Abstract
αB-crystallin, a member of the small heat shock protein family, possesses chaperone-like function. Recently, it has been shown that a missense mutation in αB-crystallin, R120G, is genetically linked to a desmin-related myopathy as well as to cataracts [Vicart, P., Caron, A., Guicheney, P., Li, A., Prevost, M.-C., Faure, A., Chateau, D., Chapon, F., Tome, F., Dupret, J.-M., et al. (1998) Nat. Genet. 20, 92–95]. By using α-lactalbumin, alcohol dehydrogenase, and insulin as target proteins, in vitro assays indicated that R120G αB-crystallin had reduced or completely lost chaperone-like function. The addition of R120G αB-crystallin to unfolding α-lactalbumin enhanced the kinetics and extent of its aggregation. R120G αB-crystallin became entangled with unfolding α-lactalbumin and was a major portion of the resulting insoluble pellet. Similarly, incubation of R120G αB-crystallin with alcohol dehydrogenase and insulin also resulted in the presence of R120G αB-crystallin in the insoluble pellets. Far and near UV CD indicate that R120G αB-crystallin has decreased β-sheet secondary structure and an altered aromatic residue environment compared with wild-type αB-crystallin. The apparent molecular mass of R120G αB-crystallin, as determined by gel filtration chromatography, is 1.4 MDa, which is more than twice the molecular mass of wild-type αB-crystallin (650 kDa). Images obtained from cryoelectron microscopy indicate that R120G αB-crystallin possesses an irregular quaternary structure with an absence of a clear central cavity. The results of this study show, through biochemical analysis, that an altered structure and defective chaperone-like function of αB-crystallin are associated with a point mutation that leads to a desmin-related myopathy and cataracts.
αB-crystallin is a major lens protein that has a structural role in maintaining the transparency of the lens. αB-crystallin has been shown to be expressed outside of the lens in a number of tissues such as skeletal and cardiac muscle and to lesser extents in skin, brain, and kidney, suggesting that it has a general cellular function (1–2). Expression of αB-crystallin confers protection to cells against thermal (3), osmotic (4), and oxidative insults (5). αB-crystallin also functions as a regulator of apoptosis in fibroblast L-929 cells (6). αB-crystallin and the related small heat shock proteins (HSP) exist as high-molecular-mass oligomeric proteins, consisting of roughly 32 subunits, with the variable assembly having a large internal cavity (7). However, the size of α-crystallin can change depending on a number of parameters, such as pH, ionic strength, as well as the age of the tissue from which it is isolated (8–11). The dynamic quaternary structure of αB-crystallin may allow the performance of different functions under varying cellular conditions.
The nonlenticular function of αB-crystallin may be related to its chaperone-like property, which allows it to bind irreversibly to denaturing proteins, preventing the formation of large light-scattering aggregates (12). In the lens, this function is of paramount importance in preventing cataracts, as proteins are subject to osmotic and oxidative stresses over the lifetime of the organism and are highly susceptible to denaturation and aggregation (13). In addition to binding a wide spectrum of denaturing proteins in vitro, α-crystallin binds and inhibits the assembly of the type III intermediate filaments vimentin and glial fibrillary acidic protein (14). It has been suggested that α-crystallin may modulate the equilibrium between soluble and insoluble type III intermediate filaments in the cell (14). Moreover, there is a specific interaction between αB-crystallin and desmin, a type III intermediate filament, in cardiac muscle cells (15). On heat shock of cells, αB-crystallin has been found to be associated with sarcomeric structures, suggesting that it may protect cytoskeletal function (16). α-Crystallin can bind and prevent the cytochalasin-induced depolymerization of actin (17). These results suggest that αB-crystallin may play a role in the remodeling of the cytoskeleton that occurs during development and cell differentiation as well as after heat shock.
Interestingly, αB-crystallin is overexpressed in a number of neurological disorders such as Alzheimer’s disease (18), diffuse Lewy body disease (19), Creutzfeldt–Jacob disease (20), and most prominently in Alexander’s disease, in which it forms the major component of Rosenthal fibers (21). In many of these neurological diseases, αB-crystallin is found to be associated with intermediate filaments in cytoplasmic inclusion bodies (21), but the role that αB-crystallin may play in these diseases is currently not known. Recently, a missense mutation in αB-crystallin, R120G, has been shown to cosegregate with a desmin-related myopathy (DRM) in a French family (22). DRMs are usually adult-onset neuromuscular diseases characterized by large accumulations of aggregates of cytoplasmic desmin in conjunction with other proteins. In the French family, these spheroid inclusion bodies were found to contain large amounts of R120G αB-crystallin with desmin. This mutation was, to our knowledge, the first example of a small HSP being linked to a disease state.
To determine the effects that the missense mutation R120G had on the structure and function of αB-crystallin, we have constructed, expressed, and purified human recombinant R120G αB-crystallin. We have characterized the chaperone function of R120G αB-crystallin by using yeast alcohol dehydrogenase, insulin, and α-lactalbumin as target proteins. In addition, we have examined the secondary and tertiary structure of R120G αB-crystallin by using both near and far UV CD and the quaternary structure of αB-crystallin by using cryoelectron microscopy (cryo-EM).
MATERIALS AND METHODS
Construction, Preparation, and Purification of Proteins.
α-Lactalbumin, yeast alcohol dehydrogenase, and insulin were obtained from Sigma. Lens α-crystallin was prepared from bovine lenses and purified as described (12). Human recombinant wild-type and R120G αB-crystallins were constructed, expressed, and purified as described (23).
Molecular Mass Determination.
Size determinations were carried out by using gel filtration chromatography with a Superose HR-6 column (Amersham Pharmacia) equilibrated in 50 mM sodium phosphate (pH 7.0) containing 100 mM NaCl. A calibration curve was generated by using γ-globulin (158 kDa), apoferritin (443 kDa), and GroEL (801.5 kDa) as molecular mass standards.
CD and Fluorescence Spectra of Wild-Type and R120G αB-Crystallin.
All CD spectra were determined by using a J-600 spectropolarimeter (Jasco, Easton, MD) interfaced with a temperature-controlled water circulator. For the near and far UV regions, 1.0-cm and 0.2-mm path-length cells were used, respectively. Fluorescence spectra measurements were performed in a Fluorolog-2 fluorometer (Spex Industries, Metuchen, NJ) by using a 1.0-cm path-length cuvette with excitation and emission slits set at 2.0 nm.
Cryo-EM of Wild-Type and R120G αB-Crystallin.
Cryo-EM was performed on a Philips CM120 transmission electron microscope (FEI, Hillsboro, OR) equipped with a Gatan (Pleasanton, CA) cryo holder at a defocus setting of −1.5 μm as described (7), and 15 cryoelectron micrographs were collected of unstained, frozen-hydrated wild-type and R120G αB-crystallin assemblies. To accentuate predominant features, 200 particle images of each assembly type were extracted and filtered to 4-nm resolution.
Assays of Chaperone-Like Activity.
The aggregation of proteins on denaturation was monitored by measuring the apparent absorption caused by light scattering (23). Assays for chaperone-like function of wild-type and R120G αB-crystallin were performed at 37°C by using insulin, α-lactalbumin, and alcohol dehydrogenase as target proteins (23). For alcohol dehydrogenase, a temperature of 42°C was used.
Analytical Methods.
Protein concentrations were determined by the method of Gill and von Hippel (24). Estimation of secondary structure parameters was performed by using the self-consistent method of analysis with the Hennessey and Johnson submethod (described in ref. 25).
RESULTS
Effect of Mutation R120G on the Molecular Mass of αB-Crystallin.
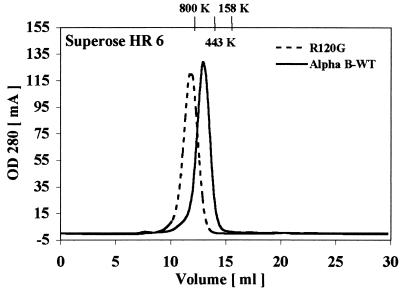
The oligomeric structure of R120G αB-crystallin was determined by using gel filtration chromatography. Consistent with previously reported results, wild-type αB-crystallin had a molecular mass of 650 kDa (26). However, the size of R120G αB-crystallin was much larger and estimated to be 1.4 MDa (Fig. 1).
Figure 1.
Gel filtration chromatography performed on a Superose HR-6 column of human wild-type and R120G αB-crystallin.
Characterization of the Secondary and Tertiary Structure of R120G αB-Crystallin.
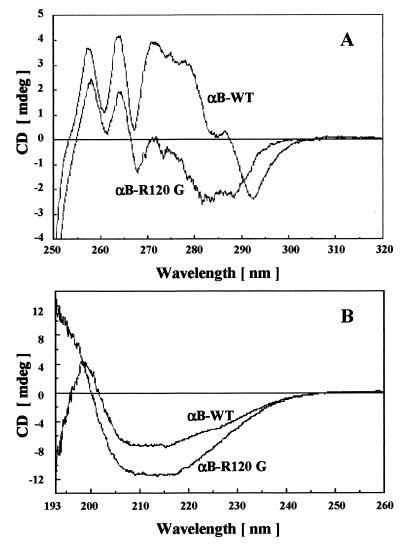
To determine the effect that mutation R120G had on secondary and tertiary structure of αB-crystallin, we have used near and far UV CD as well as tryptophan fluorescence. The far UV CD spectra of wild-type αB-crystallin had a characteristic minimum at 217 nm, indicative of a β-sheet-containing protein (Fig. 2B). Secondary structure estimates obtained from the spectra indicated that the backbone conformation of the protein consisted primarily of β-sheet with some α-helix, consistent with previously published estimates (23). Although R120G αB-crystallin also had a minimum at 217 nm, its CD spectrum was significantly more intense—between 205 nm and 225 nm—than wild-type αB-crystallin, and there were also significant differences between the two spectra in the 192- to 200-nm region (Fig. 2B). These differences in spectra were translated into differences in secondary structure, as the algorithm used for calculation showed R120G αB-crystallin possessed decreased β-sheet content with a concomitant increase in α-helix compared with wild-type αB-crystallin (see the legend of Fig. 2).
Figure 2.
Near and far UV CD of human wild-type and R120G αB-crystallin. (A) Near UV CD spectra of wild-type and R120G αB-crystallin. Each spectrum represents the average of 32 scans, and the concentration for each sample was 2.2 mg/ml. (B) Far UV CD spectra of wild-type and R120G αB-crystallin. Each spectrum represents the average of 16 scans with each sample containing a 1.0-mg/ml concentration of wild-type or R120G αB-crystallin. Estimates of secondary structure were performed as described in Materials and Methods. For wild-type αB-crystallin, the secondary structure was predicted to contain 10% α-helix, 44% β-sheet, 16% β-turns, and 30% other. For R120G αB-crystallin, the secondary structure was predicted to contain 15% α-helix, 33% β-sheet, 19% β-turns, and 33% other.
The near UV CD spectra of proteins reflect mainly the contribution of aromatic amino acids to protein tertiary structure and are very sensitive to structural perturbations. The near UV CD spectra of wild-type and R120G αB-crystallin were markedly different (Fig. 2A). Taken together, both the near and far UV CD spectra suggest that the mutation R120G introduced structural alterations into the αB-crystallin molecule. Interestingly, the tryptophan fluorescence spectra of wild-type and R120G αB-crystallin possessed similar wavelength emission maxima and intensities, suggesting that the environment of the tryptophan residues are relatively unchanged by the R120G mutation (data not shown). The near UV CD spectra are exquisitely sensitive to changes in fine structure and environment of tryptophan and tyrosine residues, and subtle differences in structure and environment not illustrated by fluorescence can be observed by CD.
Characterization of the Quaternary Structure of R120G αB-Crystallin.
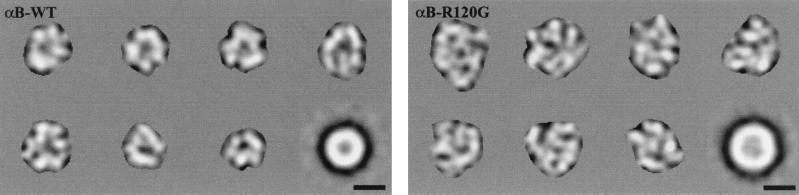
To determine the effect that mutation R120G had on the quaternary structure of αB-crystallin, we have used cryo-EM; seven representative images of wild-type and R120G αB-crystallin are shown in Fig. 3. The wild-type αB-crystallin assemblies appear in projection to have a roughly spherical protein shell with a central cavity, consistent with previous results (7). In contrast, the R120G αB-crystallin assemblies are more irregular in shape and do not appear to have a well defined central cavity. The rotationally averaged sum of 200 particle images is also presented for each assembly type. These images yield an average diameter of 13 nm for wild-type αB-crystallin and 16 nm for R120G αB-crystallin. In addition, the average image of the R120G αB-crystallin assembly has a less prominent central cavity than the average image of wild-type αB-crystallin assembly.
Figure 3.
Cryo-EM of wild-type and R120G αB-crystallin. Shown are seven representative images of wild-type and R120G αB-crystallin assemblies after low-pass filtering to 4-nm resolution, translationally aligning, and masking. The lower rightmost image of each set is a rotationally averaged sum of 200 filtered and translationally aligned images. The protein density appears white. (Bars = 10 nm.)
Effect of Mutation R120G on the Chaperone-Like Properties of Human Recombinant αB-Crystallin.
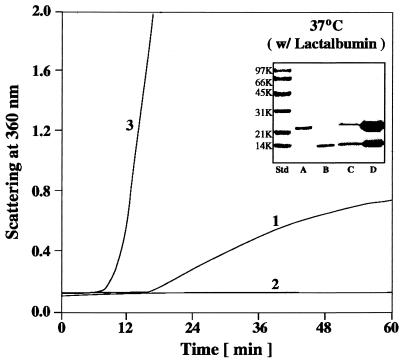
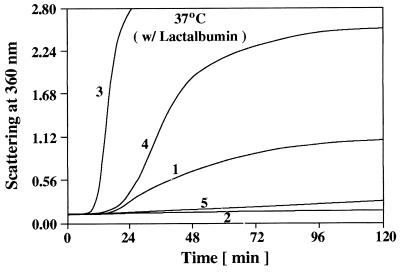
In the presence of DTT, the disulfide bonds of bovine α-lactalbumin will be reduced, and the molecule will unfold and aggregate (23, 26). The kinetics of aggregation of α-lactalbumin can be monitored by measuring the absorption caused by light scatter (Fig. 4, curve 1). At a 1:1 (wt/wt) ratio of α-lactalbumin/αB-crystallin, wild-type αB-crystallin completely suppressed the DTT-induced aggregation of α-lactalbumin at 37°C (Fig. 4, curve 2). In contrast, at a 1:1 (wt/wt) ratio of α-lactalbumin/R120G αB-crystallin, on addition of DTT, the solution rapidly became turbid (Fig. 4, curve 3). The addition of R120G αB-crystallin to α-lactalbumin under reducing conditions actually enhanced the kinetics and extent of its aggregation. In fact, R120G αB-crystallin became entangled with unfolding protein and was a major part of the resulting insoluble pellet (Fig. 4, Inset). Doubling the concentration of R120G αB-crystallin in solution with α-lactalbumin did not alter its aggregation curve (data not shown), thus R120G αB-crystallin completely lost its chaperone-like function with this substrate. It should be emphasized that DTT and heat (37°C) had no effect on the structure of wild-type or R120G αB-crystallin when in solution alone (data not shown).
Figure 4.
Aggregation of α-lactalbumin after the addition of 20 mM DTT at 37°C and pH 6.7. Curve 1, 0.25 mg of α-lactalbumin; curve 2, 0.25 mg of α-lactalbumin plus 0.25 mg of wild-type αB-crystallin; curve 3, 0.25 mg of α-lactalbumin plus 0.25 mg of R120G αB-crystallin. The Inset is an SDS/polyacrylamide gel that illustrates the particulate and soluble fractions obtained after a 60-min incubation of R120G αB-crystallin with α-lactalbumin in the presence of DTT, as shown in curve 3. Lane A, αB-crystallin; lane B, α-lactalbumin; lane C, soluble fraction of α-lactalbumin plus R120G αB-crystallin; lane D, particulate fraction of α-lactalbumin plus R120G αB-crystallin.
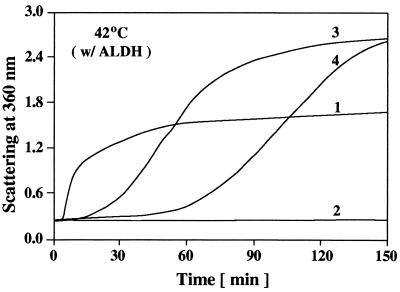
Addition of 2 mM EDTA to alcohol dehydrogenase will chelate its zinc cofactor and destabilize the enzyme, thus allowing the thermally induced aggregation of alcohol dehydrogenase (Fig. 5, curve 1) to occur at temperatures where the structure of αB-crystallin itself is not changing significantly (37–42°C). Heat (37–42°C) and EDTA had no effect on the structure of R120G αB-crystallin as judged by CD measurements (data not shown). At a 1:1 (wt/wt) ratio of alcohol dehydrogenase/αB-crystallin, wild-type αB-crystallin completely prevented the thermally induced aggregation of alcohol dehydrogenase (Fig. 5, curve 2). At the same weight ratio, R120G αB-crystallin was ineffective in suppressing the aggregation of alcohol dehydrogenase and actually enhanced the extent of its aggregation (Fig. 5, curve 3). Doubling the concentration of R120G αB-crystallin slowed the kinetics but not the extent of aggregation of unfolding alcohol dehydrogenase (Fig. 5, curve 4). On incubation of R120G αB-crystallin with unfolding alcohol dehydrogenase (Fig. 5, curves 3 and 4), R120G αB-crystallin became a major part of the resulting insoluble pellet (data not shown).
Figure 5.
Aggregation of alcohol dehydrogenase induced by heat (42°C) and 2 mM EDTA. Curve 1, 0.2 mg of alcohol dehydrogenase; curve 2, 0.2 mg of alcohol dehydrogenase plus 0.2 mg of wild-type αB-crystallin; curve 3, 0.2 mg of alcohol dehydrogenase plus 0.2 mg of R120G αB-crystallin; curve 4, 0.2 mg of alcohol dehydrogenase plus 0.4 mg of R120G αB-crystallin.
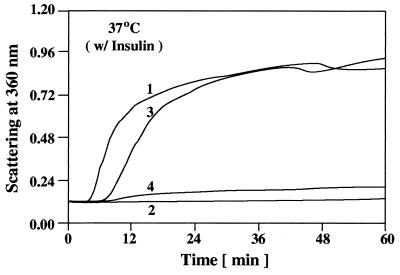
In the presence of DTT, at physiological temperatures (25–37°C), the disulfide bonds connecting the insulin A and B chains will break and the insulin B chain will unfold and aggregate (Fig. 6, curve 1). At a 1:1 (wt/wt) ratio of insulin/αB-crystallin, wild-type αB-crystallin completely prevented the aggregation of the unfolding insulin B chain (Fig. 6, curve 2). At the same weight ratio, R120G αB-crystallin was not effective in preventing the DTT-induced aggregation of the insulin B chain (Fig. 6, curve 3). SDS/PAGE analysis of the soluble and insoluble (pellet) fractions obtained from incubating R120G αB-crystallin with insulin (Fig. 6, curve 3), indicated the presence of R120G αB-crystallin as a major part of the pellet (data not shown). In contrast to α-lactalbumin and alcohol dehydrogenase, doubling the amount of R120G αB-crystallin [i.e., a 1:2 (wt/wt) ratio of insulin/R120G αB-crystallin] significantly prevented the aggregation of the insulin B chain (Fig. 6, curve 4).
Figure 6.
Aggregation of the insulin B chain after the addition of 20 mM DTT at 37°C. Curve 1, 0.1 mg of insulin; curve 2, 0.1 mg of insulin plus 0.1 mg of wild-type αB-crystallin; curve 3, 0.1 mg of insulin plus 0.1 mg of R120G αB-crystallin; curve 4, 0.1 mg of insulin plus 0.2 mg of R120G αB-crystallin.
Effect of Mixing Wild-Type αB-Crystallin with R120G αB-Crystallin on Chaperone-Like Function.
We have used both fluorescence resonance energy transfer (27) as well as gel filtration chromatography to demonstrate subunit exchange between wild-type and R120G αB-crystallin subunits. On mixing equal amounts of wild-type αB-crystallin with R120G αB-crystallin subunits for 2 h at 37°C, the two populations will integrate completely, forming high-molecular-mass heterooligomeric complexes. These heterooligomeric complexes, when chromatographed on a gel filtration system as described in Fig. 1, yield a single symmetrical peak with an apparent molecular mass of 1.2 million Da, indicating formation of a heterooligomeric complex. Patients in the family described by Vicart et al. (22), have one chromosomal copy of normal αB-crystallin and one copy of the mutant allele. This chromosomal arrangement implies that, in muscle cells of these patients, an αB-crystallin molecule consists of complexes containing an equal amount of normal and mutant αB-crystallin subunits. To understand better what may occur in vivo, we have produced complexes containing different ratios of wild-type and R120G αB-crystallin subunits and have determined their chaperone-like function. Similar to the results shown in Fig. 4, R120G αB-crystallin enhanced the kinetics and extent of aggregation of unfolding α-lactalbumin (Fig. 7, curve 3), whereas wild-type αB-crystallin completely prevented the aggregation of unfolding α-lactalbumin (Fig. 7, curve 2). A 1:1 (wt/wt) complex of wild-type and R120G αB-crystallin subunits, containing enough wild-type αB-crystallin to prevent completely the aggregation of unfolding α-lactalbumin alone, enhanced the kinetics and extent of aggregation of unfolding α-lactalbumin (Fig. 7, curve 4). However, this aggregation curve indicated a lower extent and slower kinetics of aggregation than the aggregation curve that resulted when R120G αB-crystallin alone was added to unfolding α-lactalbumin (Fig. 7, compare curves 3 and 4). A complex containing a 2:1 (wt/wt) ratio of wild-type/R120G αB-crystallin subunits, significantly prevented the aggregation of unfolding α-lactalbumin (Fig. 7, curve 5).
Figure 7.
Chaperone-like function of heterooligomers containing wild-type and R120G αB-crystallin with α-lactalbumin as a substrate. Wild-type αB-crystallin was premixed with R120G αB-crystallin for 2 h at 37°C to form complexes containing different ratios of wild-type αB-crystallin/R120G αB-crystallin subunits. Curve 1, 0.25 mg of α-lactalbumin; curve 2, 0.25 mg of α-lactalbumin plus 0.125 mg of wild-type αB-crystallin; curve 3, 0.25 mg of α-lactalbumin plus 0.25 mg of R120G αB-crystallin; curve 4, 0.25 mg of α-lactalbumin plus a complex containing 0.125 mg of wild-type αB-crystallin and 0.125 mg of R120G αB-crystallin; curve 5, 0.25 mg of α-lactalbumin plus a complex containing 0.167 mg of wild-type αB-crystallin and 0.083 mg of R120G αB-crystallin.
Effect of Mixing Native α-Crystallin with R120G αB-Crystallin on Chaperone-Like Function.
In the lens, an α-crystallin molecule consists of two highly homologous subunits, αA and αB, with the complex containing a 3:1 ratio of αA to αB subunit (12). Outside of the lens, αA- and αB-crystallin exist independently of each other. Although αA-crystallin is expressed almost exclusively in the lens, αB-crystallin has a ubiquitous tissue distribution, with particularly pronounced expression in the lens, heart, and skeletal muscle (1–2). To assess more closely what may occur in the lenses of patients described by Vicart et al. (22), we have formed complexes consisting of native α-crystallin and R120G αB-crystallin subunits and determined their chaperone-like function. A 1:1 (wt/wt) mixture of native α-crystallin and R120G αB-crystallin subunits, containing enough native α-crystallin to completely suppress aggregation of α-lactalbumin alone, showed reduced chaperone-like function (data not shown). These results were similar to what occurred when a 1:1 (wt/wt) complex between wild-type and R120G αB-crystallin subunits was incubated with unfolding α-lactalbumin (Fig. 7, curve 4). This result suggests that formation of a complex of native α-crystallin with R120G αB-crystallin subunits disrupted the chaperone-like function of native α-crystallin. Complexes containing increased amounts of α-crystallin rescued the chaperone-like function of R120G αB-crystallin (data not shown).
DISCUSSION
DRMs are inherited neuromuscular disorders that are characterized by excess accumulations of desmin, often in the form of cytoplasmic inclusion bodies (28). Cytoplasmic inclusion bodies are the hallmark of protein-folding diseases such as DRMs. Recently, Vicart et al. (22) established that a DRM was genetically linked to the point mutation R120G in αB-crystallin. Patients harboring the missense mutation R120G in αB-crystallin show symptoms of muscle fatigue, cardiomyopathy, and cataracts (22). Interestingly, αB-crystallin gene knockout mice also have muscle abnormalities (29) reminiscent of DRMs. Taken together, these studies suggest that one of the major in vivo functions of αB-crystallin is to interact with the intermediate filament desmin.
It has been shown that the point mutation R116C of αA-crystallin, the residue corresponding to R120G of αB-crystallin, is genetically linked to an autosomal dominant congenital cataract (30). This link is not surprising, because, in the eye lens, αA-crystallin is the predominant protein. In light of the fact that many investigators have found the structure and chaperone-like function of αA- and αB-crystallin to be quite refractory to point mutations (26, 31), it was somewhat surprising that single point mutations in the α-crystallins were linked to disease states. However, it has been shown that certain truncations and insertions of αA-crystallin result in a loss of its chaperone-like function. For example, C-terminal truncation of αA-crystallin at residue 157 resulted in a loss of its chaperone-like function as well as an increase in its molecular mass (32). In addition, insertion of a tryptophan residue at position 172 of αA-crystallin markedly reduced its chaperone-like function and decreased its thermal stability (33). Interestingly, in rodents, a 23-aa natural insertion of αA-crystallin, occurring between residues 63 and 64, known as αA-insert (34), and a randomly generated point mutation close to this site, D69S αA-crystallin (35), resulted in reduced chaperone-like function. Without additional structural information, it is difficult to speculate where these residues are located in relation to the conserved R116 of αA or R120 of αB-crystallin and why these truncations, insertions, and the D69S point mutation have a detrimental effect on the chaperone-like function of αA-crystallin.
How might a single point mutation in αB-crystallin lead to such a marked change in its chaperone-like function? Arg-120 of αB-crystallin lies in the most highly conserved region of the small HSP family (36). Recently, residues Y109 through L120 of αA-crystallin, which span this conserved region, were scanned sequentially with cysteine. By using site-directed spin labeling, these residues were shown to form a β-strand located near a subunit interface (37). R116 αA-crystallin was shown to exist in a buried environment, with virtually no accessibility to aqueous solvent. Charged amino acids that are buried are usually paired with a residue of opposite charge and often have functional importance (38). Mutation of R120 to glycine in αB-crystallin suggests that an unpaired negative charge may reside in the interior of this protein or at a subunit interface. It has been shown that T4 lysozyme is destabilized when an unpaired charged residue is buried within its hydrophobic core (39). Furthermore, arginine is a charged and bulky amino acid that has been replaced by the small, uncharged amino acid glycine. Large to small mutations in the core of T4 lysozyme resulted in the formation of destabilizing internal cavities (40).
The core building block of the quaternary structure of αB-crystallin and the related small HSPs is the α-crystallin domain (36). This domain is a region of ≈100 aa that contains most of the C-terminal region of αB-crystallin. Recently, the crystal structure of small HSP 16.5 from Methanococcus jannaschii, which contains an α-crystallin domain, was determined at a 2.9-Å resolution (41). The monomeric folding unit was a composite β-sandwich, in which one of the β-strands comes from a neighboring molecule. Arg-107 of HSP 16.5, the residue corresponding to R120 of αB-crystallin, exists in a highly constrained and buried environment. The guanidinium group of Arg-107 forms a hydrogen bond with the carbonyl oxygen of Gly-41 in the backbone of a loop between two β-sheets where it is thought to contribute to the integrity of the β-sheet arrangement (42). Based on our results with CD and secondary structural analysis, we conclude that R120G αB-crystallin possesses a reduced amount of β-sheet secondary structure and an altered environment of aromatic residues compared with wild-type αB-crystallin. In addition to the potentially destabilizing effects at residue 120, glycine has a poor propensity to form a β-sheet (43). The R120G mutation may have disrupted the formation of β-sheet structure. The data presented in this paper indicate that the R120G mutation in αB-crystallin significantly altered the secondary, tertiary, as well as quaternary structure of the αB-crystallin molecule.
We have used three different target proteins to analyze the chaperone-like function of R120G αB-crystallin. Incubation of R120G αB-crystallin with unfolding α-lactalbumin enhanced the kinetics and extent of its aggregation (Fig. 4). Interestingly, increasing the concentration of R120G αB-crystallin incubated with unfolding α-lactalbumin did not alter the aggregation curve. In contrast, increasing the concentration of R120G αB-crystallin incubated with unfolding alcohol dehydrogenase slowed the kinetics of its aggregation (Fig. 5). Moreover, incubation of an increased concentration of R120G αB-crystallin with insulin significantly prevented the aggregation of the insulin B chain (Fig. 6). The fact that R120G αB-crystallin possesses substrate-dependent chaperone-like function suggests that wild-type αB-crystallin may also show substrate specificity when interacting with target proteins. The mutation R120G may have altered the structure of αB-crystallin such that an underlying specificity in chaperone-like function occurs. It is also important to note that it may not be a loss of chaperone-like function of αB-crystallin, in regard to its ability to prevent the aggregation of unfolding proteins, that is the underlying cause of the DRM or cataract. The mutation of R120G in αB-crystallin may lead to faulty interactions with proteins such as desmin in their native state, which results in aggregation of desmin with R120G αB-crystallin. Further experimentation is necessary to investigate this possibility.
The experiments that were performed in this study used recombinant αB-crystallin that was expressed in Escherichia coli. The point may be raised that αB-crystallin expressed in E. coli may not undergo the same posttranslational modifications that would occur in a mammalian expression system. However, we and others have found that the structure and chaperone-like function of recombinant αA- and αB-crystallin to be similar to native lens α-crystallin (23, 44). Interestingly, αB-crystallin is phosphorylated at three different sites in a cAMP-dependent manner, yet the role that phosphorylation plays in altering its structure or chaperone-like function is unclear (45). It is possible that phosphorylation and other posttranslational modifications of αB-crystallin may have undiscovered effects on its structure and function in vivo and may play a role in DRM or cataract formation.
There are many diseases that occur as a result of protein misfolding in the cell. Many of these diseases are the result of a point mutation that leads to an inability of an essential protein to fold correctly (46). One of the many functions of molecular chaperones is to prevent misassociations and to promote proper folding. The data presented in this paper illustrate the altered structure and defective chaperone-like function of R120G αB-crystallin and provide insight into the underlying mechanisms by which this point mutation causes DRM, cataracts, and potentially other diseases.
Acknowledgments
This work was supported by National Eye Institute Grants R37-EY3897 (to J.H.) and T32-EY07026 (to M.P.B. and D.A.H.) and by National Science Foundation Grant MCB-9722353 (to P.L.S.).
ABBREVIATIONS
- DRM
desmin-related myopathy
- cryo-EM
cryoelectron microscopy
- HSP
heat shock protein
Note Added in Proof
After submission of this manuscript, we applied analytical ultracentrifugation to analyze the molecular mass of wild-type and R120G αB-crystallin. Sedimentation equilibrium analysis at 20°C indicated a molecular mass of 580 kDa and 840 kDa for wild-type and R120G αB-crystallin, respectively.
References
- 1.Srinivasan A N, Nagineni C N, Bhat S P. J Biol Chem. 1992;267:23337–23341. [PubMed] [Google Scholar]
- 2.Klemenz R, Andres A C, Frohli E, Schafer R, Aoyama A. J Cell Biol. 1993;210:639–645. doi: 10.1083/jcb.120.3.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aoyama A, Frohli E, Schafer R, Klemenz R. Mol Cell Biol. 1993;13:1824–1835. doi: 10.1128/mcb.13.3.1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dasgupta S, Hohman T C, Carper D. Exp Eye Res. 1992;54:461–470. doi: 10.1016/0014-4835(92)90058-z. [DOI] [PubMed] [Google Scholar]
- 5.Mehlen P, Preville X, Chareyron P, Briolay J, Klemenz R, Arrigo A-P. J Immunol. 1995;154:363–374. [PubMed] [Google Scholar]
- 6.Mehlen P, Schulze-Osthoff K, Arrigo A P. J Biol Chem. 1996;271:16510–16514. doi: 10.1074/jbc.271.28.16510. [DOI] [PubMed] [Google Scholar]
- 7.Haley D A, Horwitz J, Stewart P L. J Mol Biol. 1998;277:27–35. doi: 10.1006/jmbi.1997.1611. [DOI] [PubMed] [Google Scholar]
- 8.Siezen R J, Bindels J G, Hoenders H J. Exp Eye Res. 1979;28:551–567. doi: 10.1016/0014-4835(79)90043-5. [DOI] [PubMed] [Google Scholar]
- 9.Tardieu A, Laporte D, Licino P, Krop B, Delaye M. J Mol Biol. 1986;192:711–724. doi: 10.1016/0022-2836(86)90023-9. [DOI] [PubMed] [Google Scholar]
- 10.Siezen R J, Bindels J G, Hoenders H J. Eur J Biochem. 1980;111:435–444. doi: 10.1111/j.1432-1033.1980.tb04958.x. [DOI] [PubMed] [Google Scholar]
- 11.Clauwaert J, Ellerton H D, Koretz J F, Thomson K, Augusteyn R C. Curr Eye Res. 1989;8:397–403. doi: 10.3109/02713688908996387. [DOI] [PubMed] [Google Scholar]
- 12.Horwitz J. Proc Natl Acad Sci USA. 1992;89:10449–10453. doi: 10.1073/pnas.89.21.10449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rao P V, Huang Q-L, Horwitz J, Zigler J S. Biochim Biophys Acta. 1995;1245:439–447. doi: 10.1016/0304-4165(95)00125-5. [DOI] [PubMed] [Google Scholar]
- 14.Nicholl I D, Quinlan R A. EMBO J. 1994;13:945–953. doi: 10.1002/j.1460-2075.1994.tb06339.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bennardini F, Wrzosek A, Chiesi M. Circ Res. 1992;71:288–294. doi: 10.1161/01.res.71.2.288. [DOI] [PubMed] [Google Scholar]
- 16.Van de Klundert F A J M, Gijsen M L J, van den Ijssel P R L A, Snoeckx L H E H, de Jong W W. Eur J Cell Biol. 1998;75:38–45. doi: 10.1016/s0171-9335(98)80044-7. [DOI] [PubMed] [Google Scholar]
- 17.Wang K, Spector A. Eur J Biochem. 1996;242:56–66. doi: 10.1111/j.1432-1033.1996.0056r.x. [DOI] [PubMed] [Google Scholar]
- 18.Renkawek K, Voorter C E M, Bosman G J C G M, van Workum F P A, de Jong W W. Acta Neuropathol. 1994;87:155–160. doi: 10.1007/BF00296185. [DOI] [PubMed] [Google Scholar]
- 19.Lowe J, Landon M, Pike I, Spendlove I, McDermott H, Mayer R J. Lancet. 1990;336:515–516. doi: 10.1016/0140-6736(90)92075-s. [DOI] [PubMed] [Google Scholar]
- 20.Renkawek K, de Jong W W, Merck K B, Frenken C W G M, van Workum F P A, Bosman G J C G M. Acta Neuropathol. 1992;83:324–327. doi: 10.1007/BF00296796. [DOI] [PubMed] [Google Scholar]
- 21.Head M W, Corbin E, Goldman J E. Am J Pathol. 1993;143:1743–1753. [PMC free article] [PubMed] [Google Scholar]
- 22.Vicart P, Caron A, Guicheney P, Li A, Prevost M-C, Faure A, Chateau D, Chapon F, Tome F, Dupret J-M, et al. Nat Genet. 1998;20:92–95. doi: 10.1038/1765. [DOI] [PubMed] [Google Scholar]
- 23.Horwitz J, Huang Q-L, Ding L-L, Bova M P. Methods Enzymol. 1997;290:365–383. doi: 10.1016/s0076-6879(98)90032-5. [DOI] [PubMed] [Google Scholar]
- 24.Gill S C, von Hippel P H. Anal Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 25.Sreerama N, Woody R W. Anal Biochem. 1993;209:32–44. doi: 10.1006/abio.1993.1079. [DOI] [PubMed] [Google Scholar]
- 26.Horwitz J, Bova M P, Huang Q-L, Ding L-L, Yaron O, Lohman S. Int J Biol Macromol. 1998;22:263–269. doi: 10.1016/s0141-8130(98)00024-5. [DOI] [PubMed] [Google Scholar]
- 27.Bova M P, Ding L-L, Horwitz J, Fung B K-K. J Biol Chem. 1997;272:29511–29517. doi: 10.1074/jbc.272.47.29511. [DOI] [PubMed] [Google Scholar]
- 28.Goebel H H. Muscle Nerve. 1995;18:1306–1320. doi: 10.1002/mus.880181114. [DOI] [PubMed] [Google Scholar]
- 29.Wawrousek W F, Brady J P. Invest Ophthalmol Vis Sci. 1998;39:S523. (abstr.). [PubMed] [Google Scholar]
- 30.Litt M, Kramer P, LaMorticella D M, Murphey W, Lovrien E W, Weleber R G. Hum Mol Genet. 1998;7:471–474. doi: 10.1093/hmg/7.3.471. [DOI] [PubMed] [Google Scholar]
- 31.Smulders R H P H, van Boekel M A M, de Jong W W. Int J Biol Macromol. 1998;22:187–196. doi: 10.1016/s0141-8130(98)00016-6. [DOI] [PubMed] [Google Scholar]
- 32.Takemoto L, Boyle D. Int J Biol Macromol. 1998;22:331–337. doi: 10.1016/s0141-8130(98)00031-2. [DOI] [PubMed] [Google Scholar]
- 33.Smulders R H P H, Carver J A, Linder R A, van Boekel M A M, Bloemendal H, de Jong W W. J Biol Chem. 1996;271:29060–29066. doi: 10.1074/jbc.271.46.29060. [DOI] [PubMed] [Google Scholar]
- 34.Smulders R H P H, van Geel I G, Gerards W L H, Bloemendal H, de Jong W W. J Biol Chem. 1995;270:13916–13924. doi: 10.1074/jbc.270.23.13916. [DOI] [PubMed] [Google Scholar]
- 35.Smulders R H P H, Merck K B, Aendekerk J, Horwitz J, Takemoto L, Slingsby C, Bloemendal H, de Jong W W. Eur J Biochem. 1995;232:834–838. doi: 10.1111/j.1432-1033.1995.834zz.x. [DOI] [PubMed] [Google Scholar]
- 36.De Jong W W, Leunissen J A M, Voorter C E M. Mol Biol Evol. 1993;10:103–126. doi: 10.1093/oxfordjournals.molbev.a039992. [DOI] [PubMed] [Google Scholar]
- 37.Berengian A R, Bova M P, Mchaourab H S. Biochemistry. 1997;36:9951–9957. doi: 10.1021/bi9712347. [DOI] [PubMed] [Google Scholar]
- 38.Perutz M F. Science. 1978;201:1187–1191. doi: 10.1126/science.694508. [DOI] [PubMed] [Google Scholar]
- 39.Dao-pin S, Anderson D E, Baase W A, Dahlquist F W, Matthews B W. Biochemistry. 1991;30:11521–11529. doi: 10.1021/bi00113a006. [DOI] [PubMed] [Google Scholar]
- 40.Xu J, Baase W A, Baldwin E, Matthews B W. Protein Sci. 1998;7:158–177. doi: 10.1002/pro.5560070117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim K K, Kim R, Kim S-H. Nature (London) 1998;394:595–599. doi: 10.1038/29106. [DOI] [PubMed] [Google Scholar]
- 42.Van den Ijssel P, Norman D G, Quinlan R A. Curr Biol. 1999;9:R103–R105. doi: 10.1016/s0960-9822(99)80061-x. [DOI] [PubMed] [Google Scholar]
- 43.Regan L. Curr Biol. 1994;4:656–658. doi: 10.1016/s0960-9822(00)00147-0. [DOI] [PubMed] [Google Scholar]
- 44.Muchowski P J, Bassuk J A, Lubsen N H, Clark J I. J Biol Chem. 1997;272:2578–2582. doi: 10.1074/jbc.272.4.2578. [DOI] [PubMed] [Google Scholar]
- 45.Kantorow M, Piatigorsky J. Int J Biol Macromol. 1998;22:307–314. doi: 10.1016/s0141-8130(98)00028-2. [DOI] [PubMed] [Google Scholar]
- 46.Thomas P J, Qu B-H, Pedersen P L. Trends Biochem Sci. 1995;20:456–459. doi: 10.1016/s0968-0004(00)89100-8. [DOI] [PubMed] [Google Scholar]