Abstract
α-Bungarotoxin (α-bgtx) binding proteins, including certain nicotinic acetylcholine receptors and acetylcholine-binding proteins (AChBPs), are frequently characterized with radioisotope-labeled α-bgtx-binding assays. Such assays, however, preclude investigations of binding interactions in real-time and are hampered by the inconveniences associated with radioisotope-labeled reagents. We have used surface plasmon resonance-based technology (BIAcore) to investigate the binding of recombinant AChBP to CM-5 sensor chip surfaces with directly immobilized α-bgtx. We have validated our BIAcore results by comparing the same biological samples using the traditional [125I]-α-bgtx-binding assay. An α-bgtx sensor chip, as described here, enables detailed, real-time, radioisotope-free interaction studies that can greatly facilitate the characterization of novel α-bgtx-binding proteins and complexes.
Keywords: α-bungarotoxin, BIAcore, surface plasmon resonance, AChBP, acetylcholine-binding protein
The use of surface plasmon resonance (SPR)-based instrumentation for investigating protein-protein interactions has grown substantially over the past decade [1]. Instruments, such as the BIAcore T100 (GE Healthcare, Piscataway, NJ), can be used to determine binding parameters by measurement of the changes in the refractive index at the surface of a sensor chip to which a protein or peptide (ligand) is covalently bound and over which a potential binding partner (analyte) is perfused [2]. Interactions between the analyte and the ligand are detected through the increase in mass on the surface of the sensor chip [3].
The most commonly used strategies for detecting soluble α-bungarotoxin (α-bgtx)-binding proteins require radioisotope-labeled α-bgtx and are not appropriate for real-time binding analysis[4]. We have developed a radioisotope-free method for measuring the binding activity of recombinant, acetylcholine-binding protein (AChBP) that is secreted by Pichia pastoris into yeast culture medium. We utilized SPR-based technology which incorporates sensor chips directly immobilized with α-bgtx as an alternative to the traditional [125I]-α-bgtx-binding assay for AChBP detection. This radioisotope-free methodology introduces a convenient and innovative tool for the investigation of α-bgtx-binding proteins in real-time.
The ligand, α-bgtx, that we directly immobilized onto BIAcore CM-5 sensor chips, is a 74-amino acid α-neurotoxin isolated from the venom of the Taiwanese banded krait, Bungarus multicinctus [5]. In vitro and in situ studies have established that α-bgtx is a high-affinity competitive antagonist of the muscle-type nicotinic acetylcholine receptor (nAChR); of the neuronal nAChR family composed of α7, α8, α9, and α10 subunits; and also binds with high affinity to most known soluble AChBPs [6]. For the studies reported here, we used the recombinantly expressed AChBP from the mollusk, Lymnaea stagnalis, as our analyte. In recent years, AChBPs have been intensely studied as they are naturally-occurring soluble homologues of the extracellular ligand-binding domain of Cys-loop ligand-gated ion channels. Molluscan AChBPs are most closely related to the α7 homomeric subtype of nAChR in both primary structure and pharmacology [7]. The first AChBP was discovered in the central nervous system of the fresh water snail (L. stagnalis), and subsequent cloning of the AChBP gene has enabled its heterologous expression in the methylotrophic yeast Pichia pastoris [8]. For our experiments, we constructed a carboxy-terminal hexahistidine-tagged AChBP, using the cDNA subcloned by A.B. Smit into the pPIC9 vector. This plasmid encodes for an upstream Saccharomyces cerevisiae α-mating factor signal sequence, which directs the secretion of the recombinant AChBP into the culture medium. AChBP was overexpressed in Pichia pastoris strain GS115 following Invitrogen’s specifications. The cultures were grown for 48 hours in BMMY expression media (1.34% yeast nitrogenous base, 2% peptone, 100 mM potassium phosphate pH 6.0, 4 × 10-5 % biotin, 1% methanol) on an orbital shaker at 30°C and 300 revolutions per minute. The cultures were centrifuged at 4°C for 10 minutes at 6000 × g to pellet the cells and the resulting supernatant was filtered and used immediately for binding studies. Protein concentration was determined using the bicinchoninic acid assay (ThermoFisher, Waltham, MA).
α-Bgtx (Sigma-Aldrich, St. Louis, MO) was immobilized on the surface of a CM-5 sensor chip (GE Healthcare, Piscataway) via amine coupling following manufacturer’s instructions [9]. Using the BIAcore pH Scouting method provided with the instrumentation software package, we determined the pH that maximized the electrostatic attraction between α-bgtx and the carboxymethyl dextran matrix of the sensor chip. α-Bgtx was dissolved at a concentration of 50 μg/ml in 10 mM acetate buffer with pH values of 4.0, 4.5, 5.0, and 5.5. Maximal electrostatic interaction was obtained with 10 mM sodium acetate pH 5.0 (data not shown) and subsequent immobilizations were performed using α-bgtx dissolved at a concentration of 50 μg/ml in this buffer.
The surfaces of two flow cells (FC1 and FC2) on the BIAcore chip were prepared in parallel using a flow rate of 5 μl/min. FC1 served as a reference cell following mock-immobilization with buffer alone , while FC2 was immobilized with α-bgtx. We regularly obtained α-bgtx immobilization levels ranging from ~1050-1500 resonance units (RU). For example, the immobilization level of the α-bgtx flow cell used in Figure 1was 1054.5 RU, corresponding to approximately 1 ng, or 125 fmoles, of α-bgtx covalently bound to the surface of the flow cell. In comparison, the reference flow cell that was mock-immobilized in the absence of ligand gave rise to a background signal of ~150 RU.
We used the Binding Analysis Assay Wizard software to adjust assay parameters and to graph and analyze the data. For binding experiments, the BIAcore T100 instrument was operated at 25°C and the assay buffer was PT8 (150 mM NaCl, 2 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, 20 mM Tris base, 0.1% Tween-20, pH 8.0). The contact time (the period during which the analyte, AChBP, was perfused over the chip) was limited to 400 seconds and the flow rate was set at 5 μl/min. For chip surface regeneration, a solution of 1 M carbachol (a low molecular weight cholinergic ligand of the AChBP) in PT8 buffer was used to dissociate bound AChBP at the end of each experiment, while retaining surface integrity.
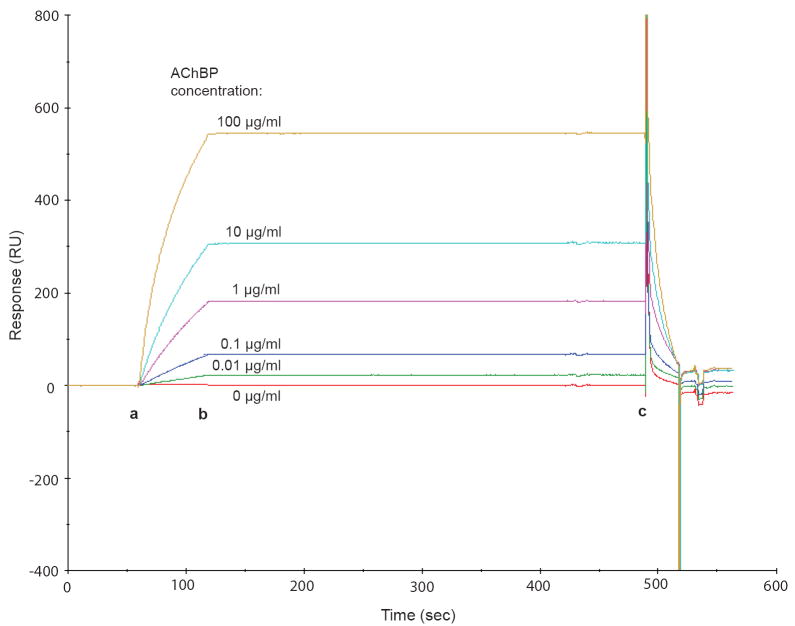
As an initial demonstration of utility, we investigated the concentration-dependence of AChBP binding by assaying five different concentrations of AChBP, ranging from 0.01 μg/ml to 100 μg/ml in order of magnitude increments, with the α-bgtx sensor chip (Fig. 1). The injection of increasing concentrations of AChBP resulted in more intense response signals (resonance units), indicating increasing levels of AChBP binding to the ligand-immobilized chip surface. In addition, we observed only minimal dissociation of AChBP from the chip prior to surface regeneration with carbachol (Fig. 1, point b to point c), which is consistent with high-affinity binding in solution between α-bgtx and AChBP [10].
Fig. 1. Concentration-dependence of signal responses due to AChBP binding to the α-bgtx sensor chip.
Purified AChBP was injected (from point a to point b) at concentrations ranging from 0 to 100 μg/ml. Point c indicates the starting point for the regeneration of the chip surface with 1 M carbachol.
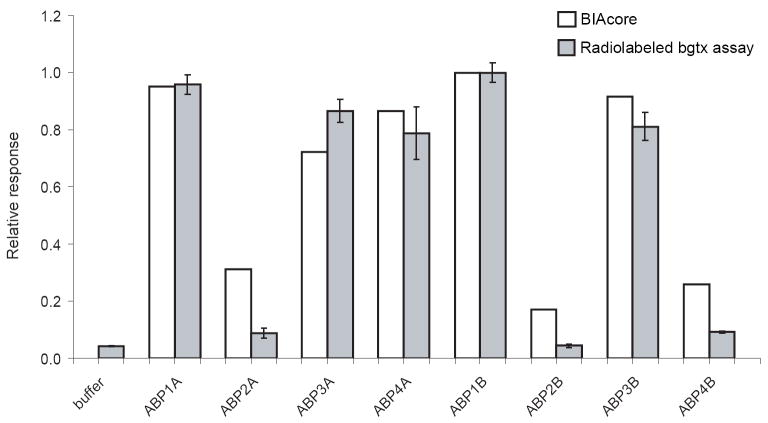
To investigate the use of the α-bgtx sensor chip in more complex samples, we screened independently isolated Pichia colonies for AChBP expression by perfusing 50 μl of filtered culture media over the α-bgtx sensor chip (5 μl/min for 10 minutes). We analyzed the culture supernatants of four colonies from each of two separate transformations (A and B) of Pichia. The exposure of the BIAcore fluidics system to the nutrient-rich media was minimized by using PT8 as the running buffer for the assay. The maximal response before surface regeneration (corresponding to point c as in Fig. 1) relative to the baseline of each sample prior to analyte injection (corresponding to point a in Fig. 1) was plotted on a bar graph (Fig. 2, white bars). Colonies ABP1A, ABP3A, ABP4A, ABP1B, and ABP3B showed the highest levels of binding activity on the α-bgtx sensor chip.
Fig. 2. Detecting AChBP expression in Pichia pastoris culture media.
This bar graph compares the assay data obtained with the BIAcore α-bgtx sensor chip (white bars) with data from an [125I]-α-bgtx-binding assay (gray bars). BIAcore and [125I]- α-bgtx-binding assay data were normalized to the value of the sample with the greatest response at point c (see Fig. 1) and the sample with the maximum radioactive counts, respectively.
We compared our BIAcore-derived results to those obtained with an [125I]-α-bgtx binding assay that is commonly used to investigate α-bgtx-binding proteins [4]. For each sample (analyzed in triplicate), 50 μl of cleared media from Pichia pastoris cultures (containing the secreted hexahistidine-tagged AChBP) was incubated with 5 nM [125I]-α-bgtx (Perkin Elmer, Shelton, CT) in 250 μl of PT8 buffer, supplemented with 1.25% BSA to decrease non-specific binding, for 1 hour at room temperature with gentle rotation. Fifty microliters of PT8-washed Talon metal-affinity bead slurry (Clontech Laboratories, Mountain View, CA) was added to the mixture which was then gently rotated for an additional 1 hour at room temperature. The samples were centrifuged for 1 minute at room temperature at 1000 × g and the supernatant was removed. The beads were washed three times with 1 ml of PT8 buffer to remove unbound [125I]-α-bgtx. The lower portion of the microcentrifuge tube retaining the bead/AChBP/[125I]-α-bgtx complexes was deposited into a scintillation vial and radioactivity was recorded using a Wallac 1275 gamma counter (Perkin Elmer, Shelton, CT).
The [125I]-α-bgtx binding assay performed with replicate aliquots of the same culture media used for the BIAcore assay revealed that samples ABP1A, ABP3A, ABP4A, ABP1B and ABP3B apparently contained greater quantities of active AChBP when compared to the other cultures tested (Fig. 2, gray bars). These results provide validation of the BIAcore-based assay. In addition, a comparison of the signals obtained with the culture medium-derived samples with the calibration curve of Fig. 1, indicates that the Pichia samples contain ~50 ng/ml of active AChBP. These findings support the use of the α-bgtx BIAcore sensor chip assay as a suitable non-radioactive alternative for detecting recombinant AChBP in complex culture media.
In prior studies, two groups had used BIAcore SA (streptavidin-coated) sensor chips to which biotinylated α-bgtx had been immobilized to screen for the binding of various peptides to α-bgtx [11; 12]. However, in crude extracts and in complex culture media, the presence of any streptavidin-binding proteins would be expected to lower instrument sensitivity [13; 14]. Direct immobilization of α-bgtx to the CM-5 chip surface eliminates potential binding artifacts and elevated backgrounds due to streptavidin. Although binding of AChBP to α-bgtx did not appear to be substantially hindered by the direct chemical immobilization of α-bgtx to the chip, caution should be taken as other protein-protein interactions may encounter steric effects adversely affecting the binding of the analyte to the ligand when using BIAcore technology [15].
In summary, we have succeeded in directly immobilizing α-bgtx onto a general-use BIAcore sensor chip, and we have used this modified sensor chip to measure the binding of AChBP in complex culture media in a concentration-dependent manner and with a lower detection limit of ~10 ng/ml (~100 pM of the pentameric AChBP). The sensitivity of the BIAcore assay is comparable to that of the [125I]-α-bgtx binding assay, as in our hands, we achieve a detection limit of ~5-7 ng/ml of AChBP using the standard binding assay with 5 nM [125I]-α-bgtx. In addition, we have demonstrated that BIAcore technology provides a non-radioactive alternative to the [125I]-α-bgtx-binding assay for the detection of heterologously-expressed AChBP directly from yeast cultures while enabling real-time monitoring of interactions during binding, washing, and dissociation. As the BIAcore T100 instrument is capable of eluant collection from the sensor chip surface, this would facilitate downstream mass spectrometric analysis and identification of novel α-bgtx-binding proteins from complex biological samples [16; 17]. Real-time detection, isolation, and identification of novel α-bgtx-binding proteins and complexes from cell cultures and tissues should now be facilitated by these advances.
Acknowledgments
This work was supported by National Institutes of Health grant #GM32629. Some of the work presented here was done in partial fulfillment of the requirements for a Ph.D. degree (J.A.P.) from Brown University. The instrumentation used in this study was made available through an NSF/EPSCoR award (#0554548). We thank A. B. Smit for the wild-type AChBP cDNA and Dr. P. Caffery for his critical reading of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Fivash M, Towler EM, Fisher RJ. BIAcore for macromolecular interaction. Curr Opin Biotechnol. 1998;9:97–101. doi: 10.1016/s0958-1669(98)80091-8. [DOI] [PubMed] [Google Scholar]
- 2.Malmqvist M. BIACORE: an affinity biosensor system for characterization of biomolecular interactions. Biochem Soc Trans. 1999;27:335–40. doi: 10.1042/bst0270335. [DOI] [PubMed] [Google Scholar]
- 3.Raghavan M, Bjorkman PJ. BIAcore: a microchip-based system for analyzing the formation of macromolecular complexes. Structure. 1995;3:331–3. doi: 10.1016/s0969-2126(01)00164-2. [DOI] [PubMed] [Google Scholar]
- 4.Salvaterra PM, Mahler HR. Nicotinic acetylcholine receptor from rat brain. Solubilization, partial purification, and characterization. J Biol Chem. 1976;251:6327–34. [PubMed] [Google Scholar]
- 5.Moise L, Zeng H, Caffery P, Rogowski RS, Hawrot E. Structure and function of alpha-bungarotoxin. Journal of Toxicology-Toxin Reviews. 2002;21:293–317. [Google Scholar]
- 6.Dutertre S, Lewis RJ. Toxin insights into nicotinic acetylcholine receptors. Biochem Pharmacol. 2006;72:661–70. doi: 10.1016/j.bcp.2006.03.027. [DOI] [PubMed] [Google Scholar]
- 7.Sine SM, Engel AG. Recent advances in Cys-loop receptor structure and function. Nature. 2006;440:448–55. doi: 10.1038/nature04708. [DOI] [PubMed] [Google Scholar]
- 8.Brejc K, van Dijk WJ, Klaassen RV, Schuurmans M, van Der Oost J, Smit AB, Sixma TK. Crystal structure of an ACh-binding protein reveals the ligand-binding domain of nicotinic receptors. Nature. 2001;411:269–76. doi: 10.1038/35077011. [DOI] [PubMed] [Google Scholar]
- 9.Johnsson B, Lofas S, Lindquist G. Immobilization of proteins to a carboxymethyldextran-modified gold surface for biospecific interaction analysis in surface plasmon resonance sensors. Anal Biochem. 1991;198:268–77. doi: 10.1016/0003-2697(91)90424-r. [DOI] [PubMed] [Google Scholar]
- 10.Smit AB, Syed NI, Schaap D, van Minnen J, Klumperman J, Kits KS, Lodder H, van der Schors RC, van Elk R, Sorgedrager B, Brejc K, Sixma TK, Geraerts WP. A glia-derived acetylcholine-binding protein that modulates synaptic transmission. Nature. 2001;411:261–8. doi: 10.1038/35077000. [DOI] [PubMed] [Google Scholar]
- 11.Bracci L, Lozzi L, Lelli B, Pini A, Neri P. Mimotopes of the nicotinic receptor binding site selected by a combinatorial peptide library. Biochemistry. 2001;40:6611–9. doi: 10.1021/bi0023201. [DOI] [PubMed] [Google Scholar]
- 12.Spiga O, Bernini A, Scarselli M, Ciutti A, Bracci L, Lozzi L, Lelli B, Di Maro D, Calamandrei D, Niccolai N. Peptide-protein interactions studied by surface plasmon and nuclear magnetic resonances. FEBS Lett. 2002;511:33–5. doi: 10.1016/s0014-5793(01)03274-4. [DOI] [PubMed] [Google Scholar]
- 13.Wang H, Pevsner J. Detection of endogenous biotin in various tissues: novel functions in the hippocampus and implications for its use in avidin-biotin technology. Cell and Tissue Research. 1999;296:511–516. doi: 10.1007/s004410051311. [DOI] [PubMed] [Google Scholar]
- 14.McKay BE, Molineux ML, Turner RW. Endogenous biotin in rat brain: implications for false-positive results with avidin-biotin and streptavidin-biotin techniques. Methods Mol Biol. 2008;418:111–28. doi: 10.1385/1-59745-579-2:111. [DOI] [PubMed] [Google Scholar]
- 15.Edwards DA. Steric hindrance effects on surface reactions: applications to BIAcore. J Math Biol. 2007;55:517–39. doi: 10.1007/s00285-007-0093-7. [DOI] [PubMed] [Google Scholar]
- 16.Nelson RW, Nedelkov D, Tubbs KA. Biomolecular Interaction Analysis Mass Spectometry. BIA/MS can detect and characterize protiens in complex biological fluids at the low- to subfemtomole level. Anal Chem. 2000;72:404A–411A. [PubMed] [Google Scholar]
- 17.Nelson RW, Nedelkov D, Tubbs KA. Biosensor chip mass spectrometry: a chip-based proteomics approach. Electrophoresis. 2000;21:1155–63. doi: 10.1002/(SICI)1522-2683(20000401)21:6<1155::AID-ELPS1155>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]