Summary
We examined the distribution of the dynein-associated protein NudE in Drosophila larval brain neuroblasts and spermatocytes, and analyzed the phenotypic consequences of a nudE null mutation. NudE can associate with kinetochores, spindles and the nuclear envelope. In nudE mutant brain mitotic cells, centrosomes are often detached from the poles. Moreover, the centrosomes of mutant primary spermatocytes do not migrate from the cell cortex to the nuclear envelope, establishing a new role for NudE. In mutant neuroblasts, chromosomes fail to congress to a tight metaphase plate, and cell division arrests because of spindle assembly checkpoint (SAC) activation. The targeting of NudE to mitotic kinetochores requires the dynein-interacting protein Lis1, and surprisingly Cenp-meta, a Drosophila CENP-E homolog. NudE is non-essential for the targeting of all mitotic kinetochore components tested. However, in the absence of NudE, the `shedding' of proteins off the kinetochore is abrogated and the SAC cannot be turned off, implying that NudE regulates dynein function at the kinetochore.
Keywords: Mitosis, Meiosis, Kinetochores, Centrosomes, Spindle assembly checkpoint, Dynein
Introduction
Kinetochores attach chromosomes to the spindle apparatus, move chromosomes along the spindle, and govern the spindle assembly checkpoint (SAC) that prevents precocious mitotic exit before the establishment of proper connections between the chromosomes and the spindle (Meraldi et al., 2006; Musacchio and Salmon, 2007). Many kinetochore components have been evolutionarily conserved between unicellular eukaryotes and metazoans, even though the homology is sometimes difficult to discern (Meraldi et al., 2006; Przewloka et al., 2007; Schittenhelm et al., 2007). However, one fundamental difference between the kinetochores of Saccharomyces cerevisiae and those of higher eukaryotes is that the latter contain the minus-end-directed motor cytoplasmic dynein, whereas yeast kinetochores do not. At least in some metazoan cell types, dynein at the kinetochores helps move the chromosomes toward the spindle poles during prometaphase and anaphase (Savoian et al., 2000; Sharp et al., 2000). Furthermore, at metaphase, dynein transports several proteins, including SAC components, off the kinetochores onto the microtubules to which they are attached, a process called `streaming' or `shedding'. This transport is thought to be responsible for turning off the SAC when chromosomes are properly bioriented (Howell et al., 2001; Wojcik et al., 2001).
Metazoan kinetochores must therefore contain molecules that attract dynein during mitosis and regulate dynein function once it has arrived at the kinetochores. The polypeptides Rough Deal, ZW10 and Zwilch form a complex (RZZ) at the kinetochore, which is required for dynein recruitment (Karess, 2005; Starr et al., 1998; Williams et al., 2003). The involvement of RZZ in dynein's association with the kinetochore is probably mediated by dynactin (a large multiprotein assembly that can activate dynein), because human ZW10 interacts with the p50 subunit of dynactin in the yeast two-hybrid system (Starr et al., 1998). A second interactor found in the same two-hybrid screen was Zwint-1, which was subsequently shown to be a human kinetochore component needed for kinetochore targeting of RZZ (Starr et al., 2000; Wang et al., 2004). The kinetochore thus contains complicated molecular machinery whose main purpose is to control the localization and regulate the function of dynein.
Yet another human ZW10 interactor identified in the same yeast two-hybrid screen was originally annotated as a homolog of Xenopus mitotic phosphoprotein 43 (MP43), whose function at that time was unknown (Starr et al., 2000). MP43 was subsequently discovered to be one of the two human homologs of Aspergillus nidulans NudE, a protein that is important for the even distribution of nuclei along the hyphae (Efimov and Morris, 2000). Several laboratories have since reported physical interactions between NudE (or the closely related Nudel) and dynein (Liu et al., 2000; Niethammer et al., 2000; Sasaki et al., 2000), and between NudE or Nudel and Lis1 (Efimov and Morris, 2000; Feng et al., 2000; Kitagawa et al., 2000; Sasaki et al., 2000; Sweeney et al., 2001), the latter being a WD40-containing, dynein-interacting protein that is involved in neuronal migration and neural stem cell division (Shu et al., 2004), and which localizes both to microtubule plus ends (Li et al., 2005) and to the kinetochores of mitotic chromosomes (Siller et al., 2005; Tai et al., 2002). These observations all suggest that NudE proteins might also be part of the dynein-related machinery at the kinetochore. However, little information currently exists about the function of NudE, particularly in terms of its roles in cell division.
In this report, we examine the properties of NudE in Drosophila. We find NudE on kinetochores, spindles and the nuclear envelope at various stages in the cell cycle or in various cell types. Furthermore, because Drosophila has only a single gene for this protein, we have been able to analyze the phenotypic consequences of nudE mutation. We observed strong defects in spindle organization and centrosome behavior in both mutant larval brain cells and spermatocytes. In mutant brains, most dividing cells are arrested in metaphase because of activation of the SAC. Despite the interaction of NudE with ZW10, the targeting of NudE to the kinetochore does not require the RZZ complex, but it does depend upon Lis1, and surprisingly, Cenp-meta, one of two Drosophila homologs of the vertebrate kinetochore-associated kinesin-like protein CENP-E (Yucel et al., 2000). We found no evidence that NudE is required for the targeting of any other SAC component to mitotic kinetochores. However, the shedding of various SAC proteins onto kinetochore microtubules (KMTs) does not occur in nudE mutants, so the SAC cannot be turned off, implying that NudE helps to regulate dynein function at the kinetochore.
Results
Localization of NudE
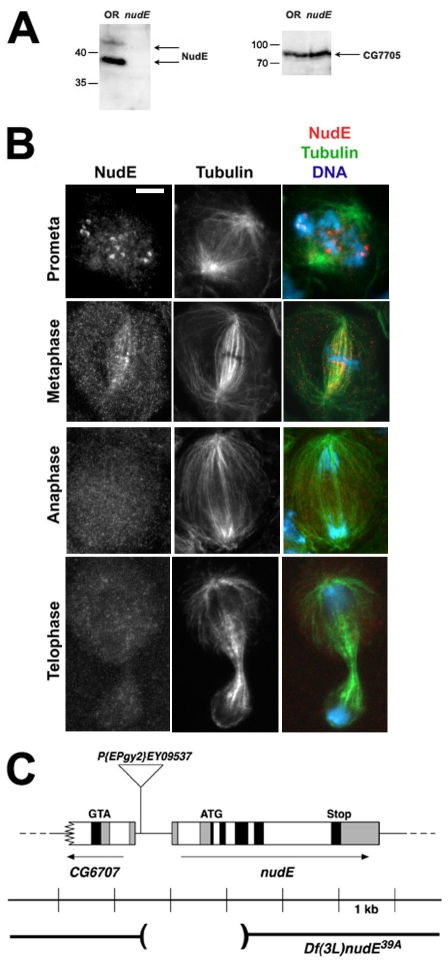
To explore the intracellular localization of NudE, we generated an antibody that recognizes this protein. Western blots of larval homogenates showed a prominent band of approximately 38 kDa in the wild type, which is close to the expected size of two isoforms of the Drosophila NudE protein annotated in FlyBase (36.2 and 37.7 kDa) (Fig. 1A). This band was not found in extracts of larvae homozygous for a null mutation in the nudE gene (nudE39A, whose genesis is described below), indicating that this band represents the NudE protein (Fig. 1A). The antibody also recognized a minor NudE-specific band of ∼45 kDa that probably corresponds to a third annotated NudE isoform of 42.6 kDa.
Fig. 1.
The nudE gene and gene product. (A) Specificity of the anti-NudE antibody. Western blot with the purified antibody shows two bands in wild-type third instar larval extracts not found in comparable extracts from nudE39A homozygous mutant larvae. These bands correspond to the molecular masses of known Drosophila NudE isoforms. As a loading control, the same blot was reprobed with antibody generated against the product of the CG7705 gene. (B) Distribution of NudE protein in wild-type larval brains. NudE is targeted to the kinetochores at prometaphase and then `streams' along KMTs during metaphase. During anaphase and telophase, NudE is not found in discrete structures. Scale bar: 5 μm. (C) Generation of a null mutation in nudE. The top row shows the genomic region that includes the nudE gene, its neighbor CG6707, and the P-element insertion P{Epgy2}EY09537. From left to right of the image corresponds to the telomere-to-centromere direction along chromosome 3L. Exons are shaded boxes; the open reading frame is in black, and the 5′ and 3′ untranslated regions are gray. The deletion mutant Df(3L)nudE39A (abbreviated in the text as nudE39A) was created by imprecise excision of P{Epgy2}EY0953; the parentheses indicate the extent of deleted DNA.
Immunofluorescence experiments using purified NudE antibody showed that the localization of NudE changed during the course of mitosis in third instar larval brains (Fig. 1B; supplementary material Fig. S1). In wild-type brains, NudE was found at discrete spots on the chromosomes during prometaphase (Fig. 1B; supplementary material Fig. S1). We interpret these spots as being the kinetochores, based on their position just distal to the centromere as defined by staining for CID, the Drosophila centromere-specific histone H3 variant (supplementary material Fig. S1) (Blower and Karpen, 2001). When the chromosomes are aligned at the metaphase plate, NudE is found either at the kinetochores or simultaneously at the kinetochores and along the spindle (Fig. 1B; supplementary material Fig. S1). From precedents established with other outer kinetochore markers including the RZZ complex (reviewed by Karess, 2005), Mad2 (Howell et al., 2000), the p50 and Glued subunits of dynactin (Siller et al., 2005; Wojcik et al., 2001), and Lis1 (Siller et al., 2005), we believe the spindle localization reflects the poleward `streaming' of NudE from the kinetochores onto the KMTs to which they are attached. However, the proportion of metaphases exhibiting streaming was much lower for the NudE protein (22% of 50 cells) than for the RZZ protein Rod (54% of 50 cells). Discrete NudE signals on chromosomes or spindles were not seen during anaphase or telophase (Fig. 1B), in contrast with Rod, which was almost always found at anaphase or telophase kinetochores (100% of 30 cells). In mitotic cells from the brains of third instar larvae homozygous for a null mutation for nudE (described below), only very weak, unlocalized signals were observed (supplementary material Fig. S1), verifying that the structures observed with our anti-NudE antibody are specific for the NudE protein.
Distribution of NudE during male meiosis
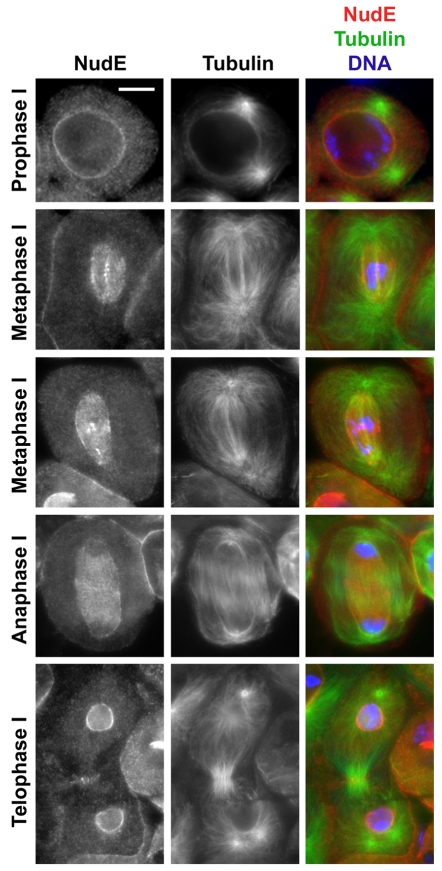
Because meiosis in Drosophila males is particularly amenable to cytological analysis (Cenci et al., 1994), we also investigated the distribution of NudE in spermatocytes (Fig. 2). During meiotic prophase, the protein is found at the nuclear envelope. Localization to kinetochores and (presumably) KMTs was seen during prometaphase or metaphase of meiosis and is similar to that seen at the equivalent mitotic stages; some diffuse spindle staining was also observed. Remarkably, the spindle staining during metaphase I and anaphase I was restricted to the `spindle envelope' and the microtubules surrounded by this structure. The spindle envelope is formed in male meiosis in many invertebrates, and consists of endoplasmic-reticulum-derived parafusorial membranes surrounding both the nuclei and part of the spindle apparatus (Fuller, 1993; Giansanti et al., 2007; Inoue et al., 2004; Wolf, 1995) (also see A. D. Tates, Cytodifferentiation during spermatogenesis in Drosophila melanogaster: an electron microscope study, PhD thesis, Rijksuniversiteit de Leiden, 1971). Little if any NudE signal was visible at the kinetochores during anaphase I. Finally, at telophase I, NudE was targeted strongly to the nuclear envelope. We also observed a NudE signal at the equator of late telophases. However, this localization is probably unrelated to cell cleavage, because nudE mutant spermatocytes are not defective in cytokinesis. We saw no staining of any discrete structure in nudE null mutant spermatocytes (data not shown), attesting to the specificity of the NudE antibody staining seen in wild-type testes.
Fig. 2.
Distribution of NudE protein in wild-type spermatocytes. NudE is mostly localized to the nuclear envelope during meiosis, except when the nuclear envelope is disrupted. NudE accumulates on the kinetochores during meiotic prometaphase or metaphase; some `streaming' along KMTs can occasionally be seen (second row). During meiotic anaphase, NudE staining is restricted to the spindle envelope region; note that the spindle itself extends beyond the spindle envelope toward the cell periphery. Scale bar: 10 μm.
Phenotypic effects of NudE depletion
To explore the consequences of the absence of NudE, we generated the null allele nudE39A by imprecise excision of a nearby P element. This deletion entirely removes the first three exons of nudE, plus part of the fourth exon (Fig. 1C). As a result, the first 37 codons of the gene, including the initiation codon, are missing, indicating that nudE39A cannot encode NudE protein. The conclusion that nudE39A is a null allele is further supported by the results of western blots of extracts from mutant larvae (Fig. 1A), and by the absence of discrete NudE immunofluorescence signals in nudE39A larval brains and testes (supplementary material Fig. S1). Animals homozygous for the nudE39A mutation die as third instar larvae or as pupae, demonstrating that nudE is an essential gene in Drosophila. We presume that nudE39A homozygous zygotes survive until late larval or pupal stages by using maternally deposited NudE protein from their heterozygous mothers, as is the case for many cell cycle genes in flies (Gatti and Goldberg, 2001).
The mitotic progression of many cells in nudE39A larval brains is arrested at prometaphase or metaphase. In orcein-stained nudE39A mutant brains, the mitotic index (the ratio of mitotic cells to microscopic fields) is substantially elevated relative to that in wild-type brains (Table 1). The large majority of mutant mitotic cells were in prometaphase or metaphase, and very few cells continue to anaphase. Chromosome overcondensation, an expected consequence of mitotic arrest (Gatti and Goldberg, 1991), was observed in most mutant brain cells (Table 1). The frequencies of all these abnormalities were roughly equivalent in the brains of nudE39A homozygotes and in the brains of larvae heterozygous for nudE39A and a chromosomal deletion [Df(3L)AC1] that removes the nudE gene (Table 1). This latter result verifies that the phenotype associated with homozygosity for the nudE39A mutation is due to lesions at the nudE gene rather than elsewhere in the genome, and further indicates that this allele fits the classical genetic definition of a null mutation.
Table 1.
Mitotic parameters of nudE mutant larval brains
| Genotype | No. fields* | Metaphase† | Anaphase‡ | Metaphase/anaphase§ | %OC¶ | MI** | Relative MI†† |
|---|---|---|---|---|---|---|---|
| OreR (wild type) | 630 | 1100 | 271 | 4.1 | 0.3 | 2.18 | 1.00 |
| nudE39A/nudE39A | 936 | 6055 | 53 | 114.2 | 89.4 | 6.53 | 3.00 |
| nudE39A/Df(3L)AC1 | 255 | 1131 | 13 | 87.0 | 96.9 | 4.49 | 2.06 |
| OreR (wild type) | 690 | 266 | 74 | 3.6 | ND | 0.49 | 1.00 |
| nudE39A/nudE39A | 227 | 434 | 6 | 72.1 | ND | 1.94 | 3.96 |
|
nudE39A zwilch1229/nudE39A
zwilch1229 |
486
|
263
|
46
|
5.7
|
ND
|
0.63
|
1.29
|
Number of microscopic fields viewed.
Number of cells in prometaphase or metaphase.
Number of cells in anaphase or telophase.
Ratio of cells in prometaphase or metaphase to those in anaphase or telophase.
Percentage of mitotic cells with overcondensed chromosomes. ND indicates not determined.
Mitotic index, the number of mitotic figures per microscopic field. The discrepancy between the MI in the two groups of samples resulted from the use of microscopes with different-sized fields.
Relative mitotic index, with wild type set equal to 1.00.
The metaphase arrest seen in nudE39A mutant larval brains probably reflects activation of the SAC, which prevents cells from initiating anaphase until all the chromosomes are properly aligned at the metaphase plate (reviewed by Malmanche et al., 2006). This conclusion arises from analysis of nudE39A zwilch1229 double mutants, where zwilch encodes an RZZ component necessary for SAC function (Williams et al., 2003). In the double mutants, the mitotic index and the proportion of mitotic figures in anaphase relative to those in prometaphase or metaphase are nearly normal (Table 1). Thus, depletion of NudE causes mitotic arrest only when the SAC is operational.
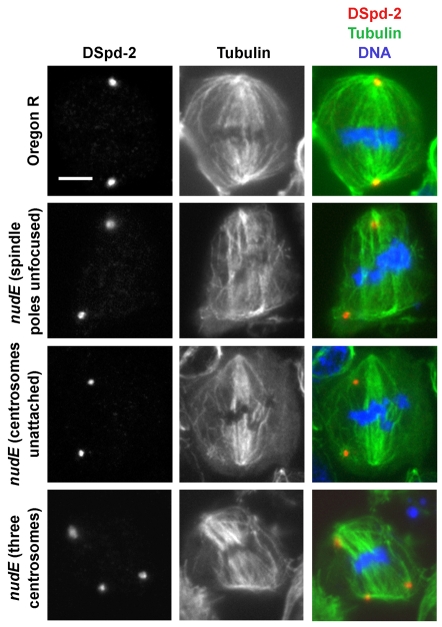
Visualization of spindles with antibody against tubulin revealed that nudE mutations also cause defects in mitotic spindle morphology and chromosome congression. In contrast to the wild type, where essentially all spindles appear to be normal, only a small minority of nudE39A cells had straight, bipolar spindles (Fig. 3; Table 2). The most frequent defects were broad, unfocused spindle poles, and, as seen by staining for the centrosomal component DSpd-2 (Giansanti et al., 2008), the failure of centrosomes to remain attached to the spindles. Smaller proportions of nudE39A brain cells showed other defects, including curved spindles, the presence of two centrosomes at one pole but none at the other pole, and supernumerary centrosomes. In the large majority of nudE39A mitoses, the chromosomes failed to form a tight metaphase plate, suggesting problems in congression (Fig. 3). By counting prometaphase or metaphase cells with chromosomes that were aligned or unaligned, we verified this idea. In wild-type controls, 42% of cells at this cell cycle stage (n=176) had uncongressed chromosomes, whereas 61% of such cells in nudE39A mutants (n=189) showed this phenotype; this difference is significant (P<0.0001).
Fig. 3.
Spindle abnormalities in nudE mutant neuroblasts. The spindles of wild-type (Oregon R) metaphase cells are bipolar, with each pole narrowly focused at a single centrosome (labeled by DSpd-2). The DNA forms a tight metaphase plate equidistant from the poles. In nudE39A homozygous mutant neuroblasts, the poles are often wide and unfocused, and the centrosomes are frequently found separated from the poles. Chromosome congression to the metaphase plate is aberrant. The presence of supernumerary centrosomes (bottom row) is also sometimes observed. Scale bar: 5 μm.
Table 2.
Mitotic aberrations in metaphase brain cells
| Genotype | Normal (bipolar, straight spindles) | Bipolar, curved spindles | Bipolar, unfocused poles | Bipolar, unattached centrosomes | Bipolar, two centrosomes on half spindle | Three spindle poles | No bipolar spindle |
|---|---|---|---|---|---|---|---|
| OreR (wild type) | 103 | 0 | 5 | 2 | 0 | 0 | 0 |
| nudE39A/nudE39A | 7 | 6 | 24* | 36 | 4 | 1 | 1 |
The number of each phenotype counted in each class is given. *Two of the 24 cells in this class had three centrosomes; the remainder had two centrosomes.
Centrosome misbehavior in nudE mutant spermatocytes
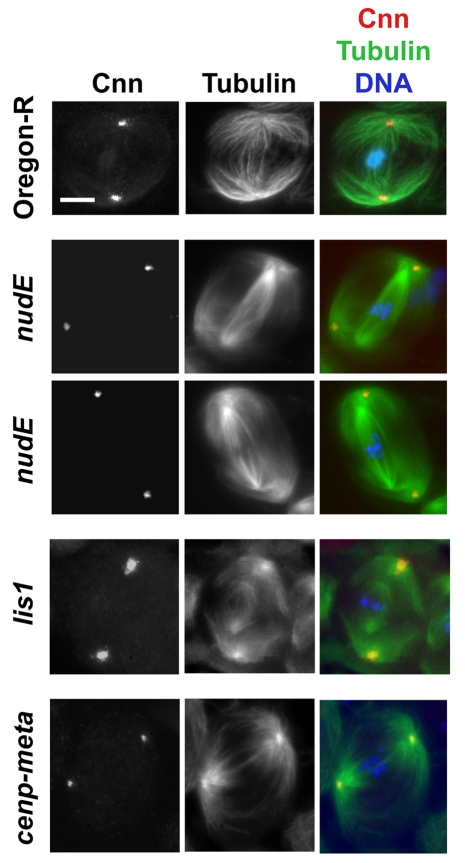
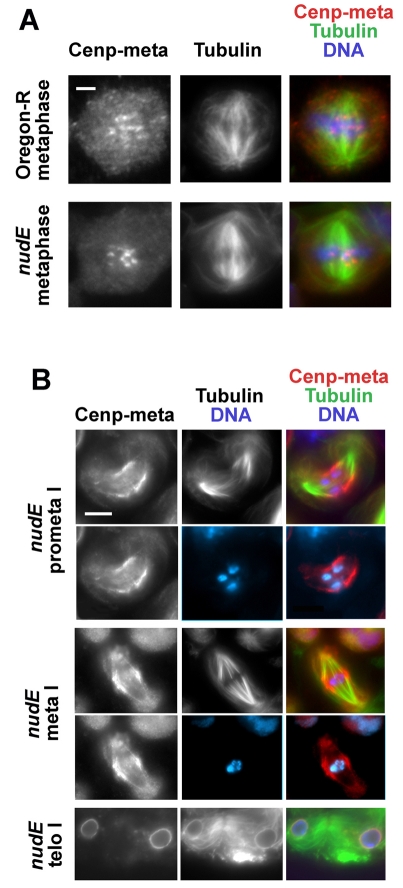
The detachment of centrosomes from the spindle is also a significant feature of meiosis in the testes of nudE39A males (Fig. 4). 88% of cells at metaphase I showed spindle-detachment defects (n=77), whereas this phenotype was not observed in any wild-type spermatocytes at the same stage (n=46). Time-lapse movies revealed the likely basis for the failure of centrosomes to associate with the spindle poles in nudE mutant spermatocytes (Fig. 5). Normally, just before the first meiotic division, the centrosomes located at one side of the spermatocyte cortex migrate towards the nuclear envelope; concomitantly they separate from each other and start to migrate to the opposite cell poles (Gunsalus et al., 1995; Rebollo et al., 2004) (also see A. D. Tates, Cytodifferentiation during spermatogenesis in Drosophila melanogaster: an electron microscope study, PhD thesis, Rijksuniversiteit de Leiden, 1971). (This migration does not occur in neuroblasts, because the centrosomes are never associated with the plasma membrane.) However, in nudE mutants, the centrosomes do not move from the cortex to the nuclear envelope. Failure of this centrosomal movement could easily produce the centrosomal detachment phenotype shown in Fig. 4, if a bipolar spindle eventually forms around the chromosomes at a location far from the cortex. Indeed, mutant metaphase I spermatocytes often exhibited spindle-like bipolar arrays of microtubules assembled around the chromosomes but not connected to the membrane-bound asters (Figs 4 and 5; supplementary material Fig. S10). This `spindle-in-the-spindle' phenotype, which has been previously observed in asp mutants (Rebollo et al., 2004), was also found in lis1, but not in cenp-meta (cmet), mutants (Fig. 4).
Fig. 4.
Prometaphase or metaphase I spindles in nudE, lis1 and cenp-meta mutant spermatocytes. Spermatocytes from nudE39A and lis1K13201 homozygous mutant larvae show centrosomes [labeled with centrosomin (Cnn)] (Li and Kaufman, 1996) detached from the meiotic spindle; by contrast, cenp-meta (cmetΔ) mutant spermatocytes form meiotic spindles with centrosomes at the poles that are indistinguishable from spindles in the wild-type (Oregon-R) controls. Scale bar: 10 μm.
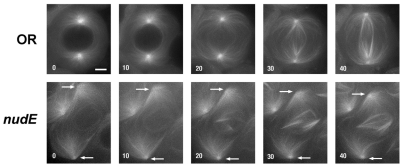
Fig. 5.
Meiotic spindle formation in wild-type and nudE mutant spermatocytes. Images are taken from time-lapse movies of control (OR) and nudE39A homozygous mutant larval spermatocytes expressing β-tubulin-GFP (time in minutes). In the wild-type panels, the centrosomes have already reached the nuclear envelope at the start of filming. By contrast, the centrosomes fail to migrate from the plasma membrane to the nuclear envelope in nudE spermatocytes. As a result, a spindle-like microtubule array forms around the chromosomes, while the asters (arrows) remain associated with the plasma membrane. Scale bar: 10 μm.
In accordance with previous findings that the SAC is relatively inactive in Drosophila spermatocytes (Basu et al., 1999; Rebollo and Gonzalez, 2000), meiotic progression was not adversely affected in nudE39A mutant testes; the proportions of cells at anaphase or telophase are normal and there is no failure in cytokinesis (data not shown). However, chromosome segregation is aberrant in many mutant cells at late anaphase and telophase, with DNA apportioned unequally to the two daughter cells, or with DNA remaining in the middle of the spindle (Table 3; supplementary material Fig. S2). Owing to these difficulties in chromosome segregation, `onion stage' spermatids displayed nuclei of different sizes. In addition, the sizes of the Nebenkern (mitochondrial derivatives) were uneven, suggesting that the cleavage furrows are not always symmetrically positioned during the male meiotic divisions (supplementary material Fig. S2).
Table 3.
DNA segregation defects in spermatocytes
| Genotype | Normal DNA segregation | Unequal DNA segregation | Unsegregated DNA at the spindle equator | DNA bridge |
|---|---|---|---|---|
| OreR (wild type) | ||||
| Meiosis I | 107 | 0 | 1 | 0 |
| Meiosis II | 74 | 0 | 0 | 0 |
| nudE39A/nudE39A | ||||
| Meiosis I | 9 | 7 | 6 | 4 |
| Meiosis II | 1 | 7 | 0 | 1 |
| cmetΔ/cmetΔ | ||||
| Meiosis I | 12 | 24 | 0 | 2 |
|
Meiosis II
|
26
|
1
|
0
|
0
|
Interactions of NudE with other kinetochore components
We used a tandem-affinity purification (TAP-tagging) approach to determine whether NudE associates with other proteins in stable, soluble complexes (Puig et al., 2001; Veraksa et al., 2005; Williams et al., 2007). Our results (supplementary material Fig. S3) clearly demonstrate a strong association between NudE and Lis1, a finding anticipated by previous results in other organisms (Efimov and Morris, 2000; Feng et al., 2000; Kitagawa et al., 2000; Sasaki et al., 2000; Sweeney et al., 2001). The complex of NudE and Lis1 formed both in normally growing tissue culture cells and in cells treated with colchicine to enrich for those in mitosis. Using mass spectrometry and western blotting, we asked whether NudE isolated by TAP-tagging copurified with any of several other candidates, including ZW10, p50 dynamitin and Glued (components of the dynactin complex), dynein heavy chain and Cenp-meta. Excepting a very weak signal seen on western blots using an antibody against p50 dynamitin, the results failed to demonstrate the association of NudE with any known kinetochore protein other than Lis1.
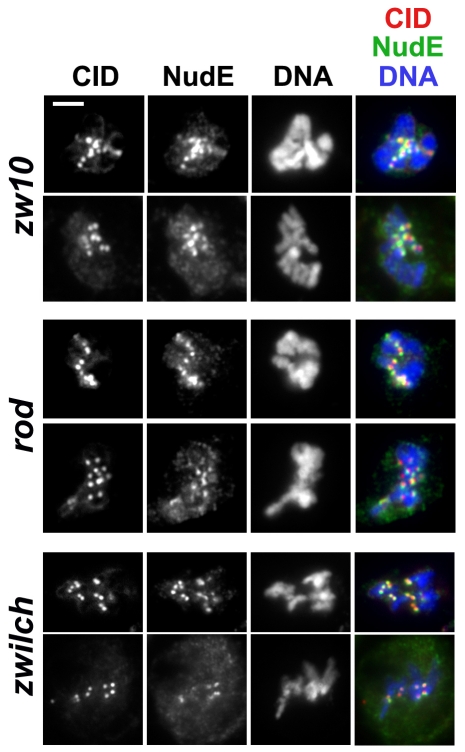
The availability of a NudE antibody allowed us to ask whether the targeting of NudE to kinetochores or to KMTs occurs in larval brain cells homozygous for loss-of-function alleles of genes encoding other outer kinetochore components. Because of the yeast two-hybrid interaction we had previously observed between NudE and ZW10 (Starr et al., 2000), we first examined NudE localization in cells lacking ZW10 or its associated RZZ subunits Rod and Zwilch. We found that NudE was targeted to the kinetochores in the larval brains of animals homozygous for mutations in zw10, rod and zwilch. However, extensive observations failed to show the `streaming' of NudE onto the KMTs at any time between prometaphase and anaphase (Fig. 6). NudE exhibited the same behavior (kinetochore targeting but lack of streaming) in mutants for dynein heavy chain (dhc64) and for NudC, a protein that forms complexes with Lis1 (Cunniff et al., 1997; Morris et al., 1998) (supplementary material Fig. S4).
Fig. 6.
NudE protein is targeted to kinetochores in RZZ mutants, but fails to stream onto KMTs. Prometaphase or metaphase cells from zw10S1, rodX5 and zwilch1229 mutant brains are shown. CID labels the centromeres. See supplementary material Fig. S1 for corresponding wild-type controls. Scale bar: 5 μm.
We obtained similar findings in mutants for the SAC component BubR1 (supplementary material Fig. S4): although NudE accumulated strongly at kinetochores during prometaphase and metaphase, we did not observe streaming in any metaphase cells (n>100). In fact, in contrast with the wild type, NudE often (63% of 30 cells) remained at kinetochores in bubR1 mutant cells during anaphase and telophase (data not shown). Because the shedding of any kinetochore component has not previously been reported to be defective in mutants for SAC components, we examined bubR1 mutant cells in more detail. Surprisingly, these cells had aberrant spindles that were very short and only thinly populated by microtubules. It is possible that these spindle defects might be responsible for the lack of NudE streaming. However, because we also found that Rod streams normally in bubR1 metaphases (data not shown), shedding per se is not blocked by the lack of BubR1.
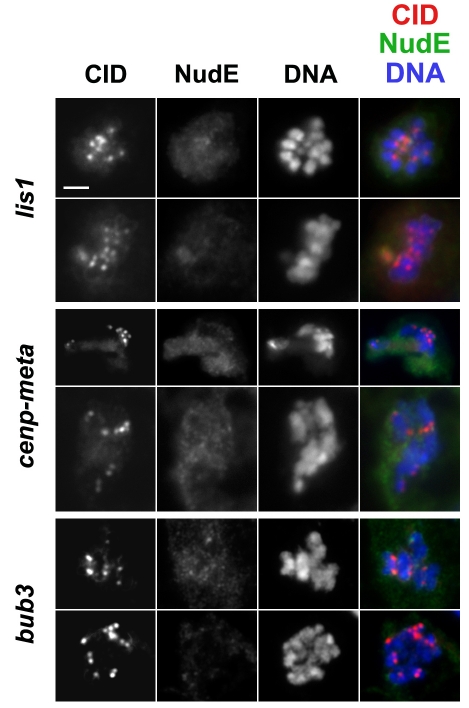
We found that mutations in three genes – bub3, lis1 and cenp-meta (cmet) – disrupted the targeting of NudE to the kinetochores and KMTs in larval neuroblasts (Fig. 7) (Lei and Warrior, 2000; Lopes et al., 2005; Yucel et al., 2000). In the lis1 and bub3 mutants, some cells in the brain exhibited weak NudE kinetochore staining, whereas other cells showed no staining. However, in both cases, the proportion of NudE-negative cells increased with larval age (to >90% in the oldest larvae for both mutants). These results indicate that the low NudE signals observed in younger third instar larvae are caused by the perdurance of maternally supplied gene products, and suggest that Lis1 and Bub3 are needed for the recruitment of NudE to kinetochores. The lack of NudE staining in cenp-meta mutants was clear-cut (even in colchicine-treated brains; data not shown) and unanticipated, because no interactions between these proteins have previously been reported.
Fig. 7.
NudE protein fails to localize to the kinetochores of chromosomes during prometaphase or metaphase in bub31, lis1K13201 and cmetΔ mutants. Very weak NudE staining in young bub3 mutant third instar larvae is visible in the top row of the bub3 figures. See supplementary material Fig. S1 for wild-type controls. Scale bar: 5 μm.
To verify that the kinetochore targeting of NudE depends upon Lis1 and Cenp-meta, we examined NudE localization in the testes of animals with mutations in these genes (supplementary material Fig. S5). NudE staining was not observed at the kinetochores in lis1 and cenp-meta mutant spermatocytes. Of particular interest, NudE was strongly redirected to the vicinity of the spindle poles in the absence of Cenp-meta (compare with the wild-type images in Fig. 2). In lis1 mutant spermatocytes, NudE accumulated anomalously in patches on the spindle envelope, particularly in the region closest to the chromosomes.
Targeting of other kinetochore proteins in nudE mutants
We examined the effects of homozygosity for the nudE39A null mutation on the localization of other outer kinetochore components.
Dynein heavy chain (Dhc), as well as the checkpoint proteins Bub3, Rod, ZW10 and Zwilch, all still associated with kinetochores in brain cells from nudE mutant third instar larvae (supplementary material Fig. S6; data not shown); the proper localization of SAC components to kinetochores is expected since the SAC is functional in nudE mutant neuroblasts (see above). However, we failed to observe the `streaming' of Rod (Fig. 8), or of ZW10 or Zwilch (data not shown) onto the KMTs, even though such movement is visible in wild-type controls, specifically during metaphase (Fig. 8) (Williams et al., 2003). Since Dhc could be visualized at the kinetochores only in the presence of colchicine (which causes spindle disassembly) (supplementary material Fig. S6), we could not assess whether Dhc streaming was also abrogated in the nudE mutant cells. In nudE39A larval brains, the kinesin-like protein Cenp-meta and the SAC component Bub3 were targeted to the kinetochores (Fig. 9A and data not shown). These proteins never moved poleward along KMTs in nudE mutant cells, but neither our own observations nor the published literature have revealed clear evidence for the streaming of Bub3 or of Cenp-meta or its mammalian homolog CENP-E during mitosis in wild-type cells (Basu et al., 1998; Yao et al., 2000; Yucel et al., 2000).
Fig. 8.
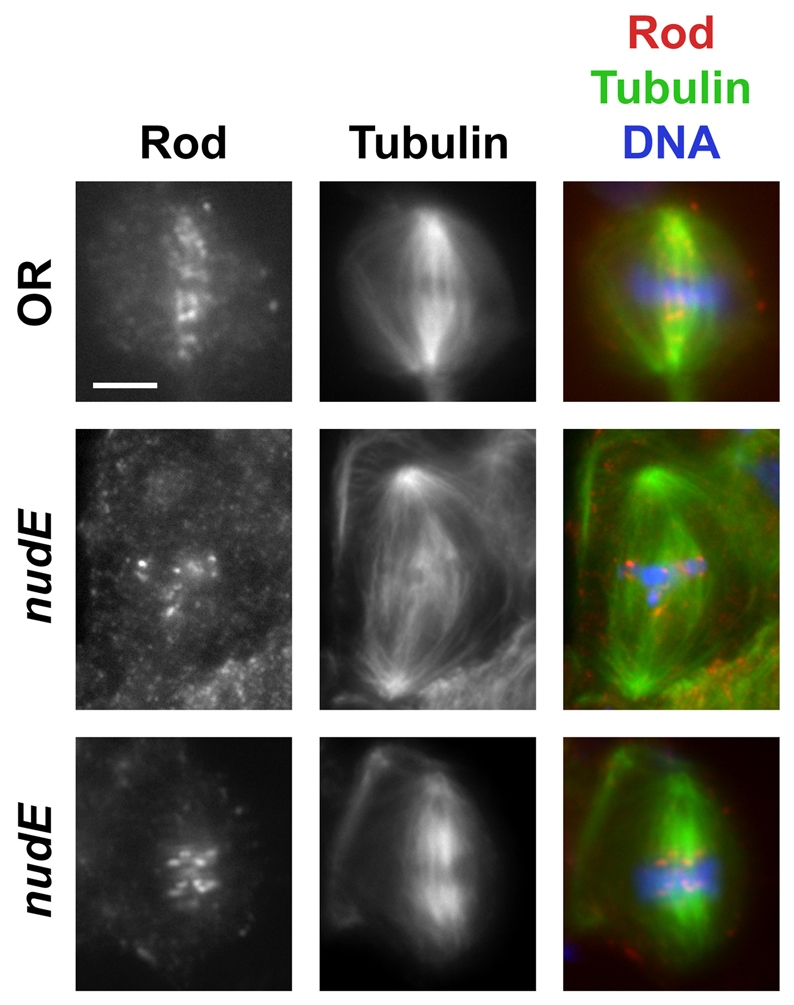
Rod is targeted to kinetochores, but fails to `stream' onto KMTs in nudE39A homozygous mutant neuroblasts. The top row shows the streaming of Rod from the kinetochores towards the poles in a wild-type (OR) metaphase cell. The two bottom rows illustrate the lack of streaming in nudE39A metaphases. Scale bar: 5 μm.
Fig. 9.
Cenp-meta behavior in nudE mutants. (A) Cenp-meta associates with kinetochores in larval neuroblasts in the absence of NudE. Streaming of Cenp-meta along the KMTs is not seen either in wild-type (Oregon-R) or in nudE mutant brain cells. Scale bar: 5 μm. (B) Cenp-meta is not targeted to kinetochores in nudE mutant spermatocytes during prometaphase I or metaphase I. Cenp-meta instead accumulates aberrantly in the middle region of the spindle envelope, similarly to NudE localization in lis1 mutant spermatocytes (compare with supplementary material Fig. S5). Merged panels without tubulin staining are provided to verify that Cenp-meta does not associate with kinetochores in nudE spermatocytes. Cenp-meta targeting to the nuclear membrane does not require NudE either during meiotic prophase (data not shown) or during telophase (bottom row). Scale bar: 10 μm.
Because the behavior of Cenp-meta in spermatocytes has not previously been described, we stained wild-type spermatocytes with antibody we generated against Cenp-meta (supplementary material Fig. S7). The localization of Cenp-meta in these meiotic cells was very similar to that shown above for NudE: Cenp-meta associated with the nuclear envelope, the kinetochores and the spindle envelope at equivalent times during meiosis. However, we did not observe streaming of Cenp-meta along the KMTs in spermatocytes during metaphase I; the signals for Cenp-meta on the kinetochores were very clear, so streaming should have been visible if it does occur. With these baseline observations, we could then investigate the behavior of Cenp-meta in nudE mutant larval testes. The behavior of Cenp-meta in spermatocytes was different from that seen in larval brains of the same animals, because it failed to localize to the kinetochores or to the KMTs (Fig. 9B). To confirm this unexpected finding, we examined spermatocytes incubated with colchicine, a treatment that causes many proteins to accumulate at abnormally high levels at centromeres. Even under these conditions, Cenp-meta was absent from kinetochores in ∼70% (n=24) of mutant cells, whereas the remaining spermatocytes displayed only very weak kinetochore signals (supplementary material Fig. S8). Remarkably, Cenp-meta is redirected in untreated nudE spermatocytes to the spindle envelope, in a pattern very similar to that noted for NudE protein in lis1 mutants (see supplementary material Fig. S5).
Asp localization in mitotic and meiotic spindles is independent of NudE function
Because the centrosomal aspects of the nudE mutant phenotype are reminiscent of aberrations reported in asp mutants (Avides and Glover, 1999; Wakefield et al., 2001; Rebollo et al., 2004), we asked whether NudE is required for proper Asp localization. We found that the Asp protein is properly localized in both mitotic and male meiotic cells of nudE mutants. Asp is enriched at the spindle poles during mitotic prophase in nudE39A neuroblasts (supplementary material Fig. S9). In mutant prometaphase or metaphase neuroblasts, Asp was still associated with microtubule minus-ends, but its distribution was more diffuse, because the spindle poles were unfocused. The spindle poles of the rare anaphases in nudE mutant brains showed normal Asp accumulation (supplementary material Fig. S9). Asp was also found at the spindle poles in nudE39A spermatocytes throughout meiosis, although the spindle poles were less compact than in the wild type (supplementary material Fig. S10). Collectively, these results indicate that the aberrant centrosome phenotype caused by nudE mutations is not due to any inability of Asp to localize to the spindle poles.
Discussion
During various stages of the cell cycle in Drosophila, NudE associates with kinetochores, KMTs and possibly other parts of the spindle apparatus, and (during meiosis) the nuclear envelope (Fig. 1B; Fig. 2; supplementary material Fig. S1). A null mutation in the nudE gene disrupts cell cycle progression and the behavior of the centrosomes, spindles and chromosomes during mitotic and meiotic divisions (Figs 3, 4 and 5; supplementary material Fig. S2).
NudE, centrosomes and the nuclear envelope
Neuroblasts and spermatocytes differ in the process of centrosome separation. During the spermatocyte growth phase, the centrosomes migrate to the periphery of the cell and associate with the plasma membrane. Shortly before the onset of the first meiotic division, the centrosomes together move towards the nucleus while nucleating astral microtubules; the centrosomes then rotate around the nucleus until they reach the opposite cell poles (Baker et al., 2004; Fuller, 1993; Rebollo et al., 2004) (also see A. D. Tates, Cytodifferentiation during spermatogenesis in Drosophila melanogaster: an electron microscope study, PhD thesis, Rijksuniversiteit de Leiden, 1971). By contrast, neuroblast centrosomes separate during interphase, and there is no evidence that these centrosomes associate with the plasma membrane. By the time the centrosomes start nucleating astral microtubules in early prophase, they have already achieved ∼70% of their final separation. Centrosome separation is then completed during late prophase or prometaphase with the formation of a bipolar spindle with a centrosome at each pole (Rebollo et al., 2004; Rusan and Peifer, 2007).
Despite these differences in the normal mechanisms of centrosome separation, centrosomes in nudE mutant neuroblasts and spermatocytes are both detached from the spindle poles (Figs 3 and 4; Table 2). Centrosome detachment from the spindle poles has been previously observed in neuroblasts from lis1, glued and dynein heavy chain (dhc64) mutants (Robinson et al., 1999; Siller et al., 2005; Wojcik et al., 2001), as well as in neuroblasts defective for Asp (Wakefield et al., 2001). In nudE mutant spermatocytes, centrosome detachment might be a secondary consequence of the tendency of centrosomes to remain associated with the plasma membrane and thus fail to migrate towards the nucleus (Figs 4 and 5). We also observe persistent association of the centrosomes with the spermatocyte plasma membrane in lis1 mutants (Fig. 4); this same phenomenon was also previously reported in asp mutant testes (Rebollo et al., 2004; Wakefield et al., 2001).
One scenario that could provide a common explanation for the problems of centrosome behavior in mutant neuroblasts and spermatocytes is that NudE and other dynein-associated proteins, as well as the microtubule-associated protein Asp, are all required to attach microtubule minus-ends to the centrosome. Roles for Drosophila Lis1 and dynactin in microtubule-centrosome interactions were proposed in a previous study (Siller et al., 2005). In addition, it has been suggested that Asp might have the same function as vertebrate NuMa (Wakefield et al., 2001), which is thought to anchor microtubule minus-ends at the centrosome (reviewed by Fant et al., 2004). Weakening of the connections between the centrosomes and the microtubule minus-ends could in theory lead to centrosome detachment from the spindle poles. Unfortunately, this attractive hypothesis is not supported by our NudE immunolocalization results, because we do not see an accumulation of NudE near centrosomes in either neuroblasts or spermatocytes. However, NudE streams along the kinetochore microtubules in both spermatocytes and neuroblasts, so the fact that NudE helps mediate interactions between the minus ends of spindle microtubules and centrosomes cannot be excluded.
The strong NudE localization we observed at the spermatocyte nuclear envelope (Fig. 2) provides another possible explanation for the nudE mutant phenotype. In spermatocytes lacking NudE, the astral microtubules could fail to interact properly with the nuclear envelope, disrupting forces that would normally pull centrosomes away from the plasma membrane towards the nucleus. A conceptually similar mechanism involving dynein-mediated microtubule interactions with the cell cortex (rather than the nuclear envelope) determines the orientation and asymmetrical localization of the mitotic spindle during the first embryonic division of Caenorhabditis elegans (Gonczy and Rose, 2005). A defective interaction between the astral microtubules and the nuclear envelope could also explain the abnormal centrosome localization in nudE mutant neuroblasts. However, since we did not observe NudE accumulation at the neuroblast nuclear envelope, the current data do not allow a precise definition of the role of NudE in centrosome positioning.
NudE, kinetochores, KMTs and the spindle checkpoint
The absence of NudE from neuroblasts results in failure of chromosomes to congress to a tightly organized metaphase plate, even though cell cycle progression is arrested at prometaphase or metaphase (Fig. 3; Table 1). Stehman and co-workers (Stehman et al., 2007) have recently reported similar findings in mammalian tissue culture cells injected with an antibody against NudE/Nudel that apparently blocks the interactions of these proteins with cytoplasmic dynein at kinetochores. Their detailed analysis showed that the majority of injected cells had one or more misoriented kinetochore pairs, and that misorientation was usually associated with the absence of microtubule attachments at these kinetochores. In accordance with such results, it seems logical to assume that the congression problems we observed result from the failure of NudE function specifically at kinetochores. It should be stressed, however, that the majority of chromosomes in both their study and ours do migrate to the metaphase plate, therefore NudE is not essential for establishing most kinetochore microtubule attachments.
Mitotic arrest in nudE mutant neuroblasts is due to persistent activation of the SAC (Table 1), therefore NudE cannot be required for SAC function. Because NudE is present at multiple locations and affects the mitotic spindle both in terms of microtubule organization and the ability of chromosomes to congress to the metaphase plate, it is difficult to pinpoint the precise reason the spindle checkpoint remains `on' in these cells. One possibility is that the checkpoint machinery detects the absence of one or more connections between kinetochores and KMTs in mutant cells, because, as mentioned above, anti-NudE/Nudel antibody appears to interfere with these connections in mammalian cells (Stehman et al., 2007). A second possibility is that generalized spindle defects or defective kinetochore connections might reduce the tension exerted across the chromosomes. In support of this idea, the average distance between sister kinetochores is shorter than normal in mammalian tissue culture cells injected with NudE/Nudel antibody (Stehman et al., 2007) and in Drosophila larval neuroblasts mutant for lis1 (Siller et al., 2005). Since only a small minority of mitotic cells in nudE mutant brains had well-defined metaphase plates, we did not make the same type of measurements, but our limited observations support the idea of reduced tension across chromosomes.
The persistence of the metaphase arrest in nudE mutant neuroblasts might also be due to a third factor: nudE mutations appear to block the `streaming' or `shedding' of outer kinetochore components along the KMTs. Current models suggest that streaming is dependent on cytoplasmic dynein activity and serves to move SAC proteins off the kinetochores, thus turning off the spindle checkpoint and allowing the initiation of anaphase (reviewed by Howell et al., 2001; Wojcik et al., 2001). In support of the idea that NudE streaming is part of the same mechanism, NudE fails to stream in neuroblasts mutant for the abnormal spindle (asp) gene (supplementary material Fig. S4). These cells display spindle defects that cause metaphase arrest, therefore SAC signaling remains on for extended periods (Basto et al., 2000). Neither our observations nor those made previously truly discriminate between these three hypotheses for the metaphase arrest seen when dynein-associated proteins are removed or inactivated. Since any of the three possibilities would be sufficient, a combination of these mechanisms might contribute to the metaphase arrest in nudE mutant brain cells.
NudE and its partners Lis1 and cytoplasmic dynein
Our TAP-tagging results indicate that NudE and Lis1 form a stable, soluble complex; furthermore, the nudE brain phenotypes described here affecting spindle structure, chromosome congression, the spindle checkpoint, and RZZ shedding are virtually identical to those previously reported for lis1 mutant Drosophila neuroblasts (Siller et al., 2005). We have observed that Lis1 is needed for the kinetochore localization of NudE; although we have not been able to detect reliably Lis1 at kinetochores using a variety of antibodies, it would not be surprising if Lis1 targeting to kinetochores reciprocally requires NudE. In our TAP-tagging experiments, we have seen no evidence for the association of NudE (or Lis1) with dynein, dynactin, NudC or ZW10, but others have reported co-immunoprecipitation or yeast two-hybrid results indicative of such interactions (Cunniff et al., 1997; Liu et al., 2000; Niethammer et al., 2000; Sasaki et al., 2000; Starr et al., 2000; Tai et al., 2002). Our inability to find other partners of NudE might be caused by the disruption of important NudE domains in the TAP-tagging construct we used, but we believe it is more likely that the interactions of NudE with these other proteins are simply weaker or less stable than its relationship with Lis1.
Our results do not agree with two recent reports in mammalian cells that NudE is required for the kinetochore localization of cytoplasmic dynein (Stehman et al., 2007; Vergnolle and Taylor, 2007). It is possible that dynein at the kinetochore is significantly reduced in Drosophila nudE mutant cells, but that we were unable to observe this because we could only visualize Dhc at the kinetochores in cells exposed to colchicine (a treatment that enhances the immunofluorescence signals of many kinetochore proteins, presumably by interfering with shedding) (supplementary material Fig. S6). Another possibility is that many intermolecular contacts are required for the kinetochore targeting of dynein and other kinetochore components, so the dependencies might vary in different cell types or in different organisms. For example, dynein does not associate with the kinetochore in flies carrying zw10 mutations (Starr et al., 1998); in a second example, the kinetochore recruitments of ZW10 and NudE are mutually independent (this paper) in spite of their apparent physical association as assayed in the yeast two-hybrid system (Starr et al., 2000). In any event, the lack of streaming observed in nudE mutant neuroblasts and spermatocytes coupled with the presence of at least some Dhc at the kinetochores in these cells implies that NudE might affect not only dynein targeting to the kinetochore, but also its function at these structures.
NudE and Cenp-meta
Our results provide clear evidence for a close but unanticipated relationship at the kinetochore between NudE and Cenp-meta, one of two Drosophila homologs of the mammalian kinesin-like protein CENP-E. NudE fails to target kinetochores in cenp-meta mutant brain cells and spermatocytes. In the reciprocal experiments, Cenp-meta associates with kinetochores in nudE mutant brain cells but not in nudE mutant spermatocytes (Figs 7 and 9; supplementary material Fig. S5), except very weakly in a small percentage of spermatocytes treated with colchicine (supplementary material Fig. S8). It is presently unclear whether these two proteins contact each other directly. Our TAP-tagging experiments (supplementary material Fig. S3) provide no evidence for a stable, soluble complex containing both NudE and Cenp-meta; however, interactions could be weak or occur only in the context of the kinetochore. One interesting possibility is that the interactions between NudE and Cenp-meta could be mediated by a Drosophila ortholog of the kinetochore protein CENP-F (also known as mitosin), since there are reports in the literature that the kinetochore targeting of NudE, Nudel and Lis1 are all dependent on CENP-F (Vergnolle and Taylor, 2007), whereas the amounts of CENP-E and cytoplasmic dynein at the kinetochores are significantly reduced in CENP-F-depleted cells (Yan et al., 2003). Unfortunately, BLAST searches do not reveal the identity of a clear-cut homolog of CENP-F in flies.
Of particular interest, the finding that NudE is not targeted to kinetochores in cenp-meta mutants raises questions about the origin of the phenotypes previously described in these mutants. The mitotic aberrations seen in larval neuroblasts that are associated with mutations in both nudE and cenp-meta are similar: Chromosomes exhibit problems in congression to the metaphase plate, and there is an increase in the mitotic index because of the accumulation of cells in prometaphase or metaphase (Maia et al., 2007; Williams et al., 2003; Yucel et al., 2000). It is quite possible that many of the problems in congression previously attributed to the lack of Cenp-meta at the kinetochore are actually caused by the absence of NudE (and thus the misregulation of cytoplasmic dynein). However, one perplexing attribute of cenp-meta mutants cannot be ascribed to the lack of NudE at the kinetochore. Although the elevated mitotic index and the delay of anaphase onset in cenp-meta mutant neuroblasts indicate activity of the SAC, depolymerization of the spindle in the mutant cells with colchicine or nocodazole leads to precocious sister chromatid separation and other events indicative of the failure of SAC signaling (Williams et al., 2003). The reason for this paradoxical behavior of the SAC in cenp-meta mutants is not currently understood, but since colchicine-treated nudE mutant cells remain arrested in prometaphase or metaphase with the sister chromatids of their chromosomes remaining attached, the failure of SAC signaling in similarly treated cenp-meta larval neuroblasts cannot be due to the absence of NudE from the kinetochores.
Materials and Methods
Genetic stocks and manipulations
Drosophila melanogaster stocks were raised on yeast-glucose-agar medium at 23±2°C in a 12 hour L:12 hour D photoperiod. Oregon R was the wild-type strain. The deletion Df(3L)AC1 (67A2-67D3) was obtained from the Drosophila stock center (Bloomington IN; stock no. 997). Strains containing the mutations zw10S1, rodX5, zwilch1229, dynein heavy chain (Dhc64C6-10), bub31, bubR11, lis1K13201, cmetΔ (a null allele of cenp-meta), and asp1 have been previously described (Basu et al., 1999; Lei and Warrior, 2000; Lopes et al., 2005; McGrail et al., 1995; Ripoll et al., 1985; Scaerou et al., 1999; Siller et al., 2005; Williams et al., 1992; Williams et al., 2003; Yucel et al., 2000). The nudC9jE8 allele was the kind gift of Rahul Warrior (University of California, Irvine, CA).
To generate a null allele of nudE, imprecise excisions were generated from the homozygous viable P element insertion P{EPgy2}EY09537 (∼600 bp upstream of the nudE gene) using standard techniques (e.g. Williams et al., 2007). We verified the extent of the deletion in the strain now known as nudE39E by direct DNA sequencing of a PCR product spanning the excision.
Immunostaining and microscopy of mitotic and meiotic figures
Third instar larval brains as well as testes from third instar larvae or early pupae were fixed using formaldehyde and stained as previously described (Bonaccorsi et al., 2000; Cenci et al., 1994; Williams et al., 1992; Williams et al., 2003). In some cases, slides were exposed to an additional incubation of goat serum before the addition of secondary antibody so as to lower background. Primary antibodies used include anti-α-tubulin (Sigma Aldrich, St Louis, MO) at a 1:1000 dilution, purified anti-NudE at 1:200 (this paper; see below), anti-DSpd-2 [a centrosomal marker (Giansanti et al., 2008)] at 1:2000, anti-Rod preparation BE40 at 1:300 [a gift from Roger Karess (Scaerou et al., 2001)]; anti-Cenp-meta at 1:100 (see below), and anti-centrosomin (CNN) at 1:1000 (Megraw et al., 1999). Secondary antibodies used include FITC-conjugated anti-rabbit IgG and IgM at a 1:20 dilution (Jackson ImmunoResearch, West Grove, PA) and Alexa Fluor 555-conjugated anti-rabbit IgG at 1:300 (Invitrogen, Carlsbad, CA).
For time-lapse imaging studies of spermatocytes, testes from third instar larvae or early pupae were prepared as described by Inoue and colleagues (Inoue et al., 2004), using flies harboring a β-tubulin-GFP construct also described in the same reference (a kind gift from Matthew Savoian, Cancer Research UK, Cambridge, UK). The imaging of living onion-stage spermatids (supplementary material Fig. S2) was according to published methods (Regan and Fuller, 1990).
To make anti-NudE, the entire coding region was PCR-amplified from the cDNA clone LD32494 (Berkeley Drosophila Genome Project) and inserted into the vector pMAL-C2 (New England Biolabs, Beverly, MA) in-frame with the gene for maltose binding protein (MBP). The resulting NudE-MBP fusion protein was purified as described (Williams et al., 2003), and then injected into rabbits (Cocalico Biologicals, Reamstown, PA). Crude serum was affinity purified against the MBP-NudE fusion protein coupled to CnBr-activated Sepharose beads. Anti-Cenp-meta was made with the same constructs and according to the same protocol described (Yucel et al., 2000); injection of the fusion protein and isolation of crude serum was performed by Cocalico Biologicals. Results obtained with this antibody on western blots (not shown) and in immunofluorescence studies (controls in Fig. 9A and data not shown) were identical to those previously reported for a different antibody made in the same way (Yucel et al., 2000; Williams et al., 2003).
Purification of NudE-containing complexes by TAP-tagging
The entire coding sequence of nudE was cloned into pMK33-NTAP (Veraksa et al., 2005), so as to fuse a TAP tag in frame to the N-terminus of NudE. This construct was transfected into Drosophila Kc tissue culture cells using Cellfectin (Invitrogen), and used to generate stable Hygromycin-resistant cell lines expressing TAP-NudE. The production of TAP-NudE was assayed on western blots using HRP-conjugated anti-Protein-A antibody (Rockland, Gilbertsville, PA). Protein complexes from one liter of TAP-NudE cells were isolated following established procedures (Puig et al., 2001), using a lysis buffer for making Drosophila extracts (Veraksa et al., 2005). After purification using IgG-Sepharose and calmodulin-Sepharose beads (GE Healthcare, Piscataway, NJ), proteins in the final eluate were precipitated with trichloroacetic acid, solubilized in Laemmli sample buffer (Bio-Rad, Hercules, CA) and subjected to SDS-PAGE. Bands were excised, digested with trypsin, and analyzed by matrix-assisted laser desorption/ionization mass spectrometry (MALDI; Cornell Bioresource Center, Ithaca, NY). Samples from the NudE complex purification were also analyzed by western blotting for the presence of Lis1, p50 dynamitin, Cenp-meta (Williams et al., 2003), Glued and ZW10 (Williams et al., 1992).
Western blotting of fractions from TAP-tagging experiments were performed using standard procedures (Williams et al., 2007) and the following diluted antibodies: purified rabbit anti-NudE at 1:1000 (this paper); rabbit anti-ZW10 1:3000 (Williams et al., 1992); rabbit anti-Cenp-meta 1:6000 (Williams et al., 2003); rabbit anti-Glued 1:2000 [(Waterman-Storer and Holzbaur, 1996); a kind gift from Erica Holzbaur (University of Pennsylvania, Philadelphia, PA)]; rat anti-Lis1 1:2000 (from R. Warrior); and rat anti-p50 dynamitin (R. Warrior).
Supplementary Material
Supplementary material available online at http://jcs.biologists.org/cgi/content/full/122/11/1747/DC1
We thank Rahul Warrior and Erica Holzbaur for generously providing antibodies and mutant strains. This work was supported by NIH grant GM48430 to M.L.G. and grants from Centro di Eccellenza di Biologia e Medicina Molecolare (BEMM) to M.G.; A.W. was supported by a European Community Training and Mobility of Researchers grant (HPRN-CT-2002-00260) to M.G. Deposited in PMC for release after 12 months.
References
- Avides, M. C. and Glover, D. M. (1999). Abnormal spindle protein, Asp, and the integrity of mitotic centrosomal microtubule organizing centers. Science 283, 1733-1735. [DOI] [PubMed] [Google Scholar]
- Baker, J. D., Adhikarakunnathu, S. and Kernan, M. J. (2004). Mechanosensory-defective, male-sterile unc mutants identify a novel basal body protein required for ciliogenesis in Drosophila. Development 131, 3411-3422. [DOI] [PubMed] [Google Scholar]
- Basto, R., Gomes, R. and Karess, R. E. (2000). Rough deal and Zw10 are required for the metaphase checkpoint in Drosophila. Nat. Cell Biol. 2, 939-943. [DOI] [PubMed] [Google Scholar]
- Basu, J., Logarinho, E., Herrmann, S., Bousbaa, H., Li, Z., Chan, G. K., Yen, T. J., Sunkel, C. E. and Goldberg, M. L. (1998). Localization of the Drosophila checkpoint control protein Bub3 to the kinetochore requires Bub1 but not Zw10 or Rod. Chromosoma 107, 376-385. [DOI] [PubMed] [Google Scholar]
- Basu, J., Bousbaa, H., Logarinho, E., Li, Z., Williams, B. C., Lopes, C., Sunkel, C. E. and Goldberg, M. L. (1999). Mutations in the essential spindle checkpoint gene bub1 cause chromosome missegregation and fail to block apoptosis in Drosophila. J. Cell Biol. 146, 13-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blower, M. D. and Karpen, G. H. (2001). The role of Drosophila CID in kinetochore formation, cell-cycle progression and heterochromatin interactions. Nat. Cell Biol. 3, 730-739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaccorsi, S., Giansanti, M. G. and Gatti, M. (2000). Spindle assembly in Drosophila neuroblasts and ganglion mother cells. Nat. Cell Biol. 2, 54-56. [DOI] [PubMed] [Google Scholar]
- Cenci, G., Bonaccorsi, S., Pisano, C., Verni, F. and Gatti, M. (1994). Chromatin and microtubule organization during premeiotic, meiotic and early postmeiotic stages of Drosophila melanogaster spermatogenesis. J. Cell Sci. 107, 3521-3534. [DOI] [PubMed] [Google Scholar]
- Cunniff, J., Chiu, Y. H., Morris, N. R. and Warrior, R. (1997). Characterization of DnudC, the Drosophila homolog of an Aspergillus gene that functions in nuclear motility. Mech. Dev. 66, 55-68. [DOI] [PubMed] [Google Scholar]
- Efimov, V. P. and Morris, N. R. (2000). The LIS1-related NUDF protein of Aspergillus nidulans interacts with the coiled-coil domain of the NUDE/RO11 protein. J. Cell Biol. 150, 681-688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fant, X., Merdes, A. and Haren, L. (2004). Cell and molecular biology of spindle poles and NuMA. Int. Rev. Cytol. 238, 1-57. [DOI] [PubMed] [Google Scholar]
- Feng, Y., Olson, E. C., Stukenberg, P. T., Flanagan, L. A., Kirschner, M. W. and Walsh, C. A. (2000). LIS1 regulates CNS lamination by interacting with mNudE, a central component of the centrosome. Neuron 28, 665-679. [DOI] [PubMed] [Google Scholar]
- Fuller, M. T. (1993). Spermatogenesis. In The Development of Drosophila Melanogaster (ed. M. Bate and A. Martinez-Arias), pp. 71-147. Cold Spring Harbor, NY: Cold Spring Harbor Press.
- Gatti, M. and Goldberg, M. L. (1991). Mutations affecting cell division in Drosophila. Methods Cell Biol. 35, 543-586. [DOI] [PubMed] [Google Scholar]
- Giansanti, M. G., Belloni, G. and Gatti, M. (2007). Rab11 is required for membrane trafficking and actomyosin ring constriction in meiotic cytokinesis of Drosophila males. Mol. Biol. Cell 18, 5034-5047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giansanti, M. G., Bucciarelli, E., Bonaccorsi, S. and Gatti, M. (2008). Drosophila SPD-2 is an essential centriole component required for PCM recruitment and astral-microtubule nucleation. Curr. Biol. 18, 303-309. [DOI] [PubMed] [Google Scholar]
- Gonczy, P. and Rose, L. S. (2005). Asymmetric cell division and axis formation in the embryo. WormBook, 1-20. [DOI] [PMC free article] [PubMed]
- Gunsalus, K. C., Bonaccorsi, S., Williams, E., Verni, F., Gatti, M. and Goldberg, M. L. (1995). Mutations in twinstar, a Drosophila gene encoding a cofilin/ADF homologue, result in defects in centrosome migration and cytokinesis. J. Cell Biol. 131, 1243-1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell, B. J., Hoffman, D. B., Fang, G., Murray, A. W. and Salmon, E. D. (2000). Visualization of Mad2 dynamics at kinetochores, along spindle fibers, and at spindle poles in living cells. J. Cell Biol. 150, 1233-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell, B. J., McEwen, B. F., Canman, J. C., Hoffman, D. B., Farrar, E. M., Rieder, C. L. and Salmon, E. D. (2001). Cytoplasmic dynein/dynactin drives kinetochore protein transport to the spindle poles and has a role in mitotic spindle checkpoint inactivation. J. Cell Biol. 155, 1159-1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue, Y. H., Savoian, M. S., Suzuki, T., Mathe, E., Yamamoto, M. T. and Glover, D. M. (2004). Mutations in orbit/mast reveal that the central spindle is comprised of two microtubule populations, those that initiate cleavage and those that propagate furrow ingression. J. Cell Biol. 166, 49-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karess, R. (2005). Rod-Zw10-Zwilch: a key player in the spindle checkpoint. Trends Cell Biol. 15, 386-392. [DOI] [PubMed] [Google Scholar]
- Kitagawa, M., Umezu, M., Aoki, J., Koizumi, H., Arai, H. and Inoue, K. (2000). Direct association of LIS1, the lissencephaly gene product, with a mammalian homologue of a fungal nuclear distribution protein, rNUDE. FEBS Lett. 479, 57-62. [DOI] [PubMed] [Google Scholar]
- Lei, Y. and Warrior, R. (2000). The Drosophila Lissencephaly1 (DLis1) gene is required for nuclear migration. Dev. Biol. 226, 57-72. [DOI] [PubMed] [Google Scholar]
- Li, J., Lee, W. L. and Cooper, J. A. (2005). NudEL targets dynein to microtubule ends through LIS1. Nat. Cell Biol. 7, 686-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, K. and Kaufman, T. C. (1996). The homeotic target gene centrosomin encodes an essential centrosomal component. Cell 85, 585-596. [DOI] [PubMed] [Google Scholar]
- Liu, Z., Steward, R. and Luo, L. (2000). Drosophila Lis1 is required for neuroblast proliferation, dendritic elaboration and axonal transport. Nat. Cell Biol. 2, 776-783. [DOI] [PubMed] [Google Scholar]
- Lopes, C. S., Sampaio, P., Williams, B., Goldberg, M. and Sunkel, C. E. (2005). The Drosophila Bub3 protein is required for the mitotic checkpoint and for normal accumulation of cyclins during G2 and early stages of mitosis. J. Cell Sci. 118, 187-198. [DOI] [PubMed] [Google Scholar]
- Maia, A. F., Lopes, C. S. and Sunkel, C. E. (2007). BubR1 and CENP-E have antagonistic effects upon the stability of microtubule-kinetochore attachments in Drosophila S2 cell mitosis. Cell Cycle 6, 1367-1378. [DOI] [PubMed] [Google Scholar]
- Malmanche, N., Maia, A. and Sunkel, C. E. (2006). The spindle assembly checkpoint: preventing chromosome mis-segregation during mitosis and meiosis. FEBS Lett. 580, 2888-2895. [DOI] [PubMed] [Google Scholar]
- McGrail, M., Gepner, J., Silvanovich, A., Ludmann, S., Serr, M. and Hays, T. S. (1995). Regulation of cytoplasmic dynein function in vivo by the Drosophila Glued complex. J. Cell Biol. 131, 411-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megraw, T. L., Li, K., Kao, L. R. and Kaufman, T. C. (1999). The centrosomin protein is required for centrosome assembly and function during cleavage in Drosophila. Development 126, 2829-2839. [DOI] [PubMed] [Google Scholar]
- Meraldi, P., McAinsh, A. D., Rheinbay, E. and Sorger, P. K. (2006). Phylogenetic and structural analysis of centromeric DNA and kinetochore proteins. Genome Biol. 7, R23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris, S. M., Albrecht, U., Reiner, O., Eichele, G. and Yu-Lee, L. Y. (1998). The lissencephaly gene product Lis1, a protein involved in neuronal migration, interacts with a nuclear movement protein, NudC. Curr. Biol. 8, 603-606. [DOI] [PubMed] [Google Scholar]
- Musacchio, A. and Salmon, E. D. (2007). The spindle-assembly checkpoint in space and time. Nat. Rev. Mol. Cell. Biol. 8, 379-393. [DOI] [PubMed] [Google Scholar]
- Niethammer, M., Smith, D. S., Ayala, R., Peng, J., Ko, J., Lee, M. S., Morabito, M. and Tsai, L. H. (2000). NUDEL is a novel Cdk5 substrate that associates with LIS1 and cytoplasmic dynein. Neuron 28, 697-711. [DOI] [PubMed] [Google Scholar]
- Przewloka, M. R., Zhang, W., Costa, P., Archambault, V., D'Avino, P. P., Lilley, K. S., Laue, E. D., McAinsh, A. D. and Glover, D. M. (2007). Molecular analysis of core kinetochore composition and assembly in Drosophila melanogaster. PLoS ONE 2, e478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig, O., Caspary, F., Rigaut, G., Rutz, B., Bouveret, E., Bragado-Nilsson, E., Wilm, M. and Seraphin, B. (2001). The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods 24, 218-229. [DOI] [PubMed] [Google Scholar]
- Rebollo, E. and Gonzalez, C. (2000). Visualizing the spindle checkpoint in Drosophila spermatocytes. EMBO Rep. 1, 65-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebollo, E., Llamazares, S., Reina, J. and Gonzalez, C. (2004). Contribution of noncentrosomal microtubules to spindle assembly in Drosophila spermatocytes. PLoS Biol. 2, E8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regan, C. L. and Fuller, M. T. (1990). Interacting genes that affect microtubule function in Drosophila melanogaster: two classes of mutation revert the failure to complement between haync2 and mutations in tubulin genes. Genetics 125, 77-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripoll, P., Pimpinelli, S., Valdivia, M. M. and Avila, J. (1985). A cell division mutant of Drosophila with a functionally abnormal spindle. Cell 41, 907-912. [DOI] [PubMed] [Google Scholar]
- Robinson, J. T., Wojcik, E. J., Sanders, M. A., McGrail, M. and Hays, T. S. (1999). Cytoplasmic dynein is required for the nuclear attachment and migration of centrosomes during mitosis in Drosophila. J. Cell Biol. 146, 597-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusan, N. M. and Peifer, M. (2007). A role for a novel centrosome cycle in asymmetric cell division. J. Cell Biol. 177, 13-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki, S., Shionoya, A., Ishida, M., Gambello, M. J., Yingling, J., Wynshaw-Boris, A. and Hirotsune, S. (2000). A LIS1/NUDEL/cytoplasmic dynein heavy chain complex in the developing and adult nervous system. Neuron 28, 681-696. [DOI] [PubMed] [Google Scholar]
- Savoian, M. S., Goldberg, M. L. and Rieder, C. L. (2000). The rate of poleward chromosome motion is attenuated in Drosophila zw10 and rod mutants. Nat. Cell Biol. 2, 948-952. [DOI] [PubMed] [Google Scholar]
- Scaerou, F., Aguilera, I., Saunders, R., Kane, N., Blottiere, L. and Karess, R. (1999). The rough deal protein is a new kinetochore component required for accurate chromosome segregation in Drosophila. J. Cell Sci. 112, 3757-3768. [DOI] [PubMed] [Google Scholar]
- Scaerou, F., Starr, D. A., Piano, F., Papoulas, O., Karess, R. E. and Goldberg, M. L. (2001). The ZW10 and Rough Deal checkpoint proteins function together in a large, evolutionarily conserved complex targeted to the kinetochore. J. Cell Sci. 114, 3103-3114. [DOI] [PubMed] [Google Scholar]
- Schittenhelm, R. B., Heeger, S., Althoff, F., Walter, A., Heidmann, S., Mechtler, K. and Lehner, C. F. (2007). Spatial organization of a ubiquitous eukaryotic kinetochore protein network in Drosophila chromosomes. Chromosoma 116, 385-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp, D. J., Rogers, G. C. and Scholey, J. M. (2000). Cytoplasmic dynein is required for poleward chromosome movement during mitosis in Drosophila embryos. Nat. Cell Biol. 2, 922-930. [DOI] [PubMed] [Google Scholar]
- Shu, T., Ayala, R., Nguyen, M. D., Xie, Z., Gleeson, J. G. and Tsai, L. H. (2004). Ndel1 operates in a common pathway with LIS1 and cytoplasmic dynein to regulate cortical neuronal positioning. Neuron 44, 263-277. [DOI] [PubMed] [Google Scholar]
- Siller, K. H., Serr, M., Steward, R., Hays, T. S. and Doe, C. Q. (2005). Live imaging of Drosophila brain neuroblasts reveals a role for Lis1/dynactin in spindle assembly and mitotic checkpoint control. Mol. Biol. Cell 16, 5127-5140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr, D. A., Williams, B. C., Hays, T. S. and Goldberg, M. L. (1998). ZW10 helps recruit dynactin and dynein to the kinetochore. J. Cell Biol. 142, 763-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr, D. A., Saffery, R., Li, Z., Simpson, A. E., Choo, K. H., Yen, T. J. and Goldberg, M. L. (2000). HZwint-1, a novel human kinetochore component that interacts with HZW10. J. Cell Sci. 113, 1939-1950. [DOI] [PubMed] [Google Scholar]
- Stehman, S. A., Chen, Y., McKenney, R. J. and Vallee, R. B. (2007). NudE and NudEL are required for mitotic progression and are involved in dynein recruitment to kinetochores. J. Cell Biol. 178, 583-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney, K. J., Prokscha, A. and Eichele, G. (2001). NudE-L, a novel Lis1-interacting protein, belongs to a family of vertebrate coiled-coil proteins. Mech. Dev. 101, 21-33. [DOI] [PubMed] [Google Scholar]
- Tai, C. Y., Dujardin, D. L., Faulkner, N. E. and Vallee, R. B. (2002). Role of dynein, dynactin, and CLIP-170 interactions in LIS1 kinetochore function. J. Cell Biol. 156, 959-968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veraksa, A., Bauer, A. and Artavanis-Tsakonas, S. (2005). Analyzing protein complexes in Drosophila with tandem affinity purification-mass spectrometry. Dev. Dyn. 232, 827-834. [DOI] [PubMed] [Google Scholar]
- Vergnolle, M. A. and Taylor, S. S. (2007). Cenp-F links kinetochores to Ndel1/Nde1/Lis1/dynein microtubule motor complexes. Curr. Biol. 17, 1173-1179. [DOI] [PubMed] [Google Scholar]
- Wakefield, J. G., Bonaccorsi, S. and Gatti, M. (2001). The Drosophila protein asp is involved in microtubule organization during spindle formation and cytokinesis. J. Cell Biol. 153, 637-648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H., Hu, X., Ding, X., Dou, Z., Yang, Z., Shaw, A. W., Teng, M., Cleveland, D. W., Goldberg, M. L., Niu, L. et al. (2004). Human Zwint-1 specifies localization of Zeste White 10 to kinetochores and is essential for mitotic checkpoint signaling. J. Biol. Chem. 279, 54590-54598. [DOI] [PubMed] [Google Scholar]
- Waterman-Storer, C. M. and Holzbaur, E. L. (1996). The product of the Drosophila gene, Glued, is the functional homologue of the p150Glued component of the vertebrate dynactin complex. J. Biol. Chem. 271, 1153-1159. [DOI] [PubMed] [Google Scholar]
- Williams, B. C., Karr, T. L., Montgomery, J. M. and Goldberg, M. L. (1992). The Drosophila l(1)zw10 gene product, required for accurate mitotic chromosome segregation, is redistributed at anaphase onset. J. Cell Biol. 118, 759-773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, B. C., Li, Z., Liu, S., Williams, E. V., Leung, G., Yen, T. J. and Goldberg, M. L. (2003). Zwilch, a new component of the ZW10/ROD complex required for kinetochore functions. Mol. Biol. Cell 14, 1379-1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, B., Leung, G., Maiato, H., Wong, A., Li, Z., Williams, E. V., Kirkpatrick, C., Aquadro, C. F., Rieder, C. L. and Goldberg, M. L. (2007). Mitch a rapidly evolving component of the Ndc80 kinetochore complex required for correct chromosome segregation in Drosophila. J. Cell Sci. 120, 3522-3533. [DOI] [PubMed] [Google Scholar]
- Wojcik, E., Basto, R., Serr, M., Scaerou, F., Karess, R. and Hays, T. (2001). Kinetochore dynein: its dynamics and role in the transport of the Rough deal checkpoint protein. Nat. Cell Biol. 3, 1001-1007. [DOI] [PubMed] [Google Scholar]
- Wolf, K. (1995). Spindle membranes and spindle architecture in invertebrates. Micron 26, 69-98. [Google Scholar]
- Yan, X., Li, F., Liang, Y., Shen, Y., Zhao, X., Huang, Q. and Zhu, X. (2003). Human Nudel and NudE as regulators of cytoplasmic dynein in poleward protein transport along the mitotic spindle. Mol. Cell. Biol. 23, 1239-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, X., Abrieu, A., Zheng, Y., Sullivan, K. F. and Cleveland, D. W. (2000). CENP-E forms a link between attachment of spindle microtubules to kinetochores and the mitotic checkpoint. Nat. Cell Biol. 2, 484-491. [DOI] [PubMed] [Google Scholar]
- Yucel, J. K., Marszalek, J. D., McIntosh, J. R., Goldstein, L. S., Cleveland, D. W. and Philp, A. V. (2000). CENP-meta, an essential kinetochore kinesin required for the maintenance of metaphase chromosome alignment in Drosophila. J. Cell Biol. 150, 1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.