Summary
Histone deacetylase 6 (HDAC6) is a tubulin-specific deacetylase that regulates microtubule-dependent cell movement. In this study, we identify the F-actin-binding protein, cortactin, as a HDAC6 substrate. We demonstrate that HDAC6 binds cortactin and that overexpression of HDAC6 leads to hypoacetylation of cortactin, while inhibition of HDAC6 activity leads to cortactin hyperacetylation. HDAC6 alters the ability of cortactin to bind F-actin by modulating a “charge patch” in its repeat region. Introduction of charge-preserving or charge-neutralizing mutations in this cortactin repeat region correlates with the gain or loss of F-actin binding ability, respectively. Cells expressing a charge-neutralizing cortactin mutant were less motile than control cells or cells expressing a charge-preserving mutant. These findings suggest that, in addition to its role in microtubule-dependent cell motility, HDAC6 influences actin-dependent cell motility by altering the acetylation status of cortactin, which, in turn, changes the F-actin binding activity of cortactin.
Introduction
Histone deacetylases (HDACs) and histone acetyltransferases (HATs) are ubiquitously expressed enzymes that primarily target core histones. The hyperacetylation of histones typically enhances gene expression, while their deacetylation usually represses gene expression. Many transcriptional co-activators are HATs and many transcriptional co-repressors are HDACs. Mammalian HDACs can be subdivided into the following three classes: class I (HDACs 1, 2, 3, and 8), class II (HDACs 4, 5, 6, 7, 9, and 10), and class III (SIRTs 1, 2, 3, 4, 5, 6, 7). HDAC11 shares homology with both class I and class II HDACs.
In addition to histones, HDACs and HATs also target non-histone proteins. Some of these non-histone targets are transcription factors such as p53, GATA-1, E2F1, YY1, and MyoD. Importantly, the reversible acetylation of these proteins modifies their activities. Other non-histone HAT and HDAC substrates include proteins that regulate cell proliferation, survival, and motility. For example, PCAF acetylates the DNA end-joining protein Ku70, leading to an attenuation of Ku70 anti-apoptotic activity. p300 acetylates the tumor suppressor Rb and prevents Rb phosphorylation by cyclin-dependent kinases and blocks cell cycle progression.
One of the most extensively studied and best characterized non-histone HDAC substrates is the cytoplasmic protein α-tubulin (Haggarty et al., 2003; Hubbert et al., 2002; Matsuyama et al., 2002; Zhang et al., 2003). HDAC6 associates with and deacetylates α-tubulin in vitro and in vivo. Cells overexpressing HDAC6 have more deacetylated α-tubulin than control cells and are more motile. Consistent with its effect on cell motility, HDAC6 is predominantly localized to the cytoplasm. Interestingly, malignant mammary epithelial cells have a more pronounced HDAC6 cytosolic localization than normal cells (Yoshida et al., 2004).
Cortactin, a protein originally identified as a substrate of the Src tyrosine kinase, also plays an important role in regulating cell motility (Wu et al., 1991). It interacts with F-actin to promote polymerization and branching. Cortactin can be found at areas of dynamic actin assembly, such as at the leading edge of migrating cells (e.g., in lamellipodia and membrane ruffles) (Kaksonen et al., 2000; Uruno et al., 2001; Weaver et al., 2001; Wu and Parsons, 1993). The translocation of cortactin to the cell periphery requires the activation of the small GTPase Rac1 and leads to the activation of the actin-nucleating complex Arp2/3 (Head et al., 2003; Uruno et al., 2001; Weaver et al., 2001; Weed et al., 1998). Overexpression of cortactin increases cell migration (Kowalski et al., 2005; Patel et al., 1998), while depletion of cortactin impairs it (Bryce et al., 2005). Cortactin contains an N-terminal acidic domain (NTA), six and a half tandem repeats of a unique 37-amino acid sequence, and a C-terminal Src homology (SH3) domain (Wu and Parsons, 1993). The repeat region of cortactin is both necessary and sufficient for F-actin binding. An α-helical structure and a proline-rich region separate the repeat and the SH3 domains. It is well-established that Src targets cortactin primarily at three residues (Tyr-421, Tyr-466, and Tyr-482 in mice), which are situated between the proline-rich region and the SH3 domain (Huang et al., 1998).
Cortactin is abnormally expressed in many human tumors, often as a result of gene amplification. EMS1, the human gene that encodes for cortactin, is amplified in head and neck squamous cell carcinomas, and the extent of amplification is inversely proportional to the survival rate (Rodrigo et al., 2000). Cortactin expression correlates with the metastatic potential of hepatocellular carcinomas (Chuma et al., 2004) and breast cancer cells injected into nude mice (Li et al., 2001). Evidence suggests that cortactin may regulate the formation of podosomes, structures that cells utilize to make contact with surfaces (Schuuring et al., 1993; Zhou et al., 2006; Tehrani et al., 2006). In invasive breast cancer cells, cortactin is present in membrane protrusions, or invadopodia, which carry proteases that digest the extracellular matrix (Bowden et al., 1999; Bowden et al., 2006). Collectively, these data demonstrate a strong link between cortactin expression and tumor invasiveness and metastasis. This link suggests the need for a greater understanding of the mechanisms by which cortactin activity is regulated.
In the present study, we show that the cortactin protein is acetylated. Importantly, we found that HDAC6 associates with and deacetylates cortactin in vitro and in vivo. Moreover, we demonstrate that hyperacetylation of cortactin prevents its translocation to the cell periphery, blocks its association with F-actin, and impairs cell motility. Together, our findings uncover a pathway by which actin-dependent cell motility can be modulated by HDAC6 and provide additional evidence for the importance of HDAC6 in the regulation of cell movement.
Results
HDAC6 Associates with the Repeat Region of Cortactin
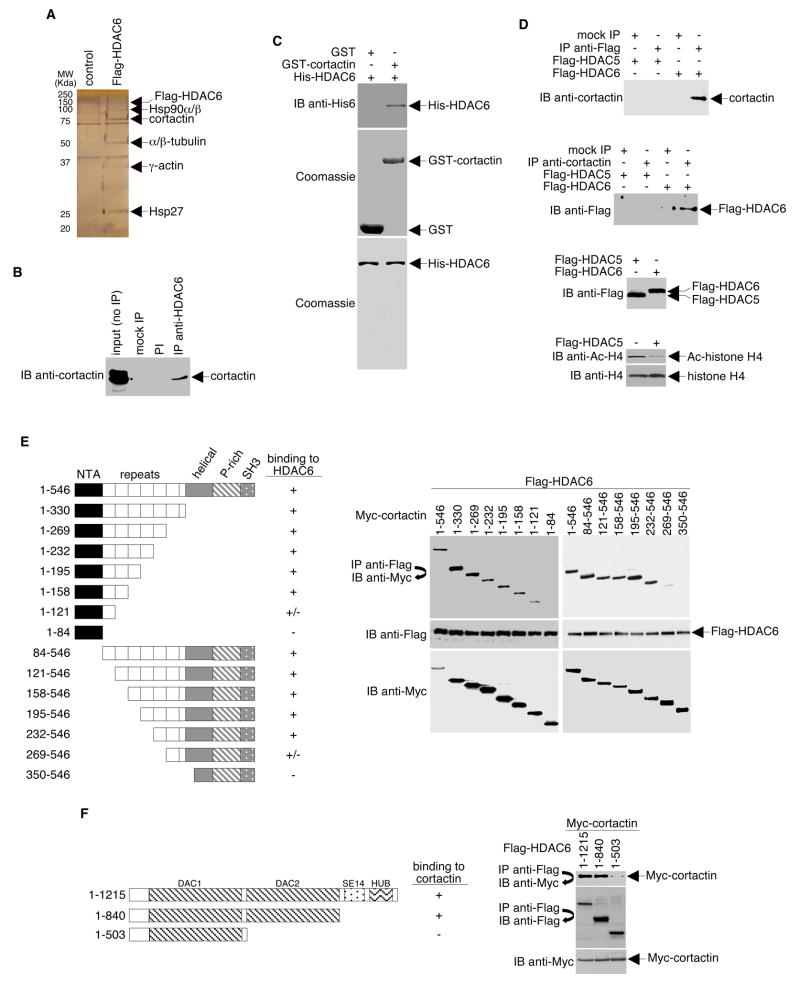
To identify HDAC6 cytoplasmic substrates, we prepared cytoplasmic extracts from a HeLa S3-derived cell line stably expressing Flag-tagged HDAC6. Flag-HDAC6 and associated proteins were purified by anti-Flag immunoaffinity chromatography. As a control, mock purifications were performed using extracts prepared from HeLa cells expressing an empty Flag vector (Figure 1A). We found at least five polypeptides that specifically associated with Flag-tagged HDAC6. Tandem mass spectrometric analysis (MS/MS) of a 80–85 kDa polypeptide yielded the amino acid sequence LPSSPVYEDAASFK, which was identified as cortactin (Accession Q14247).
Figure 1. HDAC6 Interacts with Cortactin.
(A) Silver-stained SDS-PAGE of the immunopurified Flag-tagged HDAC6-containing complexes. “Control” indicates an anti-Flag immunopurified sample prepared from HeLa cells expressing the empty Flag vector. (B) HeLa cell lysate was incubated with protein A-agarose alone (mock IP), preimmune serum (PI), or anti-HDAC6 antibody. Precipitates and total cell lysate (input) were analyzed by Western blotting using an anti-cortactin antibody. (C) GST and GST-cortactin coupled to Sepharose beads were incubated with purified His-HDAC6. The beads were washed extensively, and bound proteins were eluted and analyzed by Western blotting with anti-His6 antibodies (top panel). Coomassie blue stain gels were used to assess the quality and quantities of the purified proteins (middle and bottom panels). (D) HeLa cells were infected with adenoviruses encoding either Flag-tagged HDAC5 or Flag-tagged HDAC6. Whole cell lysates were immunoprecipitated with Flag-specific antibodies and Western blotted with cortactin antibodies (top panel) or vice versa (upper middle panel). Levels of Flag-tagged HDAC5 and Flag-tagged HDAC6 were determined by Western blot analysis of cell extracts using an anti-Flag antibody (lower middle panel). The ability of Flag-HDAC5 to deacetylate histone H4 was assessed by Western blot analysis of cell extracts using an anti-Ac-H4 antibody. (E) Left panel, a schematic diagram (not drawn to scale) of cortactin and various cortactin deletion mutants. P-rich: proline-rich. NTA: N-terminal acidic domain. For simplicity, the Myc portions of the fusion proteins are not shown. Right panel, HeLa cells were infected with adenoviruses encoding Flag-tagged HDAC6 and were transfected with plasmids encoding Myc-tagged cortactin or cortactin deletion mutants. Anti-Flag immunoprecipitates were Western blotted with antibodies specific for Myc (right top panel). Levels of Flag-tagged HDAC6 (right middle panel) and Myc-tagged cortactin (right bottom panel) were determined by Western blot analysis of cell extracts using antibodies specific for Flag or Myc, respectively. To maximize resolution of the different protein fragments, gels shown were prepared with different percentages of polyacrylamide. (F) Left panel, a schematic diagram (not drawn to scale) of HDAC6 and two HDAC6 deletion mutants. DAC: deacetylase (catalytic) domain. SE14: Ser/Glu-containing tetradecapeptide repeats. HUB: HDAC6/USP3/BRAP2-like ubiquitin-binding zinc finger. For simplicity, the Flag portions of the fusion proteins are not shown. Right panel, HeLa cells were transfected with plasmids encoding Myc-tagged cortactin and either Flag-tagged wildtype or mutant HDAC6. Anti-Flag immunoprecipitates were Western blotted with anti-Myc (top panel) or anti-Flag (middle panel). Expression levels of Myc-cortactin were assayed by Western blot with anti-Myc antibodies (bottom panel).
To determine if HDAC6 interacts with cortactin under normal physiologic conditions, co-immunoprecipitation of the endogenous proteins from a cytoplasmic extract was performed. As shown in Figure 1B, a significant fraction of cortactin could be co-precipitated with an anti-HDAC6 antibody but not with pre-immune serum. No cortactin was precipitated when the primary antibody was omitted. Using bacterially expressed and highly purified GST-cortactin and histidine-tagged HDAC6 expressed and purified from Sf9 insect cells, we found that HDAC6 interacts directly with cortactin in the absence of any other cellular proteins (Figure 1C).
To demonstrate specificity for the HDAC6-cortactin interaction, we used adenoviral infection to generate a HeLa cell line that overexpresses Flag-tagged HDAC5. HDAC5-containing protein complexes were then immunoprecipitated from HeLa extracts using a Flag-specific antibody. The presence of cortactin in these immunoprecipitates was analyzed by Western blotting. As shown in Figure 1D, the anti-Flag antibody specifically co-precipitated cortactin from cells expressing Flag-tagged HDAC6 but not from cells expressing Flag-tagged HDAC5. In a reciprocal experiment, an anti-cortactin antibody specifically co-precipitated Flag-tagged HDAC6, but not Flag-tagged HDAC5. Western blots verified the equivalent expression of the Flag-tagged HDAC6 and HDAC5 proteins in the infected cells. Further, overexpressed Flag-HDAC5 was active as determined by a comparison of acetylated histone H4 levels in Flag-HDAC5-overexpressing cells and in parental cells. Thus, Flag-HDAC5 appears to be a suitable negative control. In similar experiments, cortactin was not shown to interact with any other class II HDACs (data not shown). This finding suggests that the cortactin-HDAC6 interaction is highly specific.
To identify the HDAC6-binding region of cortactin, we performed serial deletions from the N- or C-terminus. The truncated sequences were then Myc-tagged and expressed in HeLa cells by transfection. These cells also expressed Flag-tagged HDAC6 as a result of adenoviral infection. Lysates from these cells were subjected to immunoprecipitation using a Flag-specific antibody followed by Western blotting using a Myc-specific antibody. As shown in Figure 1E, neither an N-terminal cortactin fragment (containing the acidic domain, aa 1–84) nor a C-terminal cortactin fragment (containing the α-helix, the proline-rich region, and the SH3 domain, aa 350–546) interacted with HDAC6. Results from GST pull-down assays further reveal that the repeat region alone binds HDAC6 (Figure S1). Therefore, the cortactin repeat region, a region known to be responsible for interaction with F-actin (Wu and Parsons, 1993), is necessary and sufficient for the interaction between cortactin and HDAC6.
To define the HDAC6 domain(s) required for interaction with cortactin, anti-Flag immunoprecipitates prepared from cells expressing Myc-cortactin and either Flag-tagged wildtype (1–1215) or deletion mutants were subjected to Western blot analysis using anti-Myc antibodies. As shown in Figure 1F, full-length HDAC6 and a C-terminal HDAC6 deletion mutant (1–840) bind cortactin. In contrast, an HDAC6 mutant (1–503) lacking both the C-terminal and the second deacetylase domain (DAC2) completely lost its ability to bind cortactin. Further deletion analysis indicates that DAC2 alone does not bind HDAC6 (data not shown), suggesting that both DAC1 and DAC2 are necessary, and neither domain alone is sufficient for cortactin binding.
HDAC6 Regulates the Acetylation of Cortactin in vivo
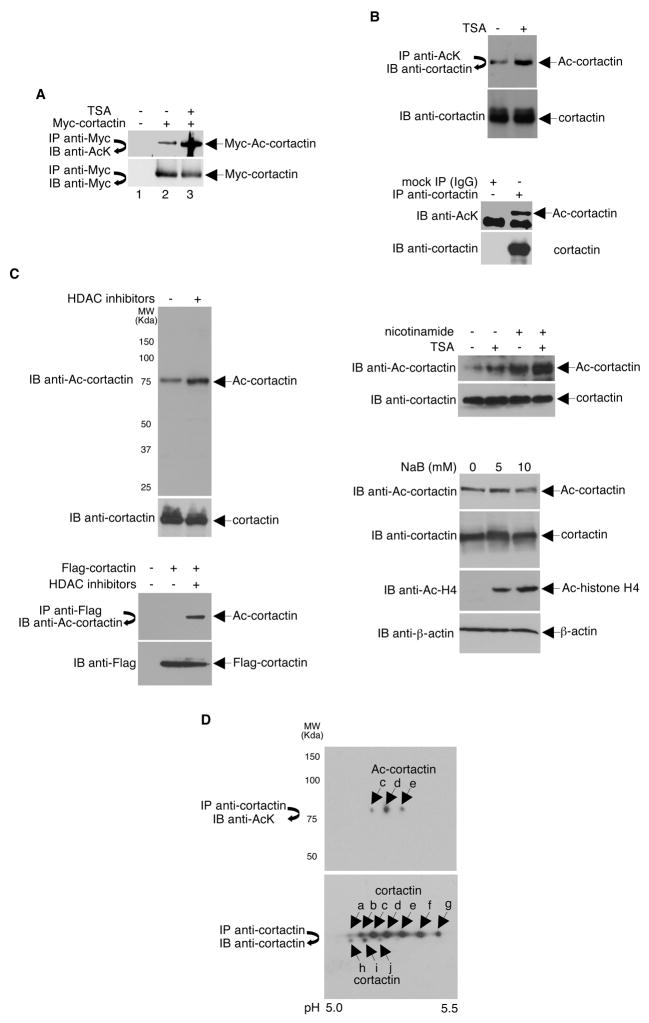
The association between cortactin and HDAC6 suggests that cortactin may be a deacetylation target of HDAC6. To determine if cortactin is acetylated, immunoprecipitated Myc-tagged cortactin was subjected to Western blotting using an anti-acetyl-lysine specific antibody. As shown in Figure 2A, Myc-cortactin is acetylated in vivo. Similarly, endogenous cortactin was also found to be acetylated as assayed by high stringency immunoprecipitation of a whole cell extract either with an anti-acetyl-lysine antibody followed by Western blotting with an anti-cortactin antibody or anti-cortactin followed by anti-acetyl-lysine antibody (Figure 2B). Treatment of cells with the class I and II HDAC inhibitor, TSA, greatly increased the level of acetylated Myc-tagged (Figure 2A) and endogenous (Figure 2B) cortactin.
Figure 2. Cortactin is Acetylated in vivo.
(A) HeLa cells transfected with either an empty vector or a vector encoding Myc-tagged cortactin were treated with ethanol (−) or 400 ng/ml TSA (+). Anti-Myc immunoprecipitates were Western blotted using antibodies specific for acetyl-lysine (AcK) or Myc. Relative level of acetylated cortactin: Ac-cortactin/cortactin = 0.33 (lane 2), 1.75 (lane 3). (B) HeLa cells were untreated (bottom panel) or treated (top panel) with either ethanol (−) or 400 ng/ml TSA (+) for 12 h. Lysates were immunoprecipitated under high stringency conditions and immunoprecipitates were Western blotted with the indicated antibodies. The same lysates were directly immunoblotted with an anti-cortactin antibody. (C) Left top panel, total extracts prepared from HeLa cells treated with ethanol (−) or 400 ng/ml TSA plus 20 mM nicotinamide (+) for 12 h were separated on SDS-PAGE and analyzed by Western blot with the anti-acetyl-cortactin antibody. The blot was stripped and re-probed with an anti-cortactin antibody. Left bottom panel, HeLa cells transfected with plasmids expressing Flag-tagged cortactin or an empty parental vector were treated with ethanol (−) or 400 ng/ml TSA plus 20 mM nicotinamide (+) for 12 h. Cell lysates were immunoprecipitated with anti-Flag and Western blotted with an anti-acetyl-cortactin antibody or directly Western blotted with a Flag-specific antibody. Right panels, extracts prepared from cells treated with nicotinamide, sodium butyrate (NaB), and/or TSA were analyzed as before. Anti-Ac-H4 blot was performed to confirm that NaB was active in this system. Anti-cortactin and anti-β-actin Western blots were done as loading controls. (D) Top panel, anti-cortactin immunoprecipitates prepared from a NIH3T3 whole cell extract was separated on a 2-dimensional gel and Western blotted with an anti-acetyl-lysine antibody. Bottom panel, the blot was stripped and re-probed with an anti-cortactin antibody.
To confirm that cortactin is acetylated, we raised a rabbit polyclonal antibody that specifically recognizes acetylated cortactin. Western blot analysis using this antibody revealed that HDAC inhibition increased acetylation of both endogenous and overexpressed cortactin (Figure 2C). Interestingly, like TSA, treatment of cells with the class III deacetylase inhibitor nicotinamide also resulted in an increase of cortactin acetylation. However, unlike TSA, cortactin acetylation was unchanged in the presence of the potent HDAC inhibitor sodium butyrate, which does not affect HDAC6 (Hubbert et al., 2002).
Next, using an alternative approach to demonstrate that cortactin is acetylated under physiologic conditions, we immunopurified cortactin from a NIH3T3 whole cell extract, separated the products on a 2-dimensional gel, and analyzed the acetylated cortactin by Western blot with an anti-acetyl-lysine antibody. As shown in Figure 2D, a significant fraction of endogenous cortactin was present in an acetylated form.
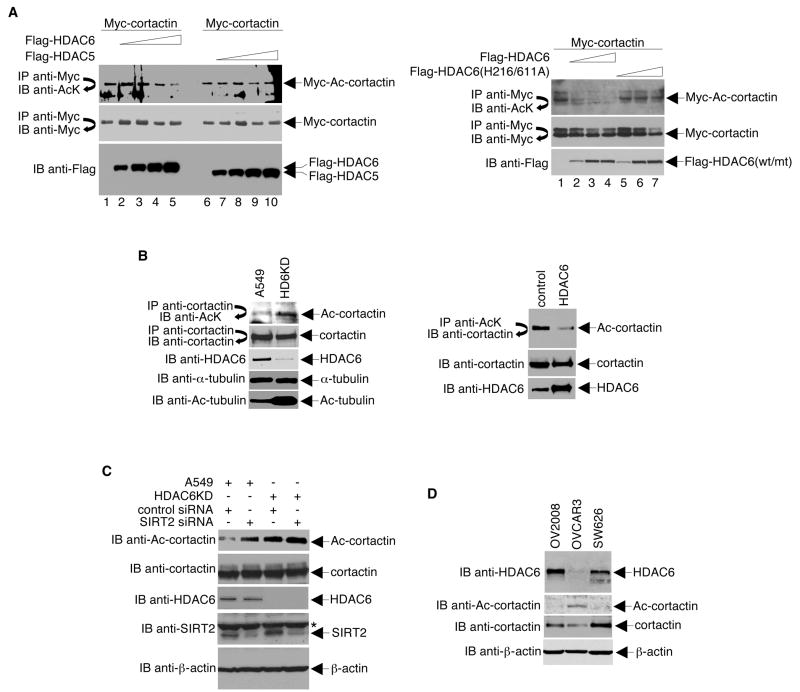
To examine the possible deacetylation of cortactin by HDAC6, we co-expressed Myc-tagged cortactin and either Flag-tagged HDAC6 or Flag-tagged HDAC5 in HeLa cells. Myc-immunoprecipitates were then analyzed by Western blot using an acetyl-lysine-specific antibody. As shown in Figure 3A, overexpression of HDAC6 resulted in a reduction in the level of acetylated cortactin. The total level of cortactin was unaffected. In contrast, the overexpression of HDAC5 or a catalytically defective HDAC6 did not alter levels of total or acetylated cortactin.
Figure 3. HDAC6 Deacetylates Cortactin in vivo.
(A) Left panel, Myc-tagged cortactin was expressed in HeLa cells by transient transfection. Various amounts of Flag-tagged HDAC6 and Flag-tagged HDAC5 were expressed in HeLa cells by adenoviral infection. Right panel, Myc-cortactin, Flag-HDAC6, and Flag-tagged HDAC6 catalytically defective mutant (H216/611A) were expressed in HeLa cells by transfection. Anti-Myc immunoprecipitates were Western blotted with antibodies specific to either acetyl-lysine (top panels) or Myc (middle panels). Cell extracts were Western blotted with a Flag-specific antibody (bottom panels). Relative level of acetylated cortactin: Ac-cortactin/cortactin = 0.83 (lane 1, left panel), 0.17 (lane 5, left panel), 0.8 (lane 6, left panel), 0.9 (lane 10, left panel), 0.65 (lane 1, right panel), 0.03 (lane 4, right panel), 0.57 (lane 7, right panel). (B) Left panel, lysates prepared from A549 cells and A549 cells stably expressing HDAC6 siRNA (HD6KD) were immunoprecipitated with a cortactin antibody, and the immunoprecipitates were Western blotted with an acetyl-lysine-specific antibody (top) or an anti-cortactin antibody (second from the top). Lysates were also directly Western blotted with antibodies specific for HDAC6, α-tubulin, and acetylated α-tubulin. Right panel, cell lysates were prepared from stably transfected HeLa cells expressing either the empty vector or high levels of HDAC6 and used either in a high stringency immunoprecipitation with anti-acetyl-lysine-specific antibodies followed by Western blotting with anti-cortactin (top) or for Western blotting with antibodies specific for either cortactin or HDAC6 (middle and bottom panels, respectively). (C) Four different sets of SIRT2 siRNA from the siGENOME SMART pool were purchased from Dharmacon and used to knock-down SIRT2 expression in A549 or HDAC6KD cells. Protein expression levels and the effects on cortactin acetylation were examined by Western blotting with the indicated antibodies. * indicates a non-specific band on the anti-SIRT2 Western blot. (D) Cell lysates prepared from three different ovarian cancer cell lines subjected to Western blot analysis with the indicated antibodies.
To verify that HDAC6 mediates the deacetylation of cortactin, we used an established A549 cell line (HD6KD) in which HDAC6 expression is specifically knocked-down by a retrovirus-mediated RNAi system (Kawaguchi et al., 2003). Consistent with previous studies (Hubbert et al., 2002), HD6KD cells contained more acetylated tubulin than control A549 cells (Figure 3B). More importantly, we found that HD6KD cells contained higher levels of acetylated cortactin than the control cells. A reciprocal experiment with a cell line stably overexpressing HDAC6 revealed that acetylated cortactin levels were markedly decreased in these cells as compared to the negative control parental cells.
Since HDAC6 has been reported to interact with SIRT2 (North et al., 2003), we tested the effect of SIRT2 on cortactin acetylation levels. As shown in Figure 3C, similar to HDAC6 knockdown, SIRT2-knockdown resulted in an increase in cortactin acetylation suggesting that, in addition to HDAC6, SIRT2 might also regulate the state of cortactin acetylation.
Previous analysis of mouse tissues has shown that HDAC6 is highly expressed in testes (Seigneurin-Berny et al., 2001). In addition, we have found that HDAC6 is highly expressed in human ovarian tumor tissues (data not shown). We examined the expression of HDAC6 in three different human ovarian cancer cell lines and found that HDAC6 expression in the OV2008 and SW626 cell lines was relatively high. On the other hand, the OVCAR3 cell line contained lower levels of HDAC6 (Figure 3D). Importantly, the acetylation level of cortactin was inversely proportional to the expression level of HDAC6. That is, endogenous cortactin was highly acetylated in the OVCAR3 cell line, but not in the OV2008 or SW626 cell lines. These data strongly suggest that HDAC6 is a chief regulator of cortactin acetylation.
Cortactin is Acetylated/Deacetylated in its Repeat Region
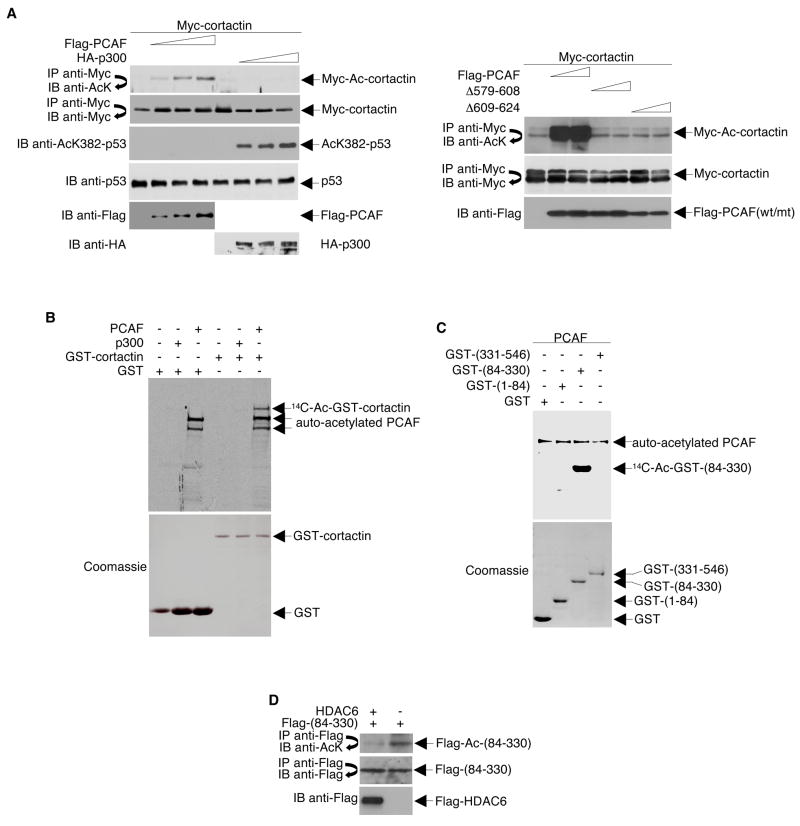
To examine the HAT-mediated acetylation of cortactin, Myc-tagged cortactin was co-expressed with either Flag-tagged PCAF or HA-tagged p300 in HeLa cells. Anti-Myc-immunoprecipitates prepared from these cells were then analyzed by Western blot using an acetyl-lysine-specific antibody. As shown in Figure 4A, PCAF expression increased the level of acetylated cortactin in a dose-dependent manner (left panel). However, overexpression of p300 or catalytic mutants of PCAF (Δ579–608 and Δ609–624) did not alter levels of acetylated cortactin.
Figure 4. Cortactin is Acetylated Primarily in its Repeat Region.
(A) HeLa cells were transfected with Myc-tagged cortactin and various amounts of Flag-tagged PCAF, Flag-tagged PCAF catalytically-dead mutants, or HA-tagged p300. Anti-Myc immunoprecipitates were Western blotted with antibodies specific for either acetyl-lysine or Myc. Levels of Flag-tagged PCAF and HA-tagged p300 were determined by Western blotting of cell extracts with antibodies specific for Flag and HA, respectively. To confirm that p300 was active in this system, a Western blot was performed using the same extracts with an antibody specific for acetylated K382 of p53. (B) GST-cortactin was incubated with 14C-acetyl CoA and recombinant PCAF or p300. Acetylation of GST-cortactin was visualized by autoradiography of SDS-PAGE gels (top panel). Levels of GST-cortactin were determined by Coomassie staining of SDS-PAGE gels (bottom panel). (C) Acetylation assays were performed using purified GST-tagged cortactin fragments and PCAF. (D) HeLa cells were infected with adenoviruses encoding either GFP (−) or Flag-tagged HDAC6 (+). Whole cell lysates were immunoprecipitated with Flag-specific antibodies and Western blotted with antibodies against acetylated lysines (top panel). The blot was stripped and re-probed with anti-Flag (middle panel). A direct Western blot was performed to assess expression of Flag-HDAC6 (bottom panel).
Although PCAF has been reported to be located both in the nucleus and cytoplasm of cells (Wong et al., 2004), most studies on PCAF so far have focused on its nuclear functions. Because cortactin is a cytoplasmic protein, to rule out the possibility that PCAF acetylation of cortactin is a result of over-expression and consequently mis-localization, we re-examined the subcellular localization of endogenous PCAF both by cellular protein fractionation and by immunostaining. As shown in Figure S2, endogenous PCAF is indeed present in both the nuclear and cytoplasm of 293T and NIH3T3 cells, confirming the possibility that cortactin could be a true physiological substrate of PCAF.
To determine if PCAF directly acetylates cortactin, an in vitro acetylation assay was performed using GST-cortactin and the catalytic domains of either PCAF or p300. Consistent with the in vivo results, these experiments showed that PCAF, but not p300, can acetylate cortactin in vitro (Figure 4B).
To map the region(s) of cortactin acetylated by PCAF in vitro, the following three GST-tagged cortactin fragments were prepared: the N-terminal acidic region (1–84), the repeat region (84–330), and the C-terminal region (331–546) (see diagram in Figure 1E). Together, these fragments cover the entire cortactin protein sequence. As determined by in vitro acetylation assays, PCAF was able to acetylate the repeat region of cortactin, but not the N-terminal acidic or the C-terminal regions (Figure 4C). These data suggest that the repeat region of cortactin is the primary site of acetylation.
To determine if the cortactin repeat region alone is sufficient to serve as a HDAC6 substrate, we infected HeLa cells that express Flag-(84–330) with adenoviruses that express either Flag-HDAC6 or GFP as control, prepared cell lysates, and assayed acetylation levels by immunoprecipitation with anti-Flag and Western blotting with anti-acetyl-lysine antibodies. As shown in Figure 4D, acetylation level of cortactin repeat region diminishes significantly in the presence of overexpressed HDAC6.
Identification of Acetylated Lysines in Cortactin
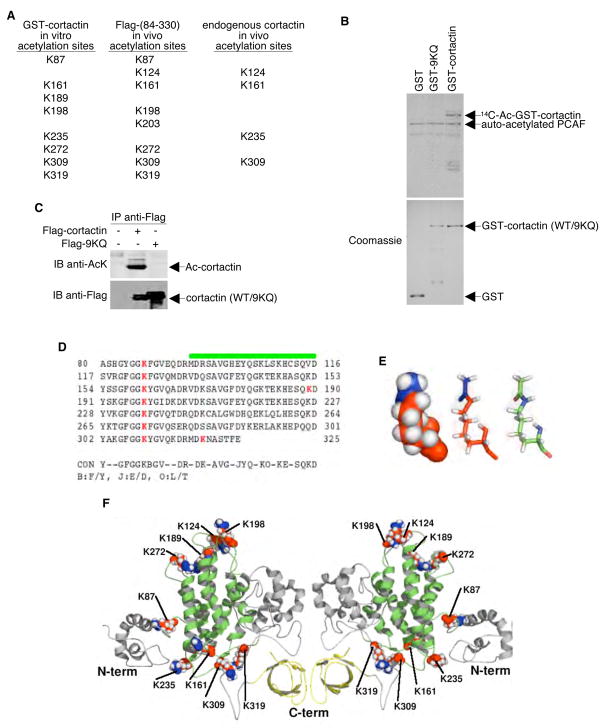
To identify the sites of acetylation on cortactin, in vitro acetylation assays were performed using GST-cortactin, the PCAF catalytic domain, and acetyl CoA. To verify cortactin acetylation, an aliquot of each reaction mixture was analyzed by Western blotting using an anti-acetyl-lysine antibody (data not shown). The remainder of the reaction mixture was resolved by SDS-PAGE, and the polypeptide band corresponding to cortactin was excised and analyzed by LC tandem mass spectrometry (LC-MS/MS). Of the 50 lysines in cortactin, 11 were found to be acetylated. Of these 11 acetyl-lysines, eight (K87, K161, K189, K198, K235, K272, K309, and K319) were present in the cortactin repeat region (Figure 5A). We also mapped the in vivo sites of acetylation on cortactin by focusing on the repeat region. For these analyses, we transfected 293T cells with a plasmid encoding the Flag-tagged repeat region of cortactin. To maximize acetylation, cells were treated with 400 ng/ml TSA for 12 h. Following this treatment, cellular extracts were prepared from these cells, and the extracts were subjected to immunoprecipitation using a Flag-specific antibody. The resulting immunoprecipitates were then resolved by SDS-PAGE, and the band corresponding to the cortactin repeat region was excised from the gel and analyzed by LC-MS/MS. Finally, using a similar strategy, we immunopurified endogenous cortactin protein using anti-cortactin antibody and subjected the purified product to LC-MS/MS analysis.
Figure 5. Identification of Acetylation Sites of Cortactin in vitro and in vivo.
(A) GST-tagged cortactin (in vitro acetylated by PCAF), Flag-tagged cortactin (expressed and purified from HeLa cells), and endogenous cortactin immunopurified with anti-cortactin antibody were cleaved with trypsin and analyzed by ion trap mass spectrometry. Positions of unambiguously identified acetylated lysine residues are listed. (B) In vitro acetylation assays performed using the catalytic domain of PCAF and GST-tagged (wildtype) or 9KQ mutant cortactin. Reaction products were resolved by SDS-PAGE. Acetylated proteins were visualized by autoradiography (top panel), and total amounts of protein were monitored by Coomassie staining (bottom panel). (C) Flag-tagged cortactin (wildtype) and Flag-tagged 9KQ mutant cortactin were expressed in HeLa cells by transient transfection. Anti-Flag immunoprecipitates were Western blotted with antibodies specific to either acetyl-lysine or Flag. (D) Alignment of the mouse cortactin repeats (residues 80 to 325). The green line represents the predicted transmembrane zone, and the red bold letters represent the potential acetylated lysine residues. (E) Representation of the acetylated residues in balls (left) and sticks (center and right). Atoms in red belong to the lysine residues and atoms in blue to the acetyl groups. The molecule on the right is colored according to atom type: carbon (green), nitrogen (blue), oxygen (red), and hydrogen (white). (F) Cartoon representations of both sides of the mouse cortactin model. The cortactin repeats are in green and the SH3 motif in the carboxyl-terminal is in yellow. The acetylated lysines are in the same color scheme described in E (left and center).
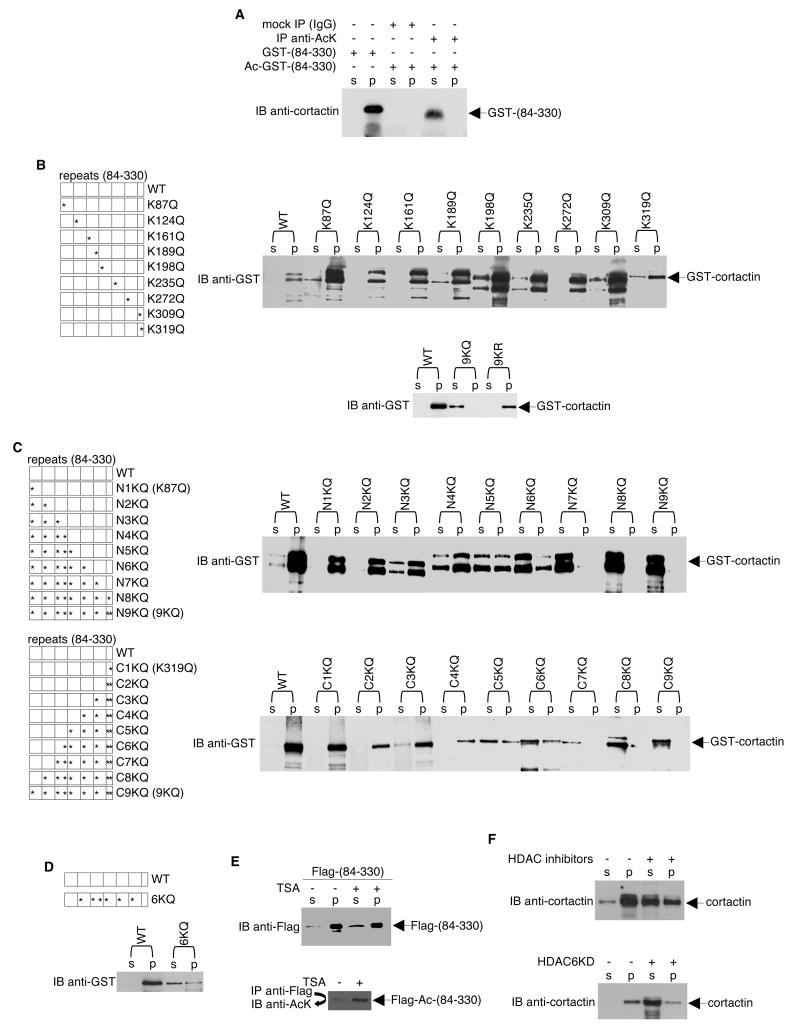
To assess their contributions to the overall acetylation status of cortactin, all eight of the lysines that were identified as the PCAF in vitro acetylation sites as well as K124 (a residue detected both in the purified Flag-tagged repeat region and endogenous cortactin) were mutated to glutamine. This mutant, referred to as 9KQ, was examined using an in vitro acetylation assay. As shown in Figure 5B, while PCAF effectively acetylated wildtype GST-cortactin, acetylation of the GST-9KQ mutant was nearly undetectable. Coomassie blue gel staining verified that similar amounts of wildtype cortactin and 9KQ were present in both assays. Furthermore, the lysine to glutamine change does not affect the binding of cortactin to HDAC6 (Figure S3). Similarly, in transiently transfected HeLa cells, the 9KQ mutant was also much less efficiently acetylated than wild-type cortactin (Figure 5C). Thus, the repeat region is most likely the primary (if not the only) cortactin region acetylated in vitro and in vivo.
The secondary structure of the individual repeats of cortactin repeat region predicts an α-helical region within the carboxyl-half of each subunit (Wu and Parsons, 1993). Previous computer modeling of this helical region suggested that highly conserved lysine residues may be positioned on the same face of the α-helices, a conformation that would contribute to a positively-charged helical surface. In our predicted model, all of the above-described acetylated lysines in the repeat domain (with the exception of K189 and K319) are in a loop rather than in the helical region (Figure 5D–F). Additionally, the acetylated lysines are present at two ends of the helices, a conformation that could result in the formation of two “charged patches”. In its deacetylated state, this charged patch (or the positively-charged loop surface) of cortactin is likely to contribute to F-actin binding.
Acetylation of Cortactin Impedes its Interaction with F-Actin
To determine the effect of cortactin acetylation status on cortactin interactions with F-actin, F-actin co-sedimentation assays were performed. For these experiments, GST-fused cortactin repeat regions were expressed in and purified from E. coli cells. This purified protein was then incubated with polymerized rabbit muscle F-actin. As shown in Figure 6A, following F-actin sedimentation, the non-acetylated GST-tagged cortactin repeat region was bound to (i.e., co-sedimented) F-actin in vitro; whereas, immunopurified, PCAF-acetylated GST-tagged cortactin repeat region was not.
Figure 6. Acetylation of Cortactin Reduces its Interaction with F-actin.
(A) GST-tagged cortactin (the repeat region) was acetylated by PCAF in vitro. Reaction mixtures were then incubated with mouse IgG or anti-acetyl-lysine antibodies. Precipitates were resolved by SDS-PAGE. The band corresponding to the repeat region was eluted from the gel with 1% SDS. Eluates were incubated with F-actin, and co-sedimentation assays were performed. Supernatants (S) and pellets (P) were subjected to Western blotting with cortactin-specific antibodies. For lanes 1 and 2, co-sedimentation assays were performed using unacetylated GST-tagged cortactin (the repeat region). (B, C, D) GST-tagged wildtype or mutant cortactin proteins were incubated with F-actin, and co-sedimentation assays were performed. Supernatants and pellets were subjected to Western blotting with an anti-GST antibody. Diagrams of the wildtype and mutant constructs are shown. (E) HeLa cells were transfected with a plasmid encoding Flag-tagged cortactin (repeat region) and treated with ethanol (vehicle control) or 400 ng/ml TSA for 12 h. Top panel, co-sedimentation assays were performed on cell lysates, and supernatants and pellets were subjected to Western blotting with an anti-Flag antibody. Bottom panel, Anti-Flag immunoprecipitates were Western blotted with an antibody specific for acetyl-lysine. (F) Similar co-sedimentation experiments were performed to assess endogenous cortactin-F-actin interactions.
To examine this effect in more detail, we prepared two sets of GST-tagged cortactin mutants. In the first set, each lysine of the cortactin repeat region was individually mutated to glutamine. Although glutamine cannot be acetylated, its neutral charge mimics the acetylation of lysine. Each of these point mutants were able to bind to F-actin as efficiently as wild-type cortactin (Figure 6B). Likewise, a charge-preserving cortactin mutant in which all nine of the repeat region lysines were mutated to arginine (9KR) was able to efficiently bind to F-actin. In sharp contrast, a charge-neutralizing cortactin mutant in which all nine of the repeat region lysine residues were mutated to glutamine (9KQ) was not able to bind F-actin. In the second set of mutants, the lysines of the cortactin repeat region were progressively mutated to glutamine beginning at either the amino or carboxyl end (Figure 6C). Mutation of less than three lysines at either terminal end did not significantly alter the ability of cortactin to bind to F-actin. However, mutation of more than four of these lysines dramatically reduced the F-actin binding activity of cortactin; the more residues mutated, the less binding detected. The effect was not limited to mutations at the N-terminal or the C-terminal end of cortactin, because mutation within the internal repeats (6KQ) resulted in similar decrease in F-actin binding when compared to N6KQ or C6KQ (Figure 6D). Thus, the acetylation of multiple lysine residues of the cortactin repeat region attenuates its actin-binding ability in vitro. This effect was also observed in vivo using cells overexpressed with the cortactin repeat region. For example, HeLa cells treated with TSA had more acetylated Flag-tagged cortactin and more cortactin in the supernatant (i.e., not bound to F-actin) than did untreated cells (Figure 6E). Consistent with this finding, inhibition of HDAC6 either by treatment of cells with HDAC inhibitors or HDAC6 siRNA decreased endogenous cortactin-F-actin association (Figure 6F).
Acetylation Causes Aberrant Cortactin Localization and Cell Motility
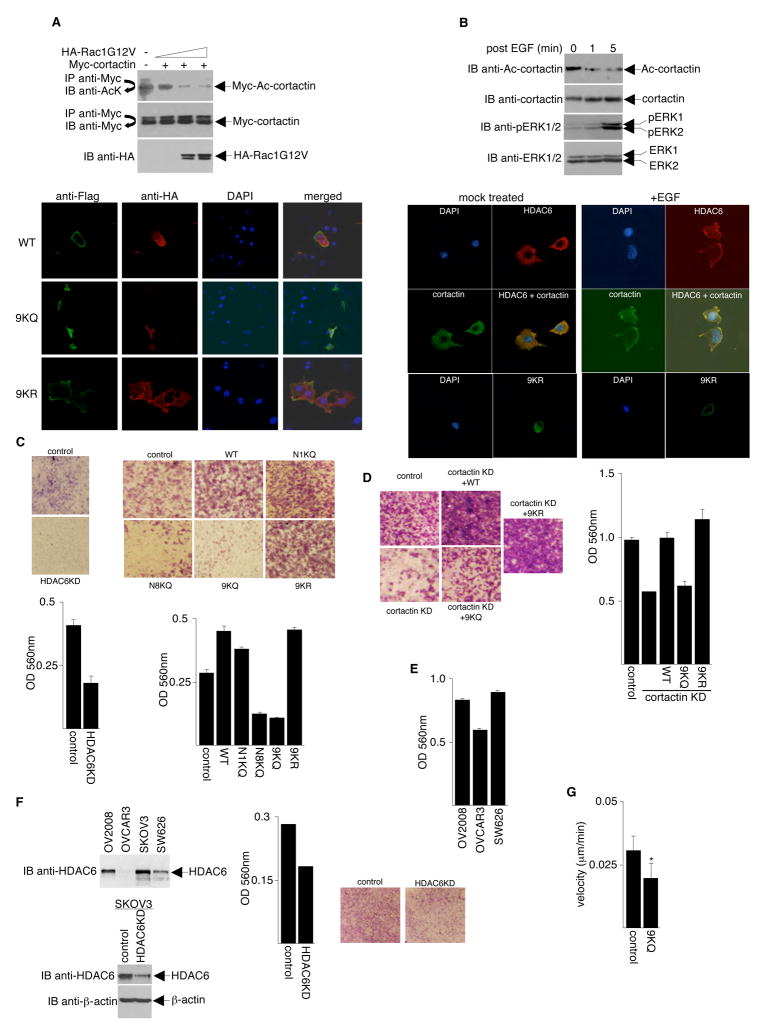
In response to growth factor stimulation or small GTPase, Rac1 activation, cortactin translocates from the cytosol to the cell periphery, where it interacts with and enhances the formation of F-actin (Weed et al., 1998; Weed et al., 2000). Because acetylation/deacetylation is a key determinate of cortactin binding to F-actin, we examined whether Rac1 has an effect on the level of cortactin acetylation. NIH3T3 cells were transfected with plasmids expressing Myc-cortactin and various amounts of constitutively active Rac1 (HA-Rac1G12V). Anti-Myc immunoprecipitates prepared from these cells were subjected to Western blot analysis using an anti-acetyl-lysine antibody. As shown in Figure 7A, consistent with our observation that deacetylated cortactin binds better to F-actin, Rac1 activation clearly resulted in cortactin deacetylation.
Figure 7. Acetylation of Cortactin Prevents its Localization to Membrane Ruffles and Inhibits Cell Motility.
(A) Top panel, NIH3T3 cells were transfected with plasmids that express Myc-cortactin and various amounts of HA-Rac1G12V. Anti-Myc immunoprecipitates prepared from these cells were assayed for cortactin acetylation by Western blotting with an anti-acetyl-lysine antibody. Immunoprecipitation efficiency and HA-Rac1G12V expression were monitored by Western blotting with with anti-Myc and anti-HA, respectively. Bottom panel, NIH 3T3 cells were transfected with plasmids encoding active Rac1 (HA-Rac1G12V) and either Flag-tagged wildtype, Flag-tagged 9KQ mutant, or Flag-tagged 9KR mutant cortactin. Twenty-four hours post-transfection, cells were immunostained with antibodies or stained with 4′,6-diamidino-2-phenyl-indole and analyzed under a confocal microscope. (B) Top panel, NIH3T3 cells were serum-starved overnight and then treated with 20 ng/ml EGF. Lysates were analyzed by Western blotting with anti-acetyl-cortactin antibodies. The blot was stripped and re-probed with anti-cortactin antibodies. For controls, anti-phospho-ERK1/2 and anti-ERK1/2 Western blots were performed to monitor the efficiency of EGF treatment. Bottom panels, NIH3T3 cells were serum-starved overnight and then either mock-treated or treated with 10 ng/ml EGF for 10 min. Endogenous HDAC6 and cortactin were detected using anti-HDAC6 and anti-cortactin specific antibodies and Alexa-594- and Alexa-488-conjugated secondary antibodies. Flag-9KR and Flag-9KQ were detected with anti-Flag antibodies. (C) Left panels, 293T cells stably expressing HDAC6 siRNA (HDAC6KD; generated by the OligoEngine retrovirus-mediated pSuper RNAi system) and 293T cells expressing control siRNA were serum-starved overnight and assayed for migratory properties. Migratory cells were stained (top) and quantified at OD 560 nm following extraction (bottom). Right panels, parental NIH 3T3 cells (control) and NIH 3T3 cells stably expressing either wildtype or cortactin mutants were serum-starved overnight and assayed for migratory properties as in left panels. (D) Plasmids expressing wildtype or mutant cortactins were transfected into HT1080 cells depleted of cortactin (cortactin KD). Stable polyclonal cell populations were selected and assayed for cell migration activity as in (C). Control cells indicate parental untransfected HT1080 cells. (E) Three different ovarian cancer cell lines were assayed for cell migration activity as in (C). (F) Top left panel, Western blot analysis of HDAC6 expression in different ovarian cancer cell lines. Bottom left panel, Western blots to assess HDAC6 knock-down in SKOV3 cells. Middle and right panels, SKOV3 cells transfected with either HDAC6 siRNA or control siRNA were assayed for cell migration activity as in (C). (G) Data showing velocity of parental MDA-MB-231 cells (control) and a pool of stable cell clones overexpressing 9KQ mutant cortactin undergoing random motility on tissue-culture dishes. For MDA-MB-231 cells, n = 64; for 9KQ stable clones, n = 76. *p < 0.0001 vs. MDA-MB-231 cells. Error bars denote the standard deviation (SD).
To further analyze whether the acetylation of cortactin affects its subcellular location, we co-transfected NIH3T3 cells with constitutively active Rac1 (HA-Rac1G12V) and either Flag-tagged wildtype, 9KQ mutant, or 9KR mutant cortactin. Cells were immunostained with anti-Flag and anti-HA antibodies followed by Alexa-594 and Alexa-488 conjugated secondary antibodies. Consistent with previous studies, wildtype cortactin was present in the membrane ruffles in cells expressing active Rac1. Similarly, the 9KR cortactin mutant, which is capable of F-actin binding, translocated to the cell periphery in the presence of active Rac1. In contrast, the 9KQ cortactin mutant, which is not capable of F-actin binding, remained cytoplasmic. Quantification of the percentage of cells expressing Rac1 and the wildtype, 9KQ, or 9KR cortactin that show a leading edge is presented in Table S1. Together, these results suggest that the acetylation of cortactin inhibits, while deacetylation may be required for, its Rac-mediated translocation to the cell periphery.
It has been reported that growth factor-induced membrane ruffling is due to activation of Rac1 (Kozma et al., 1995; Nobes and Hall, 1995). We found that the treatment of cells with EGF resulted in a decrease in cortactin acetylation (Figure 7B). To determine if HDAC6 is involved in the Rac1-mediated translocation of cortactin, we stimulated serum-starved NIH3T3 cells with EGF and monitored the subcellular localization of cortactin and HDAC6 by immunofluorescence microscopy. We found that HDAC6 was translocated to the cell periphery together with cortactin upon EGF stimulation (Figure 7B). This result suggests that HDAC6 interacts with and deacetylates cortactin at the cell periphery. Further analysis demonstrated that, upon EGF stimulation, 9KR mutant translocates to the cell periphery suggesting that the charge-preserving mutant could dislodge the HDAC6-cortactin association. Also, as can be seen in Figure S4, consistent with the notion that cortactin must be deacetylated in order to translocate to the membrane ruffle or leading edges, the localization of the 9KQ mutant is unchanged in the presence of EGF.
A previous study has shown that overexpression of HDAC6 increases the chemotactic motility of NIH3T3 cells (Hubbert et al., 2002). To confirm this result, transwell assays were performed with 293T cells expressing either HDAC6 siRNA or control siRNA. The results clearly indicate that chemotactic cell motility decreases in cells depleted of HDAC6 (Figure 7C). To examine the effect of cortactin acetylation status on chemotactic cell migration, parental NIH3T3 cells and NIH3T3 cells stably expressing either wildtype or mutated cortactins were seeded in the upper chamber of migration plates. As shown in Figure 7C, the percentage of motile cells was decreased among cultures expressing cortactin mutants that do not bind F-actin (N8KQ and 9KQ) than in cells expressing either wildtype, N1KQ mutant, or 9KR mutant with intact F-actin binding capability. Western blot analyses indicate that cortactin mutants are expressed in comparable levels as wildtype cortactin (Figure S5). These results strongly argue that cortactin deacetylation is critical for the regulation of cell migration.
In complementary experiments, we determined if cortactin mutants could rescue the phenotype caused by cortactin knockdown. Cortactin expression was assessed by a Western blot (Figure S5). As expected, a HT1080 human fibrosarcoma cell line in which cortactin protein expression was reduced by more than 95% (cortactin KD; Bryce et al., 2005) exhibited slower migration compared to parental cells (control) (Figure 7D). Interestingly, the cell motility defect in cortactin KD cells was effectively rescued by the introduction of wildtype or 9KR mutant cortactin, but not by the 9KQ mutant, underscoring the significance of deacetylation in cortactin function.
To further demonstrate that acetylation/deacetylation of cortactin affects cell motility, we performed migration assays on three different ovarian cancer cell lines that were determined to possess different levels of acetylated cortactin (Figure 3D). As shown in Figure 7E, OV2008 and SW626 cells that express high levels of HDAC6 and contain hypoacetylated cortactin show faster migration when compared to OVCAR3 that expresses low level of HDAC6 and contain hyperacetylated cortactin. Next, we used siRNA to knock-down HDAC6 expression in SKOV3 cells, an ovarian cancer cell line with high levels of HDAC6 protein (Figure 7F). We then compared the migratory properties of the HDAC6 knock-down cells with the parental cells treated with control siRNA. As expected, cells with partially depleted HDAC6 (HDAC6KD) displayed a slower migration phenotype.
Finally, using an alternative approach to assess the effects of acetylation/deacetylation of cortactin on cell motility, we established an MDA-MB-231 cell line that stably expresses the 9KQ mutant. Using a live-cell imaging technique, we then measured the actual distance and velocity these cells traveled under random motility compared to the parental cell line. As shown in Figure 7G, movement velocity was significantly decreased in cells overexpressing the 9KQ mutant. These results are consistent with the transwell assay results and unequivocally confirm that cells that express the charge-neutralizing acetylation mutant of cortactin (9KQ) travel less distance and move slower than the parental cell line.
Discussion
Results from previous studies have shown that HDAC6 associates with and regulates the acetylation of α-tubulin and Hsp90 (Bali et al., 2005; Haggarty et al., 2003; Hubbert et al., 2002; Kovacs et al., 2005; Matsuyama et al., 2002; Zhang et al., 2003). HDAC6 also functions as an adaptor linking dynein motors to their aggregated protein cargos (Kawaguchi et al., 2003). Additionally, Seigneurin-Berny et al. (2001) and Boyault et al. (2006) found that the mammalian homolog of yeast UFD3 (ubiquitin fusion degradation protein 3), as well as p97/VCP/Cdc48p, co-purifies with HDAC6 in mouse testes. They demonstrated that HDAC6 also efficiently interacts with ubiquitin and that this interaction led to the dissociation of HDAC6 from this protein complex. Furthermore, Hook et al. (2002) reported that HDAC6 binds polyubiquitin through its zinc finger and also co-purifies with de-ubiquitinating enzymes. Using multiple approaches, we have also identified an interaction between polyubiquitin and HDAC6 (data not shown). Unexpectedly, we also identified the F-actin-binding protein, cortactin, as an HDAC6-interacting protein. This protein, which co-purifies with cytoplasmic HDAC6, is a non-histone substrate of HDAC6.
Our results indicate that HDAC6 interacts with cortactin in vivo and that this interaction requires the repeat region of cortactin. Similar to the HDAC6-β-tubulin interaction (Zhang et al., 2003), the association between HDAC6 and cortactin is direct, is mediated through deacetylase domains, and does not required deacetylase activities. However, unlike the β-tubulin-HDAC6 interaction in which only one deacetylase domain is sufficient to bind HDAC6, the association of cortactin with HDAC6 requires both deacetylase domains.
We found that PCAF acetylates cortactin and HDAC6 deacetylates it. It has been shown previously that the repeat region of cortactin mediates its interaction with F-actin, and that this interaction stimulates F-actin polymerization and branching (Uruno et al., 2001; Weaver et al., 2001; Wu and Parsons, 1993). PCAF was able to acetylate cortactin in vitro and promote cortactin acetylation when overexpressed in vivo, while the related HAT, p300, was not. The cortactin repeat region was the primary site of PCAF-mediated acetylation in vitro and in vivo. Using LC tandem mass spectrometry, we identified multiple lysine residues in this region that were acetylated. Whether endogenous PCAF is solely responsible for cortactin acetylation in vivo remains to be determined. Because no significant differences in acetylation of cortactin were detected from PCAF−/− versus PCAF+/+ fibroblast cells, we currently favor the idea that besides PCAF, an additional acetyltransferase(s) may acetylate cortactin.
We showed that deacetylated cortactin accumulated in cells that overexpressed HDAC6 and that acetylated cortactin accumulated in cells depleted of HDAC6. Also, in our studies, no other ectopically-expressed class II HDAC affected the acetylation level of cortactin (data not shown). These results strongly suggest that cortactin is a physiologic substrate of HDAC6, but whether HDAC6 is the only biologically relevant cortactin deacetylase remains to be determined. HDAC6 is known to associate with SIRT2, an NAD-dependent class III HDAC and a tubulin deacetylase (North et al., 2003). Therefore, it is possible that cortactin is a substrate of, not only HDAC6, but also SIRT2. Consistent with this possibility, we found that treatment of cells with the class III inhibitor nicotinamide or knockdown of SIRT2 with siRNA further enhances the acetylation of cortactin in TSA-treated cells. The possibility of more than one class of HDACs targeting a single non-histone protein substrate is not without precedent. For example, both HDAC1 and SIRT1 deacetylate the tumor suppressor p53.
We found that PCAF-acetylated cortactin no longer binds F-actin in vitro. This loss of binding activity was dependent on the acetylation of multiple lysines in the cortactin repeat region. Our results suggest that these acetylated lysines alter a “charge patch” that reduces the affinity of cortactin for F-actin in a graded manner. Once some threshold number of lysines becomes acetylated, the F-actin-binding affinity of cortactin is decreased to a level that inhibits the cortactin-F-actin interaction. This would suggest that the acetylation of particular lysine(s) in the patch is of lesser importance than the total number of lysines that are acetylated.
Our computer model predicts two charge patches in the repeat region. The first region is comprised of K124, K189, K198, and K272, and the second is comprised of K161, K309, and K319. Studies of histones H1 and H2A. Z in Tetrahymena have demonstrated a similar mechanism in which their functions are regulated by both acetylation- and phosphorylation-generated charge patches (Dou and Gorovsky, 2000; Ren and Gorovsky, 2001; Ren and Gorovsky, 2003). The mechanism by which the charge of the cortactin repeat region affects its interaction with F-actin is not known. One possibility is that a charge-induced conformational change mediated by HDAC6-induced deactylation favors the accessibility of the cortactin repeat region for interaction with F-actin, while acetylation of this region inhibits this accessibility. A similar mechanism is observed for the ETS-1 transcription factor in which its progressive phosphorylation shifts the protein from an active, DNA-binding conformation to a folded, inactive conformation (Pufall et al., 2005). An alternative, although not mutually exclusive, possibility is that positive charges in the cortactin repeat region are required for interaction with the acidic domain of F-actin. By deacetylating cortactin, HDAC6 enhances the association between cortactin and F-actin as a result of increasing the positive charges in the cortactin repeat region. The seven GF6/Y1GGacKF4/Y3GV6I1 motifs present in the cortactin repeat region share similarity with the acetylated lysine motif in histone H4 (KGGacK). Therefore, the interaction between cortactin and F-actin may be regulated by a mechanism similar to the one that modulates the interaction between histone H4 and cellular proteins.
We found that mutation of the acetylated lysines in the cortactin repeat region prevented cortactin translocation to the cell periphery (i.e., the site of dynamic actin assembly) in response to activated Rac1. Because the phosphorylation of cortactin by Src and Src-related kinases requires the association of cortactin with F-actin (Head et al., 2003), we predict that acetylated cortactin, which is not able to efficiently interact with F-actin, will not be efficiently phosphorylated. In support of this hypothesis, the levels of Src-phosphorylated cortactin were reduced in cells ectopically expressing the 9KQ mutant cortactin relative to the levels in wild-type expressing cells (data not shown). However, further studies are needed to fully elucidate the relationship between cortactin acetylation and Src-mediated cortactin phosphorylation.
In summary, our results demonstrate that the reversible acetylation of cortactin dynamically regulates actin-dependent cell mobility. Cell motility is important for many normal cellular processes, and dysregulated cell motility contributes to vascular disease, chronic inflammatory disease, and tumor metastasis. Thus, our finding that the reversible acetylation of cortactin modulates its actin-binding activity and affects cell migration has many far-reaching clinical relevancies.
Experimental Procedures
Plasmids, Recombinant Proteins, and Antibodies
Details of all plasmid constructions and sources of recombinant proteins and antibodies are provided in the Supplementary Experimental Procedures.
Purification of HDAC6 Complexes
The Flag-tagged HDAC6 insert from pBJ-HDAC6F was excised and ligated downstream of the HA sequence in pcDNA3.1(+)HA. The resultant plasmid was transfected into HeLa S3 cells using Lipofectamine 2000 (Invitrogen), and neomycin-resistant colonies were selected in medium containing 400 μg/ml G418 for two weeks. Thirty-two colonies were screened for Flag-tagged HDAC6 expression by Western blotting with Flag- and HA-specific antibodies. One of the HDAC6-expressing clones was expanded and maintained in DMEM containing 10% calf serum and 200 μg/ml G418.
Cytoplasmic extract was prepared from HeLa cells stably expressing Flag-tagged HDAC6 using a standard protocol (Coligan et al., 1995). Affinity purification of Flag-tagged HDAC6-containing protein complexes using an anti-Flag antibody was performed according to previously published methods (Ogryzko et al., 1998). Purified complexes were concentrated, resolved by SDS-PAGE, and analyzed by silver staining. A Coomassie blue-stained sample was prepared in parallel, and bands corresponding to HDAC6-associated proteins were excised and subjected to proteolysis with trypsin. Peptides from these mixtures were sequenced by microcapillary LC-MS/MS.
Immunoprecipitation and Immunoblotting
For immunoprecipitations, cells were lysed in buffer (50 mM Tris-HCl [pH 7.5], 1 mM EDTA, 1% NP-40, and protease inhibitor cocktail) containing either 500 mM NaCl (high stringency) or 150 mM NaCl (low stringency). Lysates were incubated with the indicated primary antibodies for 12 h at 4°C. Immunocomplexes were collected, washed four times in lysis buffer, and resolved by SDS-PAGE. For immunoblotting, samples were transferred to nitrocellulose membranes that were then probed with the indicated antibodies. Bound antibodies were detected using a Chemiluminescent Detection Kit (Pierce).
GST Pull-Down Assay
GST and GST-cortactin were expressed and purified from bacteria using standard methods. Equimolar amounts of the purified proteins were conjugated to glutathione-Sepharose beads and incubated with purified histidine-tagged HDAC6 for 1 h at room temperature. After extensive washing, bound proteins were eluted and analyzed by Western blotting with anti-His6 antibodies. A full detail protocol is presented in the Supplementary Experimental Procedures.
Two-Dimensional Gel Electrophoresis
One confluent NIH3T3 culture grown in a 150 mm dish was harvested and lysed in 500 μl RIPA buffer containing a cocktail of proteinase, phosphatase, and HDAC inhibitors. Lysates were immunoprecipitated with 10 μg of anti-cortactin antibody and subjected to 2-D gel electrophoresis described in the Supplementary Experimental Procedures.
In vitro Acetylation Assay
Bacterially-expressed GST-tagged cortactin was purified using standard procedures, and 5 μg of protein were incubated with 20 μM of 14C-acetyl CoA and either a fragment of PCAF (500 ng) containing the HAT domain or the HAT fragment of p300 (5 units) in buffer comprised of 50 mM Tris (pH 8), 0.1 mM EDTA, 1 mM DTT, 10% glycerol, 10 mM sodium butyrate, and protease inhibitors. Reactions were allowed to proceed for 1 h at 30°C. Reaction products were resolved by SDS-PAGE and visualized by autoradiography.
Tandem Mass Spectrometry Acetylation Site Analysis
Gel slices containing acetylated cortactin were prepared and subjected to tandem mass spectrometry analysis as described in the Supplementary Experimental Procedures.
Three-Dimensional Modeling of Cortactin
Details of three-dimensional modeling of cortactin are provided in the Supplementary Experimental Procedures. Data are deposited in the Protein Data Bank (PDB ID 2F9X).
F-Actin Co-sedimentation Assay
Rabbit skeletal muscle actin (>99% pure; purchased from Cytoskeleton, Inc.) was polymerized to generate F-actin, according to the manufacturers’ protocol. Typically, purified actin was stored in G-buffer (20 mM Tris-HCl [pH 7.5], 0.2 mM ATP, 0.2 mM DTT, and 0.2 mM CaCl2). Actin was polymerized by adjusting the buffer condition to that of F-buffer (2 mM Tris-HCl [pH 8], 0.2 mM DTT, 0.2 mM CaCl2, 50 mM KCl, 2 mM MgCl2, and 1 mM ATP). F-actin co-sedimentation assays were carried out according to a previously published protocol (Wu and Parsons, 1993). Briefly, cell lysates or recombinant proteins were incubated with 30 μg of F-actin in 50 μl of F-actin binding buffer for 1 h at 4°C. Reaction mixtures were layered with 50 μl of 10% sucrose in F-actin binding buffer and were centrifuged at 64,000 rpm for 1 h at 4°C. Supernatants were transferred to fresh tubes, and pellets were incubated in 50 μl of G buffer for at least 1 h. Equal volumes of supernatant and pellet were analyzed by Western blotting with the indicated antibodies.
Immunostaining and Fluorescence Microscopy
Cells cultured on chamber slides (Chamber Slide System Lab-TekII) were washed with PBS and fixed in 4% paraformaldehyde for 10 min at room temperature. Fixation was terminated with 1% glycine in PBS. For permeabilization, the cells were subjected to several changes of a 1% glycine/0.5% Triton X-100 solution at room temperature for 1 h. Cells were then incubated in PBS containing 0.2% Triton X-100, 1% bovine serum albumin, and primary antibody for 1 h. After washing in PBS containing 0.1% Tween, the cells were incubated for 45 min with secondary antibody in PBS containing 1% bovine serum albumin. Finally, the cells were washed in PBS containing Tween and then PBS alone. The slides were dried and mounted with Vectashield mounting medium with DAPI.
Cell Migration Assay
Transwell migration assays were performed using the CytoSelect™ 24 (8 μm, colorimetric format) kit purchased from Cell Biolabs. A detail protocol is presented in the Supplementary Experimental Procedures.
Live-Cell Imaging
For phase-contrast microscopy, 3 × 104 cells/cm2 were plated on a MatTek coverslip and incubated overnight prior to imaging. Imaging was performed using a Nikon TE-2000-S inverted wildfield microscope equipped with a Retigia 1600 CCD camera and IP Labs 3.6 software from Scanlytics Inc. The images were taken with a 10× 0.25 NA PhI DL lens every 5 min for a total of 205 min. The Image Pro Plus v. 5.0 software was used to measure actual distance and velocity of cell motility. A 37°C re-circulating warm stage was used to maintain cell viability. To calculate the velocity, actual distance cells traveled by tracks (in pixcells) were measured. The sum of total distance was divided by cell number (64 for control and 76 for 9KQ) and time (205 min). Since 115 pixcells equal 10 μm, our final velocity was calculated in μm/cell/min. The Student t test was employed for statistical analysis.
Supplementary Material
Acknowledgments
We thank Stuart Schreiber, Alissa Weaver, Jerry Wu, and Xi Zhan for plasmids, cell lines, and antibodies; Wei Fu, Gui Gao, Michele Glozak, Mark Lloyd, Noreen Luetteke, Jason Phan, and Alejandro Villagra for helpful discussions; John Neveu and Renee Robinson for LC-MS/MS; Upstate Biotechnology (Millipore) for their help with generating the anti-Ac-cortactin antibody; and the Moffitt Cancer Center Core Facility for their technical support. This work was supported by grants to E. S. from the NIH (GM58486, CA109699) and the Kaul Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bali P, Pranpat M, Bradner J, Balasis M, Fiskus W, Guo F, Rocha K, Kumaraswamy S, Boyapalle S, Atadja P, et al. Inhibition of histone deacetylase 6 acetylates and disrupts the chaperone function of heat shock protein 90: a novel basis for antileukemia activity of histone deacetylase inhibitors. J Biol Chem. 2005;280:26729–26734. doi: 10.1074/jbc.C500186200. [DOI] [PubMed] [Google Scholar]
- Bowden ET, Barth M, Thomas D, Glazer RI, Mueller SC. An invasion-related complex of cortactin, paxillin and PKCmu associates with invadopodia at sites of extracellular matrix degradation. Oncogene. 1999;18:4440–4449. doi: 10.1038/sj.onc.1202827. [DOI] [PubMed] [Google Scholar]
- Bowden ET, Onikoyi E, Slack R, Myoui A, Yoneda T, Yamada KM, Mueller SC. Co-localization of cortactin and phosphotyrosine identifies active invadopodia in human breast cancer cells. Exp Cell Res. 2006;312:1240–1253. doi: 10.1016/j.yexcr.2005.12.012. [DOI] [PubMed] [Google Scholar]
- Boyault C, Gilquin B, Zhang Y, Rybin V, Garman E, Meyer-Klaucke W, Matthias P, Muller CW, Khochbin S. HDAC6-p97/VCP controlled polyubiquitin chain turnover. EMBO J. 2006;25:3357–3366. doi: 10.1038/sj.emboj.7601210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryce NS, Clark ES, Leysath JL, Currie JD, Webb DJ, Weaver AM. Cortactin promotes cell motility by enhancing lamellipodial persistence. Curr Biol. 2005;15:1276–1285. doi: 10.1016/j.cub.2005.06.043. [DOI] [PubMed] [Google Scholar]
- Chuma M, Sakamoto M, Yasuda J, Fujii G, Nakanishi K, Tsuchiya A, Ohta T, Asaka M, Hirohashi S. Overexpression of cortactin is involved in motility and metastasis of hepatocellular carcinoma. J Hepatol. 2004;41:629–636. doi: 10.1016/j.jhep.2004.06.018. [DOI] [PubMed] [Google Scholar]
- Coligan JE, Dunn BM, Ploegh HL, Speicher DW, Wingfield PT. Current Protocols in Protein Science. New York: John Wiley & Sons, Inc; 1995. [Google Scholar]
- Dou Y, Gorovsky MA. Phosphorylation of linker histone H1 regulates gene expression in vivo by creating a charge patch. Mol Cell. 2000;6:225–231. doi: 10.1016/s1097-2765(00)00024-1. [DOI] [PubMed] [Google Scholar]
- Haggarty SJ, Koeller KM, Wong JC, Grozinger CM, Schreiber SL. Domain-selective small-molecule inhibitor of histone deacetylase 6 (HDAC6)-mediated tubulin deacetylation. Proc Natl Acad Sci USA. 2003;100:4389–4394. doi: 10.1073/pnas.0430973100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Head JA, Jiang D, Li M, Zorn LJ, Schaefer EM, Parsons JT, Weed SA. Cortactin tyrosine phosphorylation requires Rac1 activity and association with the cortical actin cytoskeleton. Mol Biol Cell. 2003;14:3216–3229. doi: 10.1091/mbc.E02-11-0753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hook SS, Orian A, Cowley SM, Eisenman RN. Histone deacetylase 6 binds polyubiquitin through its zinc finger (PAZ domain) and copurifies with deubiquitinating enzymes. Proc Natl Acad Sci USA. 2002;99:13425–13430. doi: 10.1073/pnas.172511699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Liu J, Haudenschild CC, Zhan X. The role of tyrosine phosphorylation of cortactin in the locomotion of endothelial cells. J Biol Chem. 1998;273:25770–25776. doi: 10.1074/jbc.273.40.25770. [DOI] [PubMed] [Google Scholar]
- Hubbert C, Guardiola A, Shao R, Kawaguchi Y, Ito A, Nixon A, Yoshida M, Wang XF, Yao TP. HDAC6 is a microtubule-associated deacetylase. Nature. 2002;417:455–458. doi: 10.1038/417455a. [DOI] [PubMed] [Google Scholar]
- Kaksonen M, Peng HB, Rauvala H. Association of cortactin with dynamic actin in lamellipodia and on endosomal vesicles. J Cell Sci. 2000;113:4421–4426. doi: 10.1242/jcs.113.24.4421. [DOI] [PubMed] [Google Scholar]
- Kawaguchi Y, Kovacs JJ, McLaurin A, Vance JM, Ito A, Yao TP. The deacetylase HDAC6 regulates aggresome formation and cell viability in response to misfolded protein stress. Cell. 2003;115:727–738. doi: 10.1016/s0092-8674(03)00939-5. [DOI] [PubMed] [Google Scholar]
- Kovacs JJ, Murphy PJ, Gaillard S, Zhao X, Wu JT, Nicchitta CV, Yoshida M, Toft DO, Pratt WB, Yao TP. HDAC6 Regulates Hsp90 Acetylation and Chaperone-Dependent Activation of Glucocorticoid Receptor. Mol Cell. 2005;18:601–607. doi: 10.1016/j.molcel.2005.04.021. [DOI] [PubMed] [Google Scholar]
- Kowalski JR, Egile C, Gil S, Snapper SB, Li R, Thomas SM. Cortactin regulates cell migration through activation of N-WASP. J Cell Sci. 2005;118:79–87. doi: 10.1242/jcs.01586. [DOI] [PubMed] [Google Scholar]
- Kozma R, Ahmed S, Best A, Lim L. The Ras-related protein Cdc42Hs and bradykinin promote formation of peripheral actin microspikes and filopodia in Swiss 3T3 fibroblasts. Mol Cell Biol. 1995;15:1942–1952. doi: 10.1128/mcb.15.4.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Tondravi M, Liu J, Smith E, Haudenschild CC, Kaczmarek M, Zhan X. Cortactin potentiates bone metastasis of breast cancer cells. Cancer Res. 2001;61:6906–6911. [PubMed] [Google Scholar]
- Matsuyama A, Shimazu T, Sumida Y, Saito A, Yoshimatsu Y, Seigneurin-Berny D, Osada H, Komatsu Y, Nishino N, Khochbin S, et al. In vivo destabilization of dynamic microtubules by HDAC6-mediated deacetylation. EMBO J. 2002;21:6820–6831. doi: 10.1093/emboj/cdf682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobes CD, Hall A. Rho, rac, and cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell. 1995;81:53–62. doi: 10.1016/0092-8674(95)90370-4. [DOI] [PubMed] [Google Scholar]
- North BJ, Marshall BL, Borra MT, Denu JM, Verdin E. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol Cell. 2003;11:437–444. doi: 10.1016/s1097-2765(03)00038-8. [DOI] [PubMed] [Google Scholar]
- Ogryzko VV, Kotani T, Zhang X, Schiltz RL, Howard T, Yang XJ, Howard BH, Qin J, Nakatani Y. Histone-like TAFs within the PCAF histone acetylase complex. Cell. 1998;94:35–44. doi: 10.1016/s0092-8674(00)81219-2. [DOI] [PubMed] [Google Scholar]
- Patel AS, Schechter GL, Wasilenko WJ, Somers KD. Overexpression of EMS1/cortactin in NIH3T3 fibroblasts causes increased cell motility and invasion in vitro. Oncogene. 1998;16:3227–3232. doi: 10.1038/sj.onc.1201850. [DOI] [PubMed] [Google Scholar]
- Pufall MA, Lee GM, Nelson ML, Kang HS, Velyvis A, Kay LE, McIntosh LP, Graves BJ. Variable control of Ets-1 DNA binding by multiple phosphates in an unstructured region. Science. 2005;309:142–145. doi: 10.1126/science.1111915. [DOI] [PubMed] [Google Scholar]
- Ren Q, Gorovsky MA. Histone H2A. Z acetylation modulates an essential charge patch. Mol Cell. 2001;7:1329–1335. doi: 10.1016/s1097-2765(01)00269-6. [DOI] [PubMed] [Google Scholar]
- Ren Q, Gorovsky MA. The nonessential H2A N-terminal tail can function as an essential charge patch on the H2A. Z variant N-terminal tail. Mol Cell Biol. 2003;23:2778–2789. doi: 10.1128/MCB.23.8.2778-2789.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigo JP, Garcia LA, Ramos S, Lazo PS, Suarez C. EMS1 gene amplification correlates with poor prognosis in squamous cell carcinomas of the head and neck. Clin Cancer Res. 2000;6:3177–3182. [PubMed] [Google Scholar]
- Schuuring E, Verhoeven E, Litvinov S, Michalides RJ. The product of the EMS1 gene, amplified and overexpressed in human carcinomas, is homologous to a v-src substrate and is located in cell-substratum contact sites. Mol Cell Biol. 1993;13:2891–2898. doi: 10.1128/mcb.13.5.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seigneurin-Berny D, Verdel A, Curtet S, Lemercier C, Garin J, Rousseaux S, Khochbin S. Identification of components of the murine histone deacetylase 6 complex: link between acetylation and ubiquitination signaling pathways. Mol Cell Biol. 2001;21:8035–8044. doi: 10.1128/MCB.21.23.8035-8044.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tehrani S, Faccio R, Chandrasekar I, Ross FP, Cooper JA. Cortactin has an essential and specific role in osteoclast actin assembly. Mol Biol Cell. 2006;17:2882–2895. doi: 10.1091/mbc.E06-03-0187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uruno T, Liu J, Zhang P, Fan Y, Egile C, Li R, Mueller SC, Zhan X. Activation of Arp2/3 complex-mediated actin polymerization by cortactin. Nat Cell Biol. 2001;3:259–266. doi: 10.1038/35060051. [DOI] [PubMed] [Google Scholar]
- Weaver AM, Karginov AV, Kinley AW, Weed SA, Li Y, Parsons JT, Cooper JA. Cortactin promotes and stabilizes Arp2/3-induced actin filament network formation. Curr Biol. 2001;11:370–374. doi: 10.1016/s0960-9822(01)00098-7. [DOI] [PubMed] [Google Scholar]
- Weed SA, Du Y, Parsons JT. Translocation of cortactin to the cell periphery is mediated by the small GTPase Rac1. J Cell Sci. 1998;111:2433–2443. doi: 10.1242/jcs.111.16.2433. [DOI] [PubMed] [Google Scholar]
- Weed SA, Karginov AV, Schafer DA, Weaver AM, Kinley AW, Cooper JA, Parsons JT. Cortactin localization to sites of actin assembly in lamellipodia requires interactions with F-actin and the Arp2/3 complex. J Cell Biol. 2000;151:29–40. doi: 10.1083/jcb.151.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong K, Zhang J, Awasthi S, Sharma A, Rogers L, Matlock EF, Van Lint C, Karpova T, McNally J, Harrod R. Nerve growth factor receptor signaling induces histone acetyltransferase domain-dependent nuclear translocation of p300/CREB-binding protein-associated factor and hGCN5 acetyltransferases. J Biol Chem. 2004;279:55667–55674. doi: 10.1074/jbc.M408174200. [DOI] [PubMed] [Google Scholar]
- Wu H, Parsons JT. Cortactin, an 80/85-kilodalton pp60src substrate, is a filamentous actin-binding protein enriched in the cell cortex. J Cell Biol. 1993;120:1417–1426. doi: 10.1083/jcb.120.6.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Reynolds AB, Kanner SB, Vines RR, Parsons JT. Identification and characterization of a novel cytoskeleton-associated pp60src substrate. Mol Cell Biol. 1991;11:5113–5124. doi: 10.1128/mcb.11.10.5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida N, Omoto Y, Inoue A, Eguchi H, Kobayashi Y, Kurosumi M, Saji S, Suemasu K, Okazaki T, Nakachi K, et al. Prediction of prognosis of estrogen receptor-positive breast cancer with combination of selected estrogen-regulated genes. Cancer Sci. 2004;95:496–502. doi: 10.1111/j.1349-7006.2004.tb03239.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Li N, Caron C, Matthias G, Hess D, Khochbin S, Matthias P. HDAC-6 interacts with and deacetylates tubulin and microtubules in vivo. EMBO J. 2003;22:1168–1179. doi: 10.1093/emboj/cdg115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S, Webb BA, Eves R, Mak AS. Effects of tyrosine phosphorylation of cortactin on podosome formation in A7r5 vascular smooth muscle cells. Am J Physiol Cell Physiol. 2006;290:C463–471. doi: 10.1152/ajpcell.00350.2005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.