Abstract
Two unrelated xeroderma pigmentosum (XP) patients, with and without neurological abnormalities respectively, had identical defects in the XPC DNA nucleotide excision repair (NER) gene. Patient XP21BE, a 27 y/o woman, had developmental delay and early onset of sensorineural hearing loss. In contrast, patient XP329BE, a 13 y/o boy, had a normal neurological examination. Both patients had marked lentiginous hyperpigmentation and multiple skin cancers at an early age. Their cultured fibroblasts showed similar hypersensitivity to killing by UV and reduced repair of DNA photoproducts. Cells from both patients had a homozygous c.2T>G mutation in the XPC gene which changed the ATG initiation codon to arginine. Both had low levels of XPC message and no detectable XPC protein on Western blotting. There was no functional XPC activity in both as revealed by the failure of localization of XPC and other NER proteins at the sites of UV-induced DNA in a sensitive in vivo immunofluorescence assay. XPC cDNA containing the initiation codon mutation was functionally inactive in a post-UV host cell reactivation assay. Microsatellite markers flanking the XPC gene showed only a small region of identity (~30kBP), indicating that the patients were not closely related. Thus, the initiation codon mutation resulted in DNA repair deficiency in cells from both patients and greatly increased cancer susceptibility. The neurological abnormalities in patient XP21BE may be related to close consanguinity and simultaneous inheritance of other recessive genes or other gene modifying effects rather than the influence of XPC gene itself.
Keywords: DNA Repair, molecular genetics, sensorineural hearing loss, skin cancer, xeroderma pigmentosum
INTRODUCTION
Xeroderma pigmentosum (XP) is a rare autosomal recessive disorder caused by a defect in the nucleotide excision repair (NER) pathway [1–4] which removes a wide spectrum of structurally unrelated DNA lesions including cyclobutane pyrimidine dimers (CPD) and 6–4 photoproducts induced by ultraviolet radiation (UV) from sunlight. Cells from XP patients fall into seven genetic complementation groups XP-A through XP-G, corresponding to seven of the gene products involved in NER and a variant form with a defect in trans-lesion polymerase eta. XP patients have increased freckle-like pigmentation in response to sun exposure and a greater than 1000-fold increased incidence of UV-induced skin cancers at an early age [4, 5].
XP complementation group C (XP-C) is one of the more common forms in the United States [6]. Cells from XP-C patients have proficient transcription coupled nucleotide excision repair (TCR) but defective global genome nucleotide excision repair (GGR) of damaged DNA while cells from XP complementation groups A,B,D,F and G are defective in both GGR and TCR [3]. The XPC DNA repair gene encodes a 940 amino acid protein that forms an in vivo stable heterotrimeric complex with one of the two human orthologs of Saccharomyces cerevisiae Rad23p (RAD23A or RAD23B) and centrin 2, a component of the centrosome, and functions as a DNA-damage sensor and repair recruitment factor in GGR [3, 7, 8].
About 20% of XP patients exhibit progressive neurodegeneration [4, 9]. However, neurological symptoms are rarely seen in XP-C patients. Most of the XP patients with neurological symptoms are in XP complementation groups XP-A, XP-B, XP-D or XP-G [2, 6]. Since the development of neurologic involvement has grave clinical prognostic implications, understanding the relationship between complementation group and neurologic degeneration is extremely important. We report here two XP patients (XP21BE and XP329BE) with the same homozygous initiation codon mutation in the XPC gene. While both patients have multiple skin cancers, XP21BE has developmental delay and sensorineural hearing loss while XP329BE has no neurological abnormalities. The neurological abnormalities in XP21BE may not be related to the XPC gene defect.
MATERIALS AND METHODS
Patients
After obtaining informed consent, the XP patients were studied at the Clinical Center, NIH under protocols approved by the NCI Institutional Review Board. Both patients had thorough skin examinations and biopsy of lesions suspicious for skin cancer. Examinations included detailed ophthalmology, neurology, audiology, and other assessments as medically indicated.
Cell lines, culture conditions and DNA/RNA isolation
Fibroblast and lymphoblastoid cell cultures from two XP-C families were studied: Family A: XP21BE (GM09943, GM09942); Family B: XP329BE (2851839, JA1356), XPH395BE his father (JA1357) and XPH396BE his mother (JA1358). Normal SV40-transformed fibroblast (GM00637), normal primary skin fibroblast (AG13145) and normal lymphoblastoid (KR06057) cells and XP24BE (GM11638) and XP25BE (KR04489) fibroblasts [10] were obtained from the Human Genetic Mutant Cell Repository (Camden, NJ). SV40-transformed XP-C (XP4PA-SV-EB) cells [8, 11] were a gift from Dr. R. Legerski (M. D. Anderson Hospital, Houston, TX). Cell culture and separation of RNA and DNA was performed as described [12].
Measurement of UV sensitivity
UV sensitivity was determined using a 3-(4, 5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2 H-tetrazolium (MTS) assay (Promega) as described [13].
Measurement of UV-induced photoproducts in genomic DNA by ELISA
Cultured cells were washed with Dulbecco’s phosphate-buffered saline, UV-irradiated and incubated for various times to allow repair. Cells were harvested and genomic DNA was isolated using QIAamp Blood Kit (Qiagen, Hilden, Germany). 6–4 PP and CPD were quantified by an enzyme-linked immunosorbent assay (ELISA) using 64M-2 and TDM-2 monoclonal antibodies as described [13–15].
Post-UV HCR assay for complementation group assignment and functional analysis of mutation
The post-UV host cell reactivation (HCR) assay was used to assign XP cells to XP complementation group C as described previously [12]. A plasmid with XPC cDNA containing the c.2T>G, initiation codon mutation was assessed for function in the post-UV HCR assay. For the construction of an expression vector with the initiation codon mutation (pXPC-HAN-2T>G), the expression vector pXPC-HAN that contains full-length XPC cDNA was subjected to site-directed mutagenesis using the QuikChange Site-Directed Mutagenesis Kit (Stratagene, LaJolla, CA) as per vendor’s protocol and forward primer 5′CAGACAAGCAACAGGGCTCGGAAACGC3′ and reverse primer 5′GCGTTTCCGAGCCCTGTTGCTTGTCTG3′. The expression vector pXPC-HAN was constructed by inserting the full length wild-type XPC cDNA fragment obtained from pcDNA3/HA-XPC (a generous gift from Dr. Fumio Hanaoka, Osaka University, Osaka, Japan) into the empty expression vector pEBS7 (a generous gift from Dr. Randy Legerski, University of Texas, M.D. Anderson Cancer Center, Houston, TX).
Mutation detection by PCR amplification and nucleotide sequencing
The 16 XPC gene exons including splice donor and acceptor sites were PCR amplified using intronic primers flanking these sequences and sequenced as described previously [7].
Real time quantitative RT-PCR (QRT-PCR) for XPC message
XPC mRNA was quantified using gene specific primer pairs employing real time QRT-PCR as described [10]. The allele-specific primer pairs, oVMM-21/oVMM-22 and oCCB-331/oCCB-337 were used to measure the amount of XPC mRNA including exon 4 and exon 12, respectively. All real-time QRT-PCR data is expressed as fg of a full-length wild type XPC cDNA clone.
XPC protein detection by Western blotting
Western blotting for the XPC protein was performed as described [10] using XPC specific monoclonal (ab6264) (Abcam, Inc., Cambridge, MA) and anti-β-Actin polyclonal antibodies (Santa Cruz).
Immunocytochemistry of NER protein localization following localized UV irradiation
Fibroblasts were labeled with 0.8 μm (normal donors) or 2 μm (patients) latex beads (Polybead carboxylate microspheres; Polysciences, Warrington PA) and cultured until confluent [16]. Cells were combined, overlaid with 5 μm Millipore filters, irradiated with 100 J/m2 UV-C and double stained for immunofluorescent localization of NER proteins and photoproducts as described [13, 17–19]. Cells were visualized with a LSM 510 confocal microscope (Carl Zeiss).
Microsatellite marker analysis
Microsatellite markers flanking the XPC gene on chromosome 3 were genotyped using fluorescently labeled oligonucleotide probes as previously described [20]. PCR products were separated on ABI Prism 3100 Genetic analyzer, and were analyzed with the GeneScan, version 3.1.2, software (Perkin Elmer–Applied Biosystems).
RESULTS
Clinical Findings
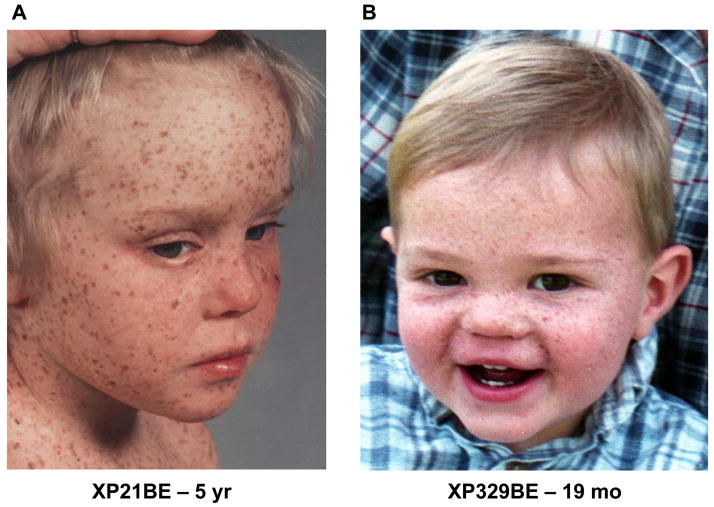
Both patient XP21BE (Figure 1A) and patient XP329BE (Figure 1B) had a history of lentiginous hyperpigmentation in sun exposed areas before the first year of age and did not have the acute photosensitivity with blistering burns after brief sun exposure which is present in some XP patients (Table 1). Both patients began to develop skin cancers by age 3 years.
Figure 1.
Xeroderma pigmentosum patients with and without neurological disease. A. Patient XP21BE, at age 5 years, had extensive lentiginous pigmentation on sun exposed portions of her face, chest and shoulders. B. Patient XP329BE, at age 19 months, had extensive freckle-like pigmentation on the sun exposed portions of his face.
TABLE 1.
Clinical Findings in Two Unrelated Patients With Identical XPC Gene Mutations Compared to Clincal Features of XP Neurological Disease and XP/CS Complex
| XP21BE | XP329BE | XP NEUROLOGICAL DISEASE | XP/CS COMPLEX | |
|---|---|---|---|---|
| Age at last observation | 27 yr | 13 yr | ||
| Acute photosensitivity | None | None | Yes | Yes |
| Onset of lentiginous hyperpigmentation of sun exposed skin | By age 10 months | By age 8 months | Early | Early |
| Skin cancers | Yes | Yes | Yes | Yes |
| Age of first skin cancer) | 3 yr | 3 yr | ||
| Basal cell carcinoma (number) | 7 | 36 | ||
| Squamous cell carcinoma (number) | 62 | None | ||
| Melanoma (number) | 13 | None | ||
| Developmental delay | Present -stable | Normal intelligence | Progressive degeneration | Progressive degeneration |
| Audiology | Sensori- neural hearing loss across entire freq range | Normal hearing | Progressive high frequency hearing loss | Progressive high frequency hearing loss |
| Cachectic dwarfism | No | No | Yes/no | Yes |
| Immature sexual development | Normal -gave birth to normal child | Normal | Normal | Yes |
| Microcephaly | Normal | Normal | Yes | Yes |
| Retinal degeneration | No | No | No | Yes |
| Ataxia | No | No | Yes | Yes |
| Primary neuronal degeneration | No | No | Yes | No |
| Demyelinating neuropathy | No | No | No | Yes |
| Cerebral atrophy | No | No | Yes | Yes |
| Brain calcification | No | No | No | Yes |
| DNA repair defect | XPC | XPC | XPA, XPD, XPG | XPB, XPD, XPG |
Patient XP21BE
By age 13 years patient XP21BE had 2 basal cell carcinomas and 33 squamous cell carcinomas. By age 27 years she had a total of 7 basal cell carcinomas, 62 squamous cell carcinomas, and 13 melanomas (Table 1). In addition, she had histopathological confirmation of 16 keratoacanthomas and multiple angiokeratomas.
Her developmental milestones were delayed; she walked at 19 months and spoke in sentences by the age of 3 years. Her mother reported hearing difficulties from 6 months of age. Upon school entrance at age 3 years, she was enrolled in regular classes with speech and hearing support. Academically she was considered learning disabled by the time she was 8 years old. At the age of 10 she developed simple partial seizures and was treated with carbamazepine which was changed to valproic acid during a two year period. At the age of 12 years she was placed into classes for the cognitively disabled. Patient XP21BE was reported to be hyperactive, impulsive and distractible as a young child, features that continued throughout her schooling. Medication for the treatment of attention deficit/hyperactivity disorder was discontinued after a tic disorder developed. As a high school student in a work-study program she was trained to monitor inventory and order supplies for the school cafeteria. However, she was physically agile and won medals in the balance beam and other gymnastic events for disabled individuals in high school. She graduated from high school at the age of 20 and took some community college courses. At 23 years of age she gave birth to a clinically normal son. At the time of her last evaluation she was working part time as a clerk while raising her 4 year old child.
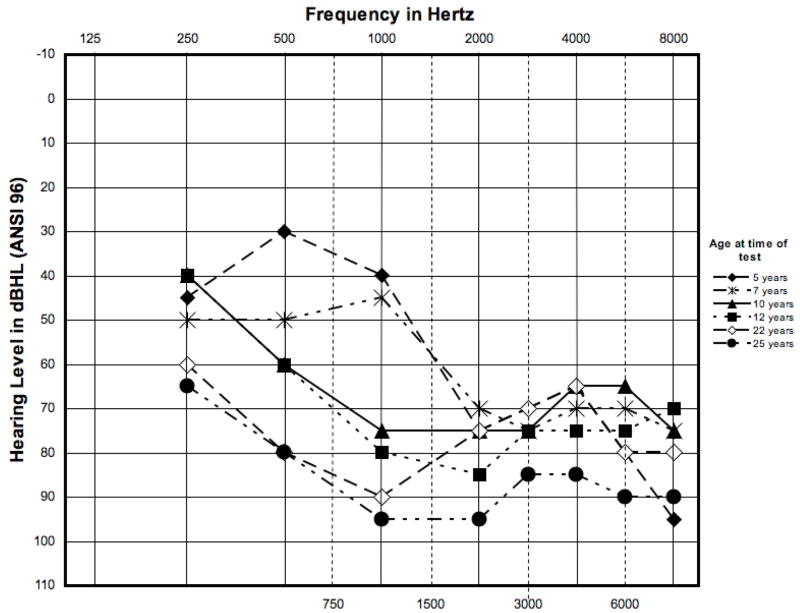
An audiogram at age 5 years showed bilateral, symmetrical sensorineural hearing loss, ranging from mild-to-profound across the entire test frequency range (250–8000 Hz). Progression to a severe-to-profound sensorineural hearing loss was documented from age 5 years to 25 years (Figure 2).
Figure 2.
Serial audiograms from the right ear of patient XP21BE. Audiograms from 250 Hz to 8000 Hz were performed at age 5 yr, 7 yr, 10 yr, 12 yr, 22 yr and 25 yr. There was a mild-to-profound sensorineural hearing loss across the entire frequency range at age 5 yr that progressed to a severe-to-profound sensorineural hearing loss at age 25 yr.
On examination at age 24 years she was cheerful and cooperative. Her speech was clear without frank dysarthria. Muscle strength was normal, tendon reflexes +1, in the upper extremities, +3 in the knees and +2 in the ankles with probable upgoing toes on plantar stimulation. Fine motor movements were normal in upper and lower extremities. Sensory examination was normal. Regular gait, hopping, heel and toe walk were normal. Nerve conduction studies performed at age 25 years showed mild sensory axonal peripherally neuropathy with decrease in the SNAP amplitudes compared to a baseline study at age 10 years. At age 26 years she was obese (BMI 33.2, weight 77.4 kg, height 152.8 cm) with head circumference 53.3 cm (17 percentile). CT and MRI of the brain were normal at age 25 and 27 years respectively.
Patient XP21BE had serial neuropsychological evaluations beginning at age 7 years using the appropriate Wechsler scale for age (Table 2). A consistent pattern was performance IQ greater than verbal IQ. The difference was always significant and ranged between 12 and 32 points which is typical of an individual with a hearing impairment. Test results at the age of 11 and 12 years were disparate from those obtained as a younger child and as an adult. This is coincident with her treatment for a seizure disorder. Compared to that of her age peers, her vocabulary was exceptionally low during those years (below the 1st percentile). At the age of 12 years there was a precipitous drop of 18–19 raw score points on Picture Arrangement and a 5 to 6 point raw score drop on Object Assembly that contributed to the 25 point deterioration in performance IQ, and by extension, the drop in full scale IQ that year. Neurologic evaluation at that time suggested that this reduction in test results might be due to either the seizure disorder or the medications used to treat the seizure disorder. She was not tested again until the age of 25 when her performance IQ returned to former levels. As an adult her vocabulary improved to the low average range but continued to be a relatively weak area. Verbal abilities were otherwise stable. Nonverbal abilities, always stronger, were also consistent, with the exception of the 12th year performance. Word recognition reading and spelling were better than math and reading comprehension. Test scores were commensurate with full scale IQ. Overall, there was no evidence of progressive intellectual deterioration.
TABLE 2.
STABILITY OF IQ OF PATIENT XP21BE
| TEST | AGE (years) | VERBAL IQ | PERFORMANCE IQ | FULL SCALE IQ |
|---|---|---|---|---|
| WISC-R | 7 | 81 | 98 | 88 |
| WISC-R | 9 | 78 | 91 | 83 |
| WISC-R | 11* | 68 | 100 | 91 |
| WISC-R | 12* | 65 | 75** | 69** |
| WAIS-III | 25 | 75 | 98 | 84 |
| WAIS-III | 27 | 83 | 102 | 91 |
Seizure disorder treated with carbamazepine followed by valproic acid. Seizures resolved and medications discontinued.
Attributed to significant decline on most tests that are timed.
Patient XP21BE was reported to be the child of a second degree consanguineous mating. Early onset of sensorineural hearing loss and developmental delay has not been reported in other XP-C patients.
Patient XP329BE
By age 13 years patient XP329BE had developed 36 histologically confirmed basal cell carcinomas (Table 1 and Figure 1B). He had normal neurological examination and normal audiogram. He is an excellent student, plays the piano and is an avid reader. Patient XP329BE was reported to be the child of a fourth degree consanguineous mating.
Elevated UV sensitivity and reduced DNA photoproduct repair
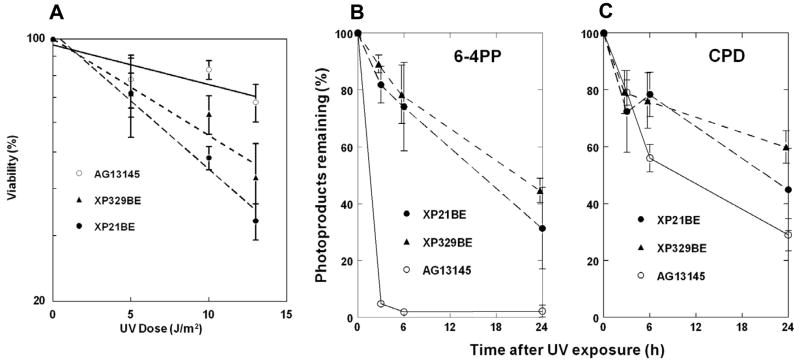
We examined the UV sensitivity cells from both patients and a normal control using an MTS assay (Figure 3A). The cells from both patients showed a higher sensitivity to UV than normal cells. After exposure to 14 J/m2 UV the XP cells had 35–45% viability while the normal cells had 70%. Though the XP patients had marked differences in their neurological status their cells showed no significant differences in UV sensitivity.
Figure 3.
Post UV cell survival and photoproduct removal in XP cells. A. Post-UV viability of normal (open circles), XP329BE (closed triangles) and XP21BE (closed circles) fibroblasts measured by MTS assay. Mean ± SEM of quadruplicate cultures. B. Removal of 6–4 photoproducts from normal (open circles), XP21BE (closed circles) and XP329BE (closed triangles) cells following 10 J/m2 UV exposure measured by ELISA assay. Mean ± SD of duplicate experiments are shown. C. Removal of CPD from normal (open circles), XP21BE (closed circles) and XP329BE (closed triangles) cells following 10 J/m2 UV exposure measured by ELISA assay. Mean ± SD of duplicate experiments are shown.
An ELISA assay indicated that normal fibroblasts had a rapid post-UV removal of 6–4 PP, with 5% remaining by 3h and 2% at 6h (Figure 3B). This is similar to previous reports for normal cells [13, 14, 21]. In contrast about 80–90% of the 6–4 PP remained in cells from both XP patients at 3h and 75–80% at 6h. This is similar to other studies for XPC cells [22]. As previously described [13, 14, 21, 22] post-UV removal of CPD photoproducts was slower in normal cells with 56% remaining at 6h and 29% remaining at 24h (Figure 3C). CPD removal was delayed in both XP-C cells with 75–80% remaining at 6h and 45–60% at 24h. This is similar to the other XP-C cells [22]. Thus, the cells from the XP-C patient with neurological abnormalities (XP21BE) and with normal neurological examinations (XP329BE) showed similar levels of reduction in the repair of photoproducts.
Assignment of XP cells to complementation group C
The post-UV host cell reactivation (HCR) assay was used to assign these cells to XP complementation groups. The UV irradiated reporter gene plasmid was transfected into the cells from the patients along with plasmids expressing wild-type, XP group A, C or D cDNA. Only co-transfection with a plasmid containing the wild-type XPC cDNA led to a markedly increased post-UV HCR in the cells from both XP patients thus assigning these cells to the XP complementation group C (data not shown).
Initiation codon mutation in the XPC gene
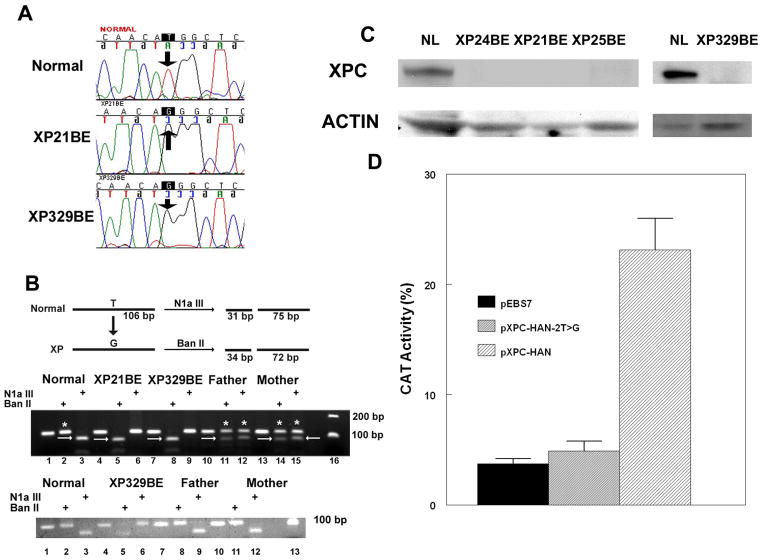
We characterized the causative mutations in these cells by nucleotide sequencing. We found a homozygous c.2T>G mutation changing the ATG initiation codon to arginine (AGG) in the XPC exon 1 in cells from both XP-C patients. This mutation was determined both in the genomic DNA (Figure 4A) and cDNA (data not shown) from the patients’ cells.
Figure 4.
XPC Initiation codon mutation in XP21BE and XP329BE cells. A. Sequence of XPC gene in genomic DNA from normal, XP21BE and XP329BE cells. The T base of the ATG initiation codon is substituted by a G in the XP cells (arrows). B. RFLP detection of c.2T>G mutation in genomic DNA and cDNA in cells from a normal donor, XP21BE, XP329BE and his father and mother. Upper gel: N1aIII digestion of a 106 bp fragment of XPC genomic DNA results in products of 31 bp and 75 bp in the normal sequence containing a T (arrows in lanes 3, 12 and 15). BanII digestion of the XPC genomic DNA fragment containing a G results in products of 34 bp and 72 bp (arrows in lanes 5, 8, 11, and 14). Undigested bands are indicated by *. Patients XP21BE and XP329BE are homozygous for the c.2T>G mutation and the parents of XP329BE are heterozygous for the mutation. Lower gel: N1aIII digestion of XPC cDNA results in products of 31 bp and 75 bp in the normal sequence containing a T (lanes 3, 9 and 12) from the normal donor and both parents. BanII digestion of the XPC cDNA fragment containing the T>G mutation results in products of 34 bp and 72 bp (lane 5) in cDNA from XP329BE. There is no detectable BanII digested cDNA in the cells from the parents indicating that only the normal allele is expressed. C. Western blot of protein extracted from cells from normal, XP24BE, XP21BE, XP25BE and XP329BE cells using XPC (upper row) and actin (lower row) antibodies. There was no detectable XPC protein in the XP cells. D. Absence of activity of XPC cDNA containing the c.2T>G mutation in a host cell reactivation assay. XP4PA-SV-EB XPC cells were co-transfected with a UV treated CAT plasmid (1000 J/m2) and a plasmid with either wild type XPC cDNA (pXPC-HAN; n=9), XPC cDNA with the c.2T>G mutation (pXPC-HAN-2T>G; n=6), or the empty vector (pEBS7; n=9). CAT activity was measured after 48 hr. The wild type XPC cDNA showed increased CAT activity (p=0.0002) while CAT activity with the XPC cDNA with the c.2T>G mutation was not significantly different from the empty vector. Mean ± SEM shown.
We developed genomic PCR and RFLP based method to characterize this new initiation codon mutation in patients and their parents. The T to G transition created a new BanII restriction site in XPC exon 1 and abolished an N1aIII restriction site (Figure 4B). Genomic DNA from the XP-C patients and their parents was digested in order to determine whether the mutations were homozygous or hemizygous. The PCR products from XP21BE and XP329BE were cut only by BanII and the PCR product from normal was cut only by N1aIII. The PCR products from parents of XP329BE were cut by both BanII and N1aIII restriction endonucleases indicating that both the parents of XP329BE were heterozygous for the same mutation in that one allele behaved like normal and the other one had the initiation codon mutation like their son. These results indicate that the XP-C patient XP329BE is homozygous for the initiation codon mutation. We do not have access to DNA from the parents of XP21BE.
XPC mRNA levels in the cells from XP-C patients
A real-time quantitative reverse transcriptase-PCR (QRT-PCR) assay [10] with allele-specific primers that detect XPC mRNAs containing either exon 4 or exon 12 (Table 3) was used to measure the levels of XPC mRNA in cells from the XP-C patients, their heterozygous parents and normal controls. Relative to the normal control, the XPC mRNA levels were 24 to 42% in cells from XP21BE and XP329BE. Interestingly, the levels of XPC mRNA in these patients with an initiation codon mutation were slightly higher compared to cells from patients with premature termination codons (PTC)[10]. This suggests that RNA harboring PTC is more likely to be degraded by nonsense mediated message decay pathway. However, the levels of XPC mRNA in the cells from the parents of XP329BE were similar to the normal cells. Restriction enzyme digestion of cDNA from patient XP329BE revealed digestion with BanII but not N1aIII indicating that all of the cDNA contained the c.2T>G mutation (Figure 4B lower gel – lanes 5 and 6). In contrast cDNA from both parents was digested by N1aIII and not BanII indicating that there was no detectable mutated cDNA expressed in these cells (Figure 4B lower gel – lanes 8,9, 11 and 12).
TABLE 3.
XPC mRNA LEVELS IN XP-C CELLS
| CELL LINE | XPC EXON 4 INCLUSION | XPC EXON 12INCLUSION | ||
|---|---|---|---|---|
| Avg (fg) | Avg (fg) | |||
| XP21BE | 57.5 | 39% | 58.5 | 42% |
| XP329BE | 35.3 | 24% | 47.4 | 34% |
| FATHER | 138 | 94% | 147 | 105% |
| MOTHER | 118 | 80% | 120 | 86% |
| NORMAL | 147 | 100% | 140 | 100% |
Reduced XPC protein levels in the cells from XP-C patients
To measure the XPC protein levels in the cells from XP-C patients and their parents, Western blot analysis was performed on total cellular extracts using XPC specific monoclonal and anti-β-actin polyclonal antibodies [10]. High levels of XPC protein were observed in the cells from the normal donors (Figure 4C). In contrast, there was no detectable XPC protein in the cells from the XP-C patients (Figure 4C). This was similar to the previously reported undectable levels of XPC protein in cells from patients XP24BE [Compound heterozygote: Intron 5.1 A to G at −2; and c.463C>T, p.Arg155*] and XP25BE [Homozygous: Intron 11 Del -1. -2 AG with Insertion of CC between −6 and −7] (Figure 4C and [10]). The parents of XP329BE, who are obligate heterozygotes, had XPC protein levels similar to that of the normal control (data not shown). These findings were similar to those previously reported in XP-C, normal and parental cells [10].
Reduced DNA repair function of initiation codon mutation
We constructed an expression vector containing the XPC cDNA with c.2T>G mutation (pXPC-HAN-2T>G) and assessed its function employing a transient post-UV HCR assay (Figure 4D) The recovery of chloramphenicol acetyl transferase (CAT) activity reflects the ability of the transfected cells to repair the UV-induced plasmid DNA damage [12]. The expression vector with wild type full-length XPC cDNA (pXPC-HAN) resulted in increased CAT activity (p=0.0002) indicating that the repair defect in XP4PA-SV-EB XP-C cells could be complemented. Transfection of pXPC-HAN-2T>G plasmid resulted in a markedly reduced CAT activity in XP4PA-SV-EB XP-C cells that was not significantly different from the CAT activity with the empty vector control (pEBS7). Thus, XPC cDNA with the initiation codon mutation was not functional in this DNA repair assay. Over-expression of XPC cDNA with the initiation codon mutation in normal GM00637 cells did not alter the normal repair capability of the cells (data not shown).
Absence of functional XPC protein in vivo
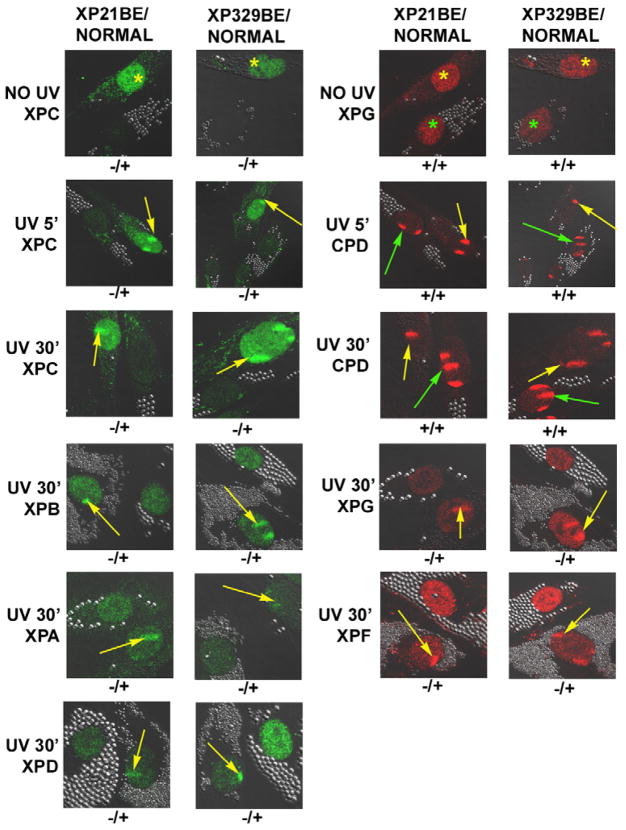
XP and normal cells were labeled by uptake of different sized polystyrene beads in their cytoplasm and then co-cultured. The cells were UV-irradiated through 5-μm diameter pores of a polycarbonate isopore membrane filter to follow localization of NER proteins to sites of UV-induced DNA damage in vivo as a measure of their DNA repair activity (Figure 5). The NER proteins were detected by immunofluorescence and confocal microscopy. Their localization to the site of DNA damage reflected their DNA repair activity. XPC protein was detected in the un-irradiated normal cells (Figure 5 – top row, left panels). In contrast, in un-irradiated XP21BE or XP329BE cells XPC protein was not detected (Figure 5 – top row, left panels), in agreement with the Western blots (Figure 4C). XPG protein was detected in the un-irradiated normal and XP cells (Figure 5 – top row, right panels). Assay within 5 min of UV irradiation produced fluorescent CPD foci in the nuclei of the normal and both the XP-C patients’ cells (Figure 5 – second row, right panels). Localization of the XPC protein (green) to the DNA damaged site was observed in the normal cells (yellow arrows) but not in the cells from the XP-C patients (Figure 5 – second row, left panels). By 30 min after UV treatment CPD were still detectable in the normal and XP-C patients’ cells (Figure 5 – third row, right panels). In the normal cells all the NER components examined (XPC, XPB, XPG, XPA, XPD, and XPF) were localized to the sites of DNA damage (yellow arrows) indicating that the NER was functionally active (Figure 5 – rows 3, 4, 5 and 6). In the absence of binding of XPC protein to the DNA damaged site, none of these NER proteins examined localized to the damaged site in the XP cells (Figure 5 – rows 3, 4, 5 and 6). These findings indicate that there was no detectible functional XPC protein activity in the cells of either patient. These findings are similar to those reported previously in other XP-C and normal cells [19].
Figure 5.
Lack of recruitment of XPC and other NER proteins to localized DNA damage in XP21BE and XP329BE cells following UV irradiation. Normal cells (AG13145) were labeled with 0.8 μm latex beads and XP21BE and XP329BE cells were labeled with 2 μm latex beads. In the absence of UV, XPC and XPG protein was stained in the normal cells (yellow *). XPG protein was present in the XP cells (green *) but XPC protein was not detected. Following 100 J/m2 UV irradiation delivered through a 5 μm filter, cells were fixed at 5 min or 30 min post-irradiation and immunostained with pairs of antibodies to simultaneously assess the location of DNA damage and NER proteins. The arrows indicate sites of localized immunostaining: normal - yellow arrows, patient – green arrows. While CPD photoproducts were detected in the XP and normal cells at 5 min and 30 min after UV exposure, the NER proteins (XPC, XPG, XPB, XPD, XPA and XPF) were only localized in the normal cells (yellow arrows). Symbols below each image indicate localization (+) or non-localization (−) of NER proteins or photoproducts in patient cells/normal cells.
Genetic marker analysis
XP21BE and XP329BE were homozygous for the same XPC initiation codon mutation (c.2T>G), suggesting the possibility that they might have a common ancestor. This can be tested by analyzing microsatellite or single nucleotide polymorphism (SNP) markers near the XPC gene on chromosome 3 [20]. Table 4 shows 9 microsatellite markers and 3 XPC SNP’s we used arranged along chromosome 3 as indicated by the maps of the human genome http://www.ncbi.nlm.nih.gov/ Examination of DNA from each of the XP patients, both of whom had a history of consanguinity, revealed the markers to be homozygous over a region of 670 kBP (Table 4). While both of these patients have the same homozygous initiation codon mutation, their alleles were different for 11 out of 12 microsatellite markers (Table 4 and data not shown). There was a common region of about 30 kBP within the XPC gene (bold type in Table 4). Thus these XP-C patients were not closely related. In addition, the markers from XP21BE were heterozygous outside of the 670 kBP homozygous region, indicating that these cells were not hemizygous.
TABLE 4.
ANALYSIS OF GENOMIC DNA FROM XP21BE AND XP329BE
| LOCATION CHR3 (BP) | XP329BE Maternal | XP329BE Paternal | XP21BE Allele I | XP21BE Allele II | UniSTS or SNP name |
|---|---|---|---|---|---|
| 1,289,933 | 1 | 2 | 4 | 5 | D3S1307 |
| 1,972,241 | 1 | 2 | 3 | 4 | D3S1297 |
| 6,853,445 | 1 | 2 | 3 | 4 | D3S1304 |
| 13,792,429 | 1 | 1 | 1 | 1 | G67699 |
| 13,875,775 | 1 | 1 | 2 | 2 | D3S1585 |
| 14,162,450 | A | A | A | A | XPC LYS939GLU |
| 14,174,889 | C | C | C | C | XPC VAL499ALA |
| 14,195,069 | G | G | G | G | XPC c.2T>G |
| 14,195,099 | C | C | C | C | XPC 5′UTR |
| 14,244,896 | 1 | 1 | 3 | 3 | G67702 |
| 14,462,499 | 1 | 1 | 3 | 3 | G67701 |
| 19,473,236 | 2 | 1 | 3 | 4 | D3S3726 |
| 30,614,544 | 1 | 2 | 4 | 5 | D3S3727 |
DISCUSSION
XPC initiation codon mutation, DNA repair and skin cancers
Most of the mutations reported in the XPC gene in cells from XP patients create premature termination codons (PTCs) [10, 20, 23–28]. PTCs can reduce the levels of XPC message and XPC protein in cells by means of nonsense-mediated mRNA decay (NMD)[29]. Only two missense mutations in the XPC gene have been reported. These are Pro334His [25] and Trp690Ser [23], which alters the stability of the encoded mutant protein [30]. Splice site mutations may result in severe or mild symptoms depending on the level of XPC mRNA. Our previous studies demonstrated that undetectable levels of normal XPC mRNA were associated with severe clinical symptoms in 2 siblings including multiple skin cancers at an early age [31]. In contrast, 3 – 5 % of normal levels of full length XPC mRNA resulted in mild symptoms in 3 affected siblings [31]. The differential expressions of XPC message in these cells were associated with different mutations in two functional lariat branch point sequences (BPS) in same XPC intron 3.
The patients in this report had a c.2T>G mutation in the ATG initiation codon. The sequences surrounding AUG codons in mammals play a role in specifying which initiation site is likely to be used during the protein translation. The sequence near the first initiation codon in the XPC gene (AGCAACaugG) matches well with the known optimal recognition sequence with an A in position −3 and G in position +4. The initiation sites are reached via a scanning mechanism. The ribosome normally scans in a 5′ to 3′ direction until it encounters the first AUG and usually initiates translation at this first AUG [32, 33]. In the cells from both XP-C patients, XPC mRNA with mutated first AUG may not be recognized by most of the ribosomes recruited at the 5′ cap-structure on the XPC mRNA thus resulting in an undectable level of XPC protein. The next in-frame unmutated AUG is located at codon 118, however, the surrounding sequence (GCTACCaugA) does not completely match with the known optimal recognition sequence. The A in position −3 is similar to the optimal recognition sequence but A in position + 4 is distinct. There are many out of frame AUGs in between the mutated initiation codon and the AUG at codon 118 which would inhibit ribosome scanning [34]. The use of this AUG would delete the NH2-terminal 117 amino acids in the XPC protein. This truncated protein has been shown to have functional activity [8, 22].
While XPC mRNA levels from XP21BE and XP329BE were slightly higher (Table 2) than in patients with XPC nonsense mutations [10],, all of the mRNA contained the mutation which rendered the allele completely inactive (Figure 4D). There was no detectable normal size XPC protein on the Western blots (Figure 4C) and no functional protein present as judged by the failure to recruit NER proteins to sites of UV damage (Figure 5). The failure to detect localization of XPC protein to UV damaged DNA is in agreement with other studies of XP cells and in vitro assays of NER protein interactions [18, 19, 35]. Thus, in the absence of XPC protein, the other core NER proteins (XPA, XPB, XPD, XPG, and XPF) did not localize to the damage site. This sensitive assay is able to detect functional activity of the 3–5% of normal full length XPC mRNA in cells from patients with XPC splice mutations (Khan, SK, Oh K-S et al unpublished data).
Genetic markers detecting the relationship between XP-C patients
Genetic analysis of the patients by use of microsatellites and SNP’s can reveal common ancestry. We previously used microsatellite markers to provide evidence of a relationship 300–500 years ago between XPC families in Turkey and in Italy [20]. Individuals that are closely related will show larger regions of identity than people who are more distant relatives. In our patients the common region was only about 30 kBP within the XPC gene (bold type in Table 4). Thus these XP-C patients were not closely related.
The presence of homozygosity of this region surrounding XPC also suggests that these patients may be homozygous for genetic changes in other regions that may be critical for their different clinical symptoms.
XPC gene mutations and neurological abnormalities
XP is characterized by sun sensitivity and early onset of lentiginous pigmentation and skin cancer in sun exposed parts of the body. About 20% XP patients have a distinct group of neurological symptoms including microcephaly, progressive neurological degeneration with sensorineural hearing loss beginning at high frequencies, loss of intellectual functions, ataxia, loss of coordination and ability to walk, and cerebral atrophy with dilated ventricles in association with primary neuronal degeneration (Table 1 and [9, 36, 37]. These patients have defects in XP complementation groups XP-A, XP-D, or XP-G. A correlation between the neurological status of the XP patients and the post-UV colony-forming ability (CFA) of their cells was reported [38]. Some XP patients have been identified with a second clinical entity: the XP/Cockayne syndrome (CS) complex (Table 1). These patients exhibit cutaneous abnormalities of XP and CS type of neurological changes including pigmentary retinal degeneration, sensorineural deafness, dysmyelination of the brain, and calcification of basal ganglia in association with progressive cachectic dwarfism [9, 36, 37]. These patients have been described in XP complementation groups XP-B, XP-D and XP-G. In contrast, the neurologic involvement in patient XP21BE differs from typical XP neurologic involvement by having normal reflexes, sensorineural deafness at many frequencies, normal CT and MRI of her brain and normal eye exam. Although she had learning disabilities, she also had good coordination and was able to win medals and compete in gymnastic events in the Special Olympics for disabled individuals. The post-UV cell survival of her cells was similar to that of patient XP329BE who had no neurological abnormalities. Similarly, her normal retina, normal height, head circumference, and normal fertility, and normal CT and MRI as well as her agility and increased body mass index distinguish patient XP21BE from patients with the XP/CS complex (Table 1). Thus, her neurological and cellular abnormalities were not typical of XP neurological disease or of the XP/CS complex.
Symptomatic neurological abnormalities have been reported in only two other XP-C patients [24, 39, 40]. Patient XP1MI was a 17 year old black woman with multiple basal cell carcinomas, scleralization of the cornea and XP associated neurological and developmental abnormalities including microcephaly (<1 %ile), mental retardation (IQ 39–48), growth retardation, and delayed sexual development (primary amenorrhea). However, unlike typical XP neurological disease, she had normal hearing and reflexes and did not have ataxia [39, 41]. In addition to XP she had systemic lupus erythematosus with joint involvement and positive serology. There was no consanguinity reported. The cells from XP1MI had a homo- or hemizygous missense mutation (c.1106C> A, p.Pro334His) in the XPC gene [25]. XP22BE was a 4 year old boy of Korean ancestry with multiple skin cancers who had neurological abnormalities not usually found with either XP or CS, including autistic features hyperactivity, normal hearing, hyperactivity, normal reflexes, and normal MRI without dilated ventricles or microcephaly along with persistent hypoglycinemia [24, 40]. He was born with a cleft palate and duplex right kidney. This patient was the product of close consanguinity. He had a homozygous XPC splice mutation (c.IVS10 +2T>G) [24]. Until additional patients are found either with the same XPC mutation or with XP and hypoglycinemia we cannot ascribe his symptoms to the defect in the XPC gene.
The hearing loss seen in XP patients is usually high frequency loss that is progressive and occurs gradually over many years [37, 41]. Clinically asymptomatic high frequency hearing loss [42] was present in one adult patient (XP1BE) with a c.1292_1293delAA, p.Lys431Argfs*6 defect in the XPC gene [10]. In contrast, the hearing loss in XP21BE was identified at an early age and spanned the entire frequency range. Patient XP21BE was a child of a close consanguineous mating with second degree relationship. In this close mating the proportion of genes shared is 25% (identical by descent), leading to the chance of homozygosity by descent of 1/8 [43]. Thus close consanguinity confers an increased probability of simultaneous occurrence of other recessive disorders. Nonsyndromic hearing loss has been reported to occur at increased frequency in association with parental consanguinity [44, 45]. We therefore looked for other genetic causes of sensorineural hearing loss. Approximately 50% of non-syndromic sensorineural hearing loss is due to mutations in GJB2 which encodes connexin 26 [46] but we were unable to identify mutations in this gene. It is likely that this hearing loss is the result of a distinct genetic abnormality. The other neurological abnormalities in patient XP21BE, as well as in patient XP22BE may be related to simultaneous inheritance of other recessive genes or other gene modifying effects rather than the influence of XPC gene itself.
Acknowledgments
This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. We are grateful to the patients and their families for their cooperation in this research. S.E. was supported in part by a grant from the Deutsche Forschungsgemeinschaft DFG (EM 63/1-1).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kraemer KH, Ruenger TM. Genome instability, DNA repair and cancer. In: Wolff K, Goldsmith LA, Katz SI, Gilchrest BA, Paller AS, Leffell DJ, editors. Fitzpatrick’s Dermatology in General Medicine. McGraw Hill; New York: 2008. pp. 977–986. [Google Scholar]
- 2.Ruenger TM, DiGiovanna JJ, Kraemer KH. Hereditary Diseases of genome instability and DNA repair. In: Wolff K, Goldsmith LA, Katz SI, Gilchrest BA, Paller AS, Leffell DJ, editors. Fitzpatrick’s Dermatology in General Medicine. McGraw Hill; New York: 2008. pp. 1311–1325. [Google Scholar]
- 3.Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis. ASM Press; Washington, D.C.: 2006. [Google Scholar]
- 4.Kraemer KH, Lee MM, Scotto J. Xeroderma pigmentosum. Cutaneous, ocular, and neurologic abnormalities in 830 published cases. Arch Dermatol. 1987;123:241–250. doi: 10.1001/archderm.123.2.241. [DOI] [PubMed] [Google Scholar]
- 5.Kraemer KH, Lee MM, Andrews AD, Lambert WC. The role of sunlight and DNA repair in melanoma and nonmelanoma skin cancer: The xeroderma pigmentosum paradigm. Arch Dermatol. 1994;130:1018–1021. [PubMed] [Google Scholar]
- 6.Moriwaki S, Kraemer KH. Xeroderma pigmentosum--bridging a gap between clinic and laboratory. Photodermatol Photoimmunol Photomed. 2001;17:47–54. doi: 10.1034/j.1600-0781.2001.017002047.x. [DOI] [PubMed] [Google Scholar]
- 7.Khan SG, Muniz-Medina V, Shahlavi T, Baker CC, Inui H, Ueda T, Emmert S, Schneider TD, Kraemer KH. The human XPC DNA repair gene: arrangement, splice site information content and influence of a single nucleotide polymorphism in a splice acceptor site on alternative splicing and function. Nucleic Acids Res. 2002;30:3624–3631. doi: 10.1093/nar/gkf469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Legerski R, Peterson C. Expression cloning of a human DNA repair gene involved in xeroderma pigmentosum group C [published erratum appears in 1992 Dec 10;360(6404):610. Nature. 1992;359:70–73. doi: 10.1038/360610b0. [DOI] [PubMed] [Google Scholar]
- 9.Kraemer KH, Patronas NJ, Schiffmann R, Brooks BP, Tamura D, DiGiovanna JJ. Xeroderma pigmentosum, trichothiodystrophy and Cockayne syndrome: A complex genotype-phenotype relationship. Neuroscience. 2007;145:1388–1396. doi: 10.1016/j.neuroscience.2006.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khan SG, Oh KS, Shahlavi T, Ueda T, Busch DB, Inui H, Emmert S, Imoto K, Muniz-Medina V, Baker CC, DiGiovanna JJ, Schmidt D, Khadavi A, Metin A, Gozukara E, Slor H, Sarasin A, Kraemer KH. Reduced XPC DNA repair gene mRNA levels in clinically normal parents of xeroderma pigmentosum patients. Carcinogenesis. 2006;27:84–94. doi: 10.1093/carcin/bgi204. [DOI] [PubMed] [Google Scholar]
- 11.Daya-Grosjean L, James MR, Drougard C, Sarasin A. An immortalized xeroderma pigmentosum, group C, cell line which replicates SV40 shuttle vectors. Mutat Res. 1987;183:185–196. doi: 10.1016/0167-8817(87)90061-7. [DOI] [PubMed] [Google Scholar]
- 12.Emmert S, Slor H, Busch DB, Batko S, Albert RB, Coleman D, Khan SG, Abu-Libdeh B, DiGiovanna JJ, Cunningham BB, Lee MM, Crollick J, Inui H, Ueda T, Hedayati M, Grossman L, Shahlavi T, Cleaver JE, Kraemer KH. Relationship of neurologic degeneration to genotype in three xeroderma pigmentosum group G patients. J Invest Dermatol. 2002;118:972–982. doi: 10.1046/j.1523-1747.2002.01782.x. [DOI] [PubMed] [Google Scholar]
- 13.Imoto K, Kobayashi N, Katsumi S, Nishiwaki Y, Iwamoto TA, Yamamoto A, Yamashina Y, Shirai T, Miyagawa S, Dohi Y, Sugiura S, Mori T. The total amount of DNA damage determines ultraviolet-radiation-induced cytotoxicity after uniformor localized irradiation of human cells. J Invest Dermatol. 2002;119:1177–1182. doi: 10.1046/j.1523-1747.2002.19514.x. [DOI] [PubMed] [Google Scholar]
- 14.Nishiwaki Y, Kobayashi N, Imoto K, Iwamoto TA, Yamamoto A, Katsumi S, Shirai T, Sugiura S, Nakamura Y, Sarasin A, Miyagawa S, Mori T. Trichothiodystrophy fibroblasts are deficient in the repair of ultraviolet-induced cyclobutane pyrimidine dimers and (6–4) photoproducts. J Invest Dermatol. 2004;122:526–532. doi: 10.1046/j.0022-202X.2004.22226.x. [DOI] [PubMed] [Google Scholar]
- 15.Nakagawa A, Kobayashi N, Muramatsu T, Yamashina Y, Shirai T, Hashimoto MW, Ikenaga M, Mori T. Three-dimensional visualization of ultraviolet-induced DNA damage and its repair in human cell nuclei. J Invest Dermatol. 1998;110:143–148. doi: 10.1046/j.1523-1747.1998.00100.x. [DOI] [PubMed] [Google Scholar]
- 16.Jaspers NG, Bootsma D. Genetic heterogeneity in ataxia-telangiectasia studied by cell fusion. Proc Natl Acad Sci U S A. 1982;79:2641–2644. doi: 10.1073/pnas.79.8.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kobayashi N, Katsumi S, Imoto K, Nakagawa A, Miyagawa S, Furumura M, Mori T. Quantitation and visualization of ultraviolet-induced DNA damage using specific antibodies: application to pigment cell biology. Pigment Cell Res. 2001;14:94–102. doi: 10.1034/j.1600-0749.2001.140204.x. [DOI] [PubMed] [Google Scholar]
- 18.Oh KS, Imoto K, Boyle J, Khan SG, Kraemer KH. Influence of XPB helicase on recruitment and redistribution of nucleotide excision repair proteins at sites of UV-induced DNA damage. DNA Repair (Amst) 2007;6:1359–1370. doi: 10.1016/j.dnarep.2007.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Volker M, Mone MJ, Karmakar P, van Hoffen A, Schul W, Vermeulen W, Hoeijmakers JH, van Driel R, van Zeeland AA, Mullenders LH. Sequential assembly of the nucleotide excision repair factors in vivo. Mol Cell. 2001;8:213–224. doi: 10.1016/s1097-2765(01)00281-7. [DOI] [PubMed] [Google Scholar]
- 20.Gozukara EM, Khan SG, Metin A, Emmert S, Busch DB, Shahlavi T, Coleman DM, Miller M, Chinsomboon N, Stefanini M, Kraemer KH. A stop codon in xeroderma pigmentosum group C families in Turkey and Italy: molecular genetic evidence for a common ancestor. J Invest Dermatol. 2001;117:197–204. doi: 10.1046/j.1523-1747.2001.01424.x. [DOI] [PubMed] [Google Scholar]
- 21.Riou L, Eveno E, van Hoffen A, van Zeeland AA, Sarasin A, Mullenders LH. Differential repair of the two major UV-induced photolesions in trichothiodystrophy fibroblasts. Cancer Res. 2004;64:889–894. doi: 10.1158/0008-5472.can-03-2070. [DOI] [PubMed] [Google Scholar]
- 22.Emmert S, Kobayashi N, Khan SG, Kraemer KH. The xeroderma pigmentosum group C gene leads to selective repair of cyclobutane pyrimidine dimers rather than 6–4 photoproducts. Proc Natl Acad Sci U S A. 2000;97:2151–2156. doi: 10.1073/pnas.040559697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chavanne F, Broughton BC, Pietra D, Nardo T, Browitt A, Lehmann AR, Stefanini M. Mutations in the XPC gene in families with xeroderma pigmentosum and consequences at the cell, protein, and transcript levels. Cancer Res. 2000;60:1974–1982. [PubMed] [Google Scholar]
- 24.Khan SG, Levy HL, Legerski R, Quackenbush E, Reardon JT, Emmert S, Sancar A, Li L, Schneider TD, Cleaver JE, Kraemer KH. Xeroderma pigmentosum group C splice mutation associated with autism and hypoglycinemia. J Invest Dermatol. 1998;111:791–796. doi: 10.1046/j.1523-1747.1998.00391.x. [DOI] [PubMed] [Google Scholar]
- 25.Li L, Bales ES, Peterson CA, Legerski RJ. Characterization of molecular defects in xeroderma pigmentosum group C. Nature Genet. 1993;5:413–417. doi: 10.1038/ng1293-413. [DOI] [PubMed] [Google Scholar]
- 26.Ridley AJ, Colley J, Wynford-Thomas D, Jones CJ. Characterisation of novel mutations in Cockayne syndrome type A and xeroderma pigmentosum group C subjects. J Hum Genet. 2005;50:151–154. doi: 10.1007/s10038-004-0228-2. [DOI] [PubMed] [Google Scholar]
- 27.Rivera-Begeman A, McDaniel LD, Schultz RA, Friedberg EC. A novel XPC pathogenic variant detected in archival material from a patient diagnosed with Xeroderma Pigmentosum: a case report and review of the genetic variants reported in XPC. DNA Repair (Amst) 2007;6:100–114. doi: 10.1016/j.dnarep.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 28.Slor H, Batko S, Khan SG, Sobe T, Emmert S, Khadavi A, Frumkin A, Busch DB, Albert RB, Kraemer KH. Clinical, Cellular, and Molecular Features of an Israeli Xeroderma Pigmentosum Family with a Frameshift Mutation in the XPC Gene: Sun Protection Prolongs Life. J Invest Dermatol. 2000;115:974–980. doi: 10.1046/j.1523-1747.2000.00190.x. [DOI] [PubMed] [Google Scholar]
- 29.Maquat LE. Nonsense-mediated mRNA decay in mammals. J Cell Sci. 2005;118:1773–1776. doi: 10.1242/jcs.01701. [DOI] [PubMed] [Google Scholar]
- 30.Yasuda G, Nishi R, Watanabe E, Mori T, Iwai S, Orioli D, Stefanini M, Hanaoka F, Sugasawa K. In vivo destabilization and functional defects of the xeroderma pigmentosum C protein caused by a pathogenic missense mutation. Mol Cell Biol. 2007;27:6606–6614. doi: 10.1128/MCB.02166-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khan SG, Metin A, Gozukara E, Inui H, Shahlavi T, Muniz-Medina V, Baker CC, Ueda T, Aiken JR, Schneider TD, Kraemer KH. Two essential splice lariat branchpoint sequences in one intron in a xeroderma pigmentosum DNA repair gene: mutations result in reduced XPC mRNA levels that correlate with cancer risk. Hum Mol Genet. 2004;13:343–352. doi: 10.1093/hmg/ddh026. [DOI] [PubMed] [Google Scholar]
- 32.Kozak M. An analysis of vertebrate mRNA sequences: intimations of translational control. J Cell Biol. 1991;115:887–903. doi: 10.1083/jcb.115.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kozak M. Initiation of translation in prokaryotes and eukaryotes. Gene. 1999;234:187–208. doi: 10.1016/s0378-1119(99)00210-3. [DOI] [PubMed] [Google Scholar]
- 34.Kozak M. Some thoughts about translational regulation: forward and backward glances. J Cell Biochem. 2007;102:280–290. doi: 10.1002/jcb.21464. [DOI] [PubMed] [Google Scholar]
- 35.Tapias A, Auriol J, Forget D, Enzlin JH, Scharer OD, Coin F, Coulombe B, Egly JM. Ordered conformational changes in damaged DNA induced by nucleotide excision repair factors. J Biol Chem. 2004;279:19074–19083. doi: 10.1074/jbc.M312611200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rapin I, Lindenbaum Y, Dickson DW, Kraemer KH, Robbins JH. Cockayne syndrome and xeroderma pigmentosum. Neurology. 2000;55:1442–1449. doi: 10.1212/wnl.55.10.1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robbins JH, Kraemer KH, Lutzner MA, Festoff BW, Coon HG. Xeroderma pigmentosum. An inherited disease with sun sensitivity, multiple cutaneous neoplasms, and abnormal DNA repair. Ann Intern Med. 1974;80:221–248. doi: 10.7326/0003-4819-80-2-221. [DOI] [PubMed] [Google Scholar]
- 38.Andrews AD, Barrett SF, Robbins JH. Xeroderma pigmentosum neurological abnormalities correlate with colony-forming ability after ultraviolet radiation. Proc Natl Acad Sci U S A. 1978;75:1984–1988. doi: 10.1073/pnas.75.4.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hananian J, Cleaver JE. Xeroderma pigmentosum exhibiting neurological disorders and systemic lupus erythematosus. Clin Genet. 1980;17:39–45. doi: 10.1111/j.1399-0004.1980.tb00112.x. [DOI] [PubMed] [Google Scholar]
- 40.Quackenbush EJ, Kraemer KH, Gahl WA, Schirch V, Whiteman DA, Levine K, Levy HL. Hypoglycinaemia and psychomotor delay in a child with xeroderma pigmentosum. J Inherit Metab Dis. 1999;22:915–924. doi: 10.1023/a:1005691424004. [DOI] [PubMed] [Google Scholar]
- 41.Robbins JH, Brumback RA, Mendiones M, Barrett SF, Carl JR, Cho S, Denckla MB, Ganges MB, Gerber LH, Guthrie RA. Neurological disease in xeroderma pigmentosum. Documentation of a late onset type of the juvenile onset form. Brain. 1991;114:1335–1361. doi: 10.1093/brain/114.3.1335. [DOI] [PubMed] [Google Scholar]
- 42.Robbins JH, Brumback RA, Moshell AN. Clinically asymptomatic xeroderma pigmentosum neurological disease in an adult: Evidence for a neurodegeneration in later life caused by defective DNA repair. Eur Neurol. 1993;33:188–190. doi: 10.1159/000116932. [DOI] [PubMed] [Google Scholar]
- 43.Harper PS. Practical Genetic Counselling. Butterworth-Heinemann, Ltd; Oxford: 1988. [Google Scholar]
- 44.Bittles AH, Sullivan SG, Zhivotovsky LA. Consanguinity, caste and deaf-mutism in Punjab, 1921. J Biosoc Sci. 2004;36:221–234. doi: 10.1017/s0021932003006230. [DOI] [PubMed] [Google Scholar]
- 45.Sajjad M, Khattak AA, Bunn JE, Mackenzie I. Causes of childhood deafness in Pukhtoonkhwa Province of Pakistan and the role of consanguinity. J Laryngol Otol. 2008:1–7. doi: 10.1017/S0022215108002235. [DOI] [PubMed] [Google Scholar]
- 46.Petersen MB, Willems PJ. Non-syndromic, autosomal-recessive deafness. Clin Genet. 2006;69:371–392. doi: 10.1111/j.1399-0004.2006.00613.x. [DOI] [PubMed] [Google Scholar]