Abstract
The nicotinic acetylcholine receptor (nAChR) is a member of the ligand-gated ion channel family and is implicated in many neurological events. Yet, the receptor is difficult to target without high-resolution structures. In contrast, the structure of the acetylcholine binding protein (AChBP) has been solved to high resolution, and it serves as a surrogate structure of the extra-cellular domain in nAChR. Here we conduct a virtual screening study of the AChBP using the relaxed-complex method, which involves a combination of molecular dynamics simulations (to achieve receptor structures) and ligand docking. The library screened through comes from the National Cancer Institute, and its ligands show great potential for binding AChBP in various manners. These ligands mimic the known binders of AChBP; a significant subset docks well against all species of the protein and some distinguish between the various structures. These novel ligands could serve as potential pharmaceuticals in the AChBP/nAChR systems.
Keywords: acetylcholine binding protein, nicotinic acetylcholine receptor, relaxed-complex, molecular dynamics, docking, virtual screening
1. Introduction
1.1. Nicotinic acetylcholine receptor and the acetylcholine binding protein
The ligand-gated ion channels (LGIC) constitute a class of membrane proteins that function in a variety of biological processes. For instance, in signal transduction, they play a key role in the transfer of information across neurological networks [1]. In the realm of disorders, these ion channels have been implicated in the onset of Alzheimer's disease and drug-addiction [2, 3]. As such, these proteins and the ligands that bind to them are of great interest in the pharmacological communities. If a binding ligand affects the activity of the channel in question, that ligand is labeled as either an agonist or antagonist. In the former case, ligand binding enhances the neurological phenomena associated with the particular ion channel. On the other hand, if an antagonist, the ligand can hinder the neurological event [4].
The nicotinic acetylcholine receptors (nAChRs) are members of the LGIC family. Upon acetylcholine (ACh) binding, the receptors assume the open-state and allow the influx of ions into the cell. This is the chemical basis of neuronal response and muscle activity [5]. As its name suggests, nicotine binds nAChR, and it does so in an agonistic fashion, leading to increased ion flow and neuronal stimulation. This agonistic behavior of nicotine leads to patient addiction by increasing the number of high-affinity nAChR in the membrane of neurons [6]. Other drugs, such as cocaine, also bind nAChR and stimulate the receptor [7]. Thus, nAChR serves as a great pharmacological target in research on smoking-cessation and drug addiction.
Yet, to be employed in pharmacology and particularly in structure-based drug discovery, the structure of the target protein must be known to a high degree of confidence. This is usually afforded by x-ray crystallography. Yet, achieving a high resolution structure is an especially difficult feat when working with membrane proteins, whose crystallization does not occur as easily as their aqueous counterparts. The lipid environment in the membrane can greatly affect the structure of the membrane protein [8], and it is difficult to capture this final structure via crystallography. To date, the highest resolution achieved for nAChR is a 4 Å result obtained by cryo-electron microscopy, by Unwin et al [9]. In this work, the pentameric structure of nAChR and the distinctive extra-cellular and transmembrane domains are resolved clearly. However, from a pharmacological standpoint, much is left to be desired regarding side chain orientations and other important conformational details.
Rather fortunately, there exists an aqueous protein that mimics the extra-cellular half of nAChR, where ligand binding occurs. This protein, the acetylcholine binding protein (AChBP), comes from three species of sea snails: Lymnaea stagnalis (Ls); Aplysia californica (Ac); and Bulinus truncates (Bt) [10]. The exact function of AChBP is unknown, although it is thought to play a role in modulating neuronal response by binding acetylcholine in synaptic clefts [11]. Experimentally, AChBP is significantly easier to work with in terms of expression and crystallization. To date, over a dozen structures of AChBP (with and without ligands) have been solved with high resolution [12, 13, 14, 15].1 Although its sequence identity to the extra-cellular domain of nAChR is only roughly 30%, AChBP bears a great resemblance with respect to its secondary structure. As shown by Brejc et al., the general shape of AChBP is that of a hollow cylinder with an outer, inner diameter and height of 80, 18, and 62 Å, respectively. AChBP is a homopentamer, each subunit consisting of 210 residues.
As in the extra-cellular domain of nAChR, ligands bind to AChBP at the interface between subunits; thus, with 5 subunits arranged in a cylinder, the maximum ratio of ligand:protein is 5:1 [16]. The ligand binds in the approximate middle region of this interfacial axis, typically behind a characteristic loop extending from one of the subunits, known as the ‘C-loop’ [11]. The chemical nature of this binding site is discussed in greater detail below. Because of its great structural similarity to the ligand-binding extra-cellular domain of nAChR, AChBP has been deemed a surrogate structure of the receptor [17]. By targeting and identifying binders of AChBP, researchers hope to gain more understanding about nAChR. Common research themes about AChBP/nAChR focus on which ligands act as agonists or antagonists, which bind universally across all subtypes of nAChR and all species of AChBP, which ligands can distinguish between the various forms, and which can serve as leads for drug discovery efforts. Although there are marked differences between the AChBP and nAChR—most obviously, the presence of the transmembrane domain in the latter—the two proteins have enough chemical and physical similarities to warrant using AChBP as a template structure of the receptor [18]. Thus, only the three species of AChBP are targeted in this study, but the implications can be far-reaching on the entire class of nicotinic acetylcholine receptors. Furthermore, what is unclear is whether or not ligand binding can occur in regions along this interfacial unit but away from the C-loop region. Can the virtual screening protocol presented here position the ligands in the traditional C-loop region as well as in novel potential binding sites? We will see that this is indeed the case.
The known ligands and various derivatives of the AChBP/nAChR systems have been extensively studied; unfortunately, many would fail as medicinal drug candidates because of their poor pharmacological properties [16]. The known small molecule ligands that do succeed in this light are harmful because of their addictive and abusive nature; these include cocaine, heroine, morphine and nicotine [19]. Thus, we are in search of new ligands that can bind AChBP and possibly play pharmacological roles in the AChBP/nAChR systems. The set through which we explore comes from the National Cancer Institute (NCI), which harbors a database of nearly 250 000 drug-like compounds. In the computational work presented here, we conduct a virtual screening study of AChBP using (as the ligand set) the National Cancer Institute Diversity Set (NCIDS)—consisting of approximately 2 000 molecules—which serves as a representative subset of the entire NCI database [20]. Indeed, the NCIDS ligands are novel to the AChBP/nAChR system (to the best of our knowledge) and they exhibit the desired pharmacological properties of small molecule drugs (low molecular weight, high solubility).
1.2 Accounting for protein flexibility: the relaxed-complex method
In any virtual screening or docking protocol, one must first determine how to obtain protein structures to dock against. One can readily employ crystal structures from the Protein Data Bank (PDB), but these static structures do not adequately depict the flexibility of the protein. Thus, docking against them does not usually reflect the true dynamical nature of most protein-ligand interactions. Various docking protocols exist that attempt to surmount this hurdle of protein flexibility. One recent application, FLIPDOCK [21], attempts to achieve protein flexibility by toggling the positions of various side chains in the active site during the docking runs. This is a useful and quick way of capturing the local protein flexibility in the binding site. Yet, because of computational expense, one can only toggle a few (3 to 5) side chains, and this can be very limiting if there are many side chains implicated in ligand binding. The technique is also unfeasible if the exact binding site is unknown, or if the user intends to explore larger sites within the protein. Moreover, the application does not capture global conformational changes of the protein, and such changes could dramatically affect the binding pocket.
An alternative method in capturing protein flexibility is the relaxed-complex method, in which a molecular dynamics (MD) simulation of the target protein is first conducted [22, 23]. If the time scale of the simulation coincides with that of the protein's functional dynamics, one can capture local (and sometimes global) conformational changes in the protein. Snapshot structures can be taken from the MD simulation and employed in the screening of the ligand set using a computational docking tool. The results from these different receptor snapshots can be compared to ascertain if/how the conformational changes in the protein affect ligand binding. In the relaxed-complex scheme, there are various simulation and post-simulation processing methods that one can employ for capturing the receptor (protein) structures. As detailed by Amaro et al., the simulation can consist of standard equilibrium MD or even non-equilibrium methods, such as steered MD. A variety of techniques such as principal component or root mean square deviation (RMSD) analysis can then be used to pick out snapshots. Exactly which simulation and analysis techniques afford the best structures for docking is the subject of much debate. The ideal method probably varies from system to system; it is thus crucial to verify any technique against experimental evidence before pursuing novel endeavors. In our case, for the AChBP, we first provide such a comparison to experimental data for validation before employing the relaxed-complex scheme on the novel set (NCIDS).
1.3 Current approach and objectives to identify novel binders
Using the relaxed-complex method, we conducted MD simulations of all three species of AChBP, selected 5 snapshots from each simulation, and screened through the NCIDS for each snapshot. Given the aforementioned knowledge of the AChBP binding site, the RMSD of the characteristic C-loop that covers the binding site was chosen as the metric for snapshot selection. Specifically, through the presented work here, we address the following questions: 1. Can our relaxed-complex scheme—focusing on just one loop region—produce comparable results with previous experimental evidence (thus implicating the usage of this technique in other AChBP/nAChR systems)? 2. Can we identify new ligand binding regions within the interface of AChBP? 3. Can the NCIDS serve as a novel source of small molecule drugs targeting AChBP (and thus possibly the entire genre of AChBP/nAChR proteins)?
The results from this virtual screening protocol agree well with previous experimental evidence, particularly with respect to the predicted poses and energetic trends of the known ligand binders of AChBP. Thus, the dynamical C-loop does serve as a sufficient metric for conformational selection. The NCIDS ligands explore almost the entire subunit interface and demonstrate their propensity to bind both the C-loop and non-C-loop regions. A good number of them dock reasonably across all species and all snapshots of AChBP, and their binding interactions resemble those of known binders. All of this suggests that the NCIDS could serve as a good set from which to pick novel ligands against AChBP/nAChR. Even more interesting is that some of the NCIDS ligands dock differently into the three species of AChBP, and these ligands could serve to distinguish between them and possibly between the various subtypes of nAChR.
2. Materials and Methods
2.1 System construction and molecular dynamics simulations
From the PDB, structures for the Ls (1I9B) [11], Ac (2BYN) [14], and Bt (2BJ0) [24] species of AChBP were obtained. All non-protein atoms were stripped from these structures. The GROMACS MD package was then used to place each protein in its own simulation box of dimensions 100 × 100 × 82 Å, to solvate and neutralize the system with counterions, and then to carry out all subsequent MD simulations [25]. Each system (3 in total) consisted of one of the AChBP species (the complete protein, as a pentamer), approximately 22 000 simple-point charge (SPC) model water molecules, and approximately 45 sodium ions. Each system totaled approximately 82 000 atoms.
Unfavorable contacts in each system were relieved by two cycles (5 000 steps each) of conjugate-gradient and steepest-descent minimizations. With the backbone of the protein restrained, each system was then heated to a temperature of 300K over a short 50 ps simulation. Pressure coupling was then added and a simulation was carried out for 1 ns with the alpha carbons of the protein restrained. The restraints were removed, and each equilibrated protein was simulated for 20 ns of production. The OPLSAA force field parameters were used for each AChBP system [26]. The simulation parameters consisted of the following: a 2 fs time-step; coordinates and velocities recorded at every 500 and 1 000 steps, respectively; bond length constraints were imposed using the LINCS method; full electrostatics were calculated using the particle mesh Ewald technique, with coulombic and van der Waals cut-offs of 9 and 14 Å, respectively; Ewald tolerance was set to 1 × 10-5; nearest-neighbor lists were updated every 10 steps, with a cut-off of 9 Å; periodic boundary conditions were employed in all directions; Berendsen temperature coupling was used across the entire system, with a coupling constant of 0.1 ps, to maintain a temperature of 300K; Berendsen isotropic pressure coupling was used across the entire system, with a reference pressure of 1.0 bar, a pressure coupling of 0.5 ps, and a compressibility of 4.5 × 10−5 bar−1. Analysis of the MD simulations was conducted using the various GROMACS tools.
2.2 Protein structure/ligand library selection and docking
For each species of AChBP, each subunit interface (there are 5 in AChBP) was examined for the greatest flexibility in the C-loop region. To reflect this flexibility, 5 snapshots of a pair of subunits forming an interface, from each species, were taken at approximately 0 (just before the production run), 5, 10, 15, and 20 ns. Thus, screening was done with 3 (species) × 5 (snapshots) = 15 AChBP receptor structures. These structures were screened against using Autodock 4.0 [27, 28] and 1 834 ligands from the NCIDS, which were previously parameterized for usage in Autodock by Chang et al [29]. These 15 × 1 834 = 27 510 Autodock jobs were performed on a cluster of 128 processors—each 3.2 GHz EM64T with hyper threading capability, 2GB RAM—at the National Biomedical Computational Resource (NBCR) [30]. Each receptor structure, consisting of 2 subunits that meet to make the binding interface, was prepared for docking using a script from the AutoDockTools kit that adds Gasteiger charges to the structure. Grid maps were generated, with the center of the grid placed in the middle of the subunit interfacial axis, at a spacing of 0.375 Å and with grid dimensions of 70 × 70 × 70 points. For each Autodock job, default parameters were used with the following exceptions: the number of energy evaluations (ga_num_evals) was set to 5 × 106; the root mean square deviation tolerance (rmstol) for clustering was set to 1.5 Å; and the number of dockings (ga_run) was set to 100. This scheme was used for both the NCIDS ligands and the experimental ligands discussed below. In the latter case, the ligand structures were obtained from the PDB codes indicated in Table 1 and docked back into their crystal structures. In the work presented here, the NCIDS ligands are referred to by their name (if available) or by their NCI index number (also known as the NSC number). The structure and pharmacological data of the NCIDS ligands were obtained using this NCI/NSC number, via the NCI Query system [31].
Table 1.
A comparison of the docked and experimental results, with respect to the pose accuracy of the ligand. The first column is the PDB ID of the ligand-AChBP complex, the second column is the full name of the ligand, followed by the ligand code (in the third column) as found in the PDB. The last column of the table shows the root mean square deviation (RMSD, in Å) of the docked ligand as compared to the crystal structure.
| PDB | Ligand Name | Code | RMSD |
|---|---|---|---|
| 2BYQ | Epibatidine | EPJ | 0.5 |
| 1UW6 | Nicotine | NCT | 0.7 |
| 2UZ6 | Alpha-Conotoxin | - | 0.9 |
| 2BJ0 | 3-Cyclo-hexyl-1-propyl-sulfonic Acid | CXS | 1.1 |
| 2BYS | Lobeline | LOB | 1.3 |
| 1UV6 | 2-[(Amino-carbonyl)oxy]-N,N,N-Trimethyl-ethanaminium | CCE | 1.4 |
| 2BR7 | 4-(2-Hydroxy-ethyl)-1-Piperazine Ethanesulfonic Acid | EPE | 1.9 |
| 1I9B | ″ | EPE | 2.8 |
| 1UX2 | ″ | EPE | 7.7 |
| 2PGZ | Cocaine | COC | 3.8 |
| 2BYN | Pentaethylene Glycol | 1PE | 6.0 |
| 2BYR | Methyllyca-conitine | MLK | 8.0 |
For any ligand, from the Autodock result, the conformation from the most populated cluster was selected as the docked ‘pose’ for that ligand; this conformation may or may not exhibit the most negative free energy of binding. Because the pose is from the most populous cluster, we take it to be the statistically-probable result. The calculated binding energy for this pose was taken to be the ‘calculated’ or ‘predicted’ free energy of binding for the ligand in question. Analysis of the Autodock results was performed using common Autodock shell scripts and several scripts written by the authors. Data was plotted using MATLAB©. Visualization and rendering of AChBP and AChBP-ligand complexes were completed using the Visual Molecular Dynamics (VMD) software [32]. Structural alignment of the AChBP species was performed using the MultiSeq module in VMD.
3. Results and Discussion
3.1 Dynamics of the acetlycholine binding protein
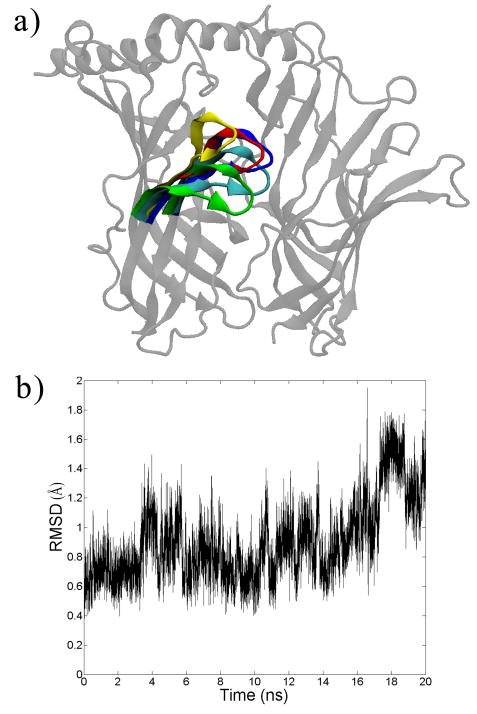
We first explore the dynamics of the binding site in AChBP, which (as mentioned above) occurs in each interfacial region between any two subunits. Following previous work, the subunit depicted on the left is known as the ‘plus’ (+) face and its characteristic feature is the ‘C-loop’, a loop in the protein consisting of residues 180-190, including the characteristic vicinal cysteine residues [33]. The subunit depicted on the right is termed the ‘minus’ (–) face. Most known binders of AChBP bind behind the C-loop in the middle of the axis in between the subunits. It has been suggested that the C-loop acts as a sort of flexible gate that covers the binding site; when a ligand binds, the C-loop becomes less flexible and covers the ligand-site complex [34, 35]. The C-loop flexibility is indeed observed in our molecular dynamics simulation as shown in Figure 1 and in the animations provided in the supporting information. At 5 ns intervals, one can readily observe the various positions of the C-loop. It should be noted though that this flexibility does not result in a complete opening of the subunit, for such an event does not occur on the present timescale of the simulation [36]. Nonetheless, one can observe that the C-loop takes on different conformations during the simulation, and the exact position of the C-loop may impact ligand binding.
Figure 1.
a) Snapshots from the molecular dynamics simulation of the acetylcholine binding protein (species Aplysia Californica, PDB: 2BYN). Only 1 of the 5 interfaces is shown here for clarity. The C-loop is colored in 5 ns increments. b) The average root mean square deviation (RMSD) of the C-loop (Cα carbons only) as a function of simulation time.
3.2 Comparison With Previous Experimental Evidence
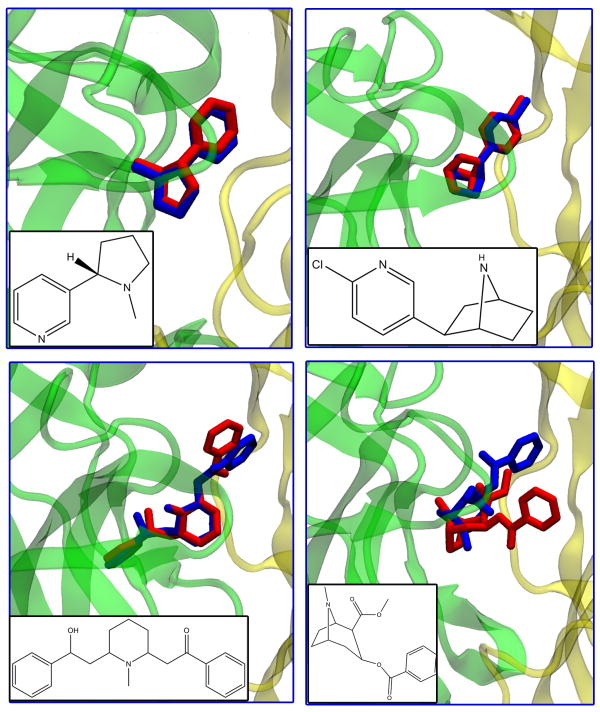
Oftentimes, the accuracy of any computational docking protocol is measured against known binding ligands of a protein. Such validation is crucial for justifying the current docking method. In our case, we seek to ensure the validity of selecting conformations based on the flexibility of one simple loop (the C-loop) in a rather large protein. Fortunately, for AChBP, there are many solved structures of ligand-receptor complexes in the PDB. The ligands of these complexes were docked back into AChBP; the results are shown in Table 1 and Figure 2. In most cases, Autodock is fairly accurate in predicting the pose of the ligand, especially in the case of the well-known binders epibatidine and nicotine, whose docked poses exhibit less than a 1 Å RMSD from the crystal structure. It is well-documented that the smaller ligands of AChBP bind more rigidly behind the C-loop in between the two interfaces [37].
Figure 2.
Docked (blue) vs. crystal (red) structures of some common ligands that bind the acetylcholine binding protein. Top Left: Nicotine, Top Right: Epibatidine, Bottom Left: Lobeline, Bottom Right: Cocaine.
As the ligands increase in size and in the number of rotatable bonds, the RMSD between docked and crystal structures increases, as can be seen in the cases of lobeline and cocaine. One suspects that the lobeline result is more accurate than the cocaine because the former exhibits a more symmetrical (or ‘butterfly’) structure, which possibly aids the lobeline in fitting along the interfacial axis between the subunits of AChBP. We discuss below ligands from the NCIDS that exhibit this same behavior. In the case of cocaine, its bulkiness and size possibly preclude a better predicted pose. In the case of the even larger ligands—pentaethylene glycol (several rotatable bonds) and methyllycaconitine (a macrocyclic structure)—the pose accuracy is even worse (> 5 Å RMSD). Such problems in dealing with large rotatable ligands are common when using Autodock and other docking codes [38]. Nonetheless, though the exact poses are off for these ligands, Autodock does place them in the general vicinity of binding (behind the C-loop) along the subunit interface, and this placement agrees well with previous experimental evidence [39].
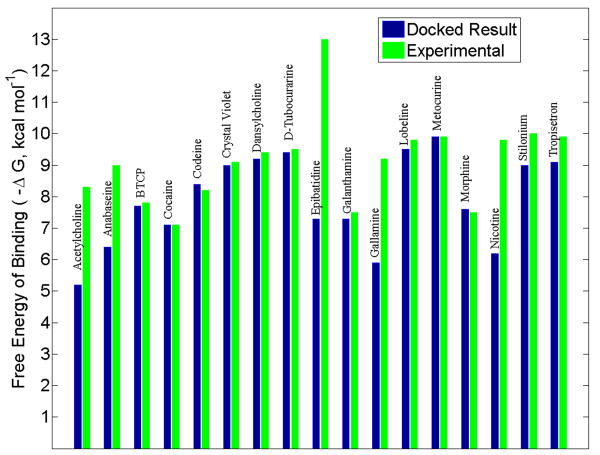
Regarding the accuracy of the calculated free energy of binding, the current docking protocol does not predict with great accuracy the actual free energy as compared to experimental evidence [14, 16, 17, 37, 40], but it does pick up on some key trends; see Figure 3. For instance, both experimental and computational results show nicotine binding better (a more negative free energy change) than the natural ligand of AChBP, acetylcholine. Although the docking does not show epibatidine to be the best binder, it does correctly show that epibatidine binds better than nicotine. The predicted free energy of binding seems to be more accurate in the larger ligands (cocaine, D-Tubocurarine, metocurine). A slight trend is also observed between the molecular weight and the calculated free energy of binding, in the sense that the larger ligands tend to bind more favorably (see Figure S1 in the Supporting Information). Given the large binding space afforded by the subunit interface, it is not surprising that the calculated energies for the larger ligands are more accurate. In any case, it stands to reason that more emphasis should be placed on the predicted trends rather than the absolute binding energy numbers. Thus, because the energetic trends and docked poses agree fairly well with previous experimental results, we have confidence that the relaxed-complex scheme presented here—where we select and dock against various conformations of the C-loop—can be employed in the search for novel ligands.
Figure 3.
A comparison of the Autodock calculated free energies of binding with experimental values, of known binders to the acetylcholine binding protein (Ls species). For the docked value, the best result was selected from the snapshots afforded by the relaxed-complex method. Experimental values for BTCP (N-[1-(2-benzothiophenyl)cyclohexyl]piperidine), codeine, crystal violet, morphine, stilonium, and tropisetron were obtained via personal correspondence with Talley. All other experimental values were obtained from references 14, 16, 17, 37, and 40.
3.3 Virtual Screening of the National Cancer Institute Diversity Set (NCIDS)
It is interesting to note that the predicted binding energies of the NCIDS ligands cluster in the same key range as the known ligands of AChBP; see Figure S2 in the Supporting Information. For the purposes of this study and because of the known calculated noise associated with Autodock [29], we will focus on those ligands with a predicted binding energy near or less than -7.0 kcal mol-1. Although we do not yet have experimental results of the NCIDS ligands against AChBP, the computational work presented here is very promising, in the sense that the novel set mimics the binding of known compounds. Furthermore, because the docking was conducted throughout the entire interfacial region, the NCIDS ligands bound to several residues along the axis between the two AChBP subunits; see Figures 4 and 5. While many docked into the traditional C-loop binding region, what is striking is that many of the NCIDS ligands explored the remaining interfacial space, suggesting the prevalence of new binding sites and modes. We discuss below both the C-loop and non-C-loop binding regions.
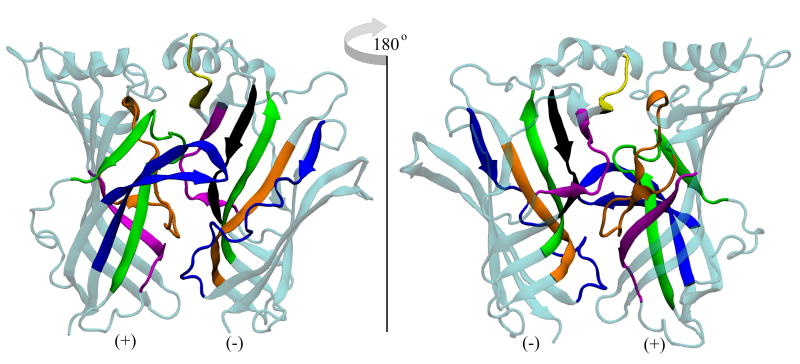
Figure 4.
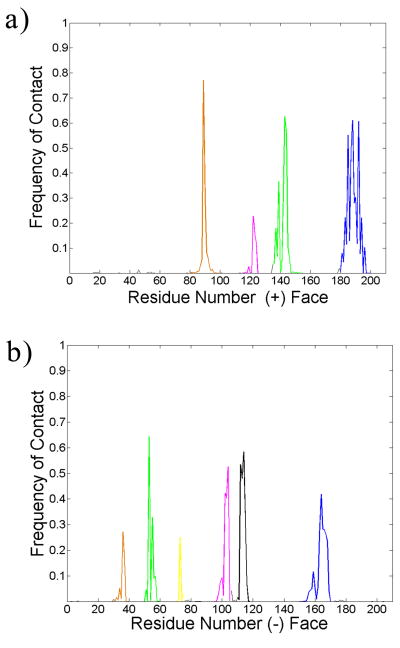
Front and rear views of the binding (interfacial) site. The subunit containing the C-loop is often termed the ‘plus’ (+) face, while the adjacent subunit is the ‘minus’ (-) face. All colored regions show significant contact with the ligands of the NCI Diversity Set. The frequency profile of contacts can be seen in Figure 5. Plus face: orange corresponds to residues 80-100, pink to 115-125, green to 135-155, blue to 180-200. Minus face: orange corresponds to residues 30-40, green to 50-60, yellow to 70-75, pink to 95-105, black to 110-118, and blue to residues 150-170.
Figure 5.
The frequency of contacts (within 5 Å of a residue) between all of the ligands of the NCI Diversity Set and the acetylcholine binding protein. Coloring scheme matches that of Figure 4.
As expected, the ligands make a significant number of contacts with and around the C-loop region of the plus face (residues 180-200). Those ligands that bind well in this region do so with respect to all three species of AChBP. Table S1 in the Supporting Information displays a list of compounds from the NCIDS that bind to all three species and across all snapshots of the protein. These ligands should not distinguish between the three species of AChBP because they all bind in a similar fashion behind the C-loop. The 50 compounds listed in Table S1 that dock across all three species and behind the C-loop exhibit the same characteristics of previous known binders of AChBP. 19 of them (almost half) bear a tryptophan-like moiety, and this structure provides a key point of stabilization in the C-loop region. In this region of the protein, an ‘aromatic cage’ stabilizes ligands that contain aromatic functionalities [41]. This cage consists of several key tryptophan and tyrosine residues, and they are observed to make stabilizing contacts with the docked NCIDS ligands as well. An example of this interaction can be seen in Figure S3 of the Supporting Information. Several of the NCIDS compounds that bind all three species and snapshots of AChBP contain amines that can bear a charge depending on the pH of the environment. In turn, these charged amines can interact with the electronic π networks of the aromatic amino acids, and these cation-π interactions also serve to stabilize the ligand in the AChBP binding pocket [15, 42]. That a significant number of the NCIDS ligands exhibit groups that can hold a charge and contain aromatic functionalities that can interact with the aromatic cage makes the NCIDS a practical pool from which to pick novel ligands against all species of AChBP.
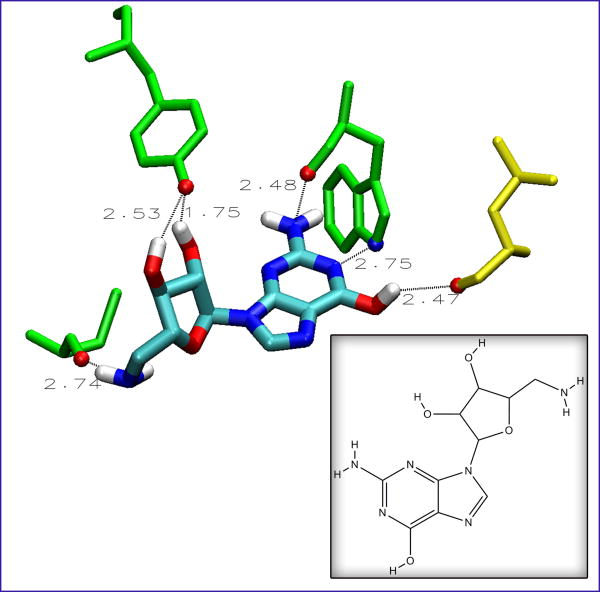
Hydrogen bonds also play a role in stabilizing the AChBP-ligand complex [35]. Backbone atoms on the protein and key side chains (such as tryptophan and tyrosine) can make hydrogen bonds with the ligand [24]. The NCIDS compounds also exhibit several hydrogen bonding moieties. Along with amines, many bear several hydroxyl groups in key positions that can help stabilize the ligand in the binding site. A prime example is NCI 108608, as shown in Figure 6. This ligand contains all of the aforementioned characteristics, including a tryptophan-like structure, amines, and several hydroxyl groups. It forms close contacts with at least two residues of the C-loop region on the plus face (CYS187 and TYR192), a third residue characteristic of the aromatic cage (TRP143), and a residue on the minus face (LEU102). These interactions coupled together could make this ligand a strong binder against all three species of AChBP.
Figure 6.
The extensive hydrogen-bonding character of NCI 108608, to the Lymnaea stagnalis species of AChBP. Numbers indicate distances in angstroms. Residues from the plus face of AChBP are shown in green, and they are (from the left) CYS187, TYR192, and TRP143. The yellow residue is LEU102 and from the minus face. The hydrogen bonding atoms on the protein residues as well as the atoms of the ligand are colored cyan, red, white, and blue, for carbon, oxygen, hydrogen, and nitrogen, respectively. Inset shows the 2D structure of the ligand.
If the ligand binds behind the C-loop in the interfacial region of AChBP, then the C-loop conformation itself plays a significant role in stabilization. For instance, in the binding of large ligands such as the conotoxins, the C-loop is usually extended outward and away from the cylindrical space of AChBP. On the other hand, when binding smaller ligands such as epibatidine, the C-loop is bent inwards and more towards the central axis of the protein [40]. Based on this knowledge and the various poses of the C-loop afforded by the relaxed-complex scheme presented here, we can qualitatively judge the fidelity of a pose for a new ligand. We see that for the NCIDS ligands that do bind in the C-loop region, the ligand pose depends significantly on the position of the C-loop; see Figure 7. It appears that as the C-loop moves away from its characteristic position, the conformation of the ligand is altered (middle and bottom panels of Figure 7a). We have not explicitly explored such dynamics here, but recent computational work by Liu et al. has shown that the dynamics of the C-loop and the ligand position are inherently tied to each other [36]. For ligands that bind well across all three species of AChBP, we take the predicted pose with the C-loop in the traditional position (as shown in the crystal structure) to be the most probable pose of that ligand.
Figure 7.
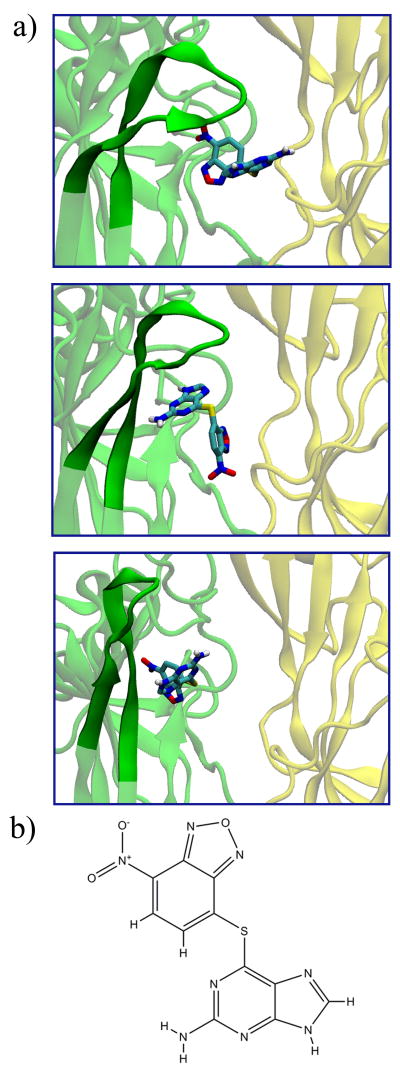
a) Binding of 2-amino-6-((7-(hydroxy(oxido)amino)-2,1,3-benzoxadiazol-4-yl)thio)-9H-purine (NCIDS 348401) to the Aplysia californica species of AChBP, at different conformations of the C-loop. The plus/minus faces of the protein are shown in a cartoon representation in green/yellow, respectively, and in a transparent fashion expect in the region of the C-loop (opaque). Ligand is shown in the licorice representations, with atoms colored cyan, red, white, and blue, for carbon, oxygen, hydrogen, and nitrogen, respectively. b) The 2D structure of the ligand.
Some of the NCIDS compounds are also reminiscent of the recently-studied neonicotinoids that bind well to AChBP [43]. NCI 348401 (shown in Figure 7), 118208, and 116805 bear a resemblance to the neonicotinoid imidacloprid, in the sense that they harbor nitro groups that can form hydrogen bonds with key residues in the C-loop of the plus face. Similar to desnitroimidacloprid, NCI 22959 contains imines and amines that can stabilize the ligand in the C-loop region. As Talley et al. point out, these nitrogenous functionalities form key hydrogen bonds and (if charged) cation-π interactions with the residues of the binding site [43]. Thus, the ligands from the NCIDS that contain these imines and amines—as well as those that exhibit properties of typical C-loop binders—may become novel AChBP-binding ligands.
As mentioned above, a significant number of the NCIDS compounds consistently bind away from the C-loop region, in all three AChBP species. By visual inspection, these compounds seem to be ‘butterfly’ structures, in the sense that they exhibit identical (or near-identical) structures on either side of a point of symmetry. Of the list in Table S1, a handful of compounds (NCI 54671, 54672, 116805, and 282034) exhibit this property. As can been seen in Figure S4 in the Supporting Information, 5-(2-(1H-tetraazol-5-yl)ethyl)-1H-tetraazole (NCI 282034) docks in a deeper pocket on the plus face, too distant to form significant contacts with the C-loop residues. These butterfly compounds can bind into other crevices along the plus/minus interface, but their effect on either the conformation or chemistry of AChBP is unclear and warrants further work. In general, such butterfly compounds are versatile and have proven of interest in other pharmacological targets, such as HIV Integrase [44]. Given the large interfacial area between the subunits of AChBP, the present docking method utilizes these butterfly compounds from the NCIDS to explore the binding space.
That the NCIDS ligands make significant contacts (see Figures 4 and 5) with parts of the plus/minus faces away from C-loop region suggests that parts of the interface above and below the C-loop may play a role in ligand binding. From the plus face, these other regions of binding are the A-loop region (residues 80 to 100), the β-strand (residues 115 to 125) following the E-loop region, and the B-loop region with accompanying β-strands (residues 135 to 155). The letter designations of the loops comes from previous work [33]. On the minus face, the various β-strands (residues 30 to 40, 50 to 60, and 110 to 118) stabilize the ligand when in the traditional binding site, behind the C-loop. Yet, we observe that some other parts of the minus face—in particular the F-loop region (residues 150 to 170)—may also provide binding residues. Work by Hibbs et al. demonstrated that agonists and antagonists of AChBP have different effects on the flexibility of the F-loop upon ligand binding, and that this flexibility may result in a conformational change that is translated down the membrane portion in the actual acetylcholine receptor [45]. Given the significant frequency of contacts with the F-loop region (see Figure 5b), the NCIDS ligands may elucidate this point further. The strands preceding the A-loop region (residues 95-105) and even a piece of the interface at the top (residues 70-75) of the minus face may also be implicated in binding. Because of the vast work on the binding in the traditional C-loop region, it is debatable how strongly ligands may bind to these other regions of the plus/minus interface. Yet, computationally, there is no reason to exclude these other interfacial regions. In fact, the work presented here suggests that it would be erroneous to discount the binding to the non-C-loop regions. Furthermore, recent experimental (unpublished) data by Talley also implicates these non-C-loop regions in ligand binding (although the exact properties of such ligands have yet to be determined).
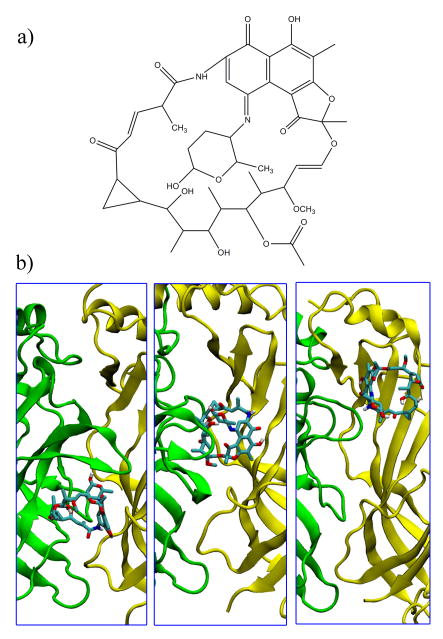
It is also interesting to note that the NCIDS ligands that do bind to other parts of the plus/minus interface bind differently amongst the three species of AChBP. Table S2 in the Supporting Information shows a subset of the NCIDS that bind differently across the 15 snapshots of AChBP (5 from each species). One such ligand, Tolypomycin (NCI 117383) is shown in Figure 8. It is a macrocyclic molecule, bearing some resemblance to the curariform antagonists that bind to AChBP in a variety of orientations [42]. In the three species, the Tolypomycin docks in the interfacial regions below, behind, and above the C-loop in the Ls, Ac, and Bt forms of AChBP, respectively. As discussed previously, because binding behind the C-loop region is the most stable form of binding, it is likely that Tolypomycin would bind better to Ac AChBP than to the other two species. In this sense, some of the ligands of the NCIDS could possibly be used to differentiate between the three species of AChBP and between the different subtypes of nAChR.
Figure 8.
a) 2D Structure of Tolypomycin (NCI 177383). b) Binding of Tolypomycin to the three species of AChBP. From left to right: Lymnaea stagnalis; Aplysia californica; Bulinus truncates. In each case, the plus/minus faces are shown in a cartoon representation in green/yellow, respectively. Ligand is shown in the licorice representations, with atoms colored cyan, red, white, and blue, for carbon, oxygen, hydrogen, and nitrogen, respectively.
Some interesting characteristics of the Tolypomycin binding are revealed by comparing the sequences of the plus/minus faces in the three species. In the plus face (Figure 9a), the sequence conservation of the C-loop region is great across all three species of AChBP. Tolypomycin makes contacts here with the conserved tyrosine-serine-cysteine-cysteine (YSCC) sequence in all three species. This again demonstrates that the C-loop region of the plus face is the ideal location for binding, and many of the NCIDS ligands resolve this point. Just upstream from the C-loop region, Tolypomycin makes contact with conserved tyrosine and aspartic acid residues. Contacts occur at some other conserved sites as well but not always across all three species of AChBP. For instance, Tolypomycin makes contact with TYR89 and TYR99 of Ls and Ac AChBP, respectively; but no such contact is made with the conserved TYR88 of Bt AChBP. The ligand also makes contacts with the conserved cysteine and aspartic acid residues of Ls AChBP but not with Bt or Ac AChBP. Slight differences in secondary structure at these conserved sites may result in differences in binding for a particular ligand. Moreover, certain strands of contact occur in some species but not in the others, such as the contacts from SER152 to GLU159 in Ac AChBP. This is even more the case in the minus face (Figure 9b), where the contact pattern differs greatly from one species to the next. Here one can see that the strands of contacts are not always aligned, and sometimes a long strand of contacts occurs in one species but to a lesser degree in the other two. These points of contact differentiation between the three species could explain the differential binding of Tolypomycin (as well as the other ligands in Table S2 of the Supporting Information) to AChBP.
Figure 9.
Structural sequence alignments of the AChBP in the a) plus face and b) minus face. The top, middle and bottom rows correspond to the species Lymnaea stagnalis (Ls), Bulinus truncates (Bt) and Aplysia californica (Ac), respectively. Asterisks delineate points of structural sequence conservation throughout all three species. Highlighted residues are those that make contacts (within 7.5 Å) with the ligand Tolypomycin, shown in Figure 8.
4. Conclusion
We have presented here a virtual screening study of the acetylcholine binding protein using the relaxed-complex method which involves a combination of MD simulation (to sample receptor structures) and docking. Regarding the ligands that bind AChBP—although the exact binding energies and poses are not always predicted—the relaxed-complex method does provide accurate results for many of the known binders and highlights key binding trends. Even the simple method of receptor structure selection based on the RMSD of a single loop can afford these results; this is likely the case for the entire realm of AChBP/nAChR proteins.
The ligands of the National Cancer Institute Diversity Set were screened through, and the results show that a number could serve as potential binders of AChBP. Like the known binders of AChBP, many of the NCIDS ligands dock behind the C-loop along the interface formed between the plus and minus subunits. This is the traditional location of ligand-binding in AChBP. The NCIDS ligands that possess aromatic functionalities are stabilized in the C-loop region by an aromatic cage. Hydrogen bonding interactions are prevalent; the ligands also feature chemical moieties that can bear a charge and thus form cation-π interactions with the residues in the binding site. Although many of the NCIDS ligands dock well across all three species and snapshots of AChBP, some appear to distinguish between the various structures. When this is the case, the ligands dock away from the C-loop region, and these molecules could serve to distinguish between the three species of AChBP. In this way, the NCIDS ligands have identified potentially-novel binding regions in the interfacial space of the protein.
Thus, the chemical space of the NCIDS provides valuable insight into ligand-binding in the acetylcholine binding protein. Because of its functional and structural similarity to the extra-cellular domain of the nicotinic acetylcholine receptor, AChBP serves as a surrogate structure in the study of the receptor. The ligands that interact significantly with AChBP could in turn bind the receptor. If this is the case, the NCIDS could be probed for ligands that bind across all of the receptor subtypes as well as those that distinguish between different structures. Such novel ligands could be helpful in pharmaceutical research on various neurological processes and disorders.
Supplementary Material
Acknowledgments
This work was supported in part by the National Science Foundation (Grants MCB-0506593 and MCA93S013), the National Institutes of Health (GM34921), the Howard Hughes Medical Institute, the National Biomedical Computing Resource, the Keck Foundation, and the Center for Theoretical and Biological Physics. We thank Dr. Rommie Amaro for assistance in setting up the virtual screening protocol. We also thank Dr. Wes Goodman and Dr. Wilfred Li at NBCR for their invaluable computing support.
Footnotes
These are some of the representative references pertaining to the solved structures of AChBP. A search in the Protein Data Bank for ‘AChBP’ or ‘acetylcholine binding protein’ will exhibit the entire body of work.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sheng M, Pak DTS. Ligand-gated ion channel interactions with cytoskeletal and signaling proteins. Annu Rev Physiol. 2000;62:755–778. doi: 10.1146/annurev.physiol.62.1.755. [DOI] [PubMed] [Google Scholar]
- 2.Whatley VJ, Harris RA. The cytoskeleton and neurotransmitter receptors. Int Rev Neurobiol. 1996;39:113–143. doi: 10.1016/s0074-7742(08)60665-0. [DOI] [PubMed] [Google Scholar]
- 3.Cami J, Farre M. Mechanisms of disease and drug addiction. N Engl J Med. 2003;349:975–986. doi: 10.1056/NEJMra023160. [DOI] [PubMed] [Google Scholar]
- 4.Tanczos AC, Palmer RA, Potter BS, Saldanha JW, Howlin BJ. Antagonist binding in the rat muscarinic receptor: a study by docking and x-ray crystallography. Comput Biol Chem. 2004;28:375–385. doi: 10.1016/j.compbiolchem.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 5.Ehrenpreis S, Kellock MG. Acetylcholine receptor protein and nerve activity. 1. Specific reaction of local anesthetics with the protein. Biochem Biophys Res Commun. 1960;2:311–313. [Google Scholar]
- 6.Buisson B, Bertrand D. Chronic exposure to nicotine upregulates the human alpha-4-beta-2 nicotinic acetylcholine receptor function. J Neurosci. 2001;21:1819–1829. doi: 10.1523/JNEUROSCI.21-06-01819.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Swanson KL, Albuquerque EX. Nicotinic acetylcholine receptor ion channel blockade by cocaine: the mechanism of synaptic action. J Pharmacol Exp Ther. 1987;243:1202–1210. [PubMed] [Google Scholar]
- 8.Ivetac A, Sansom MSP. Molecular dynamics simulations and membrane protein structure quality. Eur Biophys J. 2008;37:403–409. doi: 10.1007/s00249-007-0225-4. [DOI] [PubMed] [Google Scholar]
- 9.Unwin N. Refined structure of the nicotinic acetylcholine receptor at 4 Å resolution. J Mol Biol. 2005;346:967–989. doi: 10.1016/j.jmb.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 10.Ulens C, Hogg RC, Celie PH, Bertrand D, Tsetlin V, Smit AB, Sixma TK. Structural determinants of selective alpha-conotoxin binding to a nicotinic acetylcholine receptor homolog of AChBP. Proc Nat Acad Sci. 2006;103:3615–3620. doi: 10.1073/pnas.0507889103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brejc K, van Dijk WJ, Klaassen RV, Schuurmans M, van der Oost J, Smit AB, Sixma TK. Crystal structure of an ACh-binding protein reveals the ligand-binding domain of nicotinic receptors. Nature. 2001;411:269–276. doi: 10.1038/35077011. [DOI] [PubMed] [Google Scholar]
- 12.Celie PHN, van Rossum-Fikkert SE, van Dijk WJ, Brejc K, Smit AB, Sixma TK. Nicotine and carbamylcholine binding to nicotinic acetylcholine receptors as studied in AChBP crystal structures. Neuron. 2004;41:907–914. doi: 10.1016/s0896-6273(04)00115-1. [DOI] [PubMed] [Google Scholar]
- 13.Celie PHN, Kasheverov IE, Mordvintsev DY, Hogg RC, van Nierop P, van Elk R, van Rossum-Fikkert SE, Zhmak MN, Bertrand D, Tsetlin V, Sixma TK, Smit AB. Crystal structure of nicotinic acetylcholine receptor homolog AChBP in complex with an alpha-conotoxin PnIA variant. Nat Struct Mol Biol. 2005;12:582–588. doi: 10.1038/nsmb951. [DOI] [PubMed] [Google Scholar]
- 14.Hansen SB, Sulzenbacher G, Huxford T, Marchot P, Taylor P, Bourne Y. Structures of Aplysia AChBP complexes with nicotinic agonists and antagonists reveal distinctive binding interfaces and conformations. Eur Mol Biol Org. 2005;24:3635–3646. doi: 10.1038/sj.emboj.7600828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bourne Y, Talley TT, Hansen SB, Taylor P, Marchot P. Crystal structure of a Cbtx-AChBP complex reveals essential interactions between snake alpha-neurotoxins and nicotinic receptors. Eur Mol Biol Org. 2005;24:1512–1522. doi: 10.1038/sj.emboj.7600620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hansen SB, Radic Z, Talley TT, Molles BE, Deerinck T, Tsigelny I, Taylor P. Tryptophan fluorescence reveals conformational changes in the acetylcholine binding protein. J Biol Chem. 2002;277:41299–41302. doi: 10.1074/jbc.C200462200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Talley TT, Yalda S, Ho KY, Tor Y, Soti FS, Kem WR, Taylor P. Spectroscopic analysis of benzylidene anabaseine complexes with acetylcholine binding proteins as models for ligand-nicotinic receptor interactions. Biochemistry. 2006;45:8894–8902. doi: 10.1021/bi060534y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Talley TT, Olivera BM, Han KH, Christensen SB, Dowell C, Tsigelny I, Ho KY, Taylor P, McIntosh JM. Alpha-conotoxin OmlA is a potent ligand for the acetylcholine-binding protein as well as alpha-3-beta-2 and alpha-7 nicotinic acetylcholine receptors. J Biol Chem. 2006;281:24678–24686. doi: 10.1074/jbc.M602969200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fagen ZM, Mitchum R, Vezina P, McGehee DS. Enhanced nicotinic receptor function and drug abuse vulnerability. J Neurosci. 2007;27:8771–8778. doi: 10.1523/JNEUROSCI.2017-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.http://dtp.nci.nih.gov/index.html
- 21.http://flipdock.scripps.edu/
- 22.Lin JH, Perryman AL, Schames JR, McCammon JA. Computational drug design accommodating receptor flexibility: the relaxed complex scheme. J Am Chem Soc. 2002;124:5632–5633. doi: 10.1021/ja0260162. [DOI] [PubMed] [Google Scholar]
- 23.Amaro RE, Baron R, McCammon JA. An improved relaxed complex scheme for receptor flexibility in computer-aided drug design. J Comput Aided Mol Des. 2008;22:693–705. doi: 10.1007/s10822-007-9159-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Celie PHN, Klaassen RV, van Rossum-Fikkert SE, van Elk R, van Nierop P, Smit AB, Sixma TK. Crystal structure of acetylcholine-binding protein from bulinus truncatus reveals the conserved structural scaffold and sites of variation in nicotinic acetylcholine receptors. J Biol Chem. 2005;280:26457–26466. doi: 10.1074/jbc.M414476200. [DOI] [PubMed] [Google Scholar]
- 25.van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJ. GROMACS: fast, flexible, and free. J Comput Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 26.Kaminski GA, Friesner RA, Tirado-Reves J, Jorgensen WL. Evaluation and reparametrization of the OPLS-AA force field for proteins via comparison with accurate quantum chemical calculations on peptides. J Phys Chem B. 2001;105:6474–6487. [Google Scholar]
- 27.Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ. Automated docking using a lamarckian genetic algorithm and empirical binding free energy function. J Comput Chem. 1998;19:1639–1662. [Google Scholar]
- 28.Huey R, Morris GM, Olson AJ, Goodsell DS. A semiempirical free energy force field with charge-based desolvation. J Comput Chem. 2007;28:1145–1152. doi: 10.1002/jcc.20634. [DOI] [PubMed] [Google Scholar]
- 29.Chang MW, Lindstrom W, Olson AJ, Belew RK. Analysis of HIV wild-type and mutant structures via in-silico docking against diverse ligand libraries. J Chem Inf Model. 2007;47:1258–1262. doi: 10.1021/ci700044s. [DOI] [PubMed] [Google Scholar]
- 30.http://www.nbcr.net/
- 31.http://129.43.27.140/ncidb2/
- 32.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Molec Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 33.Cromer BA, Morton CJ, Parker MW. Anxiety over GABA(A) receptor structure relieved by AChBP. Trends Biochem Sci. 2002;27:280–287. doi: 10.1016/s0968-0004(02)02092-3. [DOI] [PubMed] [Google Scholar]
- 34.Shi J, Koeppe JR, Komives EA, Taylor P. Ligand-induced conformational changes in the acetylcholine-binding protein analyzed by hydrogen-deuterium exchange mass spectrometry. J Biol Chem. 2006;281:12170–12177. doi: 10.1074/jbc.M600154200. [DOI] [PubMed] [Google Scholar]
- 35.Gao F, Bren N, Burghardt TP, Hansen S, Henchman RH, Taylor P, McCammon JA, Sine SM. Agonist-mediated conformational changes in acetylcholine-binding protein revealed by simulation and intrinsic tryptophan fluorescence. J Biol Chem. 2005;280:8443–8451. doi: 10.1074/jbc.M412389200. [DOI] [PubMed] [Google Scholar]
- 36.Liu X, Xu Y, Wang X, Barrantes FJ, Jiang H. Unbinding of nicotine from the acetylcholine binding protein: steered molecular dynamics simulations. J Phys Chem B. 2008;112:4087–4093. doi: 10.1021/jp0716738. [DOI] [PubMed] [Google Scholar]
- 37.Hansen SB, Talley TT, Radic Z, Taylor P. Structural and ligand recognition characteristics of an acetylcholine-binding protein from aplysia californica. J Biol Chem. 2004;279:24197–24202. doi: 10.1074/jbc.M402452200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Warren GL, Andrews CW, Capelli AM, Clarke B, LaLonde J, Lambert MH, Lindvall M, Nevins N, Semus SF, Senger S, Tedesco G, Wall ID, Woolven JM, Peishoff CE, Head MS. A critical assessment of docking programs and scoring functions. J Med Chem. 2006;49:5912–5931. doi: 10.1021/jm050362n. [DOI] [PubMed] [Google Scholar]
- 39.Hibbs RE, Talley TT, Taylor P. Acrylodan-conjugated cysteine side chains reveal conformational state and ligand site locations of the acetylcholine-binding protein. J Biol Chem. 2004;279:28483–28491. doi: 10.1074/jbc.M403713200. [DOI] [PubMed] [Google Scholar]
- 40.Taylor P, Talley TT, Radic Z, Hansen SB, Hibbs RE, Shi J. Structure-guided drug design: conferring selectivity among neuronal nicotinic receptor and acetylcholine-binding protein subtypesstar. Biochem Pharmacol. 2007;74:1164–1171. doi: 10.1016/j.bcp.2007.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sixma TK, Smit AB. Acetylcholine binding protein (AChBP): a secreted glial protein that provides a high-resolution model for the extracellular domain of pentameric ligand-gated ion channels. Ann Rev Biophys Biomolec Struct. 2003;32:311–334. doi: 10.1146/annurev.biophys.32.110601.142536. [DOI] [PubMed] [Google Scholar]
- 42.Gao F, Bren N, Little A, Wang HL, Hansen SB, Talley TT, Taylor P, Sine SM. Curariform antagonists bind in different orientations to acetylcholine-binding protein. J Biol Chem. 2003;278:23020–23026. doi: 10.1074/jbc.M301151200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Talley TT, Harel M, Hibbs RE, Radic Z, Tomizawa M, Casida JE, Taylor P. Atomic interactions of neonicotinoid agonists with AChBP: molecular recognition of the distinctive electronegative pharmacophore. Proc Nat Acad Sci. 2008;105:7606–7611. doi: 10.1073/pnas.0802197105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schames JR, Henchman RH, Siegel JS, Sotriffer CA, Ni H, McCammon JA. Discovery of a novel binding trench in HIV integrase. J Med Chem. 2004;47:1879–1881. doi: 10.1021/jm0341913. [DOI] [PubMed] [Google Scholar]
- 45.Hibbs RE, Radic Z, Taylor P, Johnson DA. Influence of agonists and antagonists on the segmental motion of residues near the agonist binding pocket of the acetylcholine-binding protein. J Biol Chem. 2006;281:39708–39718. doi: 10.1074/jbc.M604752200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.