Abstract
Genetic polymorphisms in VKORC1 and CYP2C9, genes controlling vitamin K1 (VK1) epoxide reduction and (S)-warfarin metabolism, respectively, are major contributors to interindividual variability in warfarin dose. The V433M polymorphism (rs2108622) in CYP4F2 has also been associated with warfarin dose and speculatively linked to altered VK1 metabolism. Therefore, the purpose of the present study was to determine the role of CYP4F2 and the V433M polymorphism in the metabolism of VK1 by human liver. In vitro metabolic experiments with accompanying liquid chromatography-tandem mass spectrometry analysis demonstrated that recombinant CYP4F2 (Supersomes) and human liver microsomes supplemented with NADPH converted VK1 to a single product. A screen of all commercially available P450 Supersomes showed that only CYP4F2 was capable of metabolizing VK1 to this product. Steady-state kinetic analysis with recombinant CYP4F2 and with human liver microsomes revealed a substrate Km of 8 to 10 μM. Moreover, anti-CYP4F2 IgG, as well as several CYP4F2-selective chemical inhibitors, substantially attenuated the microsomal reaction. Finally, human liver microsomes genotyped for rs2108622 demonstrated reduced vitamin K1 oxidation and lower CYP4F2 protein concentrations in carriers of the 433M minor allele. These data demonstrate that CYP4F2 is a vitamin K1 oxidase and that carriers of the CYP4F2 V433M allele have a reduced capacity to metabolize VK1, secondary to an rs2108622-dependent decrease in steady-state hepatic concentrations of the enzyme. Therefore, patients with the rs2108622 polymorphism are likely to have elevated hepatic levels of VK1, necessitating a higher warfarin dose to elicit the same anticoagulant response.
The 4-hydroxycoumarin drug warfarin has been shown to be highly efficacious in the prevention of death after myocardial infarction and to greatly reduce instances of stroke in patients with atrial fibrillation (Hirsh et al., 1998; Hart et al., 2007). Warfarin is the most commonly prescribed oral anti-coagulant world-wide yet is widely believed to be underused because it has one of the highest adverse event rates of any drug on the market. The narrow therapeutic index of warfarin, coupled with the wide variability in individual patient response, poses a unique challenge to clinicians who must balance the risk of underdosing, which can result in loss of therapeutic efficacy, with that of overdosing, which may lead to potentially fatal hemorrhage (Rettie and Tai, 2006).
A number of factors are known to contribute to the considerable variability in interpatient response to coumarin anti-coagulants. Readily available patient characteristics, such as age, gender, body-mass index, and interactions due to other concomitantly administered drugs account for 20 to 30% of warfarin dose variance, and this clinical information is often integrated into initial dose calculations (Marsh and McLeod, 2006). Genetic factors have also proven to be a major determinant of warfarin dose. Polymorphisms within CYP2C9, which encodes the enzyme responsible for metabolizing the more potent S-enantiomer of warfarin, and VKORC1, the gene encoding the warfarin target receptor, together can explain approximately 30% (5-10% and 25%, respectively) of population dose variance (Rieder et al., 2005; Schwarz and Stein, 2006). Many other genes have been reported to show smaller effects on warfarin dose, although none have been fully confirmed in replication studies (Caldwell et al., 2007; Cooper et al., 2008). Still, roughly 50% of warfarin dose variance remains unexplained (Rettie and Tai, 2006; Caldwell et al., 2007). Caldwell et al. (2008) reported that polymorphism within CYP4F2, specifically rs2108622 (encoding a V433M amino acid change), led to an increase in warfarin dose requirement in a European-American cohort of patients. Subsequent replication studies were then carried out in two independent patient populations to validate these results (Caldwell et al., 2008). The same trend was found in all three patient groups, with the CYP4F2 rs2108622 variant effecting an overall increase of ∼1 mg/day of warfarin between the CC and TT genotype groups (i.e., 4-12% increase in dose per C allele).
The mechanism underlying the CYP4F2 allele effect on warfarin dose is unknown. Caldwell et al. (2008) have speculated that it might be mediated indirectly through 20-HETE production, a reaction catalyzed by CYP4F2. Alternatively, these authors also suggested that CYP4F2 might play a role in VK1 metabolism, possibly through hydroxylation of its phytyl side chain, because an analogous hydroxylation reaction of vitamin E is known to be catalyzed by CYP4F2 (Sontag and Parker, 2002, 2007; Hardwick, 2008).
Therefore, the goals of the present study were 3-fold: 1) to confirm the effect of the rs2108622 variant on warfarin dose, 2) to test the hypothesis that CYP4F2 is a VK1 oxidase, and 3) to examine molecular mechanisms for CYP4F2 genotype as a predictor of warfarin dose.
Materials and Methods
Chemicals. Sesamin and HET0016 were purchased from Cayman Chemical (Ann Arbor, MI). Troleandomycin (TAO) was obtained from BIOMOL International (Plymouth Meeting, PA). Ketoconazole (KET), VK1, VK2, zinc powder (-100 mesh), dimethyl sulfoxide, and NADPH were purchased from Sigma-Aldrich (St. Louis, MO). Zinc chloride, zinc sulfate, sodium acetate, potassium phosphate (mono and dibasic) and HPLC-grade methanol were supplied by J.T. Baker (Phillipsburg, NJ). Glacial acetic acid and isopropyl alcohol (IPA) were obtained from Thermo Fischer Scientific (Waltham, MA).
P450 Enzymes. Recombinant human P450 BD Gentest Supersomes, P450 enzymes 1A1, 1A2, 1B1, 2A6, 2B6, 2C8, 2C9, 2C18, 2C19, 2D6, 2E1, 2J2, 3A4, 3A5, 4A11, 4F2, 4F3A, 4F3B, and 4F12 were purchased from BD Biosciences (San Jose, CA) for use in the P450 screening study. The P450s were all coexpressed with P450 oxidoreductase, and most of the enzymes were also coexpressed with cytochrome b5. Only the following P450 preparations lacked b5: P450s 1A1, 1A2, 1B1, 2C18, 2D6, and 4A11.
Microsome Preparation. Human liver tissue (5-10 g dry weight) was thawed on ice in prechilled buffer 3 (100 mM Tris and 150 mM KCl, pH 7.4; 3 ml/g). Fat and connective tissue were removed, and the remaining tissue was cut into fine pieces before homogenization in a Thomas tube using a motor-driven pestle. The resultant mixture was then further homogenized on ice using a Virtishear probe. The homogenate was fractionated by centrifugation (9000g for 30 min at 4°C), and the supernatant was filtered through cotton gauze to remove residual fat. The microsomal pellet was collected upon further fractionation by ultracentrifugation (110,000g for 70 min at 4°C). The pellet was resuspended in buffer 3, rehomogenized by motor-driven pestle, and repelleted by ultracentrifugation (110,000g, 70 min). The final pellet was resuspended in a minimal amount of buffer 4 (20% glycerol, 100 mM EDTA, and 100 mM potassium phosphate, pH 7.4) and stored at -70°C. Total microsomal protein content was determined by the bicinchoninic acid assay (Smith et al., 1985).
Microsomal Incubations. Unless otherwise stated, incubations with HLM were carried out using a pool of human liver microsomes, prepared by combining equal amounts (dry weight) of eight liver samples from donors with known genotypes, obtained from the Human Liver Bank maintained in the Department of Medicinal Chemistry. Typical incubation mixtures contained 1 or 2 mg/ml microsomal protein from the HLM pool (or 50 pmol of recombinant P450), 1 mM NADPH, and 50 μM substrate (5 μl from a 5 mM VK1 stock in IPA), made up to 500-μl total volume with 100 mM potassium phosphate buffer, pH 7.4, in 1.5-ml microcentrifuge tubes. The microsomal reactions were preincubated in a water bath for 2 min at 37°C and 70 rpm before initiation with the addition of the NADPH. After incubation at 37°C and 70 rpm for 30 min, reactions were quenched with the addition of 50 μl of 15% aqueous ZnSO4. Vitamin K2 (VK2, menaquinone-4, 100 or 200 pmol) was added as internal standard, followed by 150 μl of IPA, and the reaction mixtures were extracted with hexane (2 × 650 μl). The combined hexane layers were evaporated under a N2-gas stream, and the residues were redissolved in 75 μl of IPA for analysis. Steady-state kinetic experiments for the determination of VK1 binding and turnover constants by HLM and recombinant CYP4F2 were performed at the following substrate concentrations: 1, 2, 5, 10, 20, 50, 100, and 200 μM. Kinetic analyses were carried out on Prism (ver. 4.00; GraphPad Software, San Diego, CA). Data are reported as the average values determined from triplicate runs. Error bars denote standard deviations.
HPLC-Fluorescence Assay for VK1 Oxidase Activity. HPLC was performed on a Shimadzu system equipped with two LC-10ADvp pumps, an RF-10Axl fluorescence detector, an SCL-10Avp controller, and an SIL-10ADvp or SIL-HTc autosampler (Shimadzu Scientific Instruments, Inc., Columbia, MD) using a Hypersil 5μ, 4.0 × 125-mm octadecyl silane column (Agilent Technologies, Palo Alto, CA) with a flow rate of 1.0 ml/min. A small guard column filled with zinc powder was placed in-line between the Hypersil column and the fluorescence detector. A dual-solvent gradient method was employed for metabolite isolation and quantitation; solvent A was water and solvent B was a 95:5 methanol/water mixture containing 11 mM ZnCl2, 5.5 mM NaOAc, and 5.5 mM acetic acid. The method was programmed to run at 95% solvent B for 4 min, followed by linear increases up to 97.5% B from 4 to 14 min, then up to 100% B at 15 min. Solvent B was kept at 100% from 15 to 34 min and was then brought back down to 95% by 35 min. The RF-10Axl detector wavelengths were set at 244 nm for excitation and 430 nm for emission. Under these conditions, the relative retention times for the VK1 metabolite, VK2 internal standard, and VK1 substrate were 8.5, 13, and 22 to 30 min. Data acquisition and analyses were performed on EZSTART chromatography software (ver. 7.2; Shimadzu).
LC-MS/MS MRM Assay for VK1 Metabolite Formation by HLM. LC/ESI+-MS and LC/ESI+-MS/MS multiple reaction monitoring (MRM) analyses were conducted on either a Micromass Quattro II Tandem Quadrupole Mass Spectrometer (Micromass, Ltd., Manchester, UK) connected to a Shimadzu HPLC system (similar to the system described above) or on a Micromass Quattro Premier XE Tandem Quadrupole Mass Spectrometer (Micromass, Ltd.) coupled to an ACQUITY Ultra Performance LC System with integral auto-injector (Waters Corp., Milford, MA). The Quattro II was run in electrospray positive ionization mode (ESI+) at a cone voltage of 20 V, a source block temperature of 150°C, and a desolvation temperature of 300°C. In tandem MS mode, the collision energy was set to 45 V. Separation was achieved on a Hypersil 5μ, 4.0 × 125-mm octadecyl silane column (Agilent) using an isocratic solvent system of 97:3 methanol/water with a flow rate of 0.35 ml/min. Standard assays were performed in LC-MS/MS MRM mode, monitoring the following (parent to daughter ion) mass transitions: m/z 451.7 to 187.0 (VK1 substrate); m/z 467.7 to 187.0 (VK1 metabolite), and m/z 444.7 to 187.0 (VK2 internal standard). The relative retention times for the VK1 metabolite, VK2, and VK1 were 8.8, 12.4, and 21.8 min, respectively, under these LC conditions.
The Premier XE was also run in ESI+-MS/MS MRM mode at a source temperature of 100°C and a desolvation temperature of 300°C. The cone voltage was set at 20 V, and the collision energy was set to 45 V. The same mass transitions were monitored as related above: m/z 451.7 to 187.0 (VK1 substrate); m/z 467.7 to 187.0 (VK1 metabolite), and m/z 444.7 to 187 (VK2 internal standard). Incubation product mixtures were separated on an ACQUITY Ultra Performance LC bridged ethyl hybrid 1.7 μm, 2.1 × 50-mm C8 HPLC column (Waters Corp.) with a flow rate of 0.35 ml/min and a binary solvent system of water (solvent A) and methanol (solvent B). A linear gradient was employed, whereby the methanol concentration was increased from 60 to 100% over the course of 5.1 min. Pure methanol was then run through the column until 6.0 min, followed by a return to 60% methanol by 6.5 min, with a total run time of 8.0 min. The respective retention times for the VK1 metabolite and VK2 internal standard were 4.3 and 4.6 min. Data analyses were carried out on Windows XP-based Micromass MassLynxNT software, ver. 4.1.
VK1 Metabolite Quantitation. Although quantification of the VK1 metabolite was hindered by the low turnover rate and the lack of a synthetic standard, we were able to estimate VK1 metabolite formation using the following strategy. A standard curve, using the same work-up protocol used for the VK1 metabolite assay, demonstrated that the extraction efficiency and fluorescence response of the VK1 dihydroquinone is equal to that of the reduced VK2. In determining rates for VK1 metabolite formation, it was then necessary to make the following assumptions: 1) mono-oxidation of the phytyl side chain of VK1, although slightly increasing the overall polarity of the molecule, is unlikely to significantly alter the product's extraction efficiency into hexane relative to substrate or internal standard, and 2) because oxidation occurs on the side chain of VK1, which is distantly located from its dihydroquinone derivative's fluorophore, the relative fluorescence response of the reduced VK1 metabolite should remain essentially equal to VK1 dihydroquinone and thus also to the internal standard. Therefore, the VK1 metabolite was quantified in the HPLC-fluorescence analysis assay by comparing the ratio of the peak areas of the metabolite relative to the internal standard and then accounting for the amount of VK2 that was added as internal standard.
Incubations of VK1 with HLMs resulted in the production of too little metabolite to quantify even by fluorescence. Instead, relative amounts of metabolite were determined by comparing the LC-MS/MS MRM area ratios for the VK1 metabolite transition peak versus that of the internal standard. These numbers were then compared with the MRM area ratio obtained from an incubation standard in which the amount of VK1 metabolite had already been quantified by HPLC-fluorescence analysis—we found that the VK1 metabolite consistently generates an LC-MS/MS MRM signal roughly 10 times greater than that observed for VK2. In this way, estimated turnover numbers were obtained for all metabolic reactions. A time-course study revealed that metabolite formation was essentially linear up to half an hour; all incubations were therefore run for a full 30 min to maximize the amount of metabolite produced and so improve the accuracy of analysis.
Genotyping and Association of CYP4F2 Genotype with Warfarin Dose. Liver DNA samples had been previously analyzed using the Illumina HumanHap550K genotyping platform, which includes rs2108622 (Cooper et al., 2008). To analyze genotype-dependent expression, each liver was genotyped [common allele (CC), minor allele (TT), or heterozygote (CT)], and linear regression was performed using each genotype category and assuming an additive genetic effect [coded 0 (CC), 1 (CT), or 2 (TT)] on expression levels.
VK1 Oxidase Activity of Genotyped HLM. Microsomes were prepared from a total of 12 individual human livers genotyped for CYP4F2 rs2108622. Four livers in each genotype group, CC, TT, or CT, were pooled, and microsomal protein concentrations were determined. VK1 metabolite quantitation was determined as described above by LC-MS/MS MRM analysis. All incubations were carried out in triplicate.
Immunoinhibition Assay. Polyclonal antibody raised (in rabbit) against CYP4F2 was purchased from Fitzgerald Industries International (Concord, MA), and preimmune immunoglobulin (IgG) from rabbit was obtained from ProSci Inc. (Poway, CA). Mixtures of 500 μg of protein, from HLM-CC pool (1 mg/ml), and anti-CYP4F2 (0, 50, or 150 μg from a 1 mg/ml stock) in 100 mM KPi buffer, pH 7.4 (final reaction volume, 500 μl), were preincubated on ice for 30 min to maximize antibody binding. After 2-min equilibration at 37°C and 70 rpm in a water bath, reactions were initiated by the addition of VK1 (50 μM, final) and NADPH (1 mM, final). Additional incubations containing similar amounts of blank, preimmune rabbit IgG (1 mg/ml stock) were run concurrently as negative controls. All reactions were quenched after 45-min incubation. Relative amounts of VK1 metabolite were quantified by LC-MS/MS MRM analysis. Data are reported as the mean of triplicate incubations.
Chemical Inhibition Assay. The P450 chemical inhibitors (HET0016, KET, sesamin, and TAO) were added separately to incubation mixtures containing 50 μM VK1 and 2 mg/ml microsomal protein, from the HLM-CC pool, in 100 mM KPi buffer, pH 7.4. Chemical inhibitors were added from stock solutions dissolved in methanol to give a final organic concentration of 1% in the incubation mixtures. Reactions were preincubated for 2 min before initiation with NADPH (1 mM final concentration). Incubations (30 min) were carried out in triplicate and worked up according to the standard protocol described above.
Western Blot Analysis. CYP4F2 Supersomes (1, 0.5, and 0.1 pmol/well) and HLM samples (75 μg of microsomal protein/well) were boiled in Laemmli buffer (Bio-Rad Laboratories, Hercules, CA) containing β-mercaptoethanol for 5 min. The boiled samples were resolved on a 9% Tris-glycine SDS-polyacrylamide gel. After samples transfer to nitrocellulose, the membrane was blocked overnight with Odyssey Blocking buffer (Licor Biosciences) containing 3% donkey serum. The membrane was then incubated with blocking buffer containing anti-CYP4F2 antibody (Santa Cruz Biotechnology, Inc., 1:5000 dilution) and 0.3% Tween 20 for 1 h followed by extensive washing with PBS buffer containing 0.3% Tween 20. Finally, the membrane was incubated with the blocking buffer containing antigoat IgG antibody conjugated with IR800 dye (1:30,000 dilution; Rockland Immunochemicals, Gilbertsville, PA) for 1 h. Immunochemically detected protein bands were visualized and quantitated with the Odyssey system.
CYP4F2 RNA Quantitation in Human Liver. RNA from the human liver sample repository at the University of Washington (n = 59) was extracted and normalized to ∼200 ng/μl. Using 750 ng of total RNA, whole genome expression measurement was carried out using the Illumina HumanRef-8 v.2. All liver samples were measured in duplicate (i.e., technical replicate), with each sample and replicate randomized between processed batches of 24 arrays run on different days. Raw signal intensity measurements from each sample were processed using the Illumina BeadStudio software v. 2.3.41 using the “average” normalization function. Replicate data from each liver was averaged and log2 transformed before statistical analysis according to rs2108622 genotype.
Results
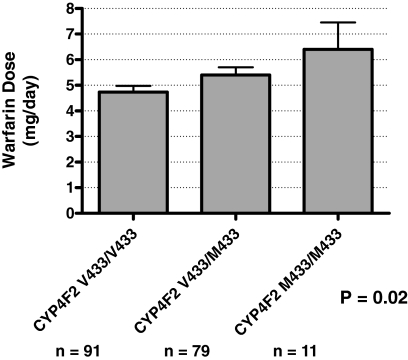
Effect of the CYP4F2 V433M Polymorphism on Warfarin Dose. The rs2108622 variant was genotyped as part of an earlier genome-wide association study conducted with DNA obtained from our cohort of 181 patients receiving warfarin (Cooper et al., 2008). As noted previously, the small size-effect (a contribution of 1-2% to the variance in warfarin dose) did not realize genome-wide statistical significance. However, when the data were closely interrogated, a similar magnitude and direction of the effect of rs2108622 on warfarin dose to that described by Caldwell et al. (2008) could be discerned (Fig. 1).
Fig. 1.
The CYP4F2 V433M polymorphism is significantly associated with the average warfarin maintenance dose in a cohort of 181 patients recruited from the University of Washington Medical Center. The P value refers to the correlation between all three genotype classes and shows that there is a statistically significant difference between the reported values (Cooper et al., 2008) for warfarin dose between data sets.
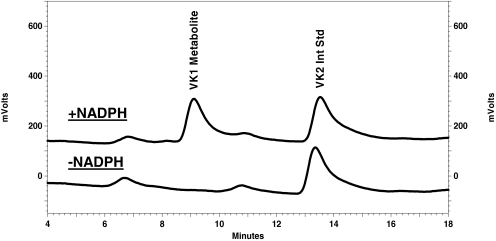
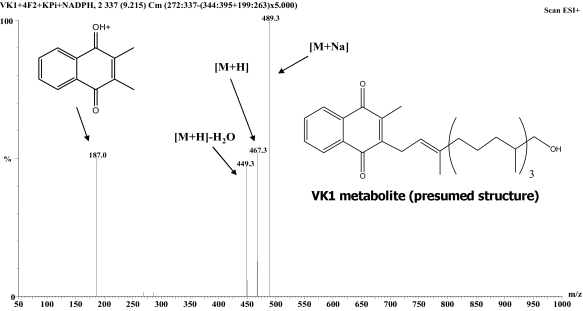
Development of HPLC-Fluorescence and HPLC-MS/MS Assays for Hydroxyvitamin K1. To test the hypothesis that CYP4F2 is a VK1 oxidase, new assay procedures were needed. These were developed by modifying a highly sensitive, fluorescence-based assay that had previously been used to analyze the reduction of VK1 epoxide (KO) to VK1 (Wang et al., 2004), a process catalyzed by the vitamin K1 epoxide reductase (VKOR) enzyme (Scheme 1). Although VK1 (phylloquinone), VK2 (menaquinone-4, used as the internal standard in our analyses) and, by extrapolation, the putative ω-hydroxyvitamin K1 metabolite, are essentially nonfluorescent, chemical reduction of the shared naphthoquinone backbone to the respective dihydroquinone derivatives results in a highly responsive fluorophore. Thus, by placing a small guard column filled with zinc powder in-line between the HPLC column and fluorescence detector, it was possible to effect separation of the metabolite, internal standard and substrate before subjecting them to chemical reduction, thereby rendering all three compounds visible by fluorescence. Using this method of analysis, we could show that incubations of VK1 with HLM or recombinant CYP4F2 generated a single NADPH-dependent product (Fig. 2). The metabolite peak was collected and identified, by ESI+-MS, as a mono-oxidized derivative of VK1 (Fig. 3). The occurrence of an m/z 187 fragment ion in both substrate and metabolite mass spectra demonstrates that the naphthoquinone portion of the substrate molecule remains unchanged, and so the site of oxidation must instead be located on the VK1 phytyl side chain.
Scheme 1.
Role for CYP4F2 in vitamin K1 homeostasis. Vitamin K1 dihydroquinone (VKH2) is the essential cofactor to γ-glutamyl carboxylase, the enzyme responsible for activating several blood factor proteins through the carboxylation of key Glu residues on those proteins, which initiates the blood coagulation cascade response. During this process, VKH2 is oxidized to the epoxide KO, which must then undergo two 2e- reductions to regenerate the active cofactor. Vitamin K1 epoxide reductase (VKORC1), the warfarin target enzyme, is known to catalyze at least the first of these reductions, generating VK1 from KO. VKORC1 is also the likely biological reductant in the conversion of VK1 back to VKH2, but it is possible that other enzymes, such as NADPH quinone oxidoreductase (NQO), might be involved in this step. CYP2C9 and CYP4F2 affect warfarin dose response by, respectively, controlling warfarin clearance and by removing VK1 from the cycle (Rettie and Tai, 2006).
Fig. 2.
VK1 metabolism by recombinant CYP4F2 Supersomes results in a single oxidative metabolite by HPLC-fluorescence analysis. Reactions contained 50 μM VK1 and 50 pmol of CYP4F2 incubated for 30 min at 37°C in 100 mM KPi buffer, pH 7.4, in the presence or absence of 1 mM NADPH cofactor. Menaquinone-4 (VK2) was added as internal standard. Reaction products were analyzed by reversed-phase HPLC with fluorescence detection as described under Materials and Methods.
Fig. 3.
ESI+-Mass spectrum of the VK1 metabolite isolated from reaction with recombinant CYP4F2. The fragment ion at m/z 187 indicates that oxidation occurred on the phytyl side chain of VK1 rather than on the menadione head group. The fragment ion at m/z 449 indicates a loss of water from the oxidized side chain of VK1.
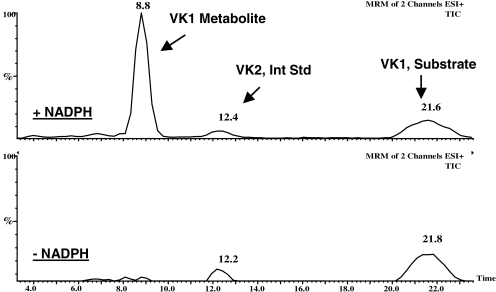
For experiments requiring an even greater degree of sensitivity than that provided by the HPLC-fluorescence assay (i.e., the various metabolic studies with HLM), analysis was performed by LC-MS/MS. The presence of a strong m/z 187 fragment ion in the ESI+-MS of the internal standard, VK2, as well as in the VK1 metabolite spectrum permitted the development of an MRM assay where the transitions from the vitamin K-related molecular ion peaks to the fragment ion at m/z 187 were followed (Fig. 4).
Fig. 4.
MRM LC/ESI+-MS/MS assay for VK1 metabolism. The figure depicts results from incubations of VK1 with recombinant CYP4F2 in the presence and absence of NADPH cofactor. The traces represent total ion chromatograms (TICs) generated from the following two mass transitions in each experiment: m/z 467.7 to 187.0 (monitored for the VK1 metabolite) and m/z 444.7 to 187.0 (monitored for the VK2 internal standard).
Screen for VK1 Metabolism by Recombinant Human P450 Supersomes. A metabolic screen was performed in which all of the commercially available recombinant human P450 enzymes (1A1, 1A2, 1B1, 2A6, 2B6, 2C8, 2C9, 2C18, 2C19, 2D6, 2E1, 2J2, 3A4, 3A5, 4A11, 4F2, 4F3A, 4F3B, and 4F12) were evaluated for their ability to metabolize VK1. Only CYP4F2 metabolized VK1 at a significant rate (∼100 pmol/min/nmol); all other P450s tested failed to generate a signal above 5 pmol/min/nmol (Fig. 5).
Fig. 5.
Metabolic screen of VK1 metabolism catalyzed by recombinant human P450 enzymes. Only CYP4F2 provided any VK1 oxidase activity above background. Reactions were run at 50 μM VK1 with 50 pmol of P450 and 1 mM NADPH unless otherwise indicated on the graph. Reported values, shown with standard errors, are averages calculated from duplicate incubations.
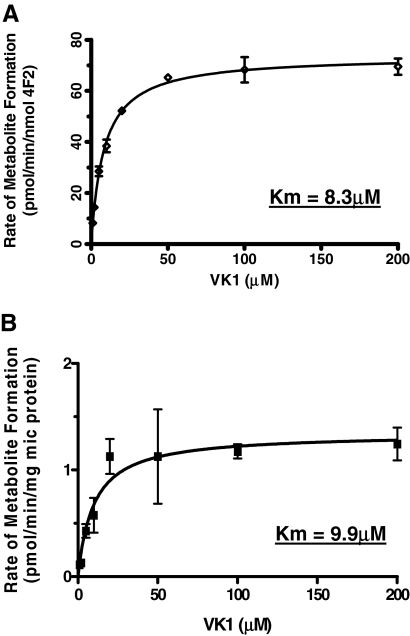
Kinetics of VK1 Metabolism. Steady-state kinetic analysis of VK1 oxidase activity was performed using both recombinant CYP4F2 and pooled HLM as enzyme sources. Recombinant CYP4F2 exhibited standard monophasic kinetics, with a Km of 8.3 μM and a Vmax of 75 pmol/min/nmol enzyme (Fig. 6A). VK1 metabolism catalyzed by HLM, also demonstrated hyperbolic kinetics, with an observed Km of 9.9 μM, similar to that obtained for recombinant CYP4F2, and a Vmax of 1.3 pmol/min/mg of microsomal protein (Fig. 6B).
Fig. 6.
Kinetic analyses of VK1 oxidase activity: A) by recombinant CYP4F2, B) by HLM. The enzyme sources both generated Km values of 8 to 10 μM as well as respective Vmax values of 75 pmol/min/nmol CYP4F2 and 1.3 pmol/min/mg microsomal protein.
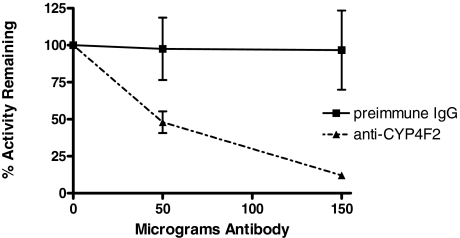
Immunoinhibition of VK1 Metabolism in HLM. A polyclonal antibody against CYP4F2 was tested for its ability to inhibit VK1 oxidase activity in pooled HLM. The anti-CYP4F2 antibody significantly inhibited VK1 metabolite formation in HLM, with metabolite formation being reduced by roughly 50 and 90%, respectively, in incubations containing 50 or 150 μg of antibody. Preimmune IgG, used as a negative control, showed essentially no inhibition of VK1 oxidase activity (<4%) over the same concentration range (Fig. 7).
Fig. 7.
VK1 metabolism in HLM is inhibited by CYP4F2 antibody. Incubations contained 500 μg of microsomal protein preincubated on ice with antibody for 30 min before initiation with the addition of saturating concentrations of VK1 and NADPH. Results represent average values obtained from triplicate incubations that were run for 45 min at 37°C. Standard deviation is denoted by error bars.
Inhibition of Microsomal VK1 Metabolism by Chemical Inhibitors. Several established chemical inhibitors of CYP4F2 were tested for their ability to inhibit VK1 oxidase activity in pooled HLM. The specific agents chosen were HET0016, which is known to be a potent inhibitor of the CYP4 family of enzymes (Miyata et al., 2001; Sato et al., 2001), and ketoconazole and sesamin, both of which have been shown to inhibit CYP4F2-mediated tocopherol (vitamin E) metabolism (Ikeda et al., 2002; Sontag and Parker, 2002; You et al., 2005). Troleandomycin was chosen as a negative control, because it is a metabolism-dependent inhibitor of CYP3A4 (Zeng et al., 1998), an enzyme that is reportedly also inhibited by ketoconazole and sesamin (von Moltke et al., 2004); however, troleandomycin does not seem to inhibit CYP4F2 (Wang et al., 2006). HET0016, ketoconazole, and sesamin all substantially inhibited VK1 oxidase activity in HLM, exhibiting a 52 to 68% reduction in metabolite formation at the substrate and inhibitor concentrations tested. Troleandomycin did not inhibit VK1 metabolite formation (Fig. 8).
Fig. 8.
VK1 metabolism in HLM is substantially attenuated by an array of selective CYP4F2 chemical inhibitors. TAO was included as a negative control. Reported values were determined from triplicate VK1 activity assays; error bars denote standard deviations.
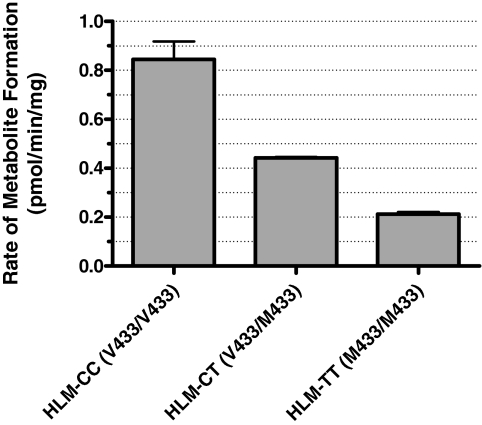
Comparison of VK1 Oxidase Activity in Genotyped HLM. The effect of the V433M polymorphism on VK1 oxidase activity was tested in pooled HLM preparations representative of each of the three genotypes: CC, TT, and CT. The HLM-CC pool (CYP4F2 Val433/Val433) exhibited the highest VK1 oxidase activity, generating metabolite at a rate of 0.85 pmol/min/mg of microsomal protein. The HLM-TT pool (CYP4F2 Met433/Met433), exhibited a 75% reduction in turnover (0.21 pmol/min/mg), whereas the HLM-CT pool displayed an intermediate activity, producing metabolite at a rate of 0.44 pmol/min/mg (Fig. 9).
Fig. 9.
VK1 metabolism in HLM is CYP4F2 genotype-dependent. HLM-CC, HLM-CT, and HLM-TT represent microsomal pools, each composed of equal amounts of microsomal protein taken from four livers of the indicated CYP4F2 genotype (single-nucleotide polymorphism rs2108622). VK1 activity assays were carried out, in triplicate, at saturating substrate and the resultant data, with standard error, is reported as the rate of metabolite formation by each HLM pool normalized to the total amount of microsomal protein used in the incubations.
CYP4F2 Immunoquantitation in Genotyped HLM. The amount of CYP4F2 in each of the three HLM pools was quantified by Western blot analysis and found to vary according to genotype and VK1 oxidase activity (Fig. 10). The HLM-CC, -CT, and -TT pools contained CYP4F2 at concentrations of 11.3, 7.2, and 2.5 pmol/mg of microsomal protein, respectively. When the VK1 oxidase activities reported in Fig. 9 were normalized to these CYP4F2 concentrations, the reaction rates of the three pools converged, generating an average maximum rate of metabolite formation of 73 pmol/min/nmol CYP4F2 (Table 1). This value is in good accord with the Vmax determined for the recombinant CYP4F2 enzyme (75-150 pmol/min/nmol).
Fig. 10.
Quantification of CYP4F2 in genotyped HLM. Samples were subjected to Western blotting with anti-CYP4F2 IgG as described under Materials and Methods. Lanes 1 and 5 contain a 55-kDa molecular mass standard. Lanes 2, 3 and 4 contain 1.0, 0.5, and 0.1 pmol of recombinant CYP4F2, respectively. The remaining lanes were each loaded with 75 μg of microsomal protein from the HLM-TT (lane 6), HLM-CT (lane 7), and HLM-CC (lane 8) pools. Marker protein was prestained with fluorescent dye having an emission maximum at 680 nm (red), whereas the CYP4F2 antibody was conjugated with fluorophore IR800, which emits between 800 and 900 nm (green).
TABLE 1.
Correlation of CYP4F2 levels with VK1 oxidase activity in genotyped HLM
Numbers in parentheses denote standard deviations.
| HLM Pool Genotype (rs2108622) | CYP4F2 Contenta | VK1 Met. Formationb | VK1 Met. Formationc |
|---|---|---|---|
| pmol/mg protein | pmol/min/mg | pmol/min/nmol | |
| CC | 11.3 | 0.85 (0.13) | 75 (11) |
| CT | 7.2 | 0.44 (0.01) | 61 (1.1) |
| TT | 2.5 | 0.21 (0.02) | 84 (6.7) |
Values determined by Western blot analysis.
VK1 oxidase activities normalized to total microsomal protein.
VK1 oxidase activities normalized to CYP4F2 content.
RNA Expression in Genotyped HLM. Finally, we determined the effect of rs2108622 on the expression of CYP4F2 mRNA in 59 human liver samples according to a method published previously (Kathiresan et al., 2008). No association was observed (p = 0.23). Testing of only the subset of livers (n = 12) that constituted the HLM pools used to evaluate genotype-dependent protein was not significant (p = 0.97).
Discussion
The main purpose of this study was to determine the mechanism of the CYP4F2 genotype-induced change in warfarin dose. In initial studies, we confirmed the previously reported CYP4F2 rs2108622 effect on average warfarin dose (Fig. 1). To analyze the metabolism of VK1 in vitro, it proved necessary to develop highly sensitive HPLC-fluorescence and LC-MS/MS assay procedures. These were then used to characterize VK1 metabolism by recombinant P450 and HLMs and to determine the contribution of CYP4F2 to human liver metabolism of VK1 using antibodies, selective chemical inhibitors, and genotyped HLM. Finally, we examined the relationship between CYP4F2 genotype, RNA expression, and CYP4F2 concentrations to provide a molecular explanation for increased warfarin dose requirements in carriers of the 433M allele.
The present studies establish that recombinant CYP4F2 catalyzes formation of a new metabolite of VK1 (Fig. 2). Indeed, a screen of all 19 commercially available recombinant human P450 Supersomes demonstrated that only CYP4F2 generates significant levels of this metabolite (Fig. 5). It should be noted that CYP3A4 and both CYP4F3A and CYP4F3B, the latter having 92% identity with CYP4F2 (Stec et al., 2007), were among the enzymes tested that produced only trace amounts of metabolite.
The ESI+-MS spectrum of the new CYP4F2-dependant metabolite shows it must be a mono-oxidized derivative of VK1. Furthermore, the presence of an m/z 187 fragment ion in the MS, resulting from the unmodified menadione nucleus, proves that oxidation occurs on the phytyl side chain, rather than on the naphthoquinone portion of the molecule (Fig. 3). Based on these analytical data, the known in vivo metabolites of VK1 (Shearer and Barkhan, 1973), and the well established product selectivity of CYP4 enzymes, we propose that this new metabolite is ω-hydroxyvitamin K1. The CYP4 family is known to ω-hydroxylate a wide variety of fatty acids, with the CYP4F subfamily showing preferential activity toward long-chain (C16-C26) fatty acids (Sanders et al., 2006; Hardwick, 2008; Sanders et al., 2008). Furthermore, CYP4F2 is the primary human enzyme responsible for ω-hydroxylating a range of tocopherols (Sontag and Parker, 2007), whose various structures all consist of a cyclized head group attached to a side chain of three linear isoprenyl units (similar to VK1), as well as phytanic acid, which is essentially the side chain of VK1 lacking the menadione head group (Xu et al., 2006). VK1, with its mostly saturated (C20) phytyl side chain composed of four linearly conjoined isoprenyl units is, therefore, a prime candidate to undergo CYP4F2-mediated ω-hydroxylation.
In vivo metabolism studies conducted with VK1 are also supportive of ω-hydroxylation. Two downstream VK1 metabolites, K acid 1 and K acid 2, are directly analogous to the known vitamin E metabolites, carboxyethyl hydroxychromans, and carboxymethyl butylhydroxychromans (Landes et al., 2003). All of these structures are consistent with a pathway of formation involving initial ω-oxidation followed by several rounds of β-oxidative cleavage. It is noteworthy that the tocopherol acid metabolites have been isolated not only from human serum and urine (Stahl et al., 1999) but also from HepG2 cells (Birringer et al., 2001; Sontag and Parker, 2002), the latter of which constitutively expresses CYP4F2 (You et al., 2005).
Hydroxyvitamin K1 generated by recombinant CYP4F2 is also produced in incubations with HLM, and we have demonstrated that CYP4F2 is the primary human liver microsomal VK1 oxidase. This conclusion is supported by the following observations: 1) recombinant CYP4F2 and HLM catalyze hydroxyvitamin K1 formation with essentially the same Km value (8-10 μM), 2) a polyclonal antibody raised against CYP4F2 inhibited microsomal metabolism by up to 90%, and 3) selective chemical inhibitors of CYP4F2 also substantially reduced microsomal metabolite formation. We note that the CYP4F2 antibody used here shows some cross-reactivity with other CYP4F proteins. Because we were unable to test recombinant CYP4F8 or CYP4F11 for activity, we cannot completely rule out the possibility that these two enzymes might also play a minor role in VK1 metabolism.
The chemical inhibitor data further strengthen the armamentarium available for discriminating between reactions catalyzed by CYP4F2 and other human P450 isoforms present in HLM. HET0016 is a strong selective inhibitor of the CYP4 family of enzymes, which has been shown to inhibit CYP4A (Miyata et al., 2001; Sato et al., 2001) and CYP4F enzymes (Miyata et al., 2001; Sato et al., 2001; Wang et al., 2006, 2007) with IC50 values ranging from 8 to 25 nM. In contrast, HET0016 is at least 200 times less potent an inhibitor of other xenobiotic metabolizing P450s, including CYP1A, CYP2C9, CYP2D6, and CYP3A4 (Miyata et al., 2001). Both ketoconazole and the lignan compound sesamin inhibit tocopherol and tocotrienol (vitamin E) metabolism, reactions known to involve CYP4F2 (Sontag and Parker, 2002; You et al., 2005; Sontag and Parker, 2007). Ketoconazole also inhibited metabolism of the antiparasitic prodrug, DB289, a process that was found to be catalyzed by CYP4F2 (Wang et al., 2006). These two inhibitors seem to be less selective than HET0016 for CYP4 isoforms, as sesamin also inhibits CYP2C and CYP3A isoforms with IC50 values of 2 to 10 μM (von Moltke et al., 2004). Ketoconazole is a submicromolar inhibitor of CYP3A4 (Eagling et al., 1998) and also inhibits CYP4F2 but not CYP4F3B-catalyzed, tocopherol hydroxylation (Wang et al., 2006). In fact, it was initially believed, erroneously, that CYP3A4 was involved in tocopherol metabolism, partly because vitamin E hydroxylase activity could be inhibited by both sesamin and ketoconazole (Parker et al., 2000). Consequently, it was important to include a selective CYP3A4 inhibitor, such as TAO, in the design of these experiments.
In further studies with HLM, we found that the V433M polymorphism leads to a pronounced genotype-dependent decrease in VK1 metabolism (Fig. 9). Western Blot analysis of HLM suggested that this effect was due to a decrease in hepatic CYP4F2 enzyme levels (Table 1). However, we observed no difference in mRNA expression levels between human liver samples with different rs2108622 genotypes (data not shown). Moreover, calculation of HLM VK1 oxidase activity based on immunochemically determined enzyme content normalized enzymatic rates for all three genotypes (Table 1). This suggests that the V433M polymorphism causes a loss of enzyme activity by affecting either translation or degradation of CYP4F2, rather than altering intrinsic catalytic activity of the enzyme. Interestingly, it was reported previously that recombinant CYP4F2 M433 metabolizes arachidonic acid to 20-HETE at approximately 35% efficiency compared with the recombinant wild-type Val433 enzyme (Stec et al., 2007). This apparent discrepancy might result from differences in the catalytic efficiencies of CYP4F2 variants for VK1 versus arachidonic acid or, perhaps, from relative differences in protein stability for CYP4F2 variants expressed recombinantly in insect cells relative to their native environment in human hepatocytes.
It was reported that lovastatin up-regulates CYP4F2 expression in human hepatocytes (Hsu et al., 2007). Therefore, the present studies might also help explain some of the drug-drug interactions that have been observed between warfarin and numerous statins (Hickmott et al., 2003; Andrus, 2004; Herman et al., 2006), which provoke an increase in INR and necessitate a decrease in warfarin dose. We speculate that up-regulation of CYP4F2, secondary to statin intake, might be expected to result in an increase in VK1 metabolism, reducing available levels of VK1 and lowering the requirement for warfarin. Although statins are generally used in the treatment of high cholesterol and act by inhibiting the enzyme HMG-CoA reductase (Blumenthal, 2000), lovastatin has also been prescribed for X-linked adrenoleukodystrophy disease, a recessive genetic disease characterized by a defect in peroxisomal β-oxidation of very long-chain fatty acids (Sanders et al., 2006). Lovastatin decreases accumulated plasma levels of very long-chain fatty acids (Singh et al., 1998), possibly by up-regulating CYP4F2. Of course, although statin-induced inhibition of warfarin metabolism through an interaction with either CYP2C9 or CYP3A4 almost certainly contributes to an increase in patient INR in specific cases (Hickmott et al., 2003), this mechanism alone does not adequately explain the full range of statin-warfarin drug interactions, based on available clinical data (Hickmott et al., 2003; Herman et al., 2006).
Finally, based on the current studies, we have modified the pathway view of the vitamin K cycle to include a role for CYP4F2 in “siphoning off” excess vitamin K1. VK1 is essential, not only for activation of clotting factors, but also for bone mineralization via γ-carboxylation of osteocalcin (Bügel, 2008). Recycling of the vitamin through the action of VKOR is clearly a major control mechanism for VK1 homeostasis. However, CYP4F2 may be an important counterpart to VKOR in limiting excessive accumulation of the vitamin. Indeed, the low basal VK1 oxidase activity of the enzyme seems consistent with such a role.
In summary, we have shown that CYP4F2 functions as a VK1 mono-oxidase, probably generating the ω-hydroxy derivative of the substrate. Carriers of the CYP4F2 V433M allele have a reduced capacity to metabolize VK1, secondary to an rs2108622-dependent decrease in steady-state hepatic concentrations of the enzyme. Therefore, patients with the rs2108622 polymorphism are likely to have elevated hepatic levels of VK1 and thus are likely to require a higher warfarin dose to elicit the same anticoagulant response (Scheme 1).
Acknowledgments
We thank Dr. Thomas F. Kalhorn and the University of Washington Mass Spectrometry Facility for technical assistance.
This work was supported by the National Institutes of Health National Institute of General Medical Sciences [Grants GM06797, GM32165] and by National Institutes of Health National Institute of Neurological Disorders and Stroke [Grant NS053646].
ABBREVIATIONS: 20-HETE, 20-hydroxyeicosatetraenoic acid; HET0016, N-hydroxy-N′-(4-n-butyl-2-methylphenyl)formamidine; TAO, troleandomycin; KET, ketoconazole; HPLC, high-performance liquid chromatography; IPA, isopropyl alcohol; P450, cytochrome P450; HLM, human liver microsomes; VK1, vitamin K1 (phylloquinone); VK2, vitamin K2 (menaquinone-4); ESI+-MS, electrospray positive ionization mass spectrometry; LC-MS/MS MRM, liquid chromatography with multiple reaction monitoring tandem mass spectrometry detection; IgG, immunoglobulin; KPi, potassium phosphate; VKORC1, vitamin K1 epoxide reductase complex 1; VKOR, vitamin K oxide reductase.
References
- Andrus MR (2004) Oral anticoagulant drug interactions with statins: case report of fluvastatin and review of the literature. Pharmacotherapy 24 285-290. [DOI] [PubMed] [Google Scholar]
- Birringer M, Drogan D, and Brigelius-Flohe R (2001) Tocopherols are metabolized in HepG2 cells by side chain omega-oxidation and consecutive beta-oxidation. Free Radic Biol Med 31 226-232. [DOI] [PubMed] [Google Scholar]
- Blumenthal RS (2000) Statins: effective antiatherosclerotic therapy. Am Heart J 139 577-583. [DOI] [PubMed] [Google Scholar]
- Bügel S (2008) Vitamin K and bone health in adult humans. Vitam Horm 78 393-416. [DOI] [PubMed] [Google Scholar]
- Caldwell MD, Awad T, Johnson JA, Gage BF, Falkowski M, Gardina P, Hubbard J, Turpaz Y, Langaee TY, Eby C, et al. (2008) CYP4F2 genetic variant alters required warfarin dose. Blood 111 4106-4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldwell MD, Berg RL, Zhang KQ, Glurich I, Schmelzer JR, Yale SH, Vidaillet HJ, and Burmester JK (2007) Evaluation of genetic factors for warfarin dose prediction. Clin Med Res 5 8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper GM, Johnson JA, Langaee TY, Feng H, Stanaway IB, Schwarz UI, Ritchie MD, Stein CM, Roden DM, Smith JD, et al. (2008) A genome-wide scan for common genetic variants with a large influence on warfarin maintenance dose. Blood 112 1022-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eagling VA, Tjia JF, and Back DJ (1998) Differential selectivity of cytochrome P450 inhibitors against probe substrates in human and rat liver microsomes. Br J Clin Pharmacol 45 107-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardwick JP (2008) Cytochrome P450 omega hydroxylase (CYP4) function in fatty acid metabolism and metabolic diseases. Biochem Pharmacol 75 2263-2275. [DOI] [PubMed] [Google Scholar]
- Hart RG, Pearce LA, and Aguilar MI (2007) Meta-analysis: antithrombotic therapy to prevent stroke in patients who have nonvalvular atrial fibrillation. Ann Intern Med 146 857-867. [DOI] [PubMed] [Google Scholar]
- Herman D, Locatelli I, Grabnar I, Peternel P, Stegnar M, Lainscak M, Mrhar A, Breskvar K, and Dolzan V (2006) The influence of co-treatment with carbamazepine, amiodarone and statins on warfarin metabolism and maintenance dose. Eur J Clin Pharmacol 62 291-296. [DOI] [PubMed] [Google Scholar]
- Hickmott H, Wynne H, and Kamali F (2003) The effect of simvastatin co-medication on warfarin anticoagulation response and dose requirements. Thromb Haemost 89 949-950. [PubMed] [Google Scholar]
- Hirsh J, Dalen JE, Anderson DR, Poller L, Bussey H, Ansell J, Deykin D, and Brandt JT (1998) Oral anticoagulants: mechanism of action, clinical effectiveness, and optimal therapeutic range. Chest 114 (5 Suppl): 445S-469S. [DOI] [PubMed] [Google Scholar]
- Hsu MH, Savas U, Griffin KJ, and Johnson EF (2007) Regulation of human cytochrome P450 4F2 expression by sterol regulatory element-binding protein and lovastatin. J Biol Chem 282 5225-5236. [DOI] [PubMed] [Google Scholar]
- Ikeda S, Tohyama T, and Yamashita K (2002) Dietary sesame seed and its lignans inhibit 2,7,8-trimethyl-2(2′-carboxyethyl)-6-hydroxychroman excretion into urine of rats fed gamma-tocopherol. J Nutr 132 961-966. [DOI] [PubMed] [Google Scholar]
- Kathiresan S, Melander O, Guiducci C, Surti A, Burtt NP, Rieder MJ, Cooper GM, Roos C, Voight BF, Havulinna AS, et al. (2008) Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat Genet 40 189-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landes N, Birringer M, and Brigelius-Flohé R (2003) Homologous metabolic and gene activating routes for vitamins E and K. Mol Aspects Med 24 337-344. [DOI] [PubMed] [Google Scholar]
- Marsh S and McLeod HL (2006) Pharmacogenomics: from bedside to clinical practice. Hum Mol Genet 15 (Spec No 1): R89-R93. [DOI] [PubMed] [Google Scholar]
- Miyata N, Taniguchi K, Seki T, Ishimoto T, Sato-Watanabe M, Yasuda Y, Doi M, Kametani S, Tomishima Y, Ueki T, et al. (2001) HET0016, a potent and selective inhibitor of 20-HETE synthesizing enzyme. Br J Pharmacol 133 325-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker RS, Sontag TJ, and Swanson JE (2000) Cytochrome P4503A-dependent metabolism of tocopherols and inhibition by sesamin. Biochem Biophys Res Commun 277 531-534. [DOI] [PubMed] [Google Scholar]
- Rettie AE and Tai G (2006) The pharmocogenomics of warfarin: closing in on personalized medicine. Mol Interv 6 223-227. [DOI] [PubMed] [Google Scholar]
- Rieder MJ, Reiner AP, Gage BF, Nickerson DA, Eby CS, McLeod HL, Blough DK, Thummel KE, Veenstra DL, and Rettie AE (2005) Effect of VKORC1 haplotypes on transcriptional regulation and warfarin dose. N Engl J Med 352 2285-2293. [DOI] [PubMed] [Google Scholar]
- Sanders RJ, Ofman R, Dacremont G, Wanders RJ, and Kemp S (2008) Characterization of the human omega-oxidation pathway for omega-hydroxy-very-long-chain fatty acids. Faseb J 22 2064-2071. [DOI] [PubMed] [Google Scholar]
- Sanders RJ, Ofman R, Duran M, Kemp S, and Wanders RJ (2006) Omega-oxidation of very long-chain fatty acids in human liver microsomes. Implications for X-linked adrenoleukodystrophy. J Biol Chem 281 13180-13187. [DOI] [PubMed] [Google Scholar]
- Sato M, Ishii T, Kobayashi-Matsunaga Y, Amada H, Taniguchi K, Miyata N, and Kameo K (2001) Discovery of a N′-hydroxyphenylformamidine derivative HET0016 as a potent and selective 20-HETE synthase inhibitor. Bioorg Med Chem Lett 11 2993-2995. [DOI] [PubMed] [Google Scholar]
- Schwarz UI and Stein CM (2006) Genetic determinants of dose and clinical outcomes in patients receiving oral anticoagulants. Clin Pharmacol Ther 80 7-12. [DOI] [PubMed] [Google Scholar]
- Shearer MJ and Barkhan P (1973) Studies on the metabolites of phylloquinone (vitamin K 1) in the urine of man. Biochim Biophys Acta 297 300-312. [DOI] [PubMed] [Google Scholar]
- Singh I, Khan M, Key L, and Pai S (1998) Lovastatin for X-linked adrenoleukodystrophy. N Engl J Med 339 702-703. [DOI] [PubMed] [Google Scholar]
- Smith PK, Krohn RI, Hermanson GT, Mallia AK, Gartner FH, Provenzano MD, Fujimoto EK, Goeke NM, Olson BJ, and Klenk DC (1985) Measurement of protein using bicinchoninic acid. Anal Biochem 150 76-85. [DOI] [PubMed] [Google Scholar]
- Sontag TJ and Parker RS (2002) Cytochrome P450 omega-hydroxylase pathway of tocopherol catabolism. Novel mechanism of regulation of vitamin E status. J Biol Chem 277 25290-25296. [DOI] [PubMed] [Google Scholar]
- Sontag TJ and Parker RS (2007) Influence of major structural features of tocopherols and tocotrienols on their omega-oxidation by tocopherol-omega-hydroxylase. J Lipid Res 48 1090-1098. [DOI] [PubMed] [Google Scholar]
- Stahl W, Graf P, Brigelius-Flohé R, Wechter W, and Sies H (1999) Quantification of the alpha- and gamma-tocopherol metabolites 2,5,7,8-tetramethyl-2-(2′-carboxyethyl)-6-hydroxychroman and 2,7, 8-trimethyl-2-(2′-carboxyethyl)-6-hydroxychroman in human serum. Anal Biochem 275 254-259. [DOI] [PubMed] [Google Scholar]
- Stec DE, Roman RJ, Flasch A, and Rieder MJ (2007) Functional polymorphism in human CYP4F2 decreases 20-HETE production. Physiol Genomics 30 74-81. [DOI] [PubMed] [Google Scholar]
- von Moltke LL, Weemhoff JL, Bedir E, Khan IA, Harmatz JS, Goldman P, and Greenblatt DJ (2004) Inhibition of human cytochromes P450 by components of Ginkgo biloba. J Pharm Pharmacol 56 1039-1044. [DOI] [PubMed] [Google Scholar]
- Wang LY, Bates CJ, Yan L, Harrington DJ, Shearer MJ, and Prentice A (2004) Determination of phylloquinone (vitamin K1) in plasma and serum by HPLC with fluorescence detection. Clin Chim Acta 347 199-207. [DOI] [PubMed] [Google Scholar]
- Wang MZ, Saulter JY, Usuki E, Cheung YL, Hall M, Bridges AS, Loewen G, Parkinson OT, Stephens CE, Allen JL, et al. (2006) CYP4F enzymes are the major enzymes in human liver microsomes that catalyze the O-demethylation of the antiparasitic prodrug DB289 [2,5-bis(4-amidinophenyl)furan-bis-O-methylamidoxime]. Drug Metab Dispos 34 1985-1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang MZ, Wu JQ, Bridges AS, Zeldin DC, Kornbluth S, Tidwell RR, Hall JE, and Paine MF (2007) Human enteric microsomal CYP4F enzymes O-demethylate the antiparasitic prodrug pafuramidine. Drug Metab Dispos 35 2067-2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F, Ng VY, Kroetz DL, and de Montellano PR (2006) CYP4 isoform specificity in the omega-hydroxylation of phytanic acid, a potential route to elimination of the causative agent of Refsum's disease. J Pharmacol Exp Ther 318 835-839. [DOI] [PubMed] [Google Scholar]
- You CS, Sontag TJ, Swanson JE, and Parker RS (2005) Long-chain carboxychromanols are the major metabolites of tocopherols and tocotrienols in A549 lung epithelial cells but not HepG2 cells. J Nutr 135 227-232. [DOI] [PubMed] [Google Scholar]
- Zeng Z, Andrew NW, Arison BH, Luffer-Atlas D, and Wang RW (1998) Identification of cytochrome P4503A4 as the major enzyme responsible for the metabolism of ivermectin by human liver microsomes. Xenobiotica 28 313-321. [DOI] [PubMed] [Google Scholar]