Abstract
Adeno-associated virus serotype 2 (AAV-2) is a leading candidate vector for gene therapy. Cell entry starts with attachment to a primary receptor, Heparan Sulfate Proteoglycan (HSPG) before binding to a co-receptor. Here, cryo-electron microscopy provides direct visualization of the virus–HSPG interactions. Single particle analysis was performed on AAV-2 complexed with a 17kDa heparin fragment at 8.3Å resolution. Heparin density covers the shoulder of spikes surrounding viral 3-fold symmetry axes. Previously implicated, positively charged residues R448/585, R451/588 and R350/487 from another subunit cluster at the center of the heparin footprint. The footprint is much more extensive than apparent through mutagenesis, including R347/484, K395/532 and K390/527 that are more conserved, but whose roles have been controversial. It also includes much of a region proposed as a co-receptor site, because prior studies had not revealed heparin interactions. Heparin density bridges over the viral 3-fold axes, indicating multivalent attachment to symmetry-related binding sites.
Keywords: Heparan sulfate, Heparin, Gene therapy, Electron microscopy, Parvovirus
Adeno-associated virus serotype 2 (AAV-2) was first discovered in tissues of children infected with adenovirus, from which its name is derived (Laughlin et al., 1983; Salo and Mayor, 1977; Salo and Mayor, 1979). Since its discovery, AAV-2 has emerged as a leading candidate vector for gene therapy. It was considered well-suited for development as a vector because the wild-type virus is non-pathogenic, elicits a low immune response, integrates itself site specifically into chromosome 19, infects both dividing and non-dividing cells and has wide cell tropism (Berns and Giraud, 1995; Bueler, 1999; Chirmule et al., 1999; Podsakoff, Wong Jr., and Chatterjee, 1994; Trempe, 1996; Wu et al., 1998; Wu et al., 2006a; Wu, Asokan, and Samulski, 2006). Recombinantly engineered AAV vectors (rAAV) differ from wild-type in that transduction is primarily episomal, and that the vector used is non-replicative and therefore not dependent on the presence of helper adenovirus (Carter, 1990; Penaud-Budloo et al., 2008). Experimental vectors have been designed to combat a variety of diseases such as AIDS, high blood pressure, cystic fibrosis, and Parkinson's disease (Chaterjee, Johnson, and Wong Jr., 1992; Chen et al., 1996; Flotte and Carter, 1998; Harster et al., 1999; Inouye et al., 1997; Mandel et al., 1997; Mochizuki et al., 2001; Phillips, 1997). However, AAV's promiscuous cell infection may also be considered a disadvantage and an obstacle towards the achievement of tailored cell-specific vectors. Structural information regarding host-cell interactions could accelerate this development.
AAV-2 is a dependovirus within the parvovirus family and is the type species. It has an ∼4.7kb ssDNA genome encapsulated by a T=1 icosahedral capsid built from three proteins, VP1, VP2, and VP3 in a respective ratio of 1:1:10 (Caspar and Klug, 1962; Xie et al., 2002). Common to all subunits is the 533 residue component, corresponding to VP3, which was seen in the crystal structure of AAV-2, determined to 3Å (Xie et al., 2002). It was infectious particles of AAV-2 that were crystallized, containing the usual complement of VP1 and VP2, but the unique parts of these minor capsid proteins were not observed in the structure which was averaged according to icosahedral symmetry (Xie et al., 2004; Xie et al., 2003). Relative to VP3, VP2 and VP1 are extended by 65 and 202 residues respectively at the N-terminus. The unique part of VP1, “VP1u” carries a phospholipase domain that is important in transport of the viral genome from endosomes and/or into the nucleus (Girod et al., 2002). Two conventions for residue numbering are well-established in the literature, one counting from the N-terminus of VP2, the other from VP1 and differing by 137 (for AAV-2). Here, to minimize ambiguity, both will be provided in the form VP2/VP3.
The common part of the VP1/2/3 subunit has a jelly-roll β-barrel motif with long-loop insertions between the core β-barell strands. These intra-strand loops compose 60% of the structure and most of the distinctive surface topology. Loops from neighboring subunits inter-digitate to form three-fold related peaks as well as the putative receptor binding site found along the shoulder of each peak. These loops have a lower sequence identity (<15%) amongst different parvovirus genera and probably determine to a large extent tissue tropism (Chapman and Rossmann, 1993). AAV-2's infection pathway is common to many viruses.
On entering many types of cells, AAV-2 first binds a low affinity attachment (primary) receptor, heparan sulfate proteoglycan (HSPG), to localize on the cell surface and then requires the presence of a co-receptor for endocystosis and infection (Summerford and Samulski, 1998). Four co-receptors have been proposed to date for AAV-2 and its close relatives: fibroblast growth factor receptor (FGFR), hepatocyte growth factor receptor (HGFR), the 36/67kDa Laminin receptor and integrins αVβ1 and αVβ5, though the latter has been questioned (Akache et al., 2006; Asokan et al., 2006; Blackburn, Steadman, and Johnson, 2005; Kashiwakura et al., 2005; Qing et al., 1999; Qiu and Brown, 1999; Smith, Collaco, and Trempe, 2004; Summerford, Bartlett, and Samulski, 1999). There are alternatives to AAV-2's HPSG-mediated cell entry, as revealed through tissue-dependent effects of mutation at heparin-binding motifs, poor correlation of infectivity with HSPG expression, and transduction by AAV vectors of low HSPG cells through CD9-mediated entry (Kern et al., 2003; Kurzeder et al., 2007; Qiu et al., 2000). Other serotypes, AAV-1, -4, -5, and -6, bind to sialic acid with different specificities for its various forms (Kaludov et al., 2001; Walters et al., 2001; Wu et al., 2006b). Thus, while cell entry is more complicated than once presumed, the consensus is that HSPG-binding plays an important, often dominant, role for at least AAV-2 & AAV-3 (Kurzeder et al., 2007).
HSPG is found on the surface of many cells. It is comprised of an integral membrane protein with an attached heparan sulfate (HS). HS, like heparin, is a linear polysaccharide glucuronic/iduronic acid and glucosamine residues. The N- and O-linked sulfates of such heparinoids impart a negative charge which is implicated in their biological interactions with growth factors, chemokines, proteases, and pathogens (Conrad, 1998). HS differs from heparin in higher diversity of disasaccharides, lower levels of sulfonation and greater ubiquity, but heparin which is more specific to mast cells is a close analog that is prepared for pharmaceutical use and is commonly used as a structural and functional analog (Conrad, 1998). Examples of viruses that use HS for cellular attachment include Foot and Mouth disease virus (FMDV), Herpes Simples Virus (HSV-1), human immunodeficiency virus (HIV-1), repiratory syncytial virus, Dengue virus, and several alphaviruses (Capila and Linhardt, 2002; Chen et al., 1997; Conrad, 1998; Jackson et al., 1996; Krusat and Streckert, 1997; Patel et al., 1993; WuDunn and Spear, 1989; Zhang et al., 2005). Atomic structures of Heparin-protein complexes have shown that interactions are often dominated by multiple ionic pairs between arginines and heparin's sulfate groups while polar interactions with sugar hydroxyls are also common (Carfi et al., 2001; Dementiev et al., 2004; DiGabriele et al., 1998; Faham et al., 1996; Lietha et al., 2001; Mulloy and Linhardt, 2001). Beyond this, there is little conservation of a binding motif between different HS-binding proteins so recognition of binding sequences is difficult (Hileman et al., 1998). In the report of the crystal structure of FMDV subtype 01, complexed with heparin, it was suggested that a single heparin fragment could bridge between multiple receptor sites on the symmetrical virus capsid (Fry et al., 1999). It was proposed that weak binding interactions of individual sites could be combined to achieve strong binding in sum.
The crystal structure of AAV-2, and calculation of the surface electrostatic potential suggested that the HS binding site might be a patch of strong positive charge on the shoulder of surface protrusions where 4 arginines and a lysine come together (Xie et al., 2002). Subsequently, three independent mutagenesis studies tested this postulate, and all concluded that basic residues on the capsid surface were crucial for HSPG binding and cell infection (Kern et al., 2003; Lochrie et al., 2006; Opie et al., 2003). There is consensus that a cluster of positively charged residues on the side of each threefold peak, specifically R448/585, R451/588 and R350/487 are involved in heparin binding. Unresolved issues include the lack of conservation of, R451/588 and R350/487 among heparin-binding serotypes, and differing experimental characterizations of mutations at more conserved sites, including R347/484, R350/487 and K395/532.
Reported here are cryo-EM reconstructions of AAV-2 with and without a 17kDa receptor analog fragment at resolutions of 8.3Å and 7.8Å respectively. Difference map analysis shows the strongest peak as an elongated ellipsoidal shape laying tangential to capsid surface directly over the basic residues R448/585 and R451/588 and extending toward R347/484, R350/487 and K395/532, in fact enclosing a footprint much larger than previously appreciated. At lower contour levels, difference density extends between 3-fold and also 2-fold related binding sites, indicating a single polysaccharide receptor bridges between multiple sites which together presumably achieve tighter attachment.
Materials and Methods
Virus was propagated in HeLa cells, and purified by cesium chloride density gradient ultracentrifugation as previously described (Xie et al., 2004). Virus was dialyzed out of the 3M CsCl salt and into a low salt buffer (125mM NaCl, 10mM Tris, and 1mM MgCl2 pH 7.5) using 50μl custom-built microdialysis cells. Quantifoil R2/1 grids were used to prepare vitrified, thin layer samples. The grids were pre-coated with carbon, but an additional continuous carbon layer was applied to enhance contrast for particle identification and strengthen the Thon rings of the power spectrum used to determine the appropriate contrast transfer function (CTF) correction. The grids were glow discharged immediately prior to sample application. For the native sample, 4μl of virus solution at 1.1 mg/ml was applied to grids, blotted to remove excess, and plunged into liquid ethane for vitrification. Grids were transferred to liquid nitrogen for long term storage. Attempts to pre-incubate virus with heparin for as little as an hour, resulted in severe aggregation, even with10-fold molar excess of heparin relative to capsid protein (600-fold excess relative to virus particles). The aggregates, containing many virus particles cross-linked by heparin, were not suitable for the preparation of thin specimens needed for high resolution EM. Thus, samples were prepared using dilute AAV-2 that had been pre-adsorbed onto EM grids, such that heparin polymers were complexed with individual viruses that were of fixed location. Note that the procedure is different from the chemical fixation used in other types of microscopy, in that the adsorption involves weak, non-specific interactions that are neither expected to induce conformational distortions or to inhibit changes induced by ligands. However, the procedure will reduce slightly the occupancy of the receptor, because a handful of the 60 symmetry-equivalent binding sites will be occluded where the virus contacts the EM grid. Heparin was purchased from Sigma Aldrich, Inc. (sodium salt, ∼17kDa fragment, catalog # H3393) and solubilized in distilled water at 10mg/ml. 4μl of virus at 1.1 mg/ml was added to the grid, as before. After blotting off excess, 4μl of receptor solution at 2.5 mg/ml was added to achieve a 10-fold molar excess relative to the capsid protein that had been applied. Thirty seconds later, the grids were again blotted and flash frozen. The grids were transferred to a pre-cooled Gatan cryotransfer holder and inserted into a Philips CM-300 FEG electron microscope. Images were collected under low dose conditions (20e−/Å2) at 300kV onto a 4K × 4K CCD camera at a magnification of 6.1 × 104, which corresponds to an effective pixel size of 2.45 Å and a Nyquist sampling limit of 4.9Å.
All 2D and 3D processing were performed with the EMAN software suite (Ludtke, Baldwin, and Chiu, 1999). CTFIT was used to assess image quality. For this, each 4k × 4k pixel image was sampled with 512 × 512 pixel sized boxes for power spectrum analysis. Images with signs of drift or astigmatism were excluded. Particles within a defocus range of 0.8 to 3.8 μm were selected automatically with the AUTOBOX option in the BOXER program, and manually checked for the presence of ice crystals, broken particles, or debris.
The refinement of parameters describing the orientation of virus particles projected in the 2-D EM images (projection alignment) requires a starting 3-D map. Convergence might have been quicker had the starting map been simulated from the known crystal structure. However, to avoid bias, the initial map for was built de novo from the EM images, assuming only that they contained icosahedral symmetry. In the STARTICOS program of EMAN, the data are searched for particles with approximate 5-, 3-, or 2-fold symmetry (in projection), indicating that the symmetry axis in the 3-D particles was approximately parallel to the projection direction. Particles exhibiting each of the symmetries were aligned and grouped into class averages. A 3-D map can then be constructed using the known angular relations of the icosahedral symmetry axes. Initial reconstructions were performed with low pass filtered (20 Å) images and refined until features of the virus began to emerge. To increase the resolution, data were corrected for the CTF and the refinement was continued. Particles were clustered into 179 classes, each corresponding to a different projection direction within the asymmetric unit. Each cycle of 3-D particle reconstruction involved 3 rounds of iterative 2-D class averaging. Individual particle images were excluded if their correlation differed by more than 0.8 standard errors from the mean. 68% and 70% of the total native and complex particles respectively were included in the final refinement. To further remove reference bias and to be sure that features in the map of the complex were real, the final 3-D map was reconstructed using 2-D projection images whose orientations were refined with reference to the native map. The resolution was assessed by Fourier Shell Correlation between even and odd maps with a cutoff of 0.5.
Complex minus native difference maps were calculated using the “DR DIFF” procedure of the SPIDER program (Frank et al., 1996). In this procedure the maps are scaled to least-squares minimize their discrepancy within an optionally masked region. Prior to scaling, each map was low-pass filtered to the same 8.3 Å resolution as the complexed particles. For scaling complex to native, the mask was a spherical shell between 79 and 125 Å, thereby including most of the capsid protein, but excluding the primary heparin site which began about 127 Å from the center (and the 3-fold capsid spikes). For difference map calculation, all maps were brought to a common scale – that of the low-pass filtered native map, so that a single estimate of the experimental noise could be used.
The significance of features in the maps, relative to noise, was assessed though analysis of a “mock” difference map, calculated between the native odd and even image half-data sets. Each data set was low-pass filtered to 8.3 Å, processed as described above and scaled to the corresponding full-data native map before calculating the differences as a measure of error. The histogram of voxels between 79 and 160 Å from the virus center (including all of the capsid protein) yielded a standard deviation that should correspond to the random error of difference maps and approximately sqrt(2) times the error of separate native and complex maps. In the discussion below, contours will be described in estimated error units (e.u.) rather than standard deviation or sigma. The latter are often cited in this context, but usually refer to the variance of the density which is a measure of both real signal in a map and noise. Note also that the noise is estimated only from regions containing virus, and is therefore not lowered by inclusion of solvent regions.
Density was compared to the atomic structure of AAV-2 (RCSB entry 1LP3) (Xie et al., 2002) using the molecular graphics program CHIMERA (Pettersen et al., 2004), after expanding the icosahedral symmetry. The electrostatic surface potential was calculated by the Poisson-Boltzmann equation using the program APBS (Baker et al., 2001), with the structure of the entire capsid, and assuming a saline like solvent at 0.15M salt.
Results & Discussion
Quality of the electron microscopic reconstructions
AAV-2 complexed with a 17kDa receptor analog fragment, together with a native control, has been visualized though cryo-EM reconstruction (Figure 1A and C) at resolutions of 8.3Å and 7.8Å respectively (Figure 2) that are among the highest that have been obtained for any parvovirus (Kaufmann et al., 2008). The reconstruction of the native virus was built from 7491 particle images.
Figure 1.
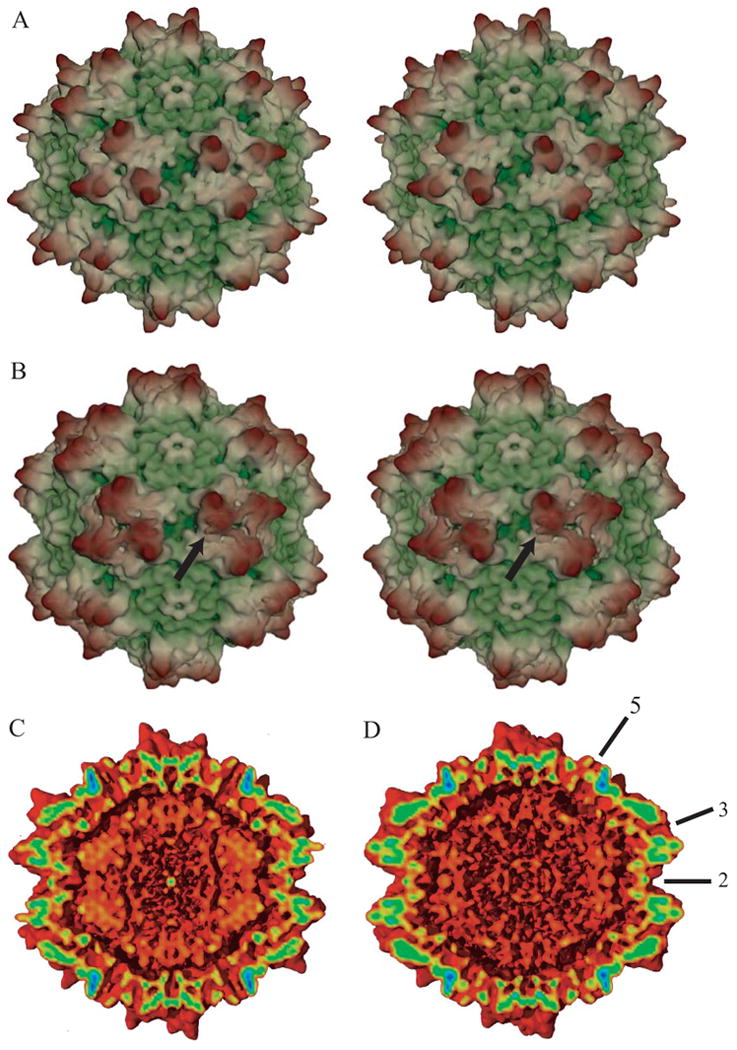
3-D electron microscope reconstructions of native (A & C) and heparin-complexed AAV-2 (B & D) at 7.8 and 8.3 Ǻ resolution respectively. Panels A & B show stereo pairs of the outer surface, while C & D show cut-aways to reveal the inner surface, all viewed in the same orientation down a 2-fold axis of symmetry. Maps were spherically masked (at 80 & 160 Ǻ) for clarity. (Note that while the particles imaged contained DNA, generally it was not seen above noise levels at these resolutions, because its structure was disordered.) In panels A & B, maps are colored according to distance from the center (from green to yellow to brown). The arrow in panel B points to the general vicinity of the bound receptor. At this contour level (4.4 e.u.), relative to the native particle in panel A, density is added to the lower surface of the 3-fold peak, and connecting density can be seen extending to a neighboring peak. In panels C & D, coloring is by strength of the density, from weakest (2.5 e.u., red) to strongest (25 e.u., blue). Some of the icosahedral symmetry axes are annotated in panel D. The differences between the complexed (D) and native (C) are subtle, and will be best visualized in difference maps in later figures. However, the map of the complex has extra density between the three-fold promixal surface peaks. The point to be emphasized from panels C & D are that the addition of the heparin makes little difference to the capsid protein structure.
Figure 2. Assessment of map quality.
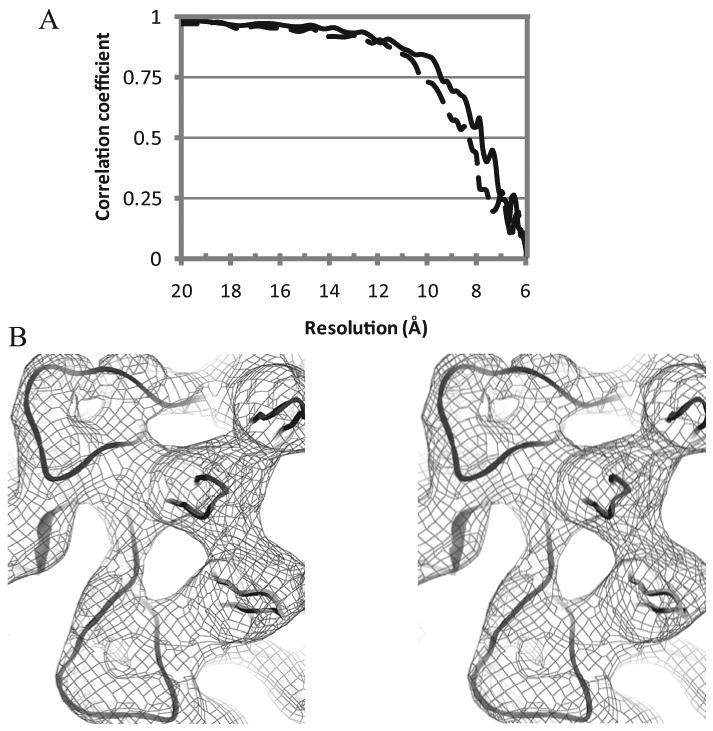
(A) Fourier shell correlation (FSC) comparing the signal from two halves of the image set as a function of resolution. The correlation for the virus alone is shown in solid line and that of the heparin complex dashed. Resolutions are estimated from the points at which the correlation coefficients drop below 0.5. (B) Example region of the 7.8 Ǻ resolution native reconstruction rendered at ∼8 e.u and overlaid on the crystallographic structure. The view is inwards, parallel to a 5-fold axis, around which 3 copies of the HI loop can be seen on the right, with two copies of the EF loop (top left & bottom). The reconstruction allows features of the virus to be resolved at the level of surface loops.
The agreement between the EM reconstruction of the native virus and the crystallographic structure is excellent, even though perfect agreement should not be expected at 8 Å resolution. All surface features characteristic of AAV-2 are apparent, including conspicuous 3-fold related peaks, a 2-fold dimple and an empty 5-fold channel. The overall consistency is remarkably good, and improved relative to the earlier EM visualizations of AAV-2 particles at ca. 10Ǻ resolution (Kronenberg et al., 2005). Several hairpin loops, comprised of just two antiparallel backbone chains are clearly visible on the surface, such as the EF loop between β-strands E & F, five symmetry-equivalent copies of which form distinctive outer surface features surrounding the 5-fold axis (Figure 2).
Interpretation was restricted to map features exceeding 3 or 4 error units. This threshold appears to be robust from several perspectives: (1) The mock difference map (odd – even images) gives highest peaks in general positions of ±3 e.u. (noting that symmetry axes can have higher noise). (2) The native EM map, contoured at 3.8 e.u. gives good correspondence to the crystallographic structure – at 4.4 e.u. some parts are outside the density, while at 1.8 e.u. all is enclosed within a possibly over-generous envelope. (3) If a single 17 kDa fragment were bound at each of 60 symmetrical sites, the appropriate volume would be enclosed by an isocontour at 2.5 e.u., 4.5 e.u. if each heparin bridges between two binding sites. Thus, three different criteria suggest that the transition from noise to real features occurs between +2.5 and +4.5 e.u..
The reconstruction of the HS-complex was built from 5950 individual virion images (Figure 1B and D). Overall, there is a good agreement with the native reconstruction. Extra density in the map of the complex covers the entire threefold area. The differences between heparin-complex and native are most apparent in the difference map which is interpreted below, but all of the significant features could also be seen in the map of the complex (Figure 1).
Visualization of the bound heparin receptor analog
In the heparin-native difference map, the strongest density at +6.9 e.u. is located just off the viral surface near a concentration of 6 positively charged residues. The closest is R448/585, about 5 Å away from the highest density (Table 1 and Figures 3 and 4). Strong density > +6 e.u. wraps around neighboring side chains, R448/585 & R451/588 (Table 1), which are on the inner shoulder of each peak surrounding the 3-fold axes. Density at +5.2 e.u extends from each of the shoulders, connecting over and around the 3-fold axes, bridging the 28 Ǻ wide valley between 3-fold-related peak shoulders. If the contour level is dropped to +3 e.u., the entire valley between 3-fold peaks is filled in, from the bottom (120 Å from the virus center) on the 3-fold to a surface at ∼150 Å from the virus center. The entire 3-fold side of each of the peaks is covered, except for the most exposed residues, P314/451 – T319/456, that protrude ∼10 Å above the heparin at the distal end of the peak, ∼160 Å from the virus center. The three peaks surrounding each 3-fold axis thereby become interconnected with heparin density. Connections between different 3-fold regions are also present, extending over the 2-fold axes, but at a lower density level (+2.5 e.u.) where the significance is borderline.
Table 1.
Basic amino acids proximal to the highest sub-peaks in the heparin-native difference map. Coordinates are given in a 222a reference frame in which icosahedral 2-folds are aligned with the principal axes. Chain “G” is a 3-fold equivalent of protomer “A”. Distances are measured to the point of maximal density, and do not account for the size of a bound heparin molecule. If the contour level were dropped to 3 e.u., all tabulated residues would contact the difference density envelope that would encompass all three sub-peaks. Distances are given in parentheses if access to the density peak would be occluded by another side chain.
| Peak height | Position | Distances (Å) | ||||||
|---|---|---|---|---|---|---|---|---|
| Chain A | Chain G | |||||||
| R347/484 Nη |
R350/487 Nη |
K390/527 Nζ |
K395/532 Nζ |
R448/585 Nη |
R451/588 Nη |
|||
| 6.9 e.u. | (2.5, 25.0, 132.8) | 13.4 | 7.3 | 8.9 | 8.8 | 4.7 | 6.9 | |
| 6.3 e.u. | (2.3, 34.8, 135.0) | (13.4) | (13.2) | (15.6) | (17.9) | 4.5 | 4.3 | |
| 6.2 e.u. | (-4.6, 37.6, 133.2) | 13.2 | (14.7) | (18.5) | (19.5) | 7.5 | 6.9 | |
| Aligned resides in serotypes: | 1/3/4/5/ 6/7/8/9 |
RRKR RRRR |
RRGG RRRR |
KKGK KKKK |
KKSN KRRR |
SSSS SAQS |
TTNT TTTA |
|
Figure 3. Differences between cryo-EM reconstructions of the heparin-complex and native AAV-2.
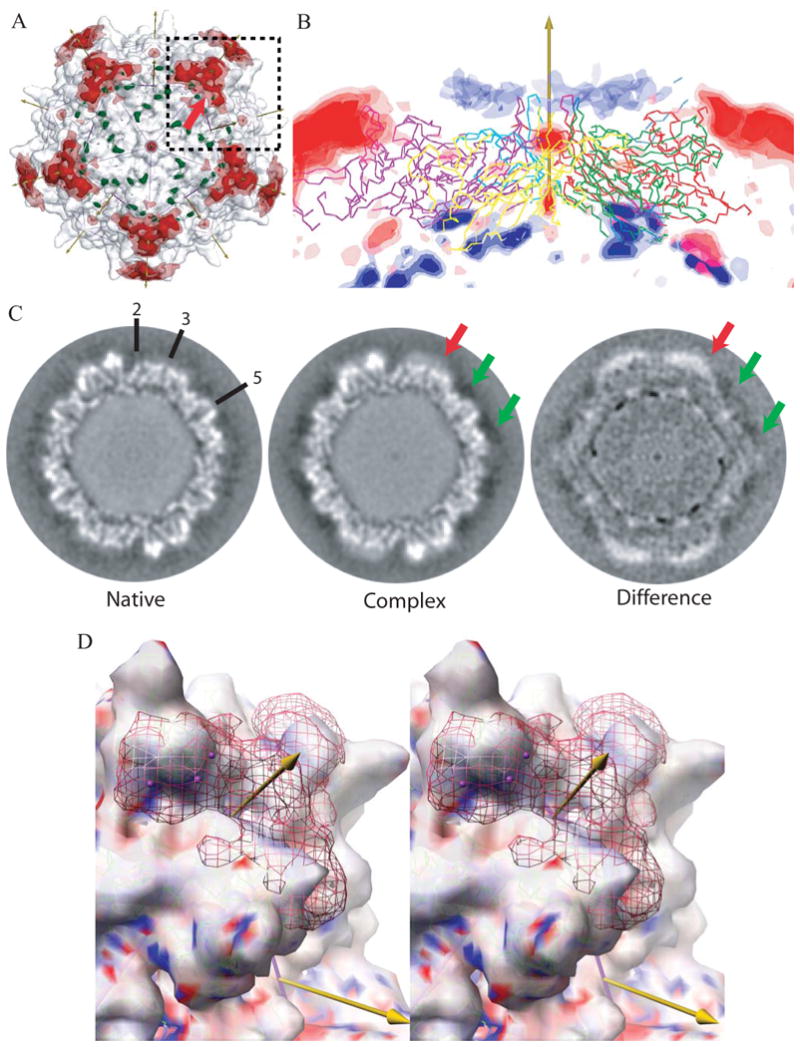
(A) Superimposed on the EM reconstruction of the native particle (gray translucent surface) are the positive (red) and negative (green) regions of the difference map. The icosahedral symmetry axes are shown with arrows (the view is down a 5-fold) and surface is divided into 60 equivalent triangular areas. The strongest positive difference densities surrounds each 3-fold axes, rising to 6.9 e.u.. Contouring of the difference density at ±4 e.u. (translucent) and ±5 e.u. (solid surface) shows that these peaks are the most massive.
(B) The difference density is shown in a cross-section with a 5-fold axis vertical. Five symmetry-related capsid proteins are shown in Cα trace. Positive density is red, negative is blue, both contoured at ±3, ±4, and ±5 e.u. The strongest negative peaks (∼-6.0 e.u.) are on the inner capsid surface, 83Å from the virus center at the boundary between protein and DNA. The next strongest negative density forms a ring of peaks at -3 to -4 e.u. surrounding each 5-fold axis. These are comparable with the experimental noise level of ∼3 e.u. (see text) and lie 11 Ǻ from the viral surface. The strong positive density at the top left and right are the 3-fold proximal heparin peaks described in panel A. The next strongest positive peaks are on the 5-fold axes of symmetry where noise levels are expected to be 2.2 × that of general positions, so these peaks are not experimentally significant.
(C) Sections through the cryo-EM reconstructions ∼2.45 Å from a plane containing multiple 2-, 3- and 5-fold axes, some of which are annotated. The grey-scale runs from dark (negative density) to white (positive). The green arrows point to the region where negative peaks (-3 to -4 e.u.) surround the 5-fold axis. At this location, 11 Ǻ from the viral surface, the density in both native and complex is negative, suggestive of experimental error or solvent differences. By contrast, the stronger density, highlighted with a red arrow in panels A & C, is clearly more positive than that in the native and is the site where heparin binds.
(D) Stereoscopic image of the most prominent difference peak. The view is similar to the boxed region in panel A, with a 3-fold axis rotated ∼17° clockwise from vertical and ∼32° forwards. The surface shows the cryo-EM reconstruction of the native virus contoured at 3.1 e.u. and colored according to the electrostatic potential of the underlying crystallographic structure (ball-&-stick, green carbons). The red net shows the EM difference density contoured at +4.7 e.u., with the strongest regions (∼6.5 e.u.) highlighted using magenta spheres. The density fills most of the valley between spikes from the bottom of the valley at 120Å to a “height” from the viral center of ∼150Å, leaving the most distal residues of the viral surface exposed. Note that the strongest density of the heparin (which carries negatively charged sulfates) lies above a part of the valley that is strongly positively charged (blue).
Figure 4. Molecular interpretation of EM results (stereoscopic images).
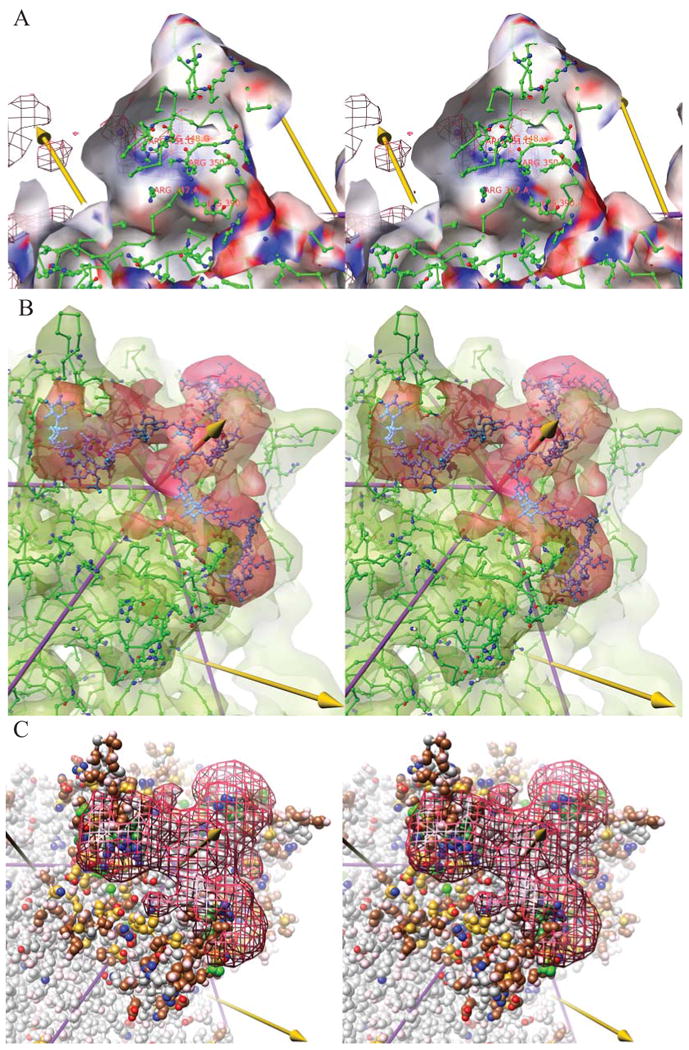
(A) The interactions of the viral protein with the heparin are best illustrated by looking outwards from inside one of the peaks surrounding a three-fold (left arrow). In this stereodiagram, the cut-away translucent surface is the experimental cryo-EM reconstruction of native, AAV-2 contoured at 3.1 e.u. and colored according to the electrostatic potential from red (most negative) to blue (most positive). The potential was calculated from the crystallographic structure (ball-&-stick). Behind the translucent surface can be seen a chicken-wire representation of the difference cryo-EM density for the heparin, contoured at a high level (5.7 e.u.) and with its strongest regions highlighted by magenta spheres. The heparin density coincides with a positively charged surface (blue strip running lower right to upper left) that is formed by the positively charged amine and guanidinium groups (blue nitrogens) of Lys390/527 and Arg350/487 from subunit A, with Arg448/585 and Arg451/588 from neighboring subunit G which together with Arg347/484 of subunit A, also in the vicinity, have been implicated in heparin-binding through structure-inspired mutagenesis (Kern et al., 2003; Opie et al., 2003; Xie et al., 2002).
(B) The difference density, contoured in red at 4.9 e.u. is commensurate in size with 3 × 9-mer saccharides, modeled in cyan ball-&-stick, surrounding a three-fold axis (central arrow). (Heparin is a heterogenous mixture – the model is not intended to represent accurately any one of the conformations averaged in the reconstruction, but only its dimensions.) The green translucent surface shows the cryo-EM reconstruction of native AAV-2 at 7.8 Ǻ resolution, contoured at 6.7 e.u. overlaid on the 3 Ǻ crystal structure(Xie et al., 2002).
(C) The heparin density, contoured at 4.7 e.u., is overlaid on the atomic structure which is colored according to atomic properties and to highlight amino acids subject to relevant mutational studies. Side chains are colored with positively changed atoms blue, negatively charged atoms red and uncharged polar atoms pink. Carbons are colored by mutation phenotype, combining the results of three studies (Kern et al., 2003; Lochrie et al., 2006; Opie et al., 2003). In green are colored residues affecting heparin interactions in any of the studies, darkly shaded for the strongest effects. Residues that did not appear to affect heparin binding are colored in brown, or in gold for those that had at least 10-fold impact upon transduction levels or infectivity. The heparin footprint has at its center the three residues with strongest impact (R448/585, R451/588 and R350/487 from), but it extends to include other charged residues for which consistent mutagenesis results have not been obtained. The footprint contains many residues (brown and gold) whose interactions were not detected previously. The gold-colored residues were previously hypothesized to affect transduction through a secondary receptor binding site proposed to be distinct from the heparin binding site, but several are within the heparin footprint.
A crude estimate of the heparin occupancy was obtained by comparing the highest peaks in the complex map in the heparin and capsid protein regions. The heparin density is about 1/3 of the height, providing an estimate of the heparin occupancy that is a lower bound to the degree that the heparin is more disordered and of weaker density than the capsid protein. The fragment size used in this study is ∼17kDa or ∼70 monosaccharide units. It was larger than the oligosaccharides used in prior preliminary experiments, and not only easier to visualize by EM, but it better represents physiological interactions. However, with a long polymer, the heparin likely exists as a population of many possible configurations. Its density will be smeared where different polysaccharide conformations are averaged by the icosahedral symmetry applied in 3-D reconstruction.
How long is the heparin polymer? Extrapolating from the structure of the fragment bound in the crystallographic study of FMDV, the 17 kDa fragment used here could be as long as ∼285 Å. In an extended configuration this corresponds to 1/3 of the virus' circumferance. At this length, a single heparin could bridge between multiple trimers. There may be many ways of winding between three-fold peaks and connecting between adjacent capsid binding sites. 60-fold averaging over the different configurations will lead to diffuse heparin density. Furthermore, some paths of the polymer will be mutually exclusive. Thus, once two neighboring sites have been connected directly over the 3-fold, the same binding configuration is not available for the third equivalent site. In summary, an occupancy of less than one represents a combination of steric exclusion, the averaging of statically disordered conformations and dynamic flexibility of the polymer chain, all of which lower the heparin density and make it more diffuse.
Each of the 3-fold symmetric parts of the heparin density can be modeled with a nonasasaccharide. The sample heterogeneity and structural disorder discussed above, together with the modest resolution mean that the density lacks the distinctive features required to discern polysaccharide sequence, chain direction or exact conformation. Thus, only a crude model was warranted, built by connecting rigidly fit monosaccharide units from RSCB entry 1SR5 using the program Coot (Emsley and Cowtan, 2004). Two of the three symmetry equivalent 9-mers can be connected through each 3-fold (Figure 4B), so the density may represent the average of possible paths of 18 of the ∼70 monosaccharide units in each sample molecule. Such a configuration allows the observable part of each heparin to wind through the strongest difference density, that is on the positively charged sides of two 3-fold proximal peaks (Figure 3D & Figure 4A). It is the regions where the heparin is mostly tightly bound that give the strongest density. The rest of the heparin is too disordered to visualize, but there are hints with density at about noise level, that the chains can also bridge between the 3-fold regions over the 2-fold axes.
Search for possible receptor-induced conformational changes
The difference map has been examined systematically for evidence of heparin-induced conformational change in the capsid (Figure 3). Protein conformational change should be apparent with adjacent negative and positive density in the difference map indicating the unbound and bound configurations. The most negative difference density is on the inside surface of the capsid where there are several peaks of ∼-6 e.u. all ∼83 Å from the virus center. Both complex and native maps have strong negative peaks, up to ∼ -20 e.u. that are about 2/3 of the strength of the maximal capsid protein positive density. The spherical shape of capsids can lead to superposition of truncation ripples and to larger-than-usual series termination effects on the inside of the capsid. These peaks correspond to modest differences between densities that are strongly negative in both complex and native, and therefore do not represent conformational change. The only other negative feature (-4.0 e.u.) in the difference map commensurate with the estimated noise is also in a location that is negative in both complex and native (Figure 3C). It also sits 11 Å off the surface beyond molecular rationalization.
The positive peaks in the difference map can similarly be analyzed. After the peak near residues R448/585 & R451/588, there is difference density on the 5-fold that is almost as strong, ∼108 Å from the virus center. The filling of the 5-fold pore, 21 Å wide at this point would be intriguing, but the expected noise on a 5-fold axis is sqrt(5)-fold higher, so this feature is not experimentally significant. The next strongest peak (3.5 e.u.) is at the noise level in a surface pocket between K528/665 and C224/361. The map of the complex covers the structure more generously than that of the native, resulting in a small peak in the center of the pocket that is not experimentally significant.
What should not be lost in this detailed discussion is that there is no evidence for large scale receptor-induced conformational changes. It is only the density near the 3-fold that rises clearly above the noise. There are several other peaks at about the noise level, but there are no paired regions of positive and negative density that would be indicative of conformational change. Furthermore, there is no connection of these peaks to the site of heparin attachment near the 3-fold.
Even though large conformational changes are not observed, there may be side chain rearrangements and small heparin-induced backbone adjustments which would only be seen at resolutions better than 8 Å. It is also possible that subtle changes are masked by the partial heparin occupancy leading to a mixture of states in the complex. It is also possible that conformational change could depend upon asymmetry between a bound site and unoccupied neighbors, which is not seen in the current multi-ligand complex, but, to the authors, this seems implausible in the absence of any observed conformational changes. In summary, with several caveats, this study cannot exclude the possibility that there are subtle changes of small magnitude, but it does rule out large receptor-induced conformational changes that would have been apparent in maps of such high quality.
Capsid-receptor interactions
Intriguingly, the contact residues for the most tightly bound part of the heparin are R448/ R585, R451/ 588 which are not conserved among AAV serotypes, even those that are closely related, such as AAV-3 & AAV-6, human serotypes that are known to bind heparin and are 89% and 85% identical in capsid sequence respectively (Negishi et al., 2004; Rutledge, Halbert, and Russell, 1998; Wu et al., 2006a) (Table 1). In the other serotypes, they are replaced, for the most part, with polar residues, Ser, Thr, Gln or Asn that are also commonly part of heparin-binding regions (Conrad, 1998), but unlikely to form associations that are as strong. The non-conserved R448/ R585, R451/ 588 form part of a contiguous positively charged surface with R347/484, R350/487, and are close to K395/532 (Xie et al., 2002) (Figure 4A), but there has not been consensus in prior studies of the importance of these residues. There are many plausible reasons for apparent discrepancies including possible differences between cell entry via heparan sulfate attachment and physical measures of the binding to the analog, heparin.
Much can be learned by integrating the current complex structure with three prior studies in which the effects of mutating many of these residues upon heparin binding were assayed (Kern et al., 2003; Lochrie et al., 2006; Opie et al., 2003). R448/585 and R451/588, which had strong impact in all three studies, are in the center of the observed heparin-binding site. R350/487 and R347/484 were found to be important in the study of Kern et al. (2003), but less so in the studies of Opie et al. (2003) and Lochrie et al. (2006). The current EM structure shows that R350/487 makes contact with the heparin density just below R448/585 while R347/484 is only slightly further removed, and still in the first shell of heparin neighbors to the side of R451/588. There has been greater discrepancy in the mutational data for K395/532, and K390/527 leading to different conclusions about their importance. The EM structure shows that both are in contact with the edge of the heparin, supporting the earlier, not later, mutational data. On the other hand, R347/484, studied by Kern et al. (2003) is buried four amino acids below, indicating that any effect upon heparin-binding would have to be collateral. The G375/512P mutant studied by Lochrie et al. (2006) is slightly removed from the heparin, but sits immediately below K390/527, so a collateral effect upon heparin-binding is plausible. The measured effects upon heparin-binding of several multi-site mutants (Lochrie et al., 2006) is more difficult to rationalize unless there are more substantial collateral effects through which buried residues have impact, or unless the modest changes in binding are within experimental error.
In summary, consensus emerged from mutagenesis studies that R448/585, R451/588 and R347/484 are critical in AAV-2/HS interactions. There has been little agreement on three other basic residues in the area: R350/487, K395/532 and K390/527 whose contact may be less intimate, but, as now seen, also fall within the binding footprint. The current EM structure shows, for the first time, the full extent of the footprint. The structure arbiters the inconsistencies between the mutagenesis studies, and suggests that some of the assays reveal the strongest interactions, while others remain undetected. This is relevant to the following point.
The mutagenesis data have also been used to infer co-receptor interactions in cases where mutation affects infectivity or transduction, but not heparin binding (Lochrie et al., 2006). Details of the interpretation need revision in light of the structure reported here. The heparin binding footprint is much larger than implicated by Lochrie et al., comprising a contiguous area of 36 amino acids (Table 2). This footprint is consistent with the prior studies in that it encloses all surface residues positively implicated in at least one of the mutagenesis studies. Within this area, however, are many additional residues whose interactions with heparin were not detected by the prior methods. Of the 36 amino acids in the footprint, 19 have previously been mutated with 6 having detectable impact in at least one of the studies. Those successfully identified were core interactions with positively charged residues. Remaining undetected were contributions from neighboring polar side chains that are also expected to contribute (Conrad, 1998). The apparent lack of heparin interaction combined with impact upon transduction led one study to conclude that a “dead zone” region on the plateau where such residues were concentrated was the likely site of co-receptor binding (Lochrie et al., 2006). However, the footprint revealed in the current work includes 10 of the 17 amino acids assigned to this site. This does not eliminate the possibility that the plateau is involved with co-receptor binding, but it undercuts a principal argument. The issue remains open pending more definitive evidence. Given the laboratory-dependent variation in assaying several mutants, it is reasonable to question whether some heparin interactions have been harder to detect than effects upon transduction. Alternatively, it is possible that some mutations within the footprint might have an indirect effect upon transduction that is greater than a direct affect upon heparin binding, if HSPG still binds, but in a conformation that is not so favorable for ternary interactions between virus, HSPG and co-receptor needed for cell entry. In fact, this is only one of many possible explanations, but speculation should be limited until it is understood whether the variation in reported mutant phenotypes reflects something more significant than experimental errors and sensitivity.
Table 2.
Amino acids at the AAV-2 heparin interface. Direct contacts with the density (contoured at 4.7 e.u.) are made by 23 amino acids. Another 13 have side chains with unobstructed access to the density through a short gap that might be filled with solvent or disordered parts of the heparin.
| Direct | Neighbor | Direct | Neighbor | Direct | Neighbor |
|---|---|---|---|---|---|
| Q320/457 | K390/527 | E437/574 | Q438/575 | ||
| R347/484 | D391/528 | T444/581 | |||
| R350/487 | D392/529 | N445/582 | |||
| A356/493 | E393/530 | L446/583 | |||
| D357/494 | K395/532 | R448/585 | |||
| N359/496 | F396/533 | G449/586 | |||
| N360/497 | S361/498 | N450/587 | |||
| T366/503 | W365/502 | R451/588 | |||
| K370/507 | Q452/589 | ||||
| H372/509 | A454/591 | ||||
| G375/512 | T455/592 | A456/593 | |||
| D377/514 | D457/594 | ||||
| N459/596 | |||||
| K569/706 |
In the current work, more questions are re-opened than answered. However, visualization of the full footprint has taken us beyond the prior mutagenic studies, resolving some of the past discrepancies. The broad extent of the footprint has provided a foundation for understanding how serotypes such as AAV-3 might bind HSPG, albeit with different affinity, even though residues previously implicated in AAV-2/heparin interactions are not conserved. The size of the footprint highlights the potential importance of ternary virus interactions involving both primary and secondary receptors. Finally, connecting heparin density between symmetry-related binding sites indicates how higher affinity multi-valent binding can be achieved.
Acknowledgments
We wish to thank Joan Hare at the Protein expression facility for virus propagation and purification and Kim Riddle and Tom Fellers at the Biological Science Imaging resource for assistance in data collection. We gratefully acknowledge the computer resources and expertise provided by the High Performance Computing center at Florida State University. Molecular graphics images were produced using the UCSF Chimera package from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIH P41 RR-01081). This work was supported by an American Heart Association pre-doctoral fellowship (J.O'D) and the National Institutes of Health (R01 GM 66875, M.S.C.).
Abbreviations and symbols
- AAV
Adenoassociated virus
- CTF
contrast transfer function
- EM
Electron Microscopy
- e.u.
error units (calculated from the map variance between two half data sets, see text)
- FMDV
Foot and mouth disease virus
- HS
heparan sulfate
- HSPG
heparan sulfate proteoglycan
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akache B, Grimm D, Pandey K, Yant SR, Xu H, Kay MA. The 37/67-kilodalton laminin receptor is a receptor for adeno-associated virus serotypes 8, 2, 3, and 9. J Virol. 2006;80(19):9831–6. doi: 10.1128/JVI.00878-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asokan A, Hamra JB, Govindasamy L, Agbandje-McKenna M, Samulski RJ. Adeno-associated virus type 2 contains an integrin alpha5beta1 binding domain essential for viral cell entry. J Virol. 2006;80(18):8961–9. doi: 10.1128/JVI.00843-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A. 2001;98(18):10037–41. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berns K, Giraud C. Adenovirus and adeno-associated virus as vectors for gene therapy. Ann N Y Acad Sci. 1995;772:95–104. doi: 10.1111/j.1749-6632.1995.tb44735.x. [DOI] [PubMed] [Google Scholar]
- Blackburn SD, Steadman RA, Johnson FB. Attachment of adeno-associated virus type 3H to fibroblast growth factor receptor 1. Arch Virol. 2005 doi: 10.1007/s00705-005-0650-6. [DOI] [PubMed] [Google Scholar]
- Bueler H. Adeno-associated viral vectors for gene transfer and gene therapy. Biol Chem. 1999;380(6):613–22. doi: 10.1515/BC.1999.078. [DOI] [PubMed] [Google Scholar]
- Capila I, Linhardt RJ. Heparin-protein interactions. Angew Chem Int Ed Engl. 2002;41(3):391–412. doi: 10.1002/1521-3773(20020201)41:3<390::aid-anie390>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- Carfi A, Willis SH, Whitbeck JC, Krummenacher C, Cohen GH, Eisenberg RJ, Wiley DC. Herpes simplex virus glycoprotein D bound to the human receptor HveA. Mol Cell. 2001;8(1):169–79. doi: 10.1016/s1097-2765(01)00298-2. [DOI] [PubMed] [Google Scholar]
- Carter BJ. Parvoviruses as Vectors. In: Tijssen P, editor. Handbook of Parvoviruses. Vol. 2. CRC Press; Boca Raton, FL: 1990. pp. 247–84. [Google Scholar]
- Caspar DLD, Klug A. Physical principles in the constructuion of regular viruses. Cold Spring Harbor Symposium in Quantitative Biology. 1962;27:1–24. doi: 10.1101/sqb.1962.027.001.005. [DOI] [PubMed] [Google Scholar]
- Chapman MS, Rossmann MG. Structure, Sequence and Function Correlations among Parvoviruses. Virology. 1993;194:491–508. doi: 10.1006/viro.1993.1288. [DOI] [PubMed] [Google Scholar]
- Chaterjee S, Johnson PR, Wong K., Jr Dual-target inhibition of HIV-1 in vitro by means of an adeno-associated virus antisense vector. Science. 1992;258:1485–8. doi: 10.1126/science.1359646. [DOI] [PubMed] [Google Scholar]
- Chen JD, Yang Q, Yang AG, Marasco WA, Chen SY. Intra- and extracellular immunization against HIV-1 infection with lymphocytes transduced with an AAV vector expressing a human anti-gp120 antibody. Human Gene Therapy. 1996;7(13):1515–1525. doi: 10.1089/hum.1996.7.13-1515. [DOI] [PubMed] [Google Scholar]
- Chen Y, Maguire T, Hileman RE, Fromm JR, Esko JD, Linhardt RJ, Marks RM. Dengue virus infectivity depends on envelope protein binding to target cell heparan sulfate. Nat Med. 1997;3(8):866–71. doi: 10.1038/nm0897-866. [DOI] [PubMed] [Google Scholar]
- Chirmule N, Propert K, Magosin S, Qian Y, Qian R, Wilson J. Immune responses to adenovirus and adeno-associated virus in humans. Gene Ther. 1999;6(9):1574–1583. doi: 10.1038/sj.gt.3300994. [DOI] [PubMed] [Google Scholar]
- Conrad HE. Heparin-Binding Proteins. 1st. Academic Press; San Diego: 1998. [Google Scholar]
- Dementiev A, Petitou M, Herbert JM, Gettins PG. The ternary complex of antithrombin-anhydrothrombin-heparin reveals the basis of inhibitor specificity. Nat Struct Mol Biol. 2004;11(9):863–7. doi: 10.1038/nsmb810. [DOI] [PubMed] [Google Scholar]
- DiGabriele AD, Lax I, Chen DI, Svahn CM, Jaye M, Schlessinger J, Hendrickson WA. Structure of a heparin-linked biologically active dimer of fibroblast growth factor. Nature. 1998;393(6687):812–7. doi: 10.1038/31741. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Faham S, Hileman RE, Fromm JR, Linhardt RJ, Rees DC. Heparin structure and interactions with basic fibroblast growth factor. Science. 1996;271(5252):1116–20. doi: 10.1126/science.271.5252.1116. [DOI] [PubMed] [Google Scholar]
- Flotte TR, Carter BJ. Adeno-associated virus vectors for gene therapy of cystic fibrosis. Methods Enzymol. 1998;292:717–32. doi: 10.1016/s0076-6879(98)92055-9. [DOI] [PubMed] [Google Scholar]
- Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116(1):190–9. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- Fry EE, Lea SM, Jackson T, Newman JW, Ellard FM, Blakemore WE, Abu-Ghazaleh R, Samuel A, King AM, Stuart DI. The structure and function of a foot-and-mouth disease virus-oligosaccharide receptor complex. Embo J. 1999;18(3):543–54. doi: 10.1093/emboj/18.3.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girod A, Wobus CE, Zadori Z, Ried M, Leike K, Tijssen P, Kleinschmidt JA, Hallek M. The VP1 capsid protein of adeno-associated virus type 2 is carrying a phospholipase A2 domain required for virus infectivity. J Gen Virol. 2002;83(Pt 5):973–8. doi: 10.1099/0022-1317-83-5-973. [DOI] [PubMed] [Google Scholar]
- Harster A, Teichmann B, Hormes R, Grimm D, Kleinschmidt J, Sczakiel G. Recombinant AAV-2 harboring gfp-antisense/ribozyme fusion sequences monitor transduction, gene expression, and show anti-HIV-1 efficacy. Gene Ther. 1999;6(7):1231–1238. doi: 10.1038/sj.gt.3300955. [DOI] [PubMed] [Google Scholar]
- Hileman RE, Fromm JR, Weiler JM, Linhardt RJ. Glycosaminoglycan-protein interactions: definition of consensus sites in glycosaminoglycan binding proteins. Bioessays. 1998;20(2):156–67. doi: 10.1002/(SICI)1521-1878(199802)20:2<156::AID-BIES8>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Inouye R, Du B, Boldt-Houle D, Ferrante A, Park I, Hammer S, Duan L, Groopman J, Pomerantz R, Terwilliger E. Potent inhibition of human immunodeficiency virus type 1 in primary T cells and alveolar macrophage by a combination anti-Rev strategy delivered in an adeno-associated virus vector. Journal of Virology. 1997;71(5):4071–4078. doi: 10.1128/jvi.71.5.4071-4078.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson T, Ellard FM, Ghazaleh RA, Brookes SM, Blakemore WE, Corteyn AH, Stuart DI, Newman JW, King AM. Efficient infection of cells in culture by type O foot-and-mouth disease virus requires binding to cell surface heparan sulfate. J Virol. 1996;70(8):5282–7. doi: 10.1128/jvi.70.8.5282-5287.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaludov N, Brown KE, Walters RW, Zabner J, Chiorini JA. Adeno-associated virus serotype 4 (AAV4) and AAV5 both require sialic acid binding for hemagglutination and efficient transduction but differ in sialic acid linkage specificity. J Virol. 2001;75(15):6884–93. doi: 10.1128/JVI.75.15.6884-6893.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashiwakura Y, Tamayose K, Iwabuchi K, Hirai Y, Shimada T, Matsumoto K, Nakamura T, Watanabe M, Oshimi K, Daida H. Hepatocyte growth factor receptor is a coreceptor for adeno-associated virus type 2 infection. J Virol. 2005;79(1):609–14. doi: 10.1128/JVI.79.1.609-614.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann B, Chipman PR, Kostynchenko VA, Modrow S, Rossmann MG. Visualization of the externalized VP2 N-termini of Infectious Human Parvovirus B19. Journal of Virology. 2008 doi: 10.1128/JVI.00512-08. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kern A, Schmidt K, Leder C, Muller OJ, Wobus CE, Bettinger K, Von der Lieth CW, King JA, Kleinschmidt JA. Identification of a heparin-binding motif on adeno-associated virus type 2 capsids. J Virol. 2003;77(20):11072–81. doi: 10.1128/JVI.77.20.11072-11081.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronenberg S, Bottcher B, von der Lieth CW, Bleker S, Kleinschmidt JA. A conformational change in the adeno-associated virus type 2 capsid leads to the exposure of hidden VP1 N termini. J Virol. 2005;79(9):5296–303. doi: 10.1128/JVI.79.9.5296-5303.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krusat T, Streckert HJ. Heparin-dependent attachment of respiratory syncytial virus (RSV) to host cells. Arch Virol. 1997;142(6):1247–54. doi: 10.1007/s007050050156. [DOI] [PubMed] [Google Scholar]
- Kurzeder C, Koppold B, Sauer G, Pabst S, Kreienberg R, Deissler H. CD9 promotes adeno-associated virus type 2 infection of mammary carcinoma cells with low cell surface expression of heparan sulphate proteoglycans. Int J Mol Med. 2007;19(2):325–33. [PubMed] [Google Scholar]
- Laughlin CA, Tratschin JD, Coon H, Carter BJ. Cloning of infectious adeno-associated virus genomes in bacterial plasmids. Gene. 1983;23(1):65–73. doi: 10.1016/0378-1119(83)90217-2. [DOI] [PubMed] [Google Scholar]
- Lietha D, Chirgadze DY, Mulloy B, Blundell TL, Gherardi E. Crystal structures of NK1-heparin complexes reveal the basis for NK1 activity and enable engineering of potent agonists of the MET receptor. Embo J. 2001;20(20):5543–55. doi: 10.1093/emboj/20.20.5543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lochrie MA, Tatsuno GP, Christie B, McDonnell JW, Zhou S, Surosky R, Pierce GF, Colosi P. Mutations on the external surfaces of adeno-associated virus type 2 capsids that affect transduction and neutralization. J Virol. 2006;80(2):821–34. doi: 10.1128/JVI.80.2.821-834.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128(1):82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- Mandel RJ, Spratt SK, Snyder RO, Leff SE. Midbrain injection of recombinant adeno-associated virus encoding rat glial cell line-derived netrotrophic factor protects nigral neurons ina progressive 6-hydroxydopamine-induced degeneraction model of Parkinson's disease in rats. Proceedings of the National Academy of Sciences, USA. 1997;94:14083–8. doi: 10.1073/pnas.94.25.14083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki H, Hayakawa H, Migita M, Shibata M, Tanaka R, Suzuki A, Shimo-Nakanishi Y, Urabe T, Yamada M, Tamayose K, Shimada T, Miura M, Mizuno Y. An AAV-derived Apaf-1 dominant negative inhibitor prevents MPTP toxicity as antiapoptotic gene therapy for Parkinson's disease. Proc Natl Acad Sci U S A. 2001;98:10918–923. doi: 10.1073/pnas.191107398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulloy B, Linhardt RJ. Order out of complexity - protein structures that interact with heparin. Current Opinion in Structural Biology. 2001;11:623–8. doi: 10.1016/s0959-440x(00)00257-8. [DOI] [PubMed] [Google Scholar]
- Negishi A, Chen JH, McCarty DM, Samulski RJ, Liu J, Superfine R. Analysis of the interaction between adeno-associated virus and heparan sulfate using atomic force microscopy. Glycobiology. 2004;14(11):969–977. doi: 10.1093/glycob/cwh118. [DOI] [PubMed] [Google Scholar]
- Opie SR, Warrington KH, Jr, Agbandje-McKenna M, Zolotukhin S, Muzyczka N. Identification of amino acid residues in the capsid proteins of adeno-associated virus type 2 that contribute to heparan sulfate proteoglycan binding. J Virol. 2003;77(12):6995–7006. doi: 10.1128/JVI.77.12.6995-7006.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel M, Yanagishita M, Roderiquez G, Bou-Habib DC, Oravecz T, Hascall VC, Norcross MA. Cell-surface heparan sulfate proteoglycan mediates HIV-1 infection of T-cell lines. AIDS Res Hum Retroviruses. 1993;9(2):167–74. doi: 10.1089/aid.1993.9.167. [DOI] [PubMed] [Google Scholar]
- Penaud-Budloo M, Le Guiner C, Nowrouzi A, Toromanoff A, Cherel Y, Chenuaud P, Schmidt M, von Kalle C, Rolling F, Moullier P, Snyder RO. Adeno-associated virus vector genomes persist as episomal chromatin in primate muscle. J Virol. 2008;82(16):7875–85. doi: 10.1128/JVI.00649-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–12. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Phillips M. Antisense inhibition and adeno-associated viral vector delivery for reducing hypertension. Hypertension. 1997;29:177–187. doi: 10.1161/01.hyp.29.1.177. [DOI] [PubMed] [Google Scholar]
- Podsakoff G, Wong KK, Jr, Chatterjee S. Efficient gene trasfer into nondividing cells by adeno-associated virus-based vectors. Journal of Virology. 1994;68:5656–66. doi: 10.1128/jvi.68.9.5656-5666.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qing K, Mah C, Hansen J, Zhou S, Dwarki V, Srivastava A. Human fibroblast growth factor receptor 1 is a co-receptor for infection by adeno-associated virus 2. Nat Med. 1999;5(1):71–7. doi: 10.1038/4758. [DOI] [PubMed] [Google Scholar]
- Qiu J, Brown KE. Integrin alphaVbeta5 is not involved in adeno-associated virus type 2 (AAV2) infection. Virology. 1999;264(2):436–40. doi: 10.1006/viro.1999.0010. [DOI] [PubMed] [Google Scholar]
- Qiu J, Handa A, Kirby M, Brown KE. The interaction of heparin sulfate and adeno-associated virus 2. Virology. 2000;269(1):137–47. doi: 10.1006/viro.2000.0205. [DOI] [PubMed] [Google Scholar]
- Rutledge E, Halbert C, Russell D. Infectious clones and vectors derived from adeno-associated virus (AAV) serotypes other than AAV type 2. Journal of Virology. 1998;72(1):309–319. doi: 10.1128/jvi.72.1.309-319.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salo RJ, Mayor HD. Structural polypeptides of parvoviruses. Virology. 1977;78(1):340–5. doi: 10.1016/0042-6822(77)90107-6. [DOI] [PubMed] [Google Scholar]
- Salo RJ, Mayor HD. Adenovirus-associated virus polypeptides synthesized in cells coinfected with either adenovirus or herpesvirus. Virology. 1979;93(1):237–45. doi: 10.1016/0042-6822(79)90291-5. [DOI] [PubMed] [Google Scholar]
- Smith A, Collaco R, Trempe JP. AAV vector delivery to cells in culture. Methods Mol Biol. 2004;246:167–77. doi: 10.1385/1-59259-650-9:167. [DOI] [PubMed] [Google Scholar]
- Summerford C, Bartlett JS, Samulski RJ. AlphaVbeta5 integrin: a co-receptor for adeno-associated virus type 2 infection. Nat Med. 1999;5(1):78–82. doi: 10.1038/4768. [DOI] [PubMed] [Google Scholar]
- Summerford C, Samulski RJ. Membrane-associated heparan sulfate proteoglycan is a receptor for adeno-associated virus type 2 virions. J Virol. 1998;72(2):1438–45. doi: 10.1128/jvi.72.2.1438-1445.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trempe JP. Packaging systems for adeno-associated virus vectors. Current Topics in Microbiology and Immunology. 1996;218:35–50. doi: 10.1007/978-3-642-80207-2_3. [DOI] [PubMed] [Google Scholar]
- Walters RW, Yi SM, Keshavjee S, Brown KE, Welsh MJ, Chiorini JA, Zabner J. Binding of adeno-associated virus type 5 to 2,3-linked sialic acid is required for gene transfer. J Biol Chem. 2001;276(23):20610–6. doi: 10.1074/jbc.M101559200. [DOI] [PubMed] [Google Scholar]
- Wu P, Phillips MI, Bui J, Terwilliger EF. Adeno-associated virus vector-mediated transgene integration into neurons and other nondividing cell targets. J Virol. 1998;72(7):5919–26. doi: 10.1128/jvi.72.7.5919-5926.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Asokan A, Grieger JC, Govindasamy L, Agbandje-McKenna M, Samulski RJ. Single Amino Acid Changes Can Influence Titer, Heparin Binding, and Tissue Tropism in Different Adeno-Associated Virus (AAV) Serotypes. J Virol. 2006a;80(22):11393–7. doi: 10.1128/JVI.01288-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Asokan A, Samulski RJ. Adeno-associated virus serotypes: vector toolkit for human gene therapy. Mol Ther. 2006;14(3):316–27. doi: 10.1016/j.ymthe.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Wu Z, Miller E, Agbandje-McKenna M, Samulski RJ. Alpha2,3 and alpha2,6 N-linked sialic acids facilitate efficient binding and transduction by adeno-associated virus types 1 and 6. J Virol. 2006b;80(18):9093–103. doi: 10.1128/JVI.00895-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WuDunn D, Spear PG. Initial interaction of herpes simplex virus with cells is binding to heparan sulfate. J Virol. 1989;63(1):52–8. doi: 10.1128/jvi.63.1.52-58.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Q, Bu W, Bhatia S, Hare J, Somasundaram T, Azzi A, Chapman MS. The atomic structure of adeno-associated virus (AAV-2), a vector for human gene therapy. Proc Natl Acad Sci U S A. 2002;99(16):10405–10. doi: 10.1073/pnas.162250899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Q, Hare J, Turnigan J, Chapman MS. Large-scale production, purification and crystallization of wild-type adeno-associated virus-2. Journal of Virological Methods. 2004;122(1):17–27. doi: 10.1016/j.jviromet.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Xie Q, Somasundaram T, Bhatia S, Bu W, Chapman MS. Structure determination of adeno-associated virus 2: three complete virus particles per asymmetric unit. Acta Crystallogr D Biol Crystallogr. 2003;59(Pt 6):959–70. doi: 10.1107/s0907444903005675. [DOI] [PubMed] [Google Scholar]
- Zhang W, Heil M, Kuhn RJ, Baker TS. Heparin binding sites on Ross River virus revealed by electron cryo-microscopy. Virology. 2005;332(2):511–8. doi: 10.1016/j.virol.2004.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]


