Summary
Lipid droplets (LDs) are cytoplasmic organelles that store neutral lipids for use as an energy supply in times of nutrient deprivation and for membrane assembly. Misregulation of LD function leads to many human diseases, including lipodystrophy, obesity and neutral lipid storage disorders. A number of proteins have been shown to localize to the surface of lipid droplets, including lipases such as adipose triglyceride lipase (ATGL) and the PAT-domain proteins ADRP (adipophilin) and TIP47, but the mechanism by which they are targeted to LDs is not known. Here we demonstrate that ATGL and ADRP, but not TIP47, are delivered to LDs by a pathway mediated by the COPI and COPII coatomer proteins and their corresponding regulators.
Keywords: ATGL, Lipid droplet, Lipid homeostasis
Introduction
Lipid droplets (LDs) are cytoplasmic organelles that serve as storage compartments for neutral lipids such as triglycerides (TGs) and cholesterol esters (CEs) in animal cells (Ducharme and Bickel, 2008; Martin and Parton, 2006; Murphy, 2001). As such, LDs participate in many important physiological processes, including the supply of lipids for energy production and for membrane assembly (Brasaemle, 2007; Ducharme and Bickel, 2008; Londos et al., 1999; Martin and Parton, 2006). LD abnormalities are associated with a range of pathologies, including lipodystrophy, obesity and neutral lipid storage disorders (Arner and Langin, 2007; Brasaemle, 2007; Ducharme and Bickel, 2008; Fischer et al., 2007; Magre et al., 2001). Despite growing appreciation of the crucial roles of LDs in health and disease, their biogenesis remains poorly understood.
LDs consist of a neutral lipid core bounded by a phospholipid monolayer and a set of associated proteins [herein referred to as LD-associated proteins (LDAPs)], including lipases [e.g. hormone-sensitive lipase (HSL; LIPE) and adipose triglyceride lipase (ATGL; PNPLA2] (Arner and Langin, 2007; Bartz et al., 2007; Brasaemle et al., 2004; Ducharme and Bickel, 2008; Liu et al., 2004; Smirnova et al., 2006; Zechner et al., 2009; Zimmermann et al., 2004) and members of the PAT-domain family [e.g. perilipin, ADRP (adipophilin; ADFP), TIP47 (M6PRBP1)] (Bartz et al., 2007; Brasaemle et al., 2004; Ducharme and Bickel, 2008; Liu et al., 2004; Targett-Adams et al., 2003). The enzymes responsible for the last steps in TG and CE synthesis reside in the endoplasmic reticulum (ER), which has led to the hypothesis that LDs arise from the ER. The prevailing view is that the neutral lipid core forms by coalescence of newly synthesized TGs and CEs between the two leaflets of the ER membrane. Eventually, nascent LDs bud from the ER to become free-standing organelles. LDAPs are thought to be synthesized by free polyribosomes (Londos et al., 1999) and then post-translationally attached either to the ER membrane prior to the budding of nascent LDs or directly to free-standing LDs (Brasaemle, 2007; Londos et al., 1999; Targett-Adams et al., 2003). The exact mechanism involved in LDAP targeting to LDs, however, is largely unknown.
In this study, we have addressed this mechanism for a prototypical LDAP, ATGL. Strikingly, we find that ATGL delivery to lipid droplets is strictly dependent on components of the ER-Golgi transport machinery, such as the small GTP-binding protein ARF1, its guanine-nucleotide exchange factor (GEF) GBF1, and its effector coatomer protein I (COPI). Other components of this machinery, such as the small GTP-binding protein SAR1 and its effector coatomer protein II (COPII), are also involved in this process. The function of these proteins in ATGL delivery to LDs occurs in the context of a close apposition of ER exit sites (ERES) and the ER-Golgi intermediate compartment (ERGIC) with LDs. On the basis of these observations, we propose that COPI, COPII and their regulators mediate segregation of ATGL into nascent lipid droplets.
Results
Tight association of ATGL with lipid droplets and membranes
To examine the mode of association of ATGL with preformed LDs, we performed fluorescence recovery after photobleaching (FRAP) analysis of cells expressing ATGL tagged with green fluorescent protein (GFP), after treatment for 19 hours with oleic acid (OA) to induce new LD formation. We observed that ATGL-GFP on LDs recovered very slowly (t1/2 ∼20 minutes) and incompletely (only ∼50% of the original fluorescence after 20 minutes) (Fig. 1A,B). This was in contrast to the fast (t1/2 ∼6 seconds) and complete (∼100% of the original fluorescence after 18 seconds) recovery of an organellar coat protein (the Golgi-associated clathrin adaptor GGA1 tagged with GFP) (Fig. 1A,B). These observations indicated that, unlike typical organellar coat proteins, ATGL is not rapidly recruited from the cytosol. In addition, subcellular fractionation on sucrose density gradients showed that most ATGL occurred in association with either floating LDs or sedimentable membrane fractions; levels in the cytosol were negligible (Fig. 1C). The membrane pool might represent a precursor compartment from which ATGL is delivered to LDs. We also found that ATGL could not be solubilized from the membrane fraction even under the harshest extraction conditions (Fig. 1C). Thus, despite not having a transmembrane domain, ATGL behaves similar to integral membrane proteins (e.g. TRAP-α; SSR1) (Fig. 1C), and is likely to require association with lipids from the earliest stages of its biosynthetic delivery to LDs.
Fig. 1.
ATGL association with LDs and membranes. (A,B) FRAP analysis of GFP-GGA1 and ATGL-GFP in HeLa cells incubated overnight with 200 μM oleic acid (OA). Boxed regions were bleached at time 0; fluorescence within these regions was monitored at 2-second (GFP-GGA1) or 30-second (ATGL-GFP) intervals. Arrows point to bleached regions. Scale bar: 5 μm. (B) Fluorescence values were plotted as a function of time. Mean and s.d. of at least three independent experiments are shown. (C) HeLa cells were treated with OA as in A, and cell lysates were fractionated into lipid droplets (LD), cytosol (C) and membranes (M) by sucrose gradient centrifugation. Membrane fractions were treated with the indicated agents or left untreated (None). Pellet (P) and supernatant (S) fractions were analyzed by SDS-PAGE and immunoblotting with antibodies to ATGL, TIP47, TRAP-α and calnexin.
Brefeldin A blocks biosynthetic delivery of ATGL to lipid droplets
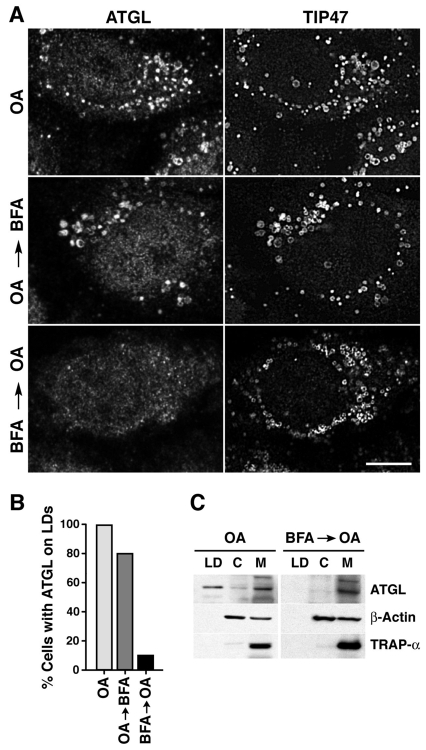
Since the lipid components of LDs arise from the ER, we examined whether the ER-Golgi transport machinery might be required for delivery of ATGL to LDs. To this end, we initially examined the effect of brefeldin A (BFA), a drug that interferes with ER-Golgi transport (Klausner et al., 1992; Lippincott-Schwartz et al., 1989). BFA acts at this stage by stabilizing an abortive complex of the GDP-bound form of the small GTPase, ARF1, with its GEF, GBF1 (Niu et al., 2005). As a consequence, ARF1 effectors such as coatomer protein I (COPI) dissociate from membranes, eventually leading to resorption of the Golgi complex into the ER and blockade of cargo export from the ER. Immunofluorescence microscopy showed that when cells were first treated with OA for 3 hours to induce ATGL synthesis (supplementary material Fig. S1) and LD formation, and then with BFA for an additional 3 hours, ATGL remained associated with LDs (Fig. 2A,B). However, when BFA was added 10 minutes prior to the 3-hour incubation with OA, ATGL was not detected on LDs (Fig. 2A,B). This indicated that BFA does not affect the pre-existing association of ATGL with LDs but prevents its delivery to newly formed LDs. By contrast, the association of TIP47 with LDs was not affected by BFA, regardless of the protocol used (Fig. 2A). This suggested the existence of at least two pathways, one BFA-sensitive (for ATGL) and the other BFA-resistant (for TIP47), for LDAP delivery to LDs. Subcellular fractionation confirmed that treatment with BFA for 19 hours blocked delivery of ATGL to LDs and additionally revealed that residual ATGL accumulated in a membrane fraction and not in the cytosol (Fig. 2C). This supports the notion that ATGL is delivered to LDs from a membrane compartment.
Fig. 2.
BFA blocks delivery of ATGL to LDs. (A) Cells were treated with 400 μM OA for 3 hours (top row), 400 μM OA for 3 hours followed by 5 μg/ml BFA for 3 hours (middle row), or 5 μg/ml BFA for 10 minutes prior to addition of 400 μM OA and 5 μg/ml BFA for 3 hours (bottom row). Cells were co-immunostained with antibodies to ATGL (left column) and TIP47 (right-hand column). Scale bar: 10 μm. (B) Quantification of results shown in A. At least 100 cells were scored for the presence or absence of ATGL on LDs, and the result plotted as a percentage of total cells examined. (C) Cells were treated with OA in the presence or absence of 1 μg/ml BFA for 19 hours, and lysates were subjected to sucrose gradient fractionation as described in Fig. 1C.
ATGL is delivered to lipid droplets through a GBF1-ARF1-COPI pathway
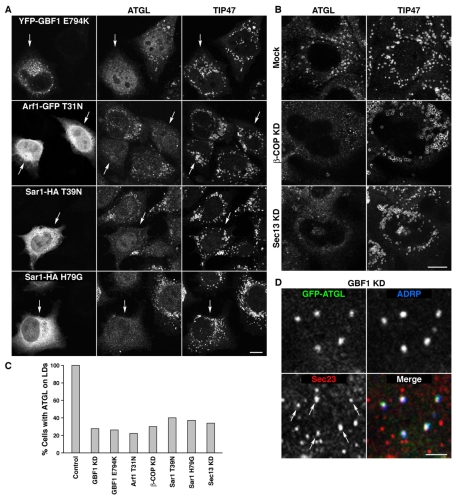
To further characterize the BFA-sensitive pathway, we examined the effect of RNAi-mediated depletion of GBF1. We found that, like BFA treatment, GBF1 depletion prevented association of ATGL, but not TIP47, with LDs (Fig. 3A) in ∼75% of the OA-treated cells (quantification shown in Fig. 4C and supplementary material Fig. S5). Moreover, GBF1 depletion decreased the total levels of ATGL in OA-treated cells (Fig. 3B) by a process that was sensitive to the proteasomal inhibitor ALLN (Fig. 3C). Therefore, impaired delivery of ATGL to LDs resulted in its degradation by the proteasome. OA-treated, GBF1-depleted cells displayed LDs that were ∼25% larger in diameter, corresponding to a ∼2-fold increase in volume (Fig. 3D), and contained higher levels of TG, CE and free cholesterol (FC), but not total phospholipids (PLs), relative to control cells (Fig. 3E). These results indicated that mistargeting of ATGL, and possibly other LDAPs, alters LD morphology and lipid homeostasis. In line with the experiments described above, expression of dominant-negative GBF1-E794K or dominant-negative ARF1-T31N mutants (tagged with YFP or GFP, respectively) (Fig. 4A), or RNAi-mediated depletion of the β-COP subunit of COPI (Fig. 4B), also precluded delivery of ATGL, but not TIP47, to LDs in 70-80% of OA-treated cells (Fig. 4C; supplementary material Fig. S5). From these experiments, we concluded that ATGL transport to LDs is mediated by a GBF1-ARF1-COPI-regulated membrane trafficking pathway. Results similar to those shown for ATGL in Figs 1, 2, 3, 4 were obtained for the PAT protein ADRP (supplementary material Figs S2-S4 and data not shown) (Jiang and Serrero, 1992; Nakamura et al., 2004; Targett-Adams et al., 2003; Xu et al., 2005), indicating that this protein is transported to LDs by the same pathway.
Fig. 3.
Depletion of GBF1 inhibits ATGL association with LDs. (A) HeLa cells were transfected with siRNA against GBF1 (knockdown, KD) or mock treated, then co-immunostained with antibodies to ATGL and TIP47. Scale bar: 10 μm. (B) Immunoblot analysis of ATGL levels upon GBF1 KD. HeLa cells were transfected as in A, then treated with 200 μM OA or left untreated, as indicated. Samples were analyzed by SDS-PAGE and immunoblotting with antibodies to ATGL and actin (loading control). Mean and s.d. of at least three independent experiments are shown. (C) Immunoblot analysis of ATGL expression in OA-treated cells incubated in the absence or presence of the proteasomal inhibitor ALLN. The same conditions were used as in B, except that ALLN was added at the same time as OA where indicated. Mean and s.d. of at least three independent experiments are shown. (D) HeLa cells were treated with siRNA against GBF1 in the presence or absence of 400 μM OA. Cells were stained with BODIPY 493/503 and LD diameters measured. Mean and s.d. of at least three independent experiments are shown. (E) Cells were treated as in D and levels of the indicated lipids quantified. Mean and s.d. are shown. In D and E, P-values for significant differences between GBF1-depleted and control cells are indicated. TG, triglyceride; CE, cholesterol ester; FC, free cholesterol; PLs, total phospholipids.
Fig. 4.
COPI, COPII and their regulators are required for ATGL delivery to LDs. (A) HeLa cells were transfected with plasmids encoding YFP-GBF1-E794K, ARF1-T31N-GFP, SAR1-T39N-GFP or SAR1-H79G-GFP. At 8 hours after transfection, 150 μM OA was added for an additional 14 hours, followed by co-immunostaining with antibodies to ATGL (middle column) and TIP47 (right-hand column). Arrows indicate cells expressing YFP- or GFP-tagged mutant constructs. (B) HeLa cells were either mock treated (top row) or transfected with siRNAs against β-COP (middle row) or SEC13 (bottom row) and incubated for a total of 72 hours, with 150 μM OA added for the last 12 hours. Cells were immunostained with antibodies to ATGL (left) or TIP47 (right). (C) Quantification of the results shown in Fig. 3A and in A,B. At least 100 cells were scored for the presence or absence of ATGL on LDs, and the result plotted as a percentage of total cells examined. (D) Co-localization of ATGL and ADRP with COPII components at ER exit sites (ERES) in GBF1-KD cells. HeLa cells were treated with GBF1 siRNAs as described in Fig. 2A, transfected with ATGL-GFP, and co-immunostained with antibodies to ADRP and SEC23. Arrows point to SEC23-positive ERES that also contain ATGL and ADRP. Scale bars: 10 μm in A,B; 2 μm in D.
Additional requirement of COPII for ATGL delivery to lipid droplets
GBF1, ARF1 and COPI are associated with various membrane structures at the ER-Golgi functional interface. These include foci adjacent to ERES, the ERGIC and cis-Golgi cisternae (Altan-Bonnet et al., 2004). Although the primary role of this protein ensemble is to retrieve proteins from the Golgi complex to the ER (Aridor et al., 1995; Letourneur et al., 1994), interference with its function also inhibits, perhaps indirectly, cargo export from the ER (Altan-Bonnet et al., 2004; Ward et al., 2001). It is thus pertinent to ask where the residual ATGL accumulates upon perturbation of the GBF1-ARF1-COPI pathway. Immunofluorescence microscopy of GBF1-depleted, OA-treated cells revealed localization of the majority of GFP-ATGL, as well as ADRP, to punctate foci that overlapped with a subset (∼40%) of ERES stained for the coatomer protein II (COPII) subunit SEC23 (Fig. 4D) (Bannykh et al., 1996).
COPII is another organellar coat protein that is specifically involved in protein export from the ER (Lee et al., 2004). This raised the possibility that ATGL could be delivered to LDs from ERES, perhaps in a COPII-dependent manner. COPII is regulated by another small GTPase, SAR1 (Lee et al., 2004). Indeed, expression of dominant-negative SAR1-T39N or constitutively active SAR1-H79G impaired the delivery of ATGL to LDs in OA-treated cells (Fig. 4A). Likewise, depletion of the SEC13 subunit of COPII inhibited transport of ATGL (Fig. 4B) and ADRP (supplementary material Fig. S4A) to LDs. The effects of perturbing SAR1-COPII, however, were quantitatively smaller than those of perturbing GBF1-ARF1-COPI (Fig. 4C; supplementary material Fig. S5), suggesting that COPI is more important than COPII for ATGL transport to LDs. In addition, unlike interference with GBF1-ARF1-COPI, interference with SAR1-COPII had no effect on LD size (supplementary material Fig. S4D), highlighting another difference in the relative importance of the two coatomer proteins in LD homeostasis.
Close apposition of ER exit sites with lipid droplets
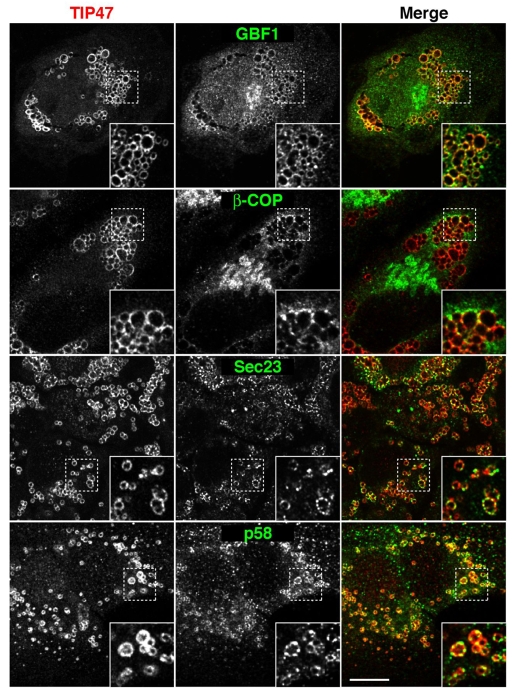
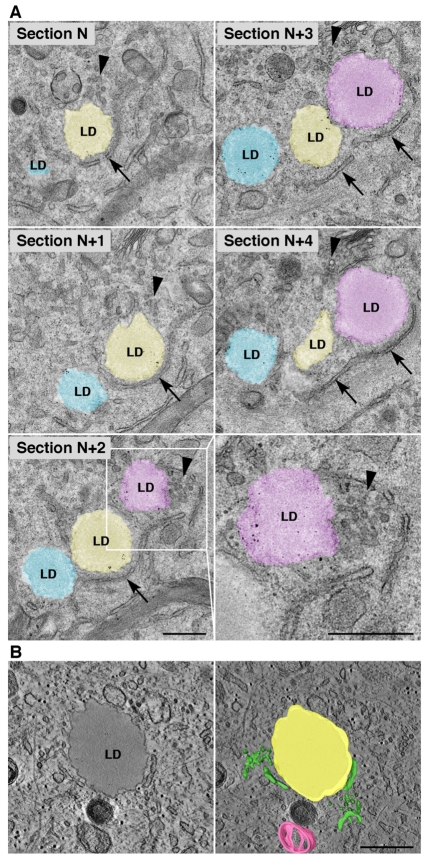
The involvement of COPI, COPII and their corresponding regulators in ATGL delivery to LDs prompted us to compare their relative distributions within cells. Double-staining immunofluorescence microscopy analyses showed that GBF1, COPI (β-COP) and COPII (SEC23) localized to discrete foci surrounding LDs after overnight incubation with OA (Fig. 5). Significantly, the transmembrane protein p58 (ERGIC53; LMAN1), which normally localizes to ERES and ERGIC (Altan-Bonnet et al., 2004), was likewise found to decorate the LD perimeter in a discontinuous fashion (Fig. 5). The presence of both peripheral coat and transmembrane proteins suggests that these foci represent a subpopulation of ERES and ERGIC structures in close apposition to LDs. By contrast, the Golgi cisternae protein GM130 (GOLGA2) remained restricted to the Golgi complex and was not found in the vicinity of LDs under these conditions (supplementary material Fig. S6A), suggesting a bifurcation of the early secretory pathway towards either LDs or the Golgi complex. Although ERES and ERGIC elements are scattered throughout the cell and could come into contact with LDs by chance, this is unlikely given the manner in which ERES and ERGIC proteins completely surround LDs in a ring. In addition, comparison of cells before and after treatment with OA indicates a clear change in the pattern of fluorescence for p58 (supplementary material Fig. S6B), as well as for the COPII and COPI components (data not shown). Prior to OA treatment the majority of p58 appears in dispersed puncta scattered throughout the cytoplasm, whereas after OA treatment a large proportion of p58 is present in rings encircling LDs (supplementary material Fig. S6B). Electron microscopy (EM) analyses of serial sections of OA-treated cells indeed revealed the presence of ER cisternae (Fig. 6A, arrows) and tubular-vesicular structures characteristic of ERES and ERGIC (Fig. 6A, arrowheads) juxtaposed to LDs. Electron tomography confirmed the close apposition of ER and ERES/ERGIC to LDs in three dimensions (Fig. 6B; supplementary material Movie 1). These observations indicate that the involvement of coatomer proteins in protein transport to LDs occurs within the framework of a close spatial association of ER export domains with LDs.
Fig. 5.
Immunofluorescence microscopy shows ERES and ER-Golgi intermediate compartment (ERGIC) membranes in close proximity to LDs. HeLa cells were treated for 20 hours with 150 μM OA and co-immunostained with antibodies to TIP47 (red channel, left column) and to GBF1, β-COP, SEC23 or p58 (green channel, middle column). Merged images are shown on the right. Insets are 3-fold magnifications of boxed areas. Scale bar: 10 μm.
Fig. 6.
Electron microscopy of ERES and ERGIC structures apposed to LDs. (A) HeLa cells were treated for 20 hours with 150 μM OA and prepared for electron microscopy as described in Materials and Methods. Five serial thin sections through one cell are shown. The bottom right panel is a magnified view of the boxed area in section N+2. Arrows, ER membranes; arrowheads, ERES/ERGIC. (B) Electron tomography showing ERES/ERGC structures in close proximity to a LD. An average of ten tomographic slices, representing a 14 nm virtual section extracted from the tomogram, shows the close proximity of ER and ERES/ERGIC structures to a LD (left). The whole tomogram is 140 nm thick and is shown in supplementary material Movie 1. The volume generated from the tomogram was merged with a single tomogram slice (right). ER and ERES/ERGIC are in green, a LD in yellow, and a mitochondrion in pink. Scale bars: 0.5 μm.
Discussion
The results of our experiments indicate the existence of a novel membrane trafficking pathway for the delivery of a subset of LDAPs, including a lipase (ATGL) and a PAT protein (ADRP), to LDs. This pathway is likely to originate from ERES/ERGIC structures that surround LDs and involves the coatomer proteins COPI and COPII and their corresponding regulators. A notable exception is the PAT protein, TIP47, which is unaffected by any of the coatomer perturbations used in this study. A possible explanation for this different behavior is that, unlike other LDAPs, TIP47 also occurs in the cytosol and on endosomal membranes, and shuttles between these two compartments (Carroll et al., 2001).
Like TIP47, neutral lipids, as detected by BODIPY 493/503 staining, are packaged into new lipid droplets independently of coatomer proteins. In fact, LDs become larger and cells accumulate more neutral lipids upon perturbation of the GBF1-ARF1-COPI pathway. These phenotypes might be due, at least in part, to impaired transport of ATGL, a lipase that was previously shown to decrease LD size through its catalytic activity (Gronke et al., 2005; Smirnova et al., 2006; Zimmermann et al., 2004). Our results provide a likely explanation for findings from a recent global RNAi screen using Drosophila S2 cells, which showed larger LDs (Guo et al., 2008) in cells depleted of ARF1 and COPI orthologs. While our manuscript was under revision, another study also showed lipid overstorage and decreased association of ATGL with LDs in cells depleted of COPI and its regulators, although not of COPII (Beller et al., 2008). Thus, the formation and budding of the neutral lipid core must depend on some other machinery. Likely components of this machinery are the recently described ER transmembrane proteins BSCL2 (seipin) (Magre et al., 2001; Szymanski et al., 2007), FIT1 and FIT2 (Kadereit et al., 2008).
How could COPI and COPII enable LDAP transport from ERES/ERGIC to LDs? One possibility is that transport between these organelles involves sequential steps mediated by COPII- and COPI-coated vesicles, as occurs in the secretory pathway. It is hard to imagine, however, how the membrane bilayer of coatomer-coated vesicles could fuse with the phospholipid monolayer that covers the surface of LDs. Moreover, GBF1-ARF1-COPI appear quantitatively more important than SAR1-COPII for ATGL delivery to LDs (Fig. 3C) and maintenance of LD size (Fig. 2D; supplementary material Fig. S4) (Beller et al., 2008; Guo et al., 2008). These differences seem incompatible with a simple model involving sequential transport by coatomer-coated vesicles, as COPII acts upstream of COPI in the secretory pathway.
Another possibility is that coatomer proteins and their regulators act directly at the level of the LD surface to enable initial association of LDAPs with the phospholipid monolayer. After this initial step, LDAPs would become stably attached to the monolayer and refractory to acute perturbation of the GBF1-ARF1-COPI pathway. In this regard, ARF1 and COPI have been detected by proteomic analyses in isolated LD preparations (Bartz et al., 2007). Given the close apposition of ER cisternae, ERES and ERGIC to LDs shown here, further studies will be needed to determine whether this truly represents direct binding to the LD surface or co-isolation of LDs with ER-derived structures.
We favor a third possibility in which the combined action of COPII and COPI directly or indirectly creates a membrane environment from which LDAPs are delivered to LDs (see model in Fig. 7). This could involve an ATGL-transfer protein that delivers ATGL to adjacent LDs, or co-segregation of LDAPs with neutral lipids on nascent LDs. This could occur within the subset of ERES or ERGIC adjacent to the LDs and involve lateral conveyance of LDAPs from COPII- and/or COPI-coated membrane bilayers to the surface of nascent LDs. Newly made LDs could then fuse with larger, pre-existing LDs.
Fig. 7.

Model for the involvement of COPII (blue) and COPI (green) in ATGL transport to LDs. ATGL associates with ER membranes and is subsequently transported to specialized ERES and ERGIC structures that are adjacent to pre-existing LDs. ATGL then co-segregates with neutral lipids into nascent LDs, which subsequently fuse with larger LDs. It is also possible that a transfer protein translocates ATGL from an ERGIC domain to LDs. Steps in this process are enabled by sequential action of SAR1-COPII and GBF1-ARF1-COPI.
LDs were long considered to be autonomous, inert lipid inclusions. However, the dependence of LDAP transport on coatomer proteins shown here, together with the finding of Rab and Arf family GTPases and SNAREs in association with LDs (Bartz et al., 2007; Bostrom et al., 2007; Liu et al., 2004), demonstrate that LDs are dynamic organelles that interface with different membrane systems in the cell.
Materials and Methods
Antibodies, cDNA constructs and siRNAs
Rabbit antiserum to full-length human ATGL (Smirnova et al., 2006) was used for immunofluorescence microscopy. Rabbit antiserum to GBF1 (Zhao et al., 2006), used for immunofluorescence experiments, was a gift from P. Melançon (Department of Cell Biology, University of Alberta, Edmonton, Canada). Rabbit anti-ATGL and mouse anti-GBF1 (Cell Signaling Technology, Danvers, MA, USA) were used for immunoblot analysis. Chicken anti-SAR1, mouse anti-β-COP, and mouse anti-β-actin were from Abcam (Cambridge, MA, USA). Guinea pig anti-TIP47 and mouse anti-ADRP were from Fitzgerald Industries International (Concord, MA, USA). Rabbit anti-SEC23 (human) was from Affinity Bioreagents (Golden, CO, USA). Antiserum to p58 was from Sigma-Aldrich (St Louis, MO, USA).
ATGL-GFP (Smirnova et al., 2006) and GFP-GGA1 (Puertollano et al., 2003) have been described previously, and GFP-ADRP was from GeneCopoeia (Germantown, MD, USA). SAR1 and HA-tagged SAR1 mutant constructs (Ward et al., 2001) were kindly provided by G. Patterson and J. Lippincott-Schwartz (Cell Biology and Metabolism Program, NICHD, NIH, Bethesda, MD, USA). GFP-tagged ARF1 constructs (wild-type and mutants) were from F. van Kuppeveld (Department of Medical Microbiology, Radboud University Nijmegen Medical Centre, Nijmegen, The Netherlands).
For RNAi experiments we used siGENOME ON-TARGETplus SMARTpool siRNA oligonucleotides (Dharmacon RNA Technologies, Lafayette, CO, USA) directed to human GBF1, human β-COP or human SEC13. The sequences of each (5′ to 3′) are as follows.
GBF1 siRNA:
sense GAUGAGGGCUUCCACAUUGUU and antisense PCAAUGUGGAAGCCCUCAUCUU;
sense CAACACACCUACUAUCUCUUU and antisense PAGAGAUAGUAGGUGUGUUGUU;
sense GAGAAGCUAGCAAUACUGAUU and antisense PUCAGUAUUGCUAGCUUCUCUU;
sense CCACUGCUGUCACUCUCUUAUU and antisense PUAGAGAGUGACAGCAGUGGUU.
Human SEC13L1 SMARTpool siRNA sequences:
sense CAUGUGAGCUGGUCCAUCAUU and antisense PUGAUGGACCAGCUCACAUGUU;
sense GGUCGUGUGUUCAUUUGGAUU and antisense PUCCAAAUGAACACACGACCUU;
sense CCAUCUCCCUGCUGACUUAUU and antisense PUAAGUCAGCAGGGAGAUGGUU;
sense GUAAUUAACACUGUGGAUAUU and antisense PUAUCCACAGUGUUAAUUACUU.
Human SMARTpool BCOP siRNA sequences:
sense CCAAGAUUGCAUUGCGCUAUU and antisense PUAGCGCAAUGCAAUCUUGGUU;
sense CAUAUAAGAAUUCGUGCAAUU and antisense PUUGCACGAAUUCUUAUAUGUU;
sense AUUAUUAAGGAGAGCGACAUU and antisense PUGUCGCUCUCCUUAAUAAUUU;
sense CCUCAUGACUUCGCAAAUAUU and antisense PUAUUUGCGAAGUCAUGAGGUU.
Preparation of oleic acid
Oleic acid (OA) complexed to bovine serum albumin (BSA) was prepared as described (Brasaemle and Wolins, 2006). Briefly, fatty-acid-free BSA (Sigma-Aldrich) was dissolved in 1 M Tris-HCl (pH 8.0) to a final concentration of 2.1 mM, then OA (Sigma-Aldrich) was added to a final concentration of 12.5 mM.
Cell culture and transfection
HeLa cells were grown in Dulbecco's Modified Eagle's Medium (DMEM) supplemented with 25 mM glucose and 1 mM sodium pyruvate, 20 mM glutamine, 100 units/ml penicillin G and 100 μg/ml streptomycin. Cells were transfected with plasmids on 24-well plates using Lipofectamine (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. For overnight OA treatment, at 8 hours post-transfection cells were treated with 150-200 μM OA and incubated for an additional 14 hours before preparation for imaging. For brefeldin A (BFA, Sigma-Aldrich) experiments, cells were either treated with 400 μM OA for 3 hours followed by treatment with 5 μg/ml BFA for an additional 3 hours, or treated with 5 μg/ml BFA for 10 minutes followed by addition of 400 μM OA and 5 μg/ml BFA for an additional 3 hours. For quantifications (Fig. 2B; Fig. 4C; supplementary material Fig. S5), at least 100 cells were scored for the presence of ATGL on LDs, and percentages calculated.
RNA interference
Approximately 24 hours before siRNA transfection, HeLa cells were seeded in antibiotic-free DMEM and transfected with an siRNA-Lipofectamine mixture. For each well of a 24-well plate, 50 pmol of siRNA was dissolved in 50 μl Optimem in one tube, and 50 μl Optimem was added to 0.5 μl Lipofectamine in a second tube. After 5 minutes, the diluted siRNA and Lipofectamine were mixed, incubated for 15 minutes, and the siRNA-Lipofectamine cocktail added in a dropwise manner to the cells in 400 μl of antibiotic-free DMEM. The next day, the medium was replaced with complete DMEM supplemented with antibiotics. For OA treatment, 60 hours after siRNA transfection cells were treated with DMEM plus 150 μM OA and incubation continued for an additional 12 hours prior to preparation of the cells for either biochemical or cell imaging experiments. N-acetyl-leucyl-leucyl-norleucinal (ALLN) (Calbiochem, Gibbstown, NJ, USA) was added at the same time as OA.
Lipomics analysis
HeLa cells were transfected with either mock or GBF1 SMARTpool siRNA, and 69 hours later treated for 3 hours with 400 μM OA. Cells were washed once with PBS, scraped from plates into PBS, spun at low speed and snap frozen in liquid nitrogen. Frozen cell pellets were sent on dry ice to Lipomics Technologies (West Sacramento, CA, USA) for lipid profiling.
Subcellular fractionation
HeLa cells were grown in DMEM containing 400 μM OA for ∼19 hours. Cells were then washed with ice-cold PBS, scraped from plates and recovered by low-speed centrifugation. Cell pellets were resuspended in an equal volume of lysis buffer (10 mM Tris-HCl pH 7.4, 500 mM sucrose, protease inhibitor cocktail) and disrupted by 30 passages through a 25-gauge needle on ice. Cell lysates were centrifuged at 1000 g for 10 minutes at 4°C, and 300 μl of post-nuclear supernatant applied to the bottom of a discontinuous gradient of 0.25, 0.5 and 1.5 M sucrose. The tubes were centrifuged at 172,000 g for 3 hours at 4°C. Three fractions containing LDs (top), cytosol (middle) and membranes (bottom) were collected. The membrane fractions were suspended in 10 mM Tris-HCl (pH 7.4) with protease inhibitor cocktail and collected by centrifugation at 172,000 g for 1 hour.
For membrane extraction experiments, the membrane fractions prepared as described above were resuspended in 10 mM Tris-HCl (pH 7.4), divided into seven equal parts, and centrifuged at 172,000 g at 4°C, for 1 hour. The membrane pellets were resuspended and incubated on ice for 1 hour with 1 M NaCl, 0.2 M Na2CO3 (pH 11.3), 2% Triton X-100, 2 M urea or with 10 mM Tris-HCl (pH 7.4). Soluble and insoluble fractions were separated by centrifugation at 172,000 g at 4°C for 1 hour. For BFA treatment, cells were incubated with 1 μg/ml BFA for 15 minutes, followed by addition of 400 μM OA, then incubated for an additional 19 hours before harvesting.
Fluorescence microscopy
For immunofluorescence microscopy, HeLa cells were grown on glass coverslips and fixed with 4% formaldehyde. The cells were permeabilized with 0.1% saponin for 10 minutes, washed with PBS, then treated with 0.2% fatty-acid-free BSA and 0.2% gelatin in PBS for 15 minutes, followed by a 30-minute wash in PBS. Cells were probed with primary and secondary antibodies for 1 hour followed by 30-minute washes in PBS. To stain the LD cores, 10 μg/ml BODIPY 493/503 was included with the secondary antibodies. Coverslips were mounted on glass slides using Fluoromount G (Southern Biotechnology Associates, Birmingham, AL, USA). Live cell imaging was carried out as previously described (Niu et al., 2005; Puertollano et al., 2003). Briefly, cells were grown on LabTek chambers (Nalge Nunc, Naperville, IL, USA), transfected with different constructs tagged with GFP or spectral variants of GFP, and transferred into culture medium buffered with 25 mM HEPES/KOH (pH 7.4). For GFP-GGA1 fluorescence recovery after photobleaching (FRAP), images were acquired at 2-second intervals; for ATGL-GFP and GFP-ADRP, images were acquired at 30-second intervals. Images were acquired using an inverted confocal laser-scanning microscope (LSM 510, Carl Zeiss) equipped with a stage heated to 37°C for live cell imaging. LD size quantifications were performed using the 510 Image Analyzer software.
Electron microscopy
Cells fixed with 4% formaldehyde were treated with reduced osmium (1:1 mixture of 2% aqueous osmium tetroxide and 3% aqueous potassium ferrocyanide) as described previously (Karnovsky, 1971), embedded in 2% agar, dehydrated in ethanol, and embedded in Epon resin. For tomography, semi-thin sections were cut to 200 nm. Sections were then stained for 2 minutes with lead citrate, and 15-nm gold fiducials were deposited on the top surface of the sections. Grids were imaged using a Tecnai T20 (FEI Company) microscope operating at 200 kV, equipped with a 2k×2k CCD camera (Gatan, Pleasanton, CA, USA). Tilt series for tomographic reconstruction were acquired using the Xplore 3D tomography software (FEI Company). The sections were tilted from –65° to +65° with images being captured at 2° intervals. Tomograms were generated using IMOD software (Kremer et al., 1996; Mastronarde, 1997). Segmentation models were generated using Amira (Mercury systems, Chelmsford, MA, USA).
Supplementary Material
Supplementary material available online at http://jcs.biologists.org/cgi/content/full/122/11/1834/DC1
We thank P. Melançon, W. Brown, F. van Kuppeveld, G. Patterson and J. Lippincott-Schwartz for kind gifts of reagents, T. Mrozek for help with 3D rendering of the tomogram, X. Zhu and N. Tsai for expert technical assistance, and R. Mattera and Y. Prabhu for critical review of the manuscript. This work was funded by the Intramural Program of NICHD. Deposited in PMC for release after 12 months.
References
- Altan-Bonnet, N., Sougrat, R. and Lippincott-Schwartz, J. (2004). Molecular basis for Golgi maintenance and biogenesis. Curr. Opin. Cell Biol. 16, 364-372. [DOI] [PubMed] [Google Scholar]
- Aridor, M., Bannykh, S. I., Rowe, T. and Balch, W. E. (1995). Sequential coupling between COPII and COPI vesicle coats in endoplasmic reticulum to Golgi transport. J. Cell Biol. 131, 875-893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arner, P. and Langin, D. (2007). The role of neutral lipases in human adipose tissue lipolysis. Curr. Opin. Lipidol. 18, 246-250. [DOI] [PubMed] [Google Scholar]
- Bannykh, S. I., Rowe, T. and Balch, W. E. (1996). The organization of endoplasmic reticulum export complexes. J. Cell Biol. 135, 19-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartz, R., Zehmer, J. K., Zhu, M., Chen, Y., Serrero, G., Zhao, Y. and Liu, P. (2007). Dynamic activity of lipid droplets: protein phosphorylation and GTP-mediated protein translocation. J. Proteome Res. 6, 3256-3265. [DOI] [PubMed] [Google Scholar]
- Beller, M., Sztalryd, C., Southall, N., Bell, M., Jackle, H., Auld, D. S. and Oliver, B. (2008). COPI complex is a regulator of lipid homeostasis. PLoS Biol. 6, e292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostrom, P., Andersson, L., Rutberg, M., Perman, J., Lidberg, U., Johansson, B. R., Fernandez-Rodriguez, J., Ericson, J., Nilsson, T., Boren, J. et al. (2007). SNARE proteins mediate fusion between cytosolic lipid droplets and are implicated in insulin sensitivity. Nat. Cell Biol. 9, 1286-1293. [DOI] [PubMed] [Google Scholar]
- Brasaemle, D. L. (2007). Thematic review series: adipocyte biology. The perilipin family of structural lipid droplet proteins: stabilization of lipid droplets and control of lipolysis. J. Lipid Res. 48, 2547-2559. [DOI] [PubMed] [Google Scholar]
- Brasaemle, D. L. and Wolins, N. E. (2006). Isolation of lipid droplets from cells by density gradient centrifugation. Curr. Protoc. Cell Biol. Chapter 3, Unit 3.15. [DOI] [PubMed]
- Brasaemle, D. L., Dolios, G., Shapiro, L. and Wang, R. (2004). Proteomic analysis of proteins associated with lipid droplets of basal and lipolytically stimulated 3T3-L1 adipocytes. J. Biol. Chem. 279, 46835-46842. [DOI] [PubMed] [Google Scholar]
- Carroll, K. S., Hanna, J., Simon, I., Krise, J., Barbero, P. and Pfeffer, S. R. (2001). Role of Rab9 GTPase in facilitating receptor recruitment by TIP47. Science 292, 1373-1376. [DOI] [PubMed] [Google Scholar]
- Ducharme, N. A. and Bickel, P. E. (2008). Lipid droplets in lipogenesis and lipolysis. Endocrinology 149, 942-949. [DOI] [PubMed] [Google Scholar]
- Fischer, J., Lefevre, C., Morava, E., Mussini, J. M., Laforet, P., Negre-Salvayre, A., Lathrop, M. and Salvayre, R. (2007). The gene encoding adipose triglyceride lipase (PNPLA2) is mutated in neutral lipid storage disease with myopathy. Nat. Genet. 39, 28-30. [DOI] [PubMed] [Google Scholar]
- Gronke, S., Mildner, A., Fellert, S., Tennagels, N., Petry, S., Muller, G., Jackle, H. and Kuhnlein, R. P. (2005). Brummer lipase is an evolutionary conserved fat storage regulator in Drosophila. Cell Metab. 1, 323-330. [DOI] [PubMed] [Google Scholar]
- Guo, Y., Walther, T. C., Rao, M., Stuurman, N., Goshima, G., Terayama, K., Wong, J. S., Vale, R. D., Walter, P. and Farese, R. V. (2008). Functional genomic screen reveals genes involved in lipid-droplet formation and utilization. Nature 453, 657-661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, H. P. and Serrero, G. (1992). Isolation and characterization of a full-length cDNA coding for an adipose differentiation-related protein. Proc. Natl. Acad. Sci. USA 89, 7856-7860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadereit, B., Kumar, P., Wang, W. J., Miranda, D., Snapp, E. L., Severina, N., Torregroza, I., Evans, T. and Silver, D. L. (2008). Evolutionarily conserved gene family important for fat storage. Proc. Natl. Acad. Sci. USA 105, 94-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnovsky, M. J. (1971). Use of ferrocyanide-reduced osmium tetroxide in electron microscopy. American Society of Cell Biology (11th Meeting) Abstr. 284, 146. [Google Scholar]
- Klausner, R. D., Donaldson, J. G. and Lippincott-Schwartz, J. (1992). Brefeldin A: insights into the control of membrane traffic and organelle structure. J. Cell Biol. 116, 1071-1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer, J. R., Mastronarde, D. N. and McIntosh, J. R. (1996). Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116, 71-76. [DOI] [PubMed] [Google Scholar]
- Lee, M. C., Miller, E. A., Goldberg, J., Orci, L. and Schekman, R. (2004). Bi-directional protein transport between the ER and Golgi. Annu. Rev. Cell Dev. Biol. 20, 87-123. [DOI] [PubMed] [Google Scholar]
- Letourneur, F., Gaynor, E. C., Hennecke, S., Demolliere, C., Duden, R., Emr, S. D., Riezman, H. and Cosson, P. (1994). Coatomer is essential for retrieval of dilysine-tagged proteins to the endoplasmic reticulum. Cell 79, 1199-1207. [DOI] [PubMed] [Google Scholar]
- Lippincott-Schwartz, J., Yuan, L. C., Bonifacino, J. S. and Klausner, R. D. (1989). Rapid redistribution of Golgi proteins into the ER in cells treated with brefeldin A: evidence for membrane cycling from Golgi to ER. Cell 56, 801-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, P., Ying, Y., Zhao, Y., Mundy, D. I., Zhu, M. and Anderson, R. G. (2004). Chinese hamster ovary K2 cell lipid droplets appear to be metabolic organelles involved in membrane traffic. J. Biol. Chem. 279, 3787-3792. [DOI] [PubMed] [Google Scholar]
- Londos, C., Brasaemle, D. L., Schultz, C. J., Segrest, J. P. and Kimmel, A. R. (1999). Perilipins, ADRP, and other proteins that associate with intracellular neutral lipid droplets in animal cells. Semin. Cell Dev. Biol. 10, 51-58. [DOI] [PubMed] [Google Scholar]
- Magre, J., Delepine, M., Khallouf, E., Gedde-Dahl, T., Jr, Van Maldergem, L., Sobel, E., Papp, J., Meier, M., Megarbane, A., Bachy, A. et al. (2001). Identification of the gene altered in Berardinelli-Seip congenital lipodystrophy on chromosome 11q13. Nat. Genet. 28, 365-370. [DOI] [PubMed] [Google Scholar]
- Martin, S. and Parton, R. G. (2006). Lipid droplets: a unified view of a dynamic organelle. Nat. Rev. Mol. Cell Biol. 7, 373-378. [DOI] [PubMed] [Google Scholar]
- Mastronarde, D. N. (1997). Dual-axis tomography: an approach with alignment methods that preserve resolution. J. Struct. Biol. 120, 343-352. [DOI] [PubMed] [Google Scholar]
- Murphy, D. J. (2001). The biogenesis and functions of lipid bodies in animals, plants and microorganisms. Prog. Lipid Res. 40, 325-438. [DOI] [PubMed] [Google Scholar]
- Nakamura, N., Akashi, T., Taneda, T., Kogo, H., Kikuchi, A. and Fujimoto, T. (2004). ADRP is dissociated from lipid droplets by ARF1-dependent mechanism. Biochem. Biophys. Res. Commun. 322, 957-965. [DOI] [PubMed] [Google Scholar]
- Niu, T. K., Pfeifer, A. C., Lippincott-Schwartz, J. and Jackson, C. L. (2005). Dynamics of GBF1, a Brefeldin A-Sensitive Arf1 Exchange Factor at the Golgi. Mol. Biol. Cell 16, 1213-1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puertollano, R., van der Wel, N. N., Greene, L. E., Eisenberg, E., Peters, P. J. and Bonifacino, J. S. (2003). Morphology and dynamics of clathrin/GGA1-coated carriers budding from the trans-Golgi network. Mol. Biol. Cell 14, 1545-1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova, E., Goldberg, E. B., Makarova, K. S., Lin, L., Brown, W. J. and Jackson, C. L. (2006). ATGL has a key role in lipid droplet/adiposome degradation in mammalian cells. EMBO Rep. 7, 106-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymanski, K. M., Binns, D., Bartz, R., Grishin, N. V., Li, W. P., Agarwal, A. K., Garg, A., Anderson, R. G. and Goodman, J. M. (2007). The lipodystrophy protein seipin is found at endoplasmic reticulum lipid droplet junctions and is important for droplet morphology. Proc. Natl. Acad. Sci. USA 104, 20890-20895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Targett-Adams, P., Chambers, D., Gledhill, S., Hope, R. G., Coy, J. F., Girod, A. and McLauchlan, J. (2003). Live cell analysis and targeting of the lipid droplet-binding adipocyte differentiation-related protein. J. Biol. Chem. 278, 15998-16007. [DOI] [PubMed] [Google Scholar]
- Ward, T. H., Polishchuk, R. S., Caplan, S., Hirschberg, K. and Lippincott-Schwartz, J. (2001). Maintenance of Golgi structure and function depends on the integrity of ER export. J. Cell Biol. 155, 557-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, G., Sztalryd, C., Lu, X., Tansey, J. T., Gan, J., Dorward, H., Kimmel, A. R. and Londos, C. (2005). Post-translational regulation of adipose differentiation-related protein by the ubiquitin/proteasome pathway. J. Biol. Chem. 280, 42841-42847. [DOI] [PubMed] [Google Scholar]
- Zechner, R., Kienesberger, P. C., Haemmerle, G., Zimmermann, R. and Lass, A. (2009). Adipose triglyceride lipase and the lipolytic catabolism of cellular fat stores. J. Lipid Res. 50, 3-21. [DOI] [PubMed] [Google Scholar]
- Zhao, X., Claude, A., Chun, J., Shields, D. J., Presley, J. F. and Melancon, P. (2006). GBF1, a cis-Golgi and VTCs-localized ARF-GEF, is implicated in ER-to-Golgi protein traffic. J. Cell Sci. 119, 3743-3753. [DOI] [PubMed] [Google Scholar]
- Zimmermann, R., Strauss, J. G., Haemmerle, G., Schoiswohl, G., Birner-Gruenberger, R., Riederer, M., Lass, A., Neuberger, G., Eisenhaber, F., Hermetter, A. et al. (2004). Fat mobilization in adipose tissue is promoted by adipose triglyceride lipase. Science 306, 1383-1386. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.