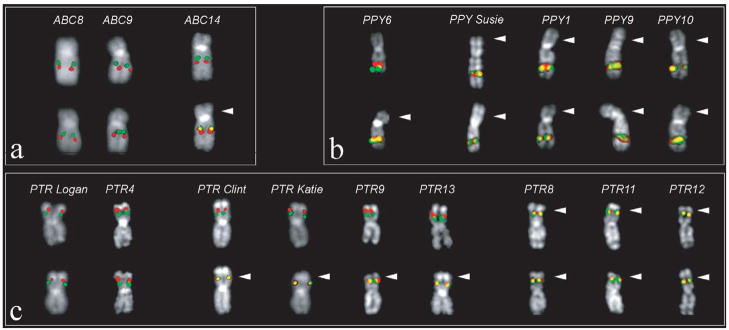
We next developed a reciprocal FISH assay to characterize the orientation of the region by taking advantage of the physical limits of metaphase chromosomes to resolve distinct signals (Supplementary Note). We tested for the presence of the inversion by examining lymphoblastoid cell lines from a diverse panel of hominoids and macaque Old World monkey species. Although the H1 and H2 haplotypes are specific to humans, for simplicity, we will refer to the H1 and H2 orientation when describing the configuration in other non-human primate species. All three macaque species tested (Macaca fascicularis, Macaca arctodies and Macaca mulatta) and orangutan showed FISH signatures consistent with the H2 orientation, suggesting that this orientation likely represents the ancestral configuration (Supplementary Note). Surprisingly, examination of a single individual from each of the two chimpanzee species (Pan paniscus and Pan troglodytes) showed that they were heterozygous for the inversion. We examined a larger population of unrelated chimpanzees (n=9, Pan troglodytes) and found the inversion to be highly polymorphic (Fig. 2). Unlike the human population, the H2 configuration represents the major allele (56% allele frequency) in chimpanzee. All Sumatran orangutans were homozygous for the H2 orientation; however, analysis of a single Bornean orangutan (PPY6) showed that it was heterozygous, indicating the inversion is likely polymorphic within this subspecies (Fig. 2b). Combined, these data argue that the H2 orientation represents the ancestral state and that this region of the genome has been subject to inversion polymorphisms for the last 12 million years of hominoid evolution.
Figure 2. Inversion polymorphism among primates.
A Metaphase FISH assay distinguishes between the H2 orientation (merged yellow) signal and H1 orientation (distinct green and red) based on proximity of two unique probes (Supplementary Note): (a) Extracted chromosome 17 from three human; (b) five orangutan and (c) nine chimpanzee lymphoblast cell lines are shown. The H2 orientation (white arrow) predominates in orangutan and chimpanzee samples. In humans, the H2 haplotype is restricted to Middle Eastern and European populations.
Sequence Analysis
Breakpoint refinement of the human inversion is complicated by extensive structural variation within the flanking duplication blocks 6,8,16. Since the current genome assembly is based on the sequence of multiple individuals, we constructed and sequenced a BAC-based assembly corresponding to the human H2 haplotype (1,481 kbp) and the human H1 haplotype (1,406 kbp) from a donor that was heterozygous for the inversion (RPCI-11). Requiring 100% sequence identity overlap between overlapping BAC clones ensured that two distinct sequence haplotypes could be constructed. A subsequent examination of 79 diagnostic SNPs confirmed that the H1 and H2 haplotypes had been successfully resolved. We also developed a BAC-based assembly of the chimpanzee (1,852 kbp) and the orangutan in H2 orientation (1,859 kbp) requiring haplotype contiguity specifically over the breakpoint regions. Due to the paucity of segmental duplications in orangutan, the WGSA and clone-based assemblies were virtually identical (see Supplementary Note for details regarding the sequence and assembly of these regions).
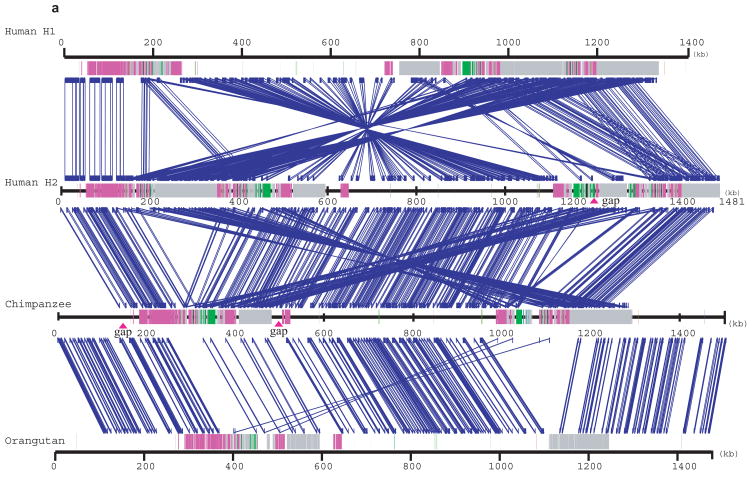
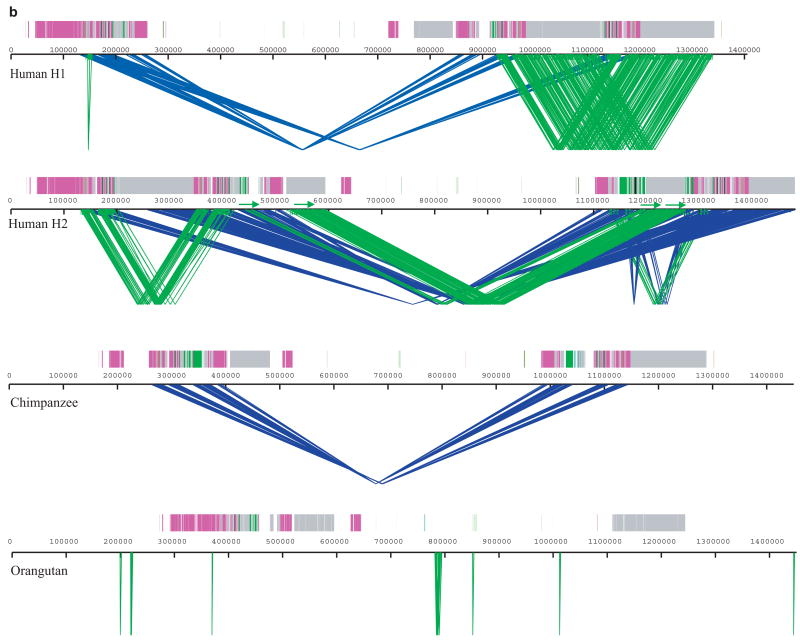
We compared the Human H1 and H2 sequence organization as well as both human haplotypes against the non-human primate sequence assemblies (Fig. 3, Supplementary Fig. 1). We identified all regions of segmental duplication based on a variety of independent analyses (Supplementary Table 1). The analysis revealed several important features. First, sequence alignments confirm an “H2 orientation” for the CRHR1-MAPT region in chimpanzees and orangutan when compared to the H1 haplotype. After the inversion, the largest genomic structural difference appears to have occurred within the shared human and chimpanzee lineage where a duplicative transposition (≥100 kb) placed two inverted copies of the core segmental duplication on either side of the inversion region (Fig. 3b). Second, although it is impossible to precisely delineate the breakpoints at the single basepair level due to the high degree of sequence identity, it was possible to identify the inversion H1-H2 “breakpoint intervals” based on alignment of flanking sequence. We estimate the inversion as ~970 kb in length and find that each of the four breakpoints map to a LRRC37 core duplication (Supplementary Table 2, Supplementary Fig. 1a).
Figure 3. A sequence comparison of the human H1, H2, chimpanzee and orangutan 17q21.31 region.
(a) BAC-based sequence assemblies of both the human inverted (H2) and non-inverted haplotype (H1) were compared using Miropeats 39 to a BAC-based assembly of the chimpanzee (PTR) and a WGS-based assembly of the orangutan (PPY, threshold –s 1000). Regions of homology are shown with blue joining-lines to the corresponding sequence above. Duplicon architecture based on human segmental duplications is overlaid as colored or grey bars. H1 shows a ~970 kbp inverted segment when compared to H2, chimpanzee (PTR) and the orangutan sequence assembly. The H2 sequence assembly shows a relocation of large (~200 kbp) high-identity duplications on either side of the unique interval when compared to chimpanzee (crisscross pattern). A comparison between orangutan and chimpanzee shows evidence of a (~100 kbp) segmental duplication from proximal to distal duplication block, which likely occurred in the common ancestor of chimpanzee and human (6–12 million years ago). (b) The extent of local direct (green) and inverted (blue) intrachromosomal SDs flanking the inversion are shown for human H1 and H2 haplotypes, chimpanzee (PTR) and orangutan (PPY) (Miropeats threshold –s 300). We examined the duplication content (WGAC) within each assembly and computed the number of non-redundant duplicated basepairs for each assembly (Supplementary Table 1). No homologous SDs (sequence identity ≥ 90%, size ≥ 1 kbp) were found in orangutan genome flanking the inversion region, while in chimpanzee and H1 haplotype, 292 kbp and 227 kbp were identified respectively. H2 shows the most extensive duplication architecture flanking the inversion including 95 kbp in direct orientation.
Sequence comparison with non-human primates shows that more extensive and complex duplication architecture has emerged in the evolutionary lineage leading to humans (Table 1, Fig. 3b). If we focus only on those duplications that align between the proximal and distal blocks (Supplementary Table 3), we find that the H1 duplication organization is slightly (59.5 kbp) larger than that of the chimpanzee. In contrast, the H2 haplotype shows the greatest duplication complexity. We find a total of 441 kbp of homologous sequence flanking either side of the inverted region in H2 when compared to only 169 kbp for H1. Similarly, we find that the average sequence identity for the H2 sequences is significantly greater (99.3%) when compared to the H1 sequences (98.3%). We constructed a series of phylogenetic trees from a multiple sequence alignment of shared duplication (40 kb) common to human H1, H2, chimpanzee and orangutan (Supplementary Note). In most cases, duplicated sequences from H2 grouped separately from H1, suggesting that the H2 segmental duplications have been significantly homogenized by gene conversion or secondary duplication events.
Table 1.
Duplication alignments flanking MAPT inversion
| Sequence | Alignments* | Length (bp) | %identity | K2M | SE |
|---|---|---|---|---|---|
| PTR | 2 | 110279 | 98.73 | 0.012814 | 0.000471 |
| H1 | 6 | 169796 | 98.34 | 0.016892 | 0.000920 |
| H2 | 8 | 441832 | 99.30 | 0.007002 | 0.000517 |
Only pairwise sequence alignments > 5 kbp mapping within the two duplication blocks flanking the inversion were considered; in H1 and PTR all pairwise alignments are in an inverted orientation with respect to one another; only within the H2 haplotype were three alignments identified in a direct orientation (corresponding to 97,301 bp with 99.53% sequence identity).
In addition to greater sequence identity, we find important differences in the orientation of the duplications. Within the sequenced H2 contig, there are 95 kbp of segmental duplication in direct orientation (Fig. 3b). This contrasts with the H1 and chimpanzee sequence where none of the alignments between the proximal and distal duplication blocks are in direct H1 orientation. Among the H2 alignments, we identify, in particular, a 73 kbp “H2-only” segmental duplication noted previously as a copy-number polymorphism in the human population 6,22. In order to test whether this large direct repeat might play a role in the predisposition of H2 to microdelete, we compared the evolutionary inversion breakpoints with the predicted microdeletion breakpoints associated with the H2 and 17q21.31 microdeletion (Supplementary Fig. 2) 16,17. We find that the inversion breakpoints and microdeletion breakpoints are not identical. Interestingly, one of the microdeletion breakpoints maps within the largest H2-specific segmental duplications, suggesting that the large direct repeats that emerged specifically within the H2 lineage predispose to rearrangement, although further experimentation will be required to define the microdeletion breakpoints more precisely.
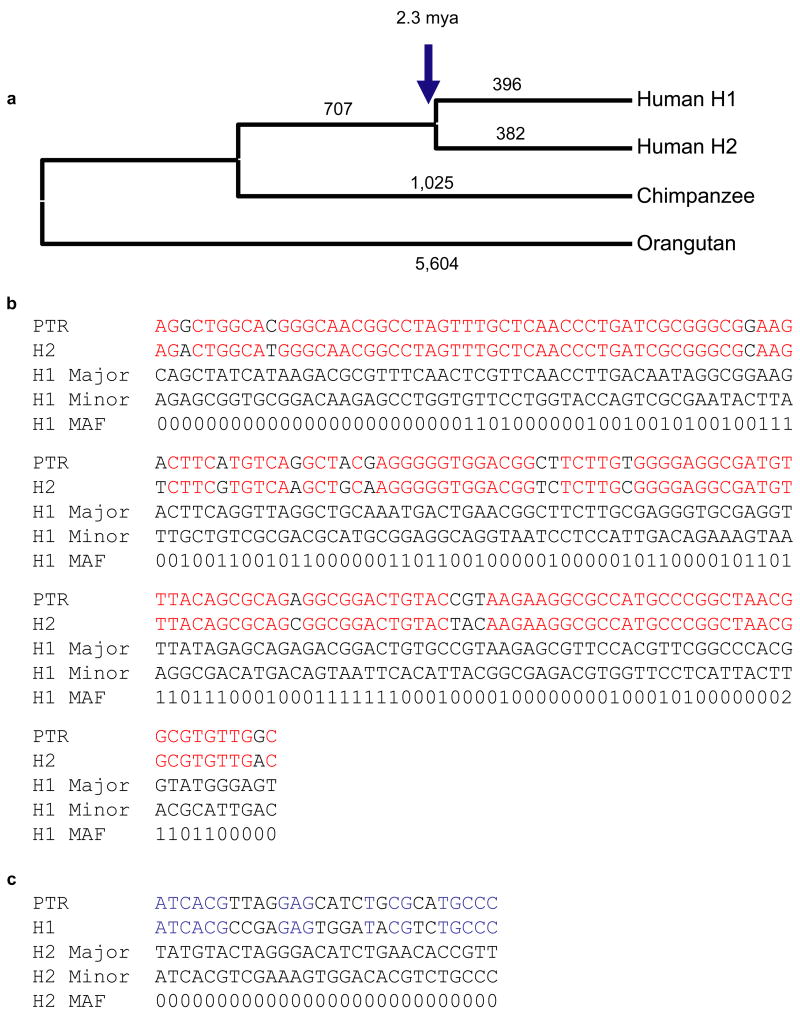
In order to determine the most likely ancestral state in humans, we constructed a 219 kbp multiple sequence alignment of human H1, H2, chimpanzee and orangutan from unique sequences mapping to the inversion interval (Fig. 4). Similar to previous analyses 6,8, the phylogenetic tree of all single nucleotide variants did not distinguish H1 or H2 as ancestral. Rather, the analysis revealed that human H1 and H2 arose from an intermediate ancestral haplotype with a large and approximately equal number of haplotype-specific single-nucleotide variants (n=382 vs. 396) mapping to both the H1 and H2 lineages.
Figure 4. Phylogenetic and SNP haplotype analysis.
(a) An unrooted neighbor-joining phylogram was constructed (MEGA pairwise deletion option, sum of branch lengths=0.0414) based on 219,165 aligned basepairs from unique sequence within the inverted region. H1 and H2 sequence taxa clustered together with 100% bootstrap support (n=500 replicates). The number of single-nucleotide variants specific for each branch in the tree is assigned above each branch. We estimate that the H1 and H2 haplotypes diverged 2.3 million years ago (Table 2). (b, c) We treated H1 and H2 haplotypes as separate populations in the analysis and identified a total of 320 SNPs that were fixed in one haplotype but polymorphic in the other. We assessed the likely ancestral state of each SNP through a comparison with the sequenced chimpanzee haplotype. For SNPs that are monomorphic among H2 haplotypes but polymorphic among the H1s (b), we found that the allele found in the H2 haplotypes matched the chimpanzee allele 90% of the time (150/166 considered positions). For SNPs that are monomorphic among H1 haplotypes but polymorphic among the H2s, the allele found in the H1 haplotypes matched the chimpanzee 60% of the time (17/28 considered positions) (c). Red indicates alleles shared between chimp and H2, while blue delineates shared alleles between H1 and chimpanzee. The major and minor alleles are denoted with the minor allele frequency represented by a single digit. For example, 4 refers to a minor allele frequency of ≥ 40%.
Assuming the chimpanzee and human lineages diverged 6 million years ago, the diversity between H1 and H2 (0.476 %) predicts that the two human haplotypes diverged 2.3 million years ago (Table 2). Furthermore, if we assume that the inversion was a unique event in human evolutionary history and that the inversion has been an effective barrier to recombination, we can treat the H1 and H2 regions as non-mixing populations. All modern copies of the derived population must be descended from a single founder, so all variants present only in the derived population must have arisen since the inversion. This means that for all SNPs segregating in the derived population, the allele found in the ancestral population would more likely match the chimpanzee variant. In order to reduce the impact of genotype error caused by paralogous sequences, we limited our consideration to HapMap SNPs that can be uniquely mapped onto both sequenced haplotypes (Supplementary Note). If we divide SNPs into those only variant within H1 haplotypes (fixed in H2) and those only variant in H2 haplotypes (fixed in H1), we find that 90% (150/166) of SNPs polymorphic in H1 have an H2 allele matching the chimpanzee allele, while for those variant only in H2, only 60% (17/28) have an H1 allele matching the chimpanzee allele (Fig. 4b, 4c, Table 3 and Supplementary Note). This significant result (p =0.0002332, Fisher’s exact test) is consistent with an ancestral H2 state in human and inconsistent with an ancestral H1 state. Interestingly, we find a small fraction of shared polymorphic sites in H1 and H2 which represent either recurrent CpG mutations or, possibly, gene flow between the H1 and H2 regions perhaps as a result of gene conversion within the inversion loop.
Table 2.
Sequence divergence of orthologous sequence
| H1 | H2 | PTR | PPY | |
|---|---|---|---|---|
| H1 | 0.000090 | 0.000140 | 0.000260 | |
| H2 | 0.004170 | 0.000140 | 0.000250 | |
| PTR | 0.010930 | 0.010890 | 0.000260 | |
| PPY | 0.034090 | 0.033920 | 0.033790 |
Kimura 2-parameter model genetic distance estimates (left diagonal) and standard error (right diagonal). 219 kbp of 4-way alignment of unique sequence within the inversion interval. Tajima’s Relative Rate test shows that the genomic sequence is evolving neutrally (p=0.22–0.81).
Table 3.
Analysis of different SNP classes
| Among H1 Chromosomes | Among H2 Chromosomes | |||||||
|---|---|---|---|---|---|---|---|---|
| Category | Number of SNPs | PTR Unk | Equal Freq | Maj=PTR | Maj != PTR | Equal Freq | Maj=PTR | Maj != PTR |
| H2-fixed, H1 polymorphic | 178 | 12 | 0 | 108 | 58 | 0 | 150 | 16 |
| H1-fixed, H2 polymorphic | 29 | 1 | 0 | 17 | 11 | 0 | 12 | 16 |
| H1-fixed, H2-fixed | 381 | 39 | 0 | 164 | 178 | 0 | 178 | 164 |
| Polymorphic among both H1and H2 | 23 | 0 | 1 | 18 | 8 | 0 | 19 | 4 |
The total number of SNPs in each category, the number of SNPs where the corresponding chimpanzee allele could not be confidently determined, and the ancestral classification among H1 and H2 chromosomes is listed. Maj=PTR: the major allele in the class matches the chimpanzee sequence, Maj != PTR, the major allele in the class is different than the chimpanzee allele; PTR Unk= SNPs where the chimp allele could not be determined because no high identity BLAT alignment could be found. In addition, we identified 23 SNPs which were polymorphic in both H1 and H2 chromosomes—most are single occurrences and likely reflect genotyping errors (Supplementary Note).
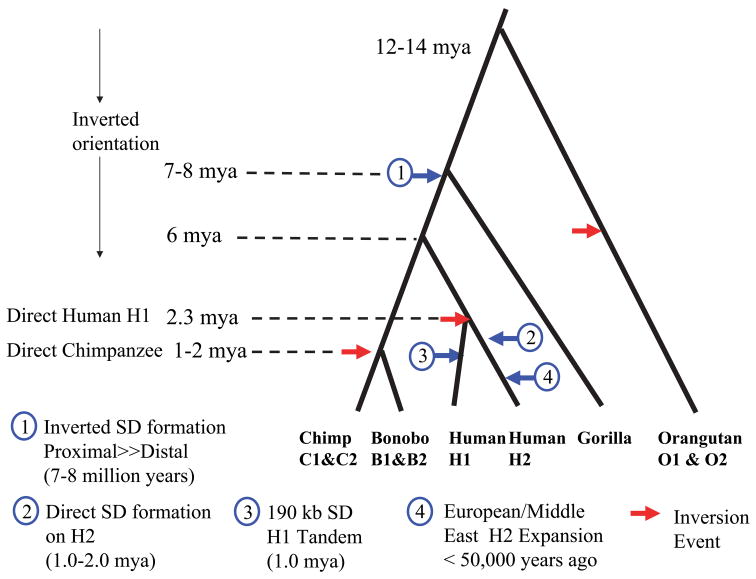
The fact that the inversion is polymorphic in human, chimpanzee, bonobo and orangutan may be the result of evolutionary recurrence 23,24 or lineage-specific sorting of an ancient polymorphism 25. In order to assess the reciprocal event within a non-human primate lineage, we took advantage of the fact that the sequenced chimpanzee (Clint) was heterozygous for the inversion (Fig. 2c). We aligned all end-sequences derived from Clint against the BAC-based chimpanzee haplotype (H2 orientation at the breakpoints) (Supplementary Note). Excluding duplicated sequences, the level of sequence divergence (1/336 bp or 0.30%) confirmed that the two chimpanzee haplotypes had emerged recently (within the last 1–2 million years of chimpanzee evolution). These values are consistent with global estimates of chimpanzee diversity 26 but slightly less than diversity between chimpanzee and bonobos (0.354%) 27. Based on sequence divergence between the human H1 and H2 haplotypes, we calculate a more ancient origin for the divergence of human H1 and H2 lineages (1.9–2.7 million years ago based on uncertainty in the chimp-human divergence which is the largest contributor to error in time estimates), but still clearly within the Homo lineage of evolution. Combined, these data strongly argue the H1 orientation has emerged independently in both lineages. Taken together with the observation of both chromosomal configurations in bonobo (Pan paniscus) and Bornean orangutan, we suggest that this particular region has been prone to recurrent inversion events within multiple primate lineages (Fig. 5). We propose that this inversion “toggling” has contributed, in part, to the complex duplication architecture that emerged over the last 12 million years of evolution 28.
Figure 5. Evolutionary Model: Inversion “toggling”, SD formation and disease susceptibility.
We estimated the evolutionary age of various duplication/gene conversion and rearrangement events by establishing a local molecular clock for single nucleotide substitution (Supplementary Table 4) and superimposed these estimates over a generally accepted hominoid phylogeny 45. Due to uncertainty in the chimp-human divergence, timing of events should only be considered an approximation. We propose that the ancestral CRHR1-MAPT region was inverted but has toggled to an H1 orientation multiple times within the evolution of different great-ape/human lineages (red arrows). Large blocks (≥100 kbp) of inverted segmental duplications were formed in the common ancestor of chimpanzee and human, further predisposing the region to recurrent inversion. An inversion of the predominant H2 allele created the H1 allele ~2.3 million years ago within the human lineage. Subsequently, larger blocks of directly oriented SDs emerged within the H2 lineage predisposing it to microdeletion and disease. As a result of this negative selection against the H2, the H1 haplotype rose in frequency and became the predominant allele in all human populations with a subsequent CNP tandem duplication occurring on some haplotypes. In the out-of-Africa European founding population, however, the H2 allele resurged in frequency due to a partial selective sweep or a population bottleneck in the founding population.
DISCUSSION
Our analysis establishes the H2 orientation as the most likely great ape/human ancestral state. Surprisingly, we find that inversion of the CRHR1-MAPT region is similarly polymorphic in other extant great ape populations where it represents the major allele. Despite the fact that the inverted configuration occurs in only 20% of European chromosomes, both SNP haplotype analysis and comparative FISH analysis point to an inverted H2-like ancestor. Previously it was assumed that H1 was the ancestral sequence because >99% of sub-Saharan African haplotypes are variants of the H1 clade 6. Based on analysis of the CEPH-HGDP sample collection 29, the few Mbuti and Biaka pygmies with an H2 allele (HGdp980, HGdp985, HGdp463, HGdp474) have a haplotype and SNP architecture identical to the European H2 allele making it difficult to distinguish an ancient origin from recent admixture 19,30. We propose that an H2-like allele was the predominant allele among ancestral populations (Homo heidelbergensis) 20, but that its frequency became subsequently reduced and nearly eliminated in ancestral African Homo sapiens populations. The lack of diversity among extant human H2 haplotypes and its apparent ancient origin (2–3 million years ago) could be the result of either a founder effect 20 or a partial selective sweep of a particular H2 haplotype within the European population, as has been posited 6.
We present evidence that the inversion has occurred independently in both chimpanzees and humans. Although the data are limited, the finding of both orientations in the Bornean orangutan argue strongly that this particular region of chromosome 17 is prone to recurrent inversions and has likely toggled multiple times between the inverted and non-inverted state during the course of hominoid evolution (Fig. 5). In humans and chimpanzees, these changes have occurred in concert with the evolution of a more complex duplication architecture flanking the inverted region in humans. These findings are strikingly reminiscent of an evolutionary survey of the human FLNA-EMDA X chromosome inversion 24. Caceres and colleagues showed that the X chromosome inversion had occurred 10 times independently in 27 Eutherian lineages, and in each case the region was flanked by duplications in an inverted orientation. If taken as a general principle of genome evolution, these data suggest extraordinary breakpoint reuse for inversions and predict that some apparently fixed inversions between species may actually be polymorphic as a result of recurrence.
In our study we similarly find large, inverted segmental duplications flanking the inversion region (in humans and chimpanzees) and show that the H1-H2 inversion breakpoints map in close proximity to the LRRC37 core duplicon sequence within these inverted segmental duplications. Cores represent some of the most abundant and rapidly evolving duplicated sequences in the human genome, perhaps because they are prone to double strand breakage and/or positive selection 23,31,32. Moreover, such sequences have been shown to be associated with recurrent evolutionary events 23. Although there are more than eleven copies of the LRRC37 core duplicon on chromosome 17, evolutionary reconstruction in primate shows that proximal (H1) 17q21.31 locus corresponds to the ancestral position 32. Comparative analyses of the mouse sequence 8 and macaque genome 33 reveal that this region of the genome has been a hotbed for multiple inversions and other rearrangements during mammalian evolution—long before most of the hominoid duplication architecture emerged. We propose that inversion toggling is a longstanding evolutionary property of the 17q21.31 region, promoted, in part, because of its association with the LRRC37 core duplicon sequence. Most of the human and chimpanzee large segmental duplications flanking the inversions are, themselves, within an inverted orientation and it is possible that such structures were created as part of the double-strand DNA repair process 28,34. Such inverted segmental duplications, once formed, would reinforce and continue to promote recurrent inversion events via non-allelic homologous recombination.
Although our analysis of the unique sequences identified a H2-like sequence as the likely ancestral allele, a detailed comparison of the genomic architecture of the segmental duplications suggests that the extant H2 sequence is much more highly derived when compared to H1 or chimpanzee (Table 1, Fig. 3b). Phylogenetic analysis of the duplicated sequences supports extensive H2-specific sequence homogenization, perhaps as a result of gene conversion between proximal and distal segmental duplication blocks. Consequently, there are three times the number of duplicated basepairs in H2 when compared to chimpanzee or H1; these duplicated bases show higher sequence identity and ~95 kbp are in direct orientation on either side of the inversion. Orientation, length and degree of sequence identity between duplicated sequences are the most important parameters for non-allelic homologous recombination 35. In the case of H2, the orientation, proportion and sequence identity would all favor microdeletion on this chromosome haplotype when compared to H1. We show that at least one of the microdeletion breakpoints associated with children with developmental delay and mental retardation corresponds to a recently evolved H2-specific segmental duplication. We propose that it is not the inversion per se that promotes microdeletion and disease, but rather the configuration and structure of the segmental duplications which favor non-allelic homologous recombination on the inverted particular haplotype. Dramatic changes in copy-number, structure and homology of flanking segmental duplications may explain why inversion haplotypes predispose to other microdeletion syndromes 36,37.
METHODS
Segmental Duplication Detection
Segmental duplication content of the CRHR1-MAPT region was initially assessed by mapping whole genome shotgun sequence assembly reads from human, chimpanzee, orangutan and macaque against human chromosome 17 (build 36, chr17:40799295-42204344) and identifying regions of excess (≥ mean +3 standard deviation) depth-of-coverage and divergence as described previously 5. For all hominoids, sequence identity alignment thresholds were set at ≥94% with the exception of macaque, where a ≥88% identity threshold was used to capture more divergent macaque sequence reads aligned to the human genome. Three independent approaches were used to analyze the segmental duplication content of each clone-based sequence assembly. A BLAST-based comparison (WGAC) method 38 identified all sequence alignments ≥1 kb and ≥90% identity 38 among the four sequence assemblies. The whole genome shotgun sequence detection approach identified regions ≥10 kb in length with a significant excess of high-quality WGS reads 21 within overlapping 5 kb windows. WSSD analysis was based on an alignment of 22,590,543 chimpanzee WGS reads and 18,355,056 orangutan WGS reads against their BAC-based sequence assemblies. Finally, we annotated all human duplications by WU-BLAST alignments of a non-redundant dataset of human duplicons 32 against each assembly. High identity sequence alignments were generated using Miropeats 39 and visualized using two-way-mirror.pl (Bailey unpublished).
FISH Inversion Assay
Metaphase spreads were obtained from lymphoblast cell lines from two human HapMap individuals (YRI NA18507 and CEU NA12156, Coriell Cell Repository, Camden, NJ), nine chimpanzees (Clint, Katie, Logan, PTR14, PTR8, PTR9, PTR11, PTR12 and PTR13), four Sumatran orangutans (Susie, PPY1, PPY9, PPY10), one Bornean orangutan (PPY6), two bonobos (PPA1, PPA2) and three subspecies of macaque: MMU (Macaca mulatta), MAR (Macaca arctoides) and MFA (Macaca fascicularis). Inversions were detected using a bi-color FISH assay (fosmid probes, WIBR2-634F12+WIBR2-1948K20) and inversion genotype status confirmed using a reciprocal assay (fosmids WIBR2-634F12 and ABC9_41289800G20). Inversion genotyping accuracy was tested by comparing FISH genotypes versus a previously designed molecular assay (Supplementary Note). 24/24 human samples (3 H2 and 45 H1 chromosomes) were concordant between FISH and molecular assays. Probes were directly labeled by nick-translation with Cy3-dUTP (Perkin-Elmer) and labeled with fluorescein-dUTP (Enzo) as described previously. Each hybridization utilized 300 ng of labeled probe, 5 μg COT1 DNA (Roche), and 3 μg sonicated salmon sperm DNA at 37°C in 10 μl 2xSSC/50% formamide/10% dextran sulphate, followed by three posthybridization washes at 60°C in 0.1xSSC. Nuclei were stained with DAPI and digital images obtained using a Leica DMRXA2 epifluorescence microscope equipped with a cooled CCD camera (Princeton Instruments).
Sequence and Assembly
We constructed, sequenced and assembled minimal tiling paths of large-insert genomic clones for both human haplotypes (H1 & H2), an H2 oriented chimpanzee chromosome and an H2 oriented orangutan chromosome (see Supplementary Note for detailed clone order, sequence assembly and annotation). In humans, this entailed disentangling existing H1 and H2 RPCI-11 BACs and generating an additional 1.7 Mb of high quality finished sequence. In chimpanzee and orangutan, a minimum tiling path of BAC clones (chimp: CHORI-251; orangutan: CHORI-276) were sequenced to derive a consensus assembly (~2 Mb) that identified BACs containing inversion breakpoints. Orangutan consensus sequence was also extracted from the draft assembly Pongo pygmaeus-2.0.2 (http://genome.wustl.edu/genome_group_index.cgi). To verify the MAPT locus orientation, analyze flanking duplication architecture and measure evolutionary distance of haplotypes in chimpanzee and orangutan, we utilized the corresponding regions (human build36 chr17:40799295-42204344) from both whole genome and BAC-based consensus sequence assemblies. Sequences were compared using Miropeats and inversion breakpoint intervals were defined based on a consistent orientation shift between the aligned sequence assemblies.
Phylogenetic and Haplotype Analyses
An unrooted neighbor-joining 40 phylogram was constructed (MEGA pairwise deletion option) 41 based on a multiple sequence alignment (ClustalW) 42 of 219,165 bp within the inverted region. Genetic distances were computed using the Kimura 2-parameter method 43 and Tajima’s relative rate test (PPY-H1-H2; PTR-H1-H2) was used to assess branch length neutrality (p=0.22–0.81). Using chimpanzee as the outgroup, an estimated local substitution rate (9.0916×10−4 substitutions per site/million years) and the uncertainty in the chimpanzee-human divergence (5–7 mya), we calculated that the human H1 and H2 haplotypes diverged 1.9–2.7 million years ago. We compared 123 chromosomes (120 CEPH HapMap chromosomes, and the sequence of the H1, H2 and PTR haplotypes) using HapMap SNPs (Phase II HapMap release 21 phased-consensus at http://hapmap.org 44). SNP genotypes were assigned to the H1, H2 and PTR sequences using BLAT and regions of segmental duplication (including H2-specific duplications) were excluded. Haplotypes were assigned to the H1 or H2 class based on two diagnostic SNPs (rs1800547 and rs9468) as described previously 6. Errors in the inferred SNP phased haplotypes were manually corrected (Supplementary Note). We assessed haplotype diversity within the chimpanzee based on alignment of Clint fosmid end-sequence pairs (ESPs) to the BAC-based chimpanzee assembly (Supplementary Table 5, Supplementary Note).
Accession Numbers
The BAC clones that were used for sequence and assembly of the MAPT region in this study have been submitted to GenBank under the following accession numbers:(Human H1 assembly) AC091132, AC126544, AC217774, AC217771, CR936218, AC217773, AC005829, AC217777, AC138645, AC217780, (Human H2 assembly) AC217778, AC217769, AC138688, AC127032, AC217772, AC217779, BX544879, AC217770, AC225613, AC217768, AC139677, AC217775, (Chimpanzee assembly), AC185328, AC185293, AC187127, AC186740, AC185975, AC186440, AC185329, AC185979, AC186439, AC187126, AC185346, AC186739, AC185985, (Orangutan assembly)AC205775, AC206340, AC206276, AC207288, AC206550, AC206558, AC205859, AC206353, AC216075, AC206444, AC207097, AC216102, AC216058, AC216103.
Supplementary Material
Acknowledgments
We thank Tomas Marques, David Reich, Arcadi Navarro, Nick Patterson, Steve McCarroll, Tonia Brown and Kari Augustyn for critical comments and valuable discussions in the preparation of this manuscript. We thank Can Alkan for providing computational assistance. We are grateful to the members of the Broad Institute Sequence Platform (BISP) and Washington University Genome Sequencing Center (WUGSC) for generating clone-based sequencing data for this project. MCZ, AA, WCW, LWH, TAG, RKW, ADR, JW, JG and the BISP and WUGSC were supported by grants from the National Human Genome Research Institute. This work was supported in part by the Intramural Research Program of the National Institute on Aging, National Institutes of Health, Department of Health and Human Services, project number Z01 AG000957-05. This work was supported by NIH grants GM058815 and HG002385 to EEE and a Rosetta Inpharmatics Fellowship (Merck Laboratories) to ZJ. EEE is an investigator of the Howard Hughes Medical Institute.
References
- 1.Iafrate AJ, et al. Detection of large-scale variation in the human genome. Nat Genet. 2004;36:949–51. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 2.Sebat J, et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–8. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 3.Tuzun E, Bailey JA, Eichler EE. Recent segmental duplications in the working draft assembly of the brown Norway rat. Genome Res. 2004;14:493–506. doi: 10.1101/gr.1907504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.CSAC. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- 5.Cheng Z, et al. A genome-wide comparison of recent chimpanzee and human segmental duplications. Nature. 2005;437:88–93. doi: 10.1038/nature04000. [DOI] [PubMed] [Google Scholar]
- 6.Stefansson H, et al. A common inversion under selection in Europeans. Nat Genet. 2005;37:129–37. doi: 10.1038/ng1508. [DOI] [PubMed] [Google Scholar]
- 7.Gijselinck I, et al. Visualization of MAPT inversion on stretched chromosomes of tau-negative frontotemporal dementia patients. Hum Mutat. 2006;27:1057–9. doi: 10.1002/humu.20391. [DOI] [PubMed] [Google Scholar]
- 8.Cruts M, et al. Genomic architecture of human 17q21 linked to frontotemporal dementia uncovers a highly homologous family of low-copy repeats in the tau region. Hum Mol Genet. 2005;14:1753–62. doi: 10.1093/hmg/ddi182. [DOI] [PubMed] [Google Scholar]
- 9.Baker M, et al. Association of an extended haplotype in the tau gene with progressive supranuclear palsy. Hum Mol Genet. 1999;8:711–5. doi: 10.1093/hmg/8.4.711. [DOI] [PubMed] [Google Scholar]
- 10.Pittman AM, et al. The structure of the tau haplotype in controls and in progressive supranuclear palsy. Hum Mol Genet. 2004;13:1267–74. doi: 10.1093/hmg/ddh138. [DOI] [PubMed] [Google Scholar]
- 11.Myers AJ, et al. A survey of genetic human cortical gene expression. Nat Genet. 2007;39:1494–9. doi: 10.1038/ng.2007.16. [DOI] [PubMed] [Google Scholar]
- 12.Conrad C, et al. Genetic evidence for the involvement of tau in progressive supranuclear palsy. Ann Neurol. 1997;41:277–81. doi: 10.1002/ana.410410222. [DOI] [PubMed] [Google Scholar]
- 13.Rademakers R, et al. High-density SNP haplotyping suggests altered regulation of tau gene expression in progressive supranuclear palsy. Hum Mol Genet. 2005;14:3281–92. doi: 10.1093/hmg/ddi361. [DOI] [PubMed] [Google Scholar]
- 14.Sundar PD, et al. Two sites in the MAPT region confer genetic risk for Guam ALS/PDC and dementia. Hum Mol Genet. 2007;16:295–306. doi: 10.1093/hmg/ddl463. [DOI] [PubMed] [Google Scholar]
- 15.Myers AJ, et al. The H1c haplotype at the MAPT locus is associated with Alzheimer’s disease. Hum Mol Genet. 2005;14:2399–404. doi: 10.1093/hmg/ddi241. [DOI] [PubMed] [Google Scholar]
- 16.Sharp AJ, et al. Discovery of previously unidentified genomic disorders from the duplication architecture of the human genome. Nat Genet. 2006 doi: 10.1038/ng1862. [DOI] [PubMed] [Google Scholar]
- 17.Koolen DA, et al. A new chromosome 17q21.31 microdeletion syndrome associated with a common inversion polymorphism. Nat Genet. 2006;38:999–1001. doi: 10.1038/ng1853. [DOI] [PubMed] [Google Scholar]
- 18.Shaw-Smith C, et al. Microdeletion encompassing MAPT at chromosome 17q21.3 is associated with develop-mental delay and learning disability. Nat Genet. 2006;38:1032–7. doi: 10.1038/ng1858. [DOI] [PubMed] [Google Scholar]
- 19.Evans W, et al. The tau H2 haplotype is almost exclusively Caucasian in origin. Neurosci Lett. 2004;369:183–5. doi: 10.1016/j.neulet.2004.05.119. [DOI] [PubMed] [Google Scholar]
- 20.Hardy J, et al. Evidence suggesting that Homo neanderthalensis contributed the H2 MAPT haplotype to Homo sapiens. Biochem Soc Trans. 2005;33:582–5. doi: 10.1042/BST0330582. [DOI] [PubMed] [Google Scholar]
- 21.Bailey JA, et al. Recent segmental duplications in the human genome. Science. 2002;297:1003–7. doi: 10.1126/science.1072047. [DOI] [PubMed] [Google Scholar]
- 22.Redon R, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–54. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson ME, et al. Recurrent duplication-driven transposition of DNA during hominoid evolution. Proc Natl Acad Sci U S A. 2006 doi: 10.1073/pnas.0605426103. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Caceres M, Sullivan RT, Thomas JW. A recurrent inversion on the eutherian X chromosome. Proc Natl Acad Sci U S A. 2007;104:18571–6. doi: 10.1073/pnas.0706604104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ebersberger I, et al. Mapping human genetic ancestry. Mol Biol Evol. 2007;24:2266–76. doi: 10.1093/molbev/msm156. [DOI] [PubMed] [Google Scholar]
- 26.Gagneux P. The genus Pan: population genetics of an endangered outgroup. Trends Genet. 2002;18:327–30. doi: 10.1016/s0168-9525(02)02695-1. [DOI] [PubMed] [Google Scholar]
- 27.Yu N, et al. Low nucleotide diversity in chimpanzees and bonobos. Genetics. 2003;164:1511–8. doi: 10.1093/genetics/164.4.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kehrer-Sawatzki H, Sandig CA, Goidts V, Hameister H. Breakpoint analysis of the pericentric inversion between chimpanzee chromosome 10 and the homologous chromosome 12 in humans. Cytogenet Genome Res. 2005;108:91–7. doi: 10.1159/000080806. [DOI] [PubMed] [Google Scholar]
- 29.Rosenberg NA, et al. Genetic structure of human populations. Science. 2002;298:2381–5. doi: 10.1126/science.1078311. [DOI] [PubMed] [Google Scholar]
- 30.Jakobsson MSS, Scheet P, Gibbs JR, VanLiere JM, Fung HC, Szpiech ZA, Degnan JH, Wang K, Guerreiro R, Bras JM, Schymick JC, Hernandez D, Traynor BJ, Simon-Sanchez J, Matarin M, Britton A, van de Leemput J, Rafferty I, Bucan M, Cann HM, Hardy JA, Rosenberg NA, Singleton AB. Genotype, haplotype, and copy number variation in worldwide human populations. Nature. 2008 doi: 10.1038/nature06742. in press. [DOI] [PubMed] [Google Scholar]
- 31.Zody MC, et al. Analysis of the DNA sequence and duplication history of human chromosome 15. Nature. 2006;440:671–5. doi: 10.1038/nature04601. [DOI] [PubMed] [Google Scholar]
- 32.Jiang Z, et al. Ancestral reconstruction of segmental duplications reveals punctuated cores of human genome evolution. Nat Genet. 2007;39:1361–8. doi: 10.1038/ng.2007.9. [DOI] [PubMed] [Google Scholar]
- 33.Cardone MF, et al. Hominoid chromosomal rearrangements on 17q map to complex regions of segmental duplication. Genome Biol. 2008;9:R28. doi: 10.1186/gb-2008-9-2-r28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Casals F, Navarro A. Chromosomal evolution: inversions: the chicken or the egg? Heredity. 2007;99:479–80. doi: 10.1038/sj.hdy.6801046. [DOI] [PubMed] [Google Scholar]
- 35.Lupski JR. Genomic disorders: structural features of the genome can lead to DNA rearrangements and human disease traits. Trends Genet. 1998;14:417–22. doi: 10.1016/s0168-9525(98)01555-8. [DOI] [PubMed] [Google Scholar]
- 36.Osborne LR, et al. A 1.5 million-base pair inversion polymorphism in families with Williams-Beuren syndrome. Nat Genet. 2001;29:321–5. doi: 10.1038/ng753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gimelli G, et al. Genomic inversions of human chromosome 15q11-q13 in mothers of Angelman syndrome patients with class II (BP2/3) deletions. Hum Mol Genet. 2003;12:849–58. doi: 10.1093/hmg/ddg101. [DOI] [PubMed] [Google Scholar]
- 38.Bailey JA, Yavor AM, Massa HF, Trask BJ, Eichler EE. Segmental duplications: organization and impact within the current human genome project assembly. Genome Res. 2001;11:1005–17. doi: 10.1101/gr.187101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parsons J. Miropeats: graphical DNA sequence comparisons. Comput Appl Biosci. 1995;11:615–619. doi: 10.1093/bioinformatics/11.6.615. [DOI] [PubMed] [Google Scholar]
- 40.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–25. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 41.Tamura K, Dudley J, Nei M, Kumar S. MEGA4:Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Molecular Biology and Evolution. 2007 doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 42.Higgins DG. CLUSTAL V: multiple alignment of DNA and protein sequences. Methods Mol Biol. 1994;25:307–18. doi: 10.1385/0-89603-276-0:307. [DOI] [PubMed] [Google Scholar]
- 43.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–20. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 44.IHMC. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–61. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goodman M. The genomic record of Humankind’s evolutionary roots. Am J Hum Genet. 1999;64:31–9. doi: 10.1086/302218. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.