Abstract
The Pax6 gene has attracted intense research interest due to its apparently important role in the development of eyes and the central nervous system (CNS) in many animal groups. Pax6 is also of interest for comparative genomics since it has not been duplicated in tetrapods, making for a direct orthology between the Ciona intestinalis gene CiPax6 and Pax6 in mammals. CiPax6 has been shown to be expressed in the anterior brain, caudal nerve cord, and in parts of the brain associated with the photoreceptive ocellus. This information was extended here using in-situ hybridization, and shows that CiPax6 transcripts mark the lateral regions of the nerve cord, remarkably similar to Pax6 expression in the mouse. As a means of dissecting the cis-regulation of CiPax6 we tested 8 kb. of sequence using transient reporter transgene assays. Three separate regions were found that work together to drive the overall CiPax6 expression pattern. A 211 bp. sequence 2 kb. upstream of the first exon was found to be a major enhancer driving expression in the sensory vesicle (the anterior portion of the ascidian brain). Other upstream sequences were shown to work with the sensory vesicle enhancer to drive expression in the remainder of the CNS. An “eye enhancer” was localized to the first intron, which controls specific expression in the central portion of the sensory vesicle, including photoreceptor cells. The fourth intron was found to repress ectopic expression of the reporter gene in middle portions of the embryonic brain. Aspects of this overall regulatory organization are similar to the organization of the Pax6 homologs in mice and Drosophila, particularly the presence of intronic elements driving expression in the eye, brain and nerve cord.
Introduction
The Pax6 gene encodes a transcription factor that has been implicated in the development of eyes throughout the animal kingdom (Gehring, 1998; Gehring, 2002). Pax6 also has important roles in other parts of the central nervous system (CNS) in both flies and vertebrates (Gruss and Walther, 1992; Simpson and Price, 2002). Pax6 is a member of the paired-box family of transcription factors, and contains both a paired domain and a homeodomain. It is of special interest in an evolutionary context since it has not been duplicated in the ascidian and tetrapod lineages, unlike many other developmental genes, and therefore the Pax6 genes across chordate phylogeny can be related as orthologs without the complication of duplicated paralogs.
Pax6 is expressed in the CNS and eyes in many, if not all, animal groups. In ascidians Pax6 is expressed strongly in the anterior and posterior portions of the brain, and in the dorsal nerve cord (Glardon et al., 1997; Mazet et al., 2003; this paper). Interestingly, its expression is absent from middle portions of the brain, reminiscent of its absence from the vertebrate midbrain, and thus it appears to be similarly deployed along the anterior-posterior axis of both ascidians and vertebrates.
The cis-regulation of Pax6 has been studied extensively in mouse (Griffin et al., 2002; Kammandel et al., 1999; Kleinjan et al., 2004; Kleinjan et al., 2006; Xu et al., 1999) and fly (Adachi et al., 2003; Hauck et al., 1999) and to a lesser extent in pufferfish (Griffin et al., 2002; Kammandel et al., 1999) and quail (Plaza et al., 1999; reviewed in (Morgan, 2004). These studies provide much material for comparison of the cis-regulation of gene homologs in disparate metazoan species.
The ascidians Ciona intestinalis and C. savignyi are emerging as important model systems for the study of chordate gene regulation and networks (Cone and Zeller, 2005; Di Gregorio and Levine, 2002; Satoh and Levine, 2005). Ascidians have a very simple version of the chordate body plan. However, several studies have shown that some major developmental gene expression patterns are conserved with those of higher chordates, such as vertebrates (e. g. (Corbo et al., 1997a; Wada et al., 1998). C. intestinalis has a very compact genome, which along with its congener C. savignyi has been completely sequenced, allowing for genomic comparisons. Often cis-regulatory elements are found quite close to the gene they regulate, which makes dissection of cis-regulation less strenuous than in many other taxa. Also, transient reporter transgenes for studying cis-regulation are easily delivered to many embryos simultaneously - a major technical advantage for this species (Zeller, 2004).
We present here an analysis of 8 kilobases of non-coding sequence upstream and in two large introns of the Pax6 gene in C. intestinalis (CiPax6) using transient reporter transgenesis. This region encompasses all the conserved non-coding sequences found in comparisons of the C. intestinalis and C. savignyi Pax6 regions. We identified major enhancers for expression in the central nervous system (CNS) upstream of the transcription start site. We also identified eye (ocellus) specific enhancers in the first intron. Finally, we found evidence that highly conserved sequences in the fourth intron act as repressors of ectopic expression in the CNS. We also examined in more detail than previously reported the transcript expression pattern of CiPax6, and discuss its similarities and differences with patterns seen in other animals.
Materials and Methods
Animals
Adult Ciona intestinalis, sp. B (Nydam and Harrison, 2007) were collected from floating docks in the Point Judith Marina at Snug Harbor, Rhode Island. Some animals used for electroporation (Ciona intestinalis, sp. A) were obtained commercially from M-Rep, Carlsbad, California. Gametes were collected by dissection and spawned in vitro. Embryos for in situ hybridization or electroporation were chemically dechorionated at spawning.
Reporter transgenes
Reporter constructs were based on a reporter vector, TV13, modified from 72-1.27 (Corbo et al., 1997b) (kindly provided by A. Di Gregorio and M. Levine) which in turn is a derivation of pPD1.27 (Fire et al., 1990). Rare-cutting restriction sites were added to 72-1.27 by cutting with PstI and XbaI and inserting a cohesive-end oligonucleotide linker with AscI and FseI sites. Subsequently, this construct was cut at PflF1 and BglII sites downstream of the reporter gene and a linker with RsrII and PacI sites was inserted to make TV13. This vector has a multiple cloning site, nuclear localization signal, lacZ sequence, SV40 polyadenylation signal, and the RsrII/PacI downstream cloning site.
Individual constructs were made using the polymerase chain reaction (PCR) to amplify desired fragments of a lambda clone (P3A) obtained from a C. intestinalis, sp. B (Rhode Island, USA population) genomic library. This library was constructed in the BlueSTAR lambda vector (Novagen, Madison, WI, U.S.A.) and a 15 kilobase clone containing the CiPax6 gene and flanking sequence was subcloned in pBlueSTAR-1 by Cre-mediated excision. PCR primers had restriction sites designed on the 5′ ends depending on the desired cloning sites in the TV13 vector.
Electroporation
Transgenes were delivered by electroporation. Fertilized eggs were dechorionated using 0.4 mg/ml Pronase E (P5147, Sigma, St. Louis, MO, USA) in 1% sodium thioglycolate in filtered sea water (FSW) pH 10.1 for 2 min. at 18°C. 150 µl of dechorionated single cell embryos in FSW (approx. 50 embryos) were transferred to the electroporation solution (50-100 ug supercoiled transgene DNA in mannitol for a final mannitol concentration of 0.5M) in a 0.4 cm electroporation cuvette. A square wave pulse was delivered using a BTX ECM 830 electroporation device (Harvard Apparatus, Holliston, MA, USA). The contents of the cuvette were immediately decanted into a 150 mm × 15 mm gelatin-coated petri dish of FSW with antibiotics (approx. 15 U penicillin and 15 µg streptomycin per milliliter) and incubated at 14-18°C to the desired stages.
β-galactosidase detection
Expression of the lacZ transgene was detected using either standard X-gal histochemistry, or immunofluorescence. For immunofluorescence, an anti-β-galactosidase monoclonal antibody, 40-1a (Developmental Studies Hybridoma Bank) was used with a goat anti-mouse secondary antibody conjugated to AlexaFluor 488 (Invitrogen).
In-situ hybridization
A 1.4 kilobase CiPax6 clone was obtained from a 9 hr. C. intestinalis, sp. B cDNA library in lambda-ZapII (kindly provided by T. Meedel). A riboprobe was made using the Maxi-Script kit (Ambion, Austin, TX, USA) with digoxygenin-UTP as label (Roche, Indianapolis, IN, USA). Whole mount in-situ hybridization was performed as previously described (Irvine, 2007; Irvine et al., 2007).
Results
Endogenous CiPax-6 transcript and expression pattern
We screened a C. intestinalis, sp. B cDNA library, and the longest clone obtained was 1.4 kb. This clone is highly similar to the Ghost cDNA Database (URL: http://ghost.zool.kyoto-u.ac.jp/indexr1.html) clone ci0100144072, being only 10 bp. shorter at the 5′ end. We found no evidence for alternate splice variants in our screen, however, the Ghost Database lists another clone from an adult cDNA library, cima822k22, which is 216 bp. longer at the 5′ end. This is a splice variant listed on the JGI v. 2 genome browser (URL: http://genome.jgi-psf.org/Cioin2/Cioin2.home.html) as estExt_fgenesh3_pg.C_chr_09q0597. Since we were examining embryonic stages only, we confined our study to the shorter embryonic 5′ variant, which will be the cDNA referred to as CiPax6.
As a means of interpreting reporter transgene assays we examined the transcript expression pattern of CiPax6 using WMISH. Portions of the CiPax6 expression pattern have been previously described by others (Mazet et al., 2003; Satou et al., 2005), ANISEED: http://crfb.univ-mrs.fr/aniseed/index.php). We first examined the gastrula stage, which exhibits one bilateral pair of cells that hybridize with the CiPax6 riboprobe in the neural plate (Fig. 1A). These are the A9.30 cells which are in the lineage contributing to brain and pigment cells (Nishida, 1987; Nicol & Meinertzhagen, 1988; Imai et al., 2004). By the neurula stage this expression has expanded to bilateral ranks of cells corresponding to the neural tube anlage, which further lengthen as the tailbud embryo develops (Fig. 1B-D). At mid-tailbud stage expression is localized to the anterior sensory vesicle, with expression conspicuously absent from the visceral ganglion (Fig. 1E-G). CiPax6 is also present in the bilateral pair of ependymal cells of the nerve cord, which consists of two lateral cells, one dorsal cell, and one ventral cell (Fig. 1G). By late tailbud stages we see expression reappearing in the posterior visceral ganglion in some specimens, but expression is always absent in the neck, anterior visceral ganglion and posterior sensory vesicle (Fig. 1H, I). This pattern is consistent with that described by Mazet et al. (2003).
Fig. 1. CiPax6 transcript expression pattern visualized by whole-mount in-situ hybridization.
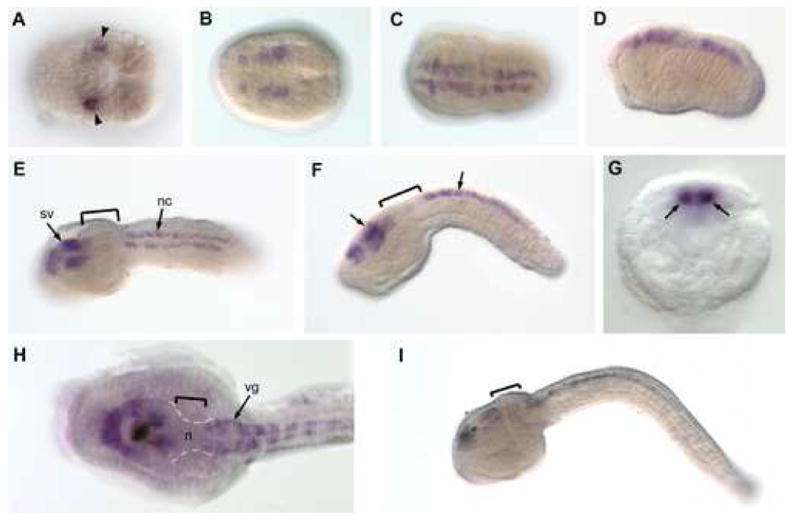
Anterior is to the left in all panels. (A) Gastrula stage dorsal view. Bilateral pair of A9.30 cells staining (arrowheads). (B) Neurula stage dorsal view with CiPax6 expression in bilateral strips of neuroectoderm. (C, D) Early tailbud embryo in dorsal and lateral views, respectively. Expression is in entire anterior-posterior length of the nascent CNS. (E, F) Mid-tailbud stage embryo in dorsal and lateral views, respectively. Expression is detected in the sensory vesicle (arrow) and nerve cord, but has become down-regulated in the forming visceral ganglion (brackets). (G) Optical section through a mid-tailbud embryo showing transcripts detected in the two lateral cell ranks of the nerve cord (arrows), which consists of only 4 ependymal cells in cross-section, one dorsal, one ventral and two lateral. (H, I) Dorsal and lateral views, respectively of late tailbud embryos, showing persistent staining in the anterior sensory vesicle and nerve cord. In the higher power view (H), staining is visible in the visceral ganglion, but absent from the neck (n, brackets) and most posterior sensory vesicle. n, neck; nc; nerve cord; sv, sensory vesicle; vg, visceral ganglion.
Comparative sequence analysis reveals conserved non-coding elements in Ciona
We performed a VISTA sequence alignment between C. intestinalis and C. savignyi Pax6 to identify conserved non-coding elements (CNEs) as candidate regulatory elements for further study (Mayor et al., 2000; URL: http://genome.lbl.gov/vista/index.shtml; C. savignyi sequence from Broad Institute database, URL: http://www.broad.mit.edu/annotation/ciona/). This method has been shown to be effective at locating functional regulatory elements in Ciona (Johnson et al., 2004). The initial analysis extended 9.5 kb. upstream of exon 1 and 4.5 kb downstream of exon 10, for a total sequence length of 24 kb (Supplementary fig. S1). 2 CNEs were identified at 2.5 kb. and 2.0 kb. upstream of the CiPax6 transcriptional start site, which have greater than 80% identity between species in places, and were identified as candidate regulatory elements (Figs. 2A & S1; labeled UB and UA respectively). Other conserved sequences outside the CiPax6 coding region correspond with predicted genes based on EST surveys (Supplementary fig. S1). Within the coding region, three candidate elements were found in Intron 1 (I1A-C), and four CNEs were located in Intron 4 (I4A-D). Since important enhancer elements have been found in the introns of mouse and fly Pax6 (refer to Fig. 6), we were especially interested in these intronic CNEs.
Fig. 2. Ciona sequence alignment, transgene diagrams and scoring.
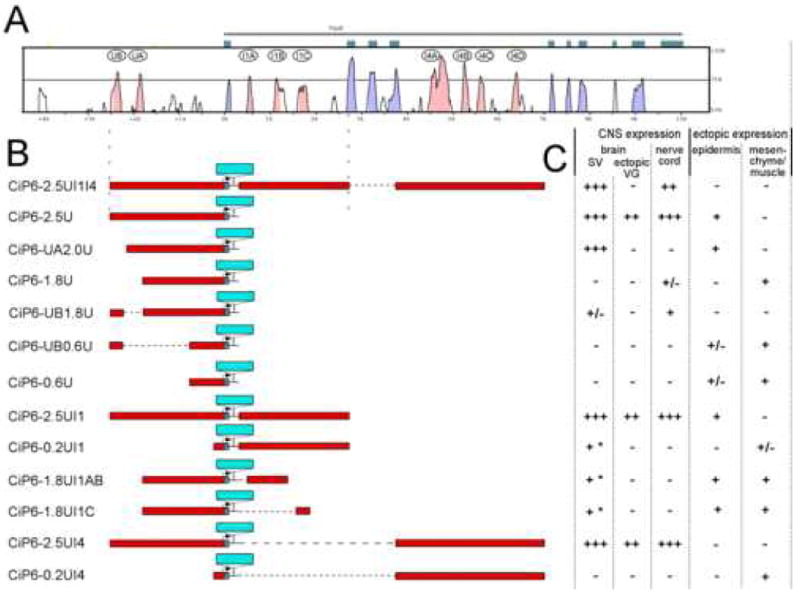
(A) mVista sequence alignment plot between C. intestinalis and C. savignyi, with CiPax6 exons shown as blue-green boxes. Curve represents levels of sequence identity in a 50 bp window. Blue-shaded peaks are in exons while pink-shaded peaks are in non-coding sequence. Major conserved non-coding sequences (CNSs) are identified by letters in ovals. (B) Diagrams of transgene constructs. Non-coding sequence is represented by red bars aligned with corresponding sequence in Vista plot above. LacZ reporter gene insertion is represented by a blue bar. Dotted horizontal lines indicate sequences not in the transgene. (In these cases the cloned sequences are connected to each other with short linkers.) (C) Scoring chart for expression driven by each transgene in C. intestinalis embryos at mid-late tailbud stages. Number of “+” symbols denotes relative intensity and penetrance of lacZ expression. “-” indicates lack of expression. “+/-” indicates that expression is present in some embryos at a low level. * refers to limited expression in central sensory vesicle only.
Fig. 6. Comparisons of Pax6 cis-regulation between mouse, Ciona and fly.
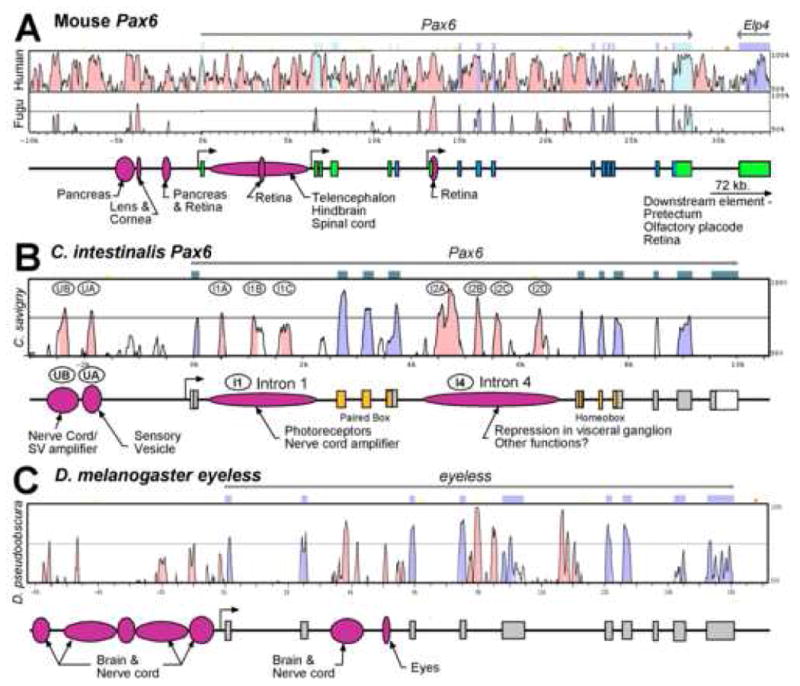
For each species a VISTA plot of the Pax6 genomic region is shown above a diagram of the exon-intron organization and experimentally verified regulatory regions. Mouse Pax6 (A) is aligned with both human and Fugu rubripes Pax6, C. intestinalis Pax6 (B) is aligned with C. savignyi Pax6, and Drosophila melanogaster eyeless (C) is aligned with the corresponding region in D. pseudoobscura. Mouse data from Kammandel et al. (1999) and Xu et al. (1999). Drosophila data from Hauck et al. (1999) and Adachi et al. (2003).
Major CNS enhancers are found upstream of the transcription start site
Since CNEs had been found upstream and in introns 1 and 4, we tested these regions from a CiPax6 genomic lambda clone for regulatory activity by inserting them into a transgene reporter vector and electroporating it into single-cell embryos. We also constructed various combinations of parts of this larger construct and tested them in like fashion, to dissect the regulatory activity of various parts of the CiPax6 non-coding regions (Fig. 2B). We found that the upstream plus introns 1 and 4 construct (CiP6-2.5UI1I4) reproduced most of the endogenous expression pattern we found in the WMISH specimens (Fig. 1), as visualized in β-galactosidase (β-gal) histochemistry (Fig. 3A, B). The important exceptions to this statement were i) that ectopic β-gal expression was present in the visceral ganglion and neck region, and ii) that ectopic β-gal expression was usually seen in two bilateral pairs of muscle cells in the caudal tail. In the first case, since the β-gal expression pattern might not reflect the transcript expression pattern of the WMISH work, we performed WMISH using a lacZ riboprobe to examine the transcript expression pattern. The reporter gene transcript expression pattern much more faithfully reproduced the endogenous mRNA pattern in the brain. (See below for more on WMISH results). Regarding point ii) above, ectopic expression in tail muscle, the expressing cells are derived from the A8.16 lineage (Nishida, 1987), which gives rise to both lateral nerve cord, which endogenously expresses CiPax6, and these 4 muscle cells, which do not. Apparently a regulatory element required to repress expression in the tail muscle subset of A8.16 descendents is not present in the CiP6-2.5UI1I4 construct.
Fig. 3. Major CNS enhancers are located upstream of CiPax6 coding sequence.
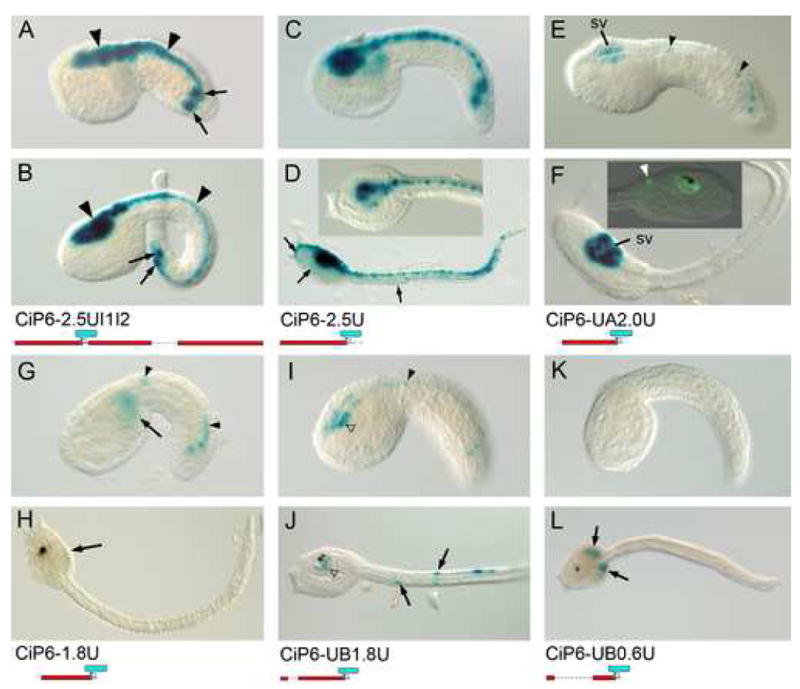
Photomicrographs of β-galactosidase (β-gal) histochemical assays of lacZ expression driven by various reporter transgenes. Mid-tail (upper panels, A, C, E, G, I, K) and late tail (lower panels, B, D, F, H, J, L) embryos in lateral view with anterior to the left, except as noted. Transgene name and diagram are located below each embryo pair. (A, B) Expression from upstream, intron 1, and intron 4 fragments together. β-gal staining is seen in the entire CNS (arrowheads). Expression is also seen in 2 muscle cells on each side of the trunk, derived from the A8.16 lineage (arrows, see text). These cells also stain with the CiP6-2.5U, -2.0U, and -1.8U constructs. (C, D) 2.5 kb of upstream sequence alone drives expression in the entire CNS, as well as ectopic expression in epidermis (arrows) (D inset) dorsal detailed view of trunk of late tail CiP6-2.5U embryo. (E, F) Specific expression in the sensory vesicle (sv) is driven by a construct with 1986 bp. of upstream sequence. At mid tailbud stage sporadic expression is also seen in the caudal nerve cord (arrowheads) (F inset) Right lateral view of late tail embryo with CiP6-2.0U transgene expression detected with an anti-lacZ antibody. Expression can be seen in the sensory vesicle surrounding the pigmented otolith and ocellus. A cell in the caudal visceral ganglion (arrowhead) also reacts with the lacZ antibody (white arrowhead). (G, H) Deletion of 211 bp. containing the UA CNE results in loss of most CNS expression except for slight expression in the caudal nerve cord at mid tailbud stage (arrowheads). Ectopic expression in mesenchyme appears in this truncated construct (arrows). (I, J) Addition of the distal sequence containing the UB CNE to the -1.8U construct results in gain of some sensory vesicle expression (white arrowheads) in addition to the low level expression in the nerve cord (arrowhead). This construct exhibits some ectopic expression in tail muscle cells at late stages (arrows). (K, L) A construct with only 0.6 kb. of upstream sequence shows only ectopic β-gal expression in trunk mesenchyme at late stages (arrows).
In order to test the regulatory activity of the upstream region alone, we made a series of transgenes including various fractions of the upstream sequence. A construct with 2436 bp. of upstream sequence (CiP6-2.5U, Fig. 3C, D) is able to drive expression in the entire CNS. This fragment contains both the upstream CNEs, UB and UA. Deletion of the distal 452 bp. containing the UB element results in a construct that drives expression specifically in the sensory vesicle (CiP6-2.0U, Fig. 3E, F). Deletion of a further 211 bp. containing the UA element (CiP6-1.8U, Fig. 3G, H) results in nearly complete loss of CNS expression, except for slight nerve cord expression, indicating that the UA element is required for anterior brain expression, and that the UB element is required to gain expression in the nerve cord.
A construct with the UA element deleted from the entire upstream sequence (CiP6-UB1.8U, Fig. 3I, J) shows only sporadic expression in the CNS, indicating a cooperative effect between the UA element and the remainder of the upstream sequence. The UB element, on the other hand, in combination with a short 600 bp. segment of upstream sequence including the basal promoter (CiP6-UB0.6U, Fig. 3K, L) is inactive in the CNS, exhibiting only ectopic expression indistinguishable from that driven by the upstream 600 bp. fragment alone (CiP6-0.6U, data not shown). Therefore, the UB element alone does not have enhancer activity without the sequence between 0.6 kb. up to the UA element. When the UA element itself is added, the expression is increased radically throughout the CNS.
Intron 1 contains a photoreceptor enhancer
To examine the possible enhancer activity of Intron 1 sequence we made a transgene, CiP6-2.5UI1, which adds Intron 1 to the upstream sequence already shown to drive expression throughout the CNS. This reporter showed β-gal expression in the same regions as the upstream construct (compare Figs. 3C & D with 4A & B). However, the expression in the visceral ganglion and caudal nerve cord is elevated in the transgene with Intron 1, indicating that this intron contains sequences that can amplify the enhancer activity of upstream sequences. This effect is also seen at the transcript level in WMISH experiments (Fig. 5C, D explained in the next section).
Fig. 4. Intron 1 contains elements driving expression in photoreceptors.
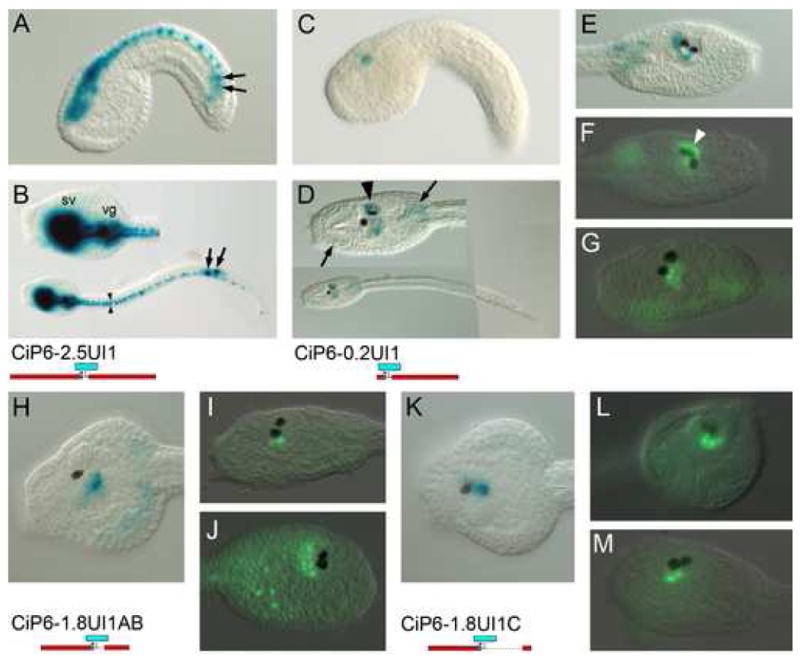
β-gal histochemistry (A-E, H, K) and anti-β-gal immunofluorescent detection (F, G, I, J, L, M) of lacZ reporter transgenes. Constructs are identified below and to the left of corresponding images. (A, C) are mid-tailbud stage in lateral view. Others are late tailbud stages in lateral views, except B, D and K, which are dorsal views, and L which is frontal. (A, B) Construct with 2.5 kb. upstream region and entire intron 1. Expression is in entire CNS, as well as ectopic expression in lineage A8.16 muscle cells (arrows) as seen in Fig. 3. Note expression in bilateral ranks of caudal nerve cord cells (arrowheads) (C-G) Intron 1 fragment connected with 200 bp. of basal promoter sequence. Expression is in the central sensory vesicle (arrowheads). Ectopic expression is also in trunk mesenchyme (arrows). In (F) note expression in photoreceptor array associated with the ocellus (white arrowhead).(H-J) Expression driven from the proximal 1 kb. of intron 1 sequence containing the I1A and I1B CNEs. Variable β-gal signal is seen in ventral and caudal portions of the central sensory vesicle. (K-L) Expression driven from the distal 300 bp. of intron 1 containing the I1C CNE. Variable β-gal signal is in similar regions to that driven by the proximal intron 1 fragment.
Fig. 5. Intron 4 sequence downregulates expression in the neck and visceral ganglion.
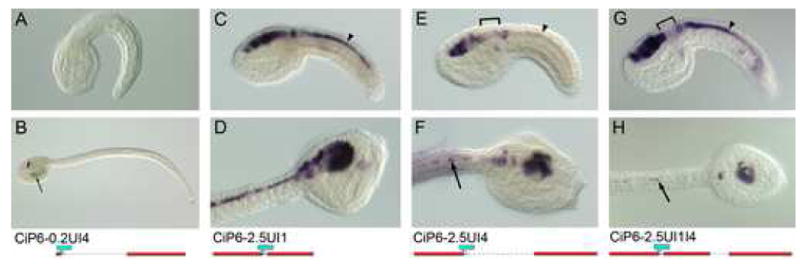
Whole mount in-situ hybridization using a lacZ antisense riboprobe, except A and B, which are β-gal histochemical staining. Transgene name and diagram are located below each embryo pair. Top row (A, C, E, G) are mid-tailbud stages, and second row are lateral (B, D, H), or dorsolateral (F) views of late tailbud stages. (A, B) Intron 4 alone drives only ectopic expression in trunk mesenchyme at late stages (arrow). (C-H) LacZ transcript expression is shown for reporter transgenes with upstream and Intron 1 sequence (C, D), upstream and Intron 4 sequence (E, F), and upstream and Intron 1 and Intron 4 sequence (G, H). Note that expression becomes downregulated in the neck and anterior visceral ganglion in the transgenes incorporating intron 4 sequence (brackets). Expression in the caudal nerve cord is reduced in E, F compared with constructs incorporating Intron 1 (C, D & G, H) (arrowheads). However at late stages (H) Intron 4 sequence represses nerve cord expression even in the presence of Intron 1 (arrows). Refer to Table 1 for scoring of multiple embryos for each transgene.
To examine the enhancer activity of Intron 1 by itself, a construct containing Intron 1 along with only 200 bp. of basal promoter sequence (CiP6-0.2UI1; Fig. 4C-G) reveals that the intronic sequence drives specific expression immediately dorsal, ventral and caudal to the pigment cell sensory organs of the sensory vesicle. This expression is obscured by the stronger or overlapping intense CNS expression driven from the 2.5 kb upstream sequence. Immunofluorescent detection of β-gal helps distinguish more specifically the reporter gene expression pattern. In some embryos, e.g. Fig. 4F, an arc of cells dorsal and left of the ocellus show reporter expression. These have been identified as photoreceptor cells by Horie et al. (2005) which is consistent in location with data in Imai and Meinertzhagen (2007). Other embryos show cells at the ventral and caudal parts of the sensory vesicle with β-gal expression. The mosaic nature of expression suggests that at least two different lineages give rise to the cells which are capable of expressing the CiP6-0.2UI1 transgene.
We attempted to determine which portion of the intron 1 sequence is responsible for sensory vesicle expression activation by testing reporter transgenes with either the proximal or distal portions of the intron in transgenes with 1.8 kb of upstream sequence (CiP6-1.8UI1A and B; Fig. 4H-M). Both constructs drove expression in the region encompassed by the CiP6-0.2UI1 pattern. However, given the mosaic character of the expression patterns we were not able to distinguish a separate specific expression domain for the two transgenes. Rather, both constructs appeared to drive expression in the same areas when the patterns from a number of transgenic embryos for each transcript were added together. Further work, such as use of two different reporter genes would be required to resolve whether different parts of the intronic sequence control the same or different expression domains.
Intron 4 may contain repressors of ectopic visceral ganglion expression
The non-coding sequences most highly conserved between C. intestinalis and C. savignyi are found in Intron 4 (I4A-I4D, Fig. 2A). Because of this we were surprised that Intron 4 sequence in combination with a minimal promoter fails to have any activation effect in the β-gal reporter, except for a low level of ectopic expression in mesenchyme (CiP6-0.2UI4; Fig. 5A, B). Likewise, the addition of Intron 4 sequence to transgenes with upstream and Intron 1 sequences in CiP6-2.5UI1I4 (Fig. 3A, B) has no discernable effect on the β-gal signal, as compared with transgenes lacking Intron 4, such as CiP6-2.5UI1 (Fig. 4A, B). Notably, all these constructs exhibit ectopic expression in the visceral ganglion, as visualized by β-gal protein expression. This led us to the hypothesis that the Intron 4 sequence may contain repressors of ectopic visceral ganglion expression at the transcriptional level, but its action is hidden because of the perdurance of the β-gal protein in the transgene assays, such as those of CiP6-2.5UI1I4 (Fig. 3A, B).
To test this hypothesis we performed WMISH using a riboprobe complementary to lacZ mRNA on embryos electroporated with upstream sequence in combination with either Intron 1, Intron 4 or both (Fig. 5C-H). In this way we could see if the reporter gene was reproducing the endogenous expression pattern at the transcriptional level. The results are rather variable, either due to the mosaic nature of the transient transgenic assay, or due to variability in the regulatory effects of the Intron 4 sequence in the transgene. Scoring of numerous embryos from the various reporter experiments are summarized in Table 1. From the endogenous expression pattern the expected results would show much lower levels of expression in the visceral ganglion than in either the sensory vesicle or nerve cord. The data indicates that both upstream sequence alone (CiP6-2.5U) and upstream plus Intron 1 sequence (CiP6-2.5UI1; Fig. 5C, D) have high levels of ectopic visceral ganglion transcript expression. On the other hand, the upstream sequence plus Intron 4 construct (CiP6-2.5UI4; Fig. 5E, F) and upstream plus Intron 1 plus Intron 4 construct (CiP6-2.5UI1I4; Fig. 5G, H) both have much lower levels of expression in the visceral ganglion. Put together, these data suggest that Intron 4 is responsible, at least in part, for repression of expression in the neck and anterior visceral ganglion, as seen in the endogenous expression patterns shown in Fig. 1.
Table 1. Region-specific mRNA expression due to various intron plus upstream non-coding sequence reporter transgene combinations, as assessed by WMISH.
| Stage | Anatomical region | % of embryos with reporter expression in region listed | |||
|---|---|---|---|---|---|
| CiP6-2.5U | CiP6-2.5UI1 | CiP6-2.5UI4 | CiP6-2.5UI1I4 | ||
| Mid tailbud stage | Sensory vesicle | 100 | 100 | 100 | 100 |
| Visceral ganglion (ectopic) | 58 | 87 | 20 | 16 | |
| Nerve cord | 67 | 100 | 7 | 42 | |
| n=12 | n=23 | n=30 | n=19 | ||
| Late tailbud stage | Sensory vesicle | 100 | 100 | 100 | 90 |
| Visceral ganglion (ectopic) | 30 | 80 | 7 | 10 | |
| Nerve cord | 20 | 80 | 13 | 40 | |
| n=10 | n=10 | n=15 | n=10 | ||
Refer to Fig. 2B for diagrams describing the reporter transgene constructs.
As summarized in Table 1, in the case of CiP6-2.5UI4, expression also drops off in the nerve cord, but this expression is largely recovered when Intron 1 sequence is present in CiP6-2.5UI1I4. Likewise lower levels of expression are seen in the nerve cord for upstream sequence alone (CiP6-2.5U) than for the reporter transgene with upstream and Intron 1 sequence (CiP6-2.5UI1). These effects are consistent with the result of β-gal histochemical detection of CiP6-2.5UI1 activity commented on above, which appeared elevated in visceral ganglion and caudal nerve cord compared with the construct with upstream sequence alone (CIP6-2.5U). These comparisons suggest that Intron 1, while not driving nerve cord expression alone, increases the level of expression in combination with upstream sequence.
Discussion
Pax6 expression marks the lateral neural tube in C. intestinalis, as it does in vertebrates
Cañestro et al. (2005) compared the Pax6 expression patterns of urochordates, based on the ascidians Phallusia mammilata (Glardon et al., 1997) and C. intestinalis (Imai et al., 2004; Mazet et al., 2003), and the larvacean Oikopleura dioica, with those found in vertebrates, particularly zebrafish. This comparison showed that in urochordates Pax6 is expressed in the brain anterior to the Pax2/5/8 and engrailed expression domains, which in vertebrates mark the midbrain. In C. intestinalis this Pax6 expressing region corresponds to most of the sensory vesicle, except for the most caudal part. In zebrafish Pax6 expression is also anterior to the zone of Pax2/Pax5/Pax8 and engrailed. This Pax6 expression region is the vertebrate prosencephalon, which has been previously proposed to be homologous to the ascidian sensory vesicle, based on Otx, Hox1 and Pax2/5/8 expression (Wada et al., 1998). CiPax6 is also expressed in the caudal visceral ganglion, and in vertebrates in the hindbrain, adding evidence to the proposed homology between those two structures (Dufour et al., 2006). In vertebrates a gap in Pax6 expression corresponds fairly precisely with the midbrain. However, although CiPax6 also has a gap in expression in the middle portion of the brain, recent studies of markers of the vertebrate midbrain-hindbrain boundary suggest that urochordates lack a homolog of the vertebrate midbrain. Rather the region lacking CiPax6 expression, which contains the neck region, the caudal sensory vesicle and the rostral visceral ganglion, is the zone giving rise to most of the CNS neurons of the post-metamorphic ascidian brain. The presence of MHB markers in this region may be related to functions in neuronal specification and differentiation rather than indicating a homology with the vertebrate midbrain (Canestro et al., 2005; Lacalli, 2006).
It has been shown previously that certain aspects of dorso-ventral patterning in the C. intestinalis neural tube are conserved with the pattern in vertebrates (Corbo et al., 1997a). In particular, the Ciona forkhead/HNF3β homolog Ci-FoxAa marks the ventral cell of the neural tube, which consists of just four ependymal cells in cross-section, one ventral, one dorsal and a bilateral pair. This pattern corresponds to the expression of HNF3β in the floor plate of the mouse neural tube. Similarly, the expression of Ci-sna, the Ciona homolog of the mouse snail gene, is expressed in the lateral pair of neural tube cells, reflecting the lateral expression of snail in mouse. Another marker of the neural tube at the intermediate dorso-ventral level in vertebrates is Pax6 (Burrill et al., 1997; Walther and Gruss, 1991) Our data adds to this picture by showing that CiPax6 is expressed specifically in the lateral pair of neural tube cells (Fig. 1G). Thus, like the expression of HNF3β, and snail, Pax6 is expressed at a similar dorso-ventral level of the neural tube, reinforcing the notion that patterning of the neural tube is conserved between vertebrates and urochordates.
CiPax6 expression is controlled by several positive and negative cis-regulatory elements
The genomic regulation of the CiPax6 gene is previously undescribed. Our work has identified three regions of cis-regulatory activity in the CiPax6 gene - two activating regions and one with repressive effects. 2 kb. upstream of the embryonic transcription start site is found a 211 bp region which is responsible for driving strong expression of a reporter transgene in the sensory vesicle (CiP6-2.0U, Fig. 3E, F), since when this is deleted from a 2 kb. upstream reporter all specific expression is lost (CiP6-1.8U, Fig. 3G, H) This 211 bp. region corresponds with a stretch of sequence conserved between C. intestinalis and C. savignyi (UA, Figs. 2A, 6B). Addition of a further 452 bp. of upstream sequence, which includes another stretch of conserved sequence (UB) gives expression along the entire CNS (CiP6-2.5U, Fig. 3C, D). The UA and UB regions appear to work synergistically to drive expression outside of the sensory vesicle, as evidenced by the loss of nerve cord and posterior brain expression in construct CiP6-UB1.8U, which has a deletion of the UA element (Fig. 3I, J). There is also some effect of sequence more proximal to the transcription start site than UA, as shown by the difference between the expression patterns driven by the CiP6-UB1.8U and CiP6-UB0.6U reporter transgenes (Fig. 3I-L).
Although the UA element is a major enhancer for sensory vesicle expression, sequence in the first intron drives specific expression in portions of the sensory vesicle, some of which are photoreceptor neurons (Fig. 4C-M) (Horie et al., 2005; Imai and Meinertzhagen, 2007; Nicol and Meinertzhagen, 1991). The photoreceptor cell expression driven by the Intron 1 sequence is most clearly shown in Fig. 4F, where an arc of cell nuclei reactive to β-gal antibody are seen covering the pigment cup cell of the ocellus, which Horie et al. (2005) have shown to be photoreceptors. In addition to the photoreceptor cells, other cells surrounding the ventral and caudal walls of the sensory vesicle cavity also show expression driven by the Intron 1 reporter gene. We did not determine if these cells were a subset of the expression pattern driven by the upstream sensory vesicle enhancer, or if their CiPax6 expression is solely due to the Intron 1 sequences. We did examine portions of the Intron 1 sequence in an attempt to localize the activity of specific enhancer sequences. However, both proximal and distal fragments of the intron drove expression in variable and overlapping regions around the wall of the sensory vesicle cavity. This variability may be due either to the mosaic nature of the transient transgene reporter assay, or to variable properties of these particular reporter transgenes in vivo. On the other hand, there may be specificity to the different fragments that is hidden by the lack of complete cellular level resolution in these whole mount assays, which would require extensive confocal section analysis to resolve.
The most highly conserved elements in the region of CiPax6 that we examined are located in the large fourth intron (Fig. 2A). However, reporters comprised of this intron and a native CiPax6 promoter failed to drive any enhancer activity (CiP6-0.2UI4, Fig. 5A, B). We hypothesized that this intron might function as a repressor of ectopic expression. In particular, we thought repressor action from this intron might downregulate transcription in the visceral gangion at mid and late tailbud stages, a part of the expression pattern that the β-gal reporters had failed to reproduce. To test this idea we used WMISH to the lacZ transcript to see if visceral ganglion, or other expression, was downregulated by the Intron 4 sequence. Indeed, Intron 4 had a repressive effect on expression in the posterior brain (compare Figs. 5C, D to 5E-H), and embryos with the combination of upstream, Intron 1, and Intron 4 sequence (Fig. 5G,H) were the most successful at reproducing the endogenous expression patterns of Fig. 1, as shown in Table 1. These experiments also showed that Intron 1 had an amplifying effect on expression in the nerve cord, which is attenuated at late tailbud stage by the addition of Intron 4 sequence (cf. Fig. 5D & H).
These repressive effects of Intron 4 are subtle, and we have not determined which part or parts of the intron are required for repression. It is possible that other functions are served by parts of the Intron 4 sequence, especially the highly conserved portions, such as matrix attachment, nucleosome formation, or modulation of mRNA stability (Shabalina and Spiridonov, 2004).
Combinatorial modular organization of CiPax6 cis-regulation
If the CiPax6 regulatory elements described here are looked at as modules (Davidson, 2006), the combinatorial function of these modules in driving the full tailbud stage expression pattern can be seen. (In Fig. 2A and 6B the CNEs denoted in ovals – UB, UA, I1A, etc. - can be seen as putative modules.) We have experimentally verified cis-regulatory activity for the UB and UA modules. We have also verified cis-regulatory activity for portions of introns 1 and 4, denoted as Modules I1 and I4 in Fig. 6, each of which may actually be composed of multiple separable sub-modules. The following summarizes the overall organization of CiPax6 regulation. Module UA drives expression in the sensory vesicle, the largest portion of the ascidian brain. Module UB, in combination with UA drives expression in the nerve cord and amplifies sensory vesicle expression. Module I1 drives specific expression in photoreceptors, and also amplifies nerve cord expression in combination with the upstream modules. Finally, Module I4 serves to attenuate expression in the visceral ganglion. Thus, at tailbud stages all the conserved non-coding sequences within 3 kb. upstream and in the introns, ie. all the cis-regulatory modules identified here, are required in combination to recapitulate the endogenous CiPax6 expression pattern.
Comparison of Pax6 regulation in Ciona, flies, and vertebrates
Fig. 6 summarizes published information on Pax6 regulation in mouse and Drosophila, along with the data in this paper for C. intestinalis. Comparison of the three reveals a similar level of complexity in cis-regulatory elements on a gross scale, especially given that the eyes and nervous systems in flies and mice are much more complex than those of the ascidian larva. Some other intriguing similarities may be seen upon examination of these diagrams. All three species have elements in a large intron close to the 5′ end of the gene that drive transcription in photoreceptors, brain, and nerve cord. Both Drosophila and Ciona have major enhancers for brain and nerve cord expression upstream of the transcription start site. Interestingly, the mouse has enhancers grouped upstream of the first exon driving expression in vertebrate-specific organs, namely pancreas and the vertebrate eye structures, lens and cornea. Long-range enhancers have also been found in mouse 72 kb. downstream of the coding region controlling expression in brain, olfactory placode and retina.
We attempted to align the confirmed enhancer regions between these three disparate species. No sequence similarity was apparent using BLAST alignment programs. In spite of this, the similarities in genomic organization of regulatory elements, especially for the intronic eye, brain and nerve cord enhancers, suggests that some of these might be descended from an ancestral Pax6 gene, which would have been expressed in the eyes, brain and nerve cord of the common ancestor of flies, mice and ascidians. In this case any sequence similarity would have been long since obscured by binding site turnover and rearrangement, which has been shown to occur relatively rapidly (Ludwig et al., 1998; Oda-Ishii et al., 2005; Takahashi et al., 1999; Wray et al., 2003). However, even though the actual enhancer sequence has evolved beyond recognition, there may be constraints in the cis-regulatory machinery for Pax6 maintaining certain eye, brain and nerve cord enhancer elements in a close downstream relationship to the basal promoter in all three animal groups. One hint that this constrained enhancer site relationship might be the case is the presence of Pax6 autoregulatory binding sites in the intronic enhancer regions of all three taxa (Morgan, 2004). As sequence analysis techniques for detecting cis-regulatory motifs advance, it may become possible to relate the functional elements of enhancers in different species despite extensive rearrangement and turnover. A future application of the present work is as a basis for comparison with ongoing study of Pax6 regulation in other ascidians. This work is intended to provide pictures of cis-regulatory organization in homologous genes at selected phylogenetic distances for the analysis of tempo and mode in cis-regulatory evolution.
Supplementary Material
VISTA program was used to align 24 kb. of C. intestinalis genomic sequence from the JGI database (Ciona v. 2) with the corresponding region of the C. savignyi genome (URL: http://www.broad.mit.edu/annotation/ciona/). CiPax6 regulatory module designations within ovals. Conserved non-coding sequence peaks are colored pink, while conserved coding sequences are colored blue. Genes flanking CiPax6 (derived from EST screens) are identified by ANISEED gene model numbers. Dashed line upstream of CiPax6 encompasses predicted exons of adult stage transcript (refer to text for explanation).
Acknowledgments
We warmly acknowledge the support of Frank H. Ruddle in the initial stages of this work. We also thank the Point Judith Marina for allowing collection of animals from their docks and Thomas Meedel, Robert Zeller, and Mark Q. Martindale for technical advice. We also benefited from discussion with Anna DiGregorio, Brad Davidson, Jean-Francois Brunet, and Cristian Cañestro. This work was supported by National Institutes of Health Grant 1R15GM07373701 from the National Center for General Medical Sciences, and Grant P20 RR016457 from the BRIN/INBRE Program of the National Center for Research Resources. Additional support was from a National Science Foundation/Alfred P. Sloan Postdoctoral Fellowship in Molecular Evolution to SQI.
Grant support: National Institutes of Health Grant 1R15GM07373701 from the National Center for General Medical Sciences, and Grant P20 RR016457 from the BRIN/INBRE Program of the National Center for Research Resources; National Science Foundation/Alfred P. Sloan Postdoctoral Fellowship in Molecular Evolution to SQI.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication.As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proofbefore it is published in its final citable form. Please note that during the productionprocess errorsmaybe discovered which could affect the content, and all legal disclaimersthat apply to the journal pertain.
References
- Adachi Y, Hauck B, Clements J, Kawauchi H, Kurusu M, Totani Y, Kang YY, Eggert T, Walldorf U, Furukubo-Tokunaga K, Callaerts P. Conserved cis-regulatory modules mediate complex neural expression patterns of the eyeless gene in the Drosophila brain. Mech Dev. 2003;120:1113–26. doi: 10.1016/j.mod.2003.08.007. [DOI] [PubMed] [Google Scholar]
- Burrill JD, Moran L, Goulding MD, Saueressig H. PAX2 is expressed in multiple spinal cord interneurons, including a population of EN1+ interneurons that require PAX6 for their development. Development. 1997;124:4493–503. doi: 10.1242/dev.124.22.4493. [DOI] [PubMed] [Google Scholar]
- Cañestro C, Bassham S, Postlethwait J. Development of the central nervous system in the larvacean Oikopleura dioica and the evolution of the chordate brain. Dev Biol. 2005;285:298–315. doi: 10.1016/j.ydbio.2005.06.039. [DOI] [PubMed] [Google Scholar]
- Cone AC, Zeller RW. Using ascidian embryos to study the evolution of developmental gene regulatory networks. Can J Zool. 2005;83:75–89. [Google Scholar]
- Corbo JC, Erives A, Di Gregorio A, Chang A, Levine M. Dorsoventral patterning of the vertebrate neural tube is conserved in a protochordate. Development. 1997a;124:2335–44. doi: 10.1242/dev.124.12.2335. [DOI] [PubMed] [Google Scholar]
- Corbo JC, Levine M, Zeller RW. Characterization of a notochord-specific enhancer from the Brachyury promoter region of the ascidian, Ciona intestinalis. Development. 1997b;124:589–602. doi: 10.1242/dev.124.3.589. [DOI] [PubMed] [Google Scholar]
- Davidson EH. The Regulatory Genome. Academic Press; San Diego: 2006. [Google Scholar]
- Di Gregorio A, Levine M. Analyzing gene regulation in ascidian embryos: new tools for new perspectives. Differentiation. 2002;70:132–9. doi: 10.1046/j.1432-0436.2002.700402.x. [DOI] [PubMed] [Google Scholar]
- Dufour HD, Chettouh Z, Deyts C, de Rosa R, Goridis C, Joly JS, Brunet JF. Precraniate origin of cranial motoneurons. Proc Natl Acad Sci USA. 2006;103:8727–8732. doi: 10.1073/pnas.0600805103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A, Harrison SW, Dixon D. A modular set of lacZ fusion vectors for studying gene expression in Caenorhabditis elegans. Gene. 1990;93:189–198. doi: 10.1016/0378-1119(90)90224-f. [DOI] [PubMed] [Google Scholar]
- Gehring WJ. Master Control Genes in Development and Evolution: The Homeobox Story. Yale, New Haven: 1998. [Google Scholar]
- Gehring WJ. The genetic control of eye development and its implications for the evolution of the various eye-types. Int J Dev Biol. 2002;46:65–73. [PubMed] [Google Scholar]
- Glardon S, Callaerts P, Halder G, Gehring WJ. Conservation of Pax-6 in a lower chordate, the ascidian Phallusia mammillata. Development. 1997;124:817–25. doi: 10.1242/dev.124.4.817. [DOI] [PubMed] [Google Scholar]
- Griffin C, Kleinjan DA, Doe B, van Heyningen V. New 3′ elements control Pax6 expression in the developing pretectum, neural retina and olfactory region. Mech Dev. 2002;112:89–100. doi: 10.1016/s0925-4773(01)00646-3. [DOI] [PubMed] [Google Scholar]
- Gruss P, Walther C. Pax in development. Cell. 1992;69:719–22. doi: 10.1016/0092-8674(92)90281-g. Review. [DOI] [PubMed] [Google Scholar]
- Hauck B, Gehring WJ, Walldorf U. Functional analysis of an eye specific enhancer of the eyeless gene in Drosophila. Proc Natl Acad Sci USA. 1999;96:564–9. doi: 10.1073/pnas.96.2.564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie T, Orii H, Nakagawa M. Structure of ocellus photoreceptors in the ascidian Ciona intestinalis larva as revealed by an anti-arrestin antibody. J Neurobiology. 2005;65:241–250. doi: 10.1002/neu.20197. [DOI] [PubMed] [Google Scholar]
- Imai JH, Meinertzhagen IA. Neurons of the ascidian larval nervous system in Ciona intestinalis: I. Central nervous system. J Comp Neurology. 2007;501:316–334. doi: 10.1002/cne.21246. [DOI] [PubMed] [Google Scholar]
- Imai KS, Hino K, Yagi K, Satoh N, Satou Y. Gene expression profiles of transcription factors and signaling molecules in the ascidian embryo: towards a comprehensive understanding of gene networks. Development. 2004;131:4047–4058. doi: 10.1242/dev.01270. [DOI] [PubMed] [Google Scholar]
- Irvine SQ. Whole-mount in-situ hybridization using laboratory mini-columns. BioTechniques. 2007;43:764–768. doi: 10.2144/000112617. [DOI] [PubMed] [Google Scholar]
- Irvine SQ, Cangiano MC, Millette BJ, Gutter ES. Non-overlapping expression patterns of the clustered DllA/B genes in the ascidian Ciona intestinalis. J Exp Zool (Mol Dev Evol) 2007;308B:428–441. doi: 10.1002/jez.b.21169. [DOI] [PubMed] [Google Scholar]
- Johnson DS, Davidson B, Brown CD, Smith WC, Sidow A. Noncoding regulatory sequences of Ciona exhibit strong correspondence between evolutionary constraint and functional importance. Genome Res. 2004;14:2448–2456. doi: 10.1101/gr.2964504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kammandel B, Chowdhury K, Stoykova A, Aparicio S, Brenner S, Gruss P. Distinct cis-essential modules direct the time-space pattern of the Pax6 gene activity. Dev Biol. 1999;205:79–97. doi: 10.1006/dbio.1998.9128. [DOI] [PubMed] [Google Scholar]
- Kleinjan D, Seawright A, Childs A, van Heyningen V. Conserved elements in Pax6 intron 7 involved in (auto)regulation and alternative transcription. Dev Biol. 2004;265:462–477. doi: 10.1016/j.ydbio.2003.09.011. [DOI] [PubMed] [Google Scholar]
- Kleinjan D, Seawright A, Mella S, Carr C, Tyas D, Simpson T, Mason J, Price D, van Heyningen V. Long-range downstream enhancers are essential for Pax6 expression. Dev Biol. 2006;299:563–581. doi: 10.1016/j.ydbio.2006.08.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacalli TC. Prospective protochordate homologs of vertebrate midbrain and MHB, with some thoughts on MHB origins. Int J Dev Biol. 2006;2:104–109. doi: 10.7150/ijbs.2.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig MZ, Patel NH, Kreitman M. Functional analysis of eve stripe 2 enhancer evolution in Drosophila: rules governing conservation and change. Development. 1998;125:949–958. doi: 10.1242/dev.125.5.949. [DOI] [PubMed] [Google Scholar]
- Mayor C, Brudno M, Schwartz JR, Poliakov A, Rubin EM, Frazer KA, Pachter LS, Dubchak I. VISTA: Visualizing Global DNA Sequence Alignments of Arbitrary Length. Bioinformatics. 2000;1046 doi: 10.1093/bioinformatics/16.11.1046. [DOI] [PubMed] [Google Scholar]
- Mazet F, Hutt JA, Millard J, Shimeld SM. Pax gene expression in the developing central nervous system of Ciona intestinalis. Gene Expr Patterns. 2003;3:743–745. doi: 10.1016/s1567-133x(03)00137-6. [DOI] [PubMed] [Google Scholar]
- Morgan R. Conservation of sequence and function in the Pax6 regulatory elements. Trends Genet. 2004;20:283–7. doi: 10.1016/j.tig.2004.04.009. [DOI] [PubMed] [Google Scholar]
- Nicol D, Meinertzhagen IA. Cell counts and maps in the larval central nervous system of the ascidian Ciona intestinalis. J Comp Neurol. 1991;309:415–429. doi: 10.1002/cne.903090402. [DOI] [PubMed] [Google Scholar]
- Nishida H. Cell lineage analysis in ascidian embryos by intracellular injection of a tracer enzyme: III. up to the tissue restricted stage. Dev Biol. 1987;121:526–541. doi: 10.1016/0012-1606(87)90188-6. [DOI] [PubMed] [Google Scholar]
- Nydam ML, Harrison RG. Geneological relationships within and among shallow-water Ciona species (Ascidiacea) Mar Biol. 2007;151:1839–1847. [Google Scholar]
- Oda-Ishii I, Bertrand V, Matsuo I, Lemaire P, Saiga H. Making very similar embryos with divergent genomes: conservation of regulatory mechanisms of Otx between the ascidians Halocynthia roretzi and Ciona intestinalis. Development. 2005;132:1663–1674. doi: 10.1242/dev.01707. [DOI] [PubMed] [Google Scholar]
- Satoh N, Levine M. Surfing with the tunicates into the post-genome era. Genes & Development. 2005;19:2407–2411. doi: 10.1101/gad.1365805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satou YTK, Shoguchi E, Nakayama A, Satoh N. An integrated database of the ascidian, Ciona intestinalis: towards functional genomics. Zool Sci. 2005;22:837–843. doi: 10.2108/zsj.22.837. [DOI] [PubMed] [Google Scholar]
- Shabalina SA, Spiridonov NA. The mammalian transcriptome and the function of non-coding DNA sequences. Genome Biol. 2004;5:105. doi: 10.1186/gb-2004-5-4-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson TI, Price DJ. Pax6; a pleiotropic player in development. BioEssays. 2002;24:1041–51. doi: 10.1002/bies.10174. [DOI] [PubMed] [Google Scholar]
- Takahashi H, Mitani Y, Satoh G, Satoh N. Evolutionary alterations of the minimal promoter for notochord-specific Brachyury expression in ascidian embryos. Development. 1999;126:3725–3734. doi: 10.1242/dev.126.17.3725. [DOI] [PubMed] [Google Scholar]
- Wada H, Saiga H, Satoh N, Holland PWH. Tripartite organization of the ancestral chordate brain and the antiquity of placodes: insights from ascidian Pax-2/5/8, Hox and Otx genes. Development. 1998;125:1113–1122. doi: 10.1242/dev.125.6.1113. [DOI] [PubMed] [Google Scholar]
- Walther C, Gruss P. Pax-6, a murine paired box gene, is expressed in the developing CNS. Development. 1991;113:1435–1449. doi: 10.1242/dev.113.4.1435. [DOI] [PubMed] [Google Scholar]
- Wray GA, Hahn MW, Abouheif E, Balhoff JP, Pizer M, Rockman MV, Romano LA. The evolution of transcriptional regulation in eukaryotes. Mol Biol Evol. 2003;20:1377–1419. doi: 10.1093/molbev/msg140. [DOI] [PubMed] [Google Scholar]
- Xu PX, Zhang X, Heaney S, Yoon A, Michelson AM, Maas RL. Regulation of Pax6 expression is conserved between mice and flies. Development. 1999;126:383–95. doi: 10.1242/dev.126.2.383. [DOI] [PubMed] [Google Scholar]
- Zeller RW. Generation and use of transgenic ascidian embryos. Development of Sea Urchins, Ascidians, and Other Invertebrate Deuterostomes: Experimental Approaches. Meth Cell Biol. 2004;74:713–730. doi: 10.1016/s0091-679x(04)74029-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
VISTA program was used to align 24 kb. of C. intestinalis genomic sequence from the JGI database (Ciona v. 2) with the corresponding region of the C. savignyi genome (URL: http://www.broad.mit.edu/annotation/ciona/). CiPax6 regulatory module designations within ovals. Conserved non-coding sequence peaks are colored pink, while conserved coding sequences are colored blue. Genes flanking CiPax6 (derived from EST screens) are identified by ANISEED gene model numbers. Dashed line upstream of CiPax6 encompasses predicted exons of adult stage transcript (refer to text for explanation).


