Abstract
Point mutations and molecular modeling have been used to study the activation of the M1 muscarinic acetylcholine receptor (mAChR) by the functionally selective agonists 4-n-butyl-1-[4-(2-methylphenyl)-4-oxo-1-butyl]-piperidine (AC-42), and 1-[3-(4-butyl-1-piperidinyl)propyl]-3,4-dihydro-2(1H)-quinolinone (77-LH-28-1), comparing them with N-desmethylclozapine (NDMC) and acetylcholine (ACh). Unlike NDMC and ACh, the activities of AC-42 and 77-LH-28-1 were undiminished by mutations of Tyr404 and Cys407 (transmembrane helix 7), although they were reduced by mutations of Tyr408. Signaling by AC-42, 77-LH-28-1, and NDMC was reduced by L102A and abolished by D105E, suggesting that all three may interact with transmembrane helix 3 at or near the binding site Asp105 to activate the M1 mAChR. In striking contrast to NDMC and ACh, the affinities of AC-42 and 77-LH-28-1 were increased 100-fold by W101A, and their signaling activities were abolished by Y82A. Tyr82 and Leu102 contact the indole ring of Trp101 in a structural model of the M1 mAChR. We suggest the hypothesis that the side chain of Trp101 undergoes conformational isomerization, opening a novel binding site for the aromatic side chain of the AC-42 analogs. This may allow the positively charged piperidine nitrogen of the ligands to access the neighboring Asp105 carboxylate to activate signaling following a vector within the binding site that is distinct from that of acetylcholine. NDMC does not seem to use this mechanism. Subtype-specific differences in the free energy of rotation of the side chain and indole ring of Trp101 might underlie the M1 selectivity of the AC-42 analogs. Tryptophan conformational isomerization may open up new avenues in selective muscarinic receptor drug design.
The M1 muscarinic receptor (mAChR) is an attractive drug target to treat Alzheimer's disease and schizophrenia (Langmead et al., 2008b). The amino acid side chains that contact acetylcholine are modeled on the inward-facing segments of transmembrane (TM) helices 3, 4, 5, 6, and 7 within the outer leaflet of the lipid bilayer (Hulme et al., 2003). Their conservation between the five receptor subtypes has restricted the discovery of selective agonists.
AC-42 is a novel ligand that activates M1 mAChRs selectively (Spalding et al., 2002). Activation is little affected by alanine substitution of the orthosteric binding pocket residues Tyr381 (6.51; Ballesteros and Weinstein, 1995) and Asn382 (6.52) but requires sequences in the N terminus and TM1, extracellular loop 3, and the outer part of TM7. Thus AC-42 may activate M1 mAChRs from a distinct “ectopic” site (Spalding et al., 2002). Supporting this, AC-42 can display allosteric interactions with N-methyl scopolamine (NMS) and atropine (Langmead et al., 2006). A study of TM3 showed that activation of M1 mAChRs by AC-42 analogs was unaffected by alanine substitution of Tyr106 (3.33) neighboring the ACh counter-ion Asp105 (3.32) or Ser109 (3.36) one helical turn below but was inhibited by L102A (3.29) and paradoxically very strongly potentiated by W101A (3.28), one helical turn above (Spalding et al., 2006). The compounds did not activate D105A, but ACh itself is also completely inactive at this mutant.
N-desmethylclozapine (NDMC) is a metabolite of the antipsychotic drug clozapine that has agonist activity at M1, M3, and M5 but not M2 and M4 mAChRs (Davies et al., 2005). Unlike ACh, its activity is potentiated by Y381A (Sur et al., 2003) and preserved by Y106A and S109A (Spalding et al., 2006). Unlike AC-42, its potency is not enhanced by W101A, suggesting a variant mode of interaction.
TM2 does not participate in binding ACh or NMS (Bee and Hulme, 2007). However, in a range of receptors, TM2, TM3, and TM7 define a subsidiary binding pocket (Flanagan et al., 2000; Stitham et al., 2003; Leonardi et al., 2004; de Mendonca et al., 2005; Jensen et al., 2007). An aromatic cluster between TM2 and TM3 may contribute to the activation of CCR5 by chemokines (Govaerts et al., 2003) and may be a determinant of D2 versus D4 receptor selectivity (Simpson et al., 1999; Schetz et al., 2000).
Optimized molecular models of the M1 mAChR (Goodwin et al., 2007) also show a well packed microdomain enriched in hydrophobic and aromatic amino acids at the top of TM2, TM3, and TM7. The side chains of Tyr82 (2.61) and Leu102 may be close to the ring of Trp101, whereas Met79 (2.58) may help to buttress the ring of Tyr408 (7.43) (Bee and Hulme, 2007). It is noteworthy that the hydroxyl group of the homolog of Tyr408 is hydrogen-bonded to the binding site aspartic acid in the crystal structure of the β2-adrenergic receptor (Cherezov et al., 2007).
TM3 and TM7 define the frontier between the major and subsidiary pockets. Asp105 (Lu and Hulme, 1999), Tyr404 (7.39), and Cys407 (7.42) (Lu et al., 2001) lie within the major orthosteric pocket. Met79 and Tyr82 are better classified as belonging to the subsidiary pocket. This is consistent with the limited effects of the Y82A and M79A mutations on ACh-mediated activation of the M1 mAChR (Lee et al., 1996; Bee and Hulme, 2007). Trp101 and Leu102 are in the “second shell” of the ACh binding site (Lu and Hulme, 1999). However, certain residues, particularly Tyr408, may be at the interface of the two pockets and able to contribute to both.
In this study, we have investigated the effect of mutations of these residues on the binding and activation of M1 mAChRs by AC-42, its analog 77-LH-28-1 (Langmead et al., 2008a), and NDMC (for convenience, collectively designated as novel selective agonists, NSAs) comparing them with ACh. We have included a control residue, Leu100, believed to face toward the lipid bilayer. To evaluate the importance of the binding site aspartic acid, we used the D105E mutant. This strongly reduces the potency of activation of the M1 mAChR by ACh without abolishing the signaling response (Page et al., 1995), providing a more discriminating probe than D105A. The results suggest that the AC-42 analogs may activate the M1 mAChR by a novel conformational trapping mechanism, whereby a moiety of the ligand replaces the indole ring of Trp101 within the structure of the receptor after rotation of the side chain. This may allow the presentation of the charged piperidine head group of the ligands to the Asp105 counter-ion following a vector distinct from that of ACh.
Materials and Methods
Materials. (-)-N-[3H]Methylscopolamine (82, 84 Ci/mmol) and myo-d-[3H]inositol (80 Ci/mmol) were purchased from GE Healthcare (Chalfont St. Giles, Buckinghamshire, UK). Unlabeled ligands were from Sigma-Aldrich (Gillingham, UK). AC-42 and 77-LH-28-1 were synthesized as described previously (Langmead et al., 2006, 2008a). Stock solutions (10-2 M) were prepared in DMSO and stored at -20°C. Ligand dilutions were prepared in DMSO.
DNA and Expression. With the exception of Y82A, the rat M1 mutant receptors used in this work have been described previously (Page et al., 1995; Lu and Hulme, 1999; Lu et al., 2001; Bee and Hulme, 2007). They were cloned in the pCD expression vector and were transiently expressed in COS-7 cells grown in minimal essential medium-α supplemented with 10% newborn calf serum and 1% glutamate at 37°C in 5% CO2. Approximately 2 × 107 cells (1 cuvette) were transfected by electroporation with 15 μg of DNA using a Bio-Rad Gene Pulser (260 V, 960 μF; Bio-Rad, Hercules, CA). The transfected cells were resuspended in 30 ml of warm supplemented media and plated into three dishes (10 ml; ≈6.5 × 106 cells/plate) for the binding experiments and into 96-well plates (200 μl/well; ≈1.5 × 105 cells/well) for the phosphoinositide turnover assays. Twenty-four hours after the transfection, cells were washed twice in warmed phosphate-buffered saline (PBS) and new minimal essential medium-α media added. When necessary, the media were supplemented with atropine (10-6 M) to rescue the low-expressing mutant receptors (M79A, Y404A, and Y408A) as described previously (Lu et al., 2001; Bee and Hulme, 2007). Y82A was constructed by the QuikChange (Stratagene, La Jolla, CA) procedure and verified by dideoxy nucleotide sequencing.
Cell Membrane Preparation. Membrane preparations were made 72 h after transient transfection. The COS-7-transfected cells were washed twice with PBS. Cells were harvested in 20 mM sodium-HEPES and 10 mM EDTA, pH 7.5, at 4°C (1 ml/dish) and homogenized by using a mechanical glass homogenizer. Membranes were pelleted by centrifugation for 30 min at 30,000g and were resuspended in storage buffer composed of 20 mM sodium-HEPES and 1 mM EDTA, pH 7.5, and then homogenized using a Polytron PT 3100 (twice for 15 s at 12,000 rpm; Kinematica AG, Lucerne, Switzerland). Aliquots (1 ml) were snap-frozen on dry ice and stored at -80°C.
Radioligand Binding Assays. Binding experiments were carried out in 96-well plates (2 ml deep, polystyrene plates) using three different concentrations of [3H]NMS that were equal, where possible, to 0.1, 2, and 10 times the Kd value of [3H]NMS for the particular mutant. Each agonist concentration was performed in quadruplicate. Membrane preparations were spun at 14,000 rpm (17,000g) and then resuspended in binding buffer (20 mM sodium-HEPES, 0.1 M NaCl, and 1 mM MgCl2, pH 7.5). The membrane suspension was homogenized with the Polytron PT 3100 (15 s at 12,000 rpm). Diluted compounds (10 μl) in DMSO and 10 μl of [3H]NMS diluted in binding buffer were mixed with 980 μl of membrane preparation (7-30 μg membrane protein/ml), and the plates were covered with a plastic seal and incubated for 2 h (4 h for the W101A mutant) at 30°C. Binding was stopped by filtration onto Wallac 96-Printed GF/B filter mats (PerkinElmer Wallac, Gaithersburg, MD) using a TOMTEC Harvester 96, MACH III M (TOMTEC, Hamden, CT). The filter mats were washed three times with 3 ml of cold double-distilled H2O. The filters were dried at 55°C for approximately 1 h. A solid Melt-On Scintillator Sheet (Meltilex B/HS, Wallac, Turku, Finland) was melted onto the filter mat containing the harvested membranes. Radioactivity was counted on a normalized, calibrated 1450 Microbeta Trilux counter (5 min per sample). The expression level of the wild-type receptor was 2.92 ± 0.80 pmol/mg membrane protein (mean ± S.D.). This corresponds to ∼2 × 105 [3H]NMS binding sites per cell (Bee and Hulme, 2007). Expression levels of the mutants (percentage of wild type) were as follows: M79A, 96%; Y82A, 36%; L100A, 102%; L102A, 76%; W101A, 47%; D105E, 46%; Y404A, 109%; Y404F, 125%; C407A, 107%; Y408A, 162%; and Y408F, 99%.
Phosphoinositide Functional Response. Measurement of G-protein-coupled receptor activation was carried out using a scintillation proximity assay of inositol phosphates in cell extracts (Brandish et al., 2003) with some modifications. Transfected cells were grown in 96-well plates at 37°C, 5% CO2. After 24 h, the cells were washed twice in warm PBS and labeled with 2.5 μCi/ml myo-d-[3H]inositol in medium for 48 h. After 48 h, myo-d-[3H]inositol was removed, the cells were washed twice in warm PBS and once in Krebs' solution containing 10 mM LiCl, and finally they were incubated for 30 min in Krebs/Li at 37°C, 5% CO2. Cells were then incubated with 100 μl of agonist (diluted in Krebs/Li solution to the final concentration required) for 1 h at 37°C, 5% CO2. The reaction was terminated by the removal of the agonist dilution and the addition of 200 μl of 0.1 M formic acid. The cell solubilization was allowed to continue for 1 h at room temperature, after which 20-μl samples of the lysates were mixed with 80 μl of RNA binding yttrium silicate scintillation proximity assay beads (12.5 mg/ml) in 96-well isoplates. Plates were agitated for 1 h, and then the beads were left to settle for 2 h, protected from the light. The accumulation of negatively charged [3H]inositol phosphates was determined using a 1450 Microbeta Trilux counter (5 min per sample). In each experiment, atropine (10-6 M) was added to one triplicate set of assays to control for atropine-sensitive ACh-independent constitutive activity. This was maximally 8.5% for the wild-type receptor.
Data Analysis. Binding curves and phosphoinositide (PhI) dose-response curves were fitted using SigmaPlot (version 10; Systat Software, San Jose, CA). Binding curves for [3H]NMS in the presence of agonists were fitted to an allosteric ternary complex model of binding in which the apparent affinity constant for the radioligand is given by the expression Kapp = KL · (1 + α (KA · [A])nH)/(1 + KI · [I] + (KA · [A])nH).
This analysis yields an affinity constant (-log M) for NMS (KL), for the compounds (KA), an expression level value of the receptor (RT), and in the case of mutants displaying allosteric behavior, a cooperativity factor value (α) and a slope factor (nH). Nonspecific binding was determined with 10-6 M unlabeled NMS (designated [I]). The equations were written to compensate for any radioligand depletion (less than 10%). The three data sets for a particular combination of mutant and unlabeled ligand, acquired using [3H]NMS concentrations equivalent to 0.1, 2, and 10 times the Kd value for the particular mutant wherever possible, were first analyzed simultaneously providing estimates of the affinity constants and the cooperativity factor in a logarithmic form. Fits were compared using the F statistic. The cooperativity factor is quoted only when the increase in the goodness-of-fit compared with the competitive model (α = 0) was significant at the 1% level. The three data sets for each compound were then reanalyzed as independent experiments using the Hill equation. The pIC50 values were corrected for the Cheng-Prusoff shift, using the mean affinity constant for [3H]NMS from 5 independent measurements, to calculate individual pKd values. These were used in statistical analysis by one-way analysis of variance as detailed below. The pKd values from the two methods of analysis were not significantly different (mean difference = 0.044 ± 0.044; mean ± S.D.) except in the case of AC-42 at Y82A (pKd, allosteric ternary complex model = 6.25; pKd Hill analysis = 5.75). This gave the highest cooperativity factor (0.12) of any ligand-mutant combination, rendering Hill analysis inappropriate. Thus, all of the values given are from global fits to the allosteric ternary complex model. Values are tabulated as pKd ± S.E.M.
PhI dose-response curves were fitted to a four-parameter logistic function, yielding EMax, EBasal, and pEC50 values for each NSA at the particular mutant under study. Slope factors were close to 1.0. EMax values for the NSAs, corrected for basal signaling, were expressed relative to the corresponding values for ACh in the same experiment as follows: EMaxN = (EMaxNSA - EBasal)/(EMaxACh - EBasal). EBasal was the measurement obtained in the presence of atropine, except in the case of L102A, D105E, and Y404A, where the values were not different. pEC50 and EMaxN values are tabulated. The values are mean ± S.E.M. of three to four separate measurements.
Statistical Analysis. One-way analysis of variance followed by Dunnett's post hoc test was used to determine the levels of significance of differences between the means.
Construction of the M1 mAChR Model. The initial model of the TM domain of human M1 receptor was constructed by homology with the published high-resolution X-ray crystal structure of the β2-adrenergic receptor (Cherezov et al., 2007). Alignment between the M1 sequence and β2 was based on the “classic” motifs found in each TM region, the asparagine in TM1, the aspartate in TM2, the “DRY” motif of TM3, the tryptophan in TM4, and the conserved prolines in TM5, TM6, and TM7. These alignments were used with the standard homology modeling tools in the Quanta program to construct the seven helical bundle domain of M1. The extracellular loop regions were subsequently added using a procedure developed within Glaxo-SmithKline (Harlow, Essex, UK), which makes use of a combined distance geometry sampling and molecular dynamics simulation (Blaney et al., 2001) The side chains of this model were then refined using the Karplus standard rotamer library (Dunbrack and Karplus, 1993). The final model was optimized fully (500 steps of Steepest Descent followed by 5000 steps of Adopted Basis Newton Raphson) with the CHARMM force field (Brooks et al., 1983) using helical distance constraints between the ith and i + 4th residues (except proline) within a range of 1.8 to 2.5Å to maintain the backbone hydrogen bonds of the helix bundle. A distance-dependent dielectric was used by selecting the CHARMM keyword RDIE, believed to be more appropriate when relaxing a structure.
A variant of the model was constructed in which the χ1 angle of Trp101 before the final minimization step was manually adjusted from trans to gauche+ (χ1 = -60°). This enabled the energy difference between the two different states of the receptor to be calculated. The calculated energy difference is given in kilocalories per mole.
Ligand Docking Studies. Ligand molecules were built and optimized within Spartan (SPARTAN SGI, version 5.1.3; Wavefunction, Inc., Irvine, CA) using an AM1 Hamiltonian. The atom-centered charges for the ligands, used in the docking studies, were natural atomic orbital charges calculated at the Hartree-Fock level within Spartan using a 6-31G* basis set.
Ligands were docked into the receptor model manually using a variety of low-energy starting conformations. The starting conformation of the Trp101 side chain was gauche+ (χ1 = -60°). Adjustments of the receptor protein side chains were made where necessary, always ensuring that these side chains were only in allowed rotameric states (Dunbrack and Karplus, 1993). Once again, full optimization of the receptor-ligand complexes was performed using CHARMM, the only constraints used being those that maintained the hydrogen bonding pattern of the helical bundle and the charge-charge interaction between the basic nitrogen of our ligand species and the acidic head group of Asp105 on TM3. This procedure allows full relaxation of both the ligand and the whole protein, something that is not possible with automated docking procedures.
After energy minimization, the interaction energies of the ligand 77-LH-28-1 and the indole component of the Trp101 side chain with the nearest neighbor residues Tyr82 and Leu102 were calculated by energy deconvolution. These represent the sum of the electrostatic and the Van der Waals energy contributions from all of the atoms in these two residues that interact either with the ligand or with the indole moiety of the Trp101 side chain. The force field used was CHARMM. Energies are given in kilocalories per mole. A value for the universal gas constant of 1.986 cal/°/mol was used to calculate the free energies corresponding to changes in the ligand affinity constants, measured at the assay temperature of 303 K.
Results
Ligand Binding Studies. The binding of the unlabeled ligands to wild-type and mutant M1 mAChRs was investigated by measuring inhibition of the binding of three concentrations of [3H]NMS, equal (where possible) to 0.1, 2, and 10 times the Kd value of the radioligand for the particular mutant. The inhibition curves were analyzed simultaneously using an allosteric ternary complex model of binding. This allowed three parameters to be estimated for each ligand: 1) the affinity of the ligand for the unoccupied receptor, measured by its pKd; 2) its cooperativity with the tracer ligand NMS, measured by the cooperativity factor α; and 3) a slope factor, usually approximately 0.9. In addition, the experimental design provided a measure of the expression level of the mutant receptor and a three-point estimate of its affinity for [3H]NMS, providing a useful set of data for comparison with previous studies in the laboratory. The mutants were expressed at levels ranging from 36 to 162% of the wild-type (see Materials and Methods). The pKd value and slope factor for ACh binding to the wild-type receptor were 4.97 ± 0.07 and 0.87 ± 0.03, respectively. These are in good agreement with published values, as are the affinity constants of NMS for the mutant receptors (Table 1) (Page et al., 1995; Lu and Hulme, 1999; Lu et al., 2001; Bee and Hulme, 2007).
TABLE 1.
Binding affinities of N-methylscopolamine and novel selective agonists for M1 mAChR mutants
The affinity constants pKd (−log M) for [3H]NMS, NDMC, AC-42, and 77-LH-28-1 are tabulated. A cooperativity factor value α is specified where its inclusion significantly improved the global fit to an allosteric ternary complex model of ligand-[3H]NMS interaction (see Materials and Methods). Otherwise, the interactions seemed competitive. pKd,NMS values were estimated using three different NMS concentrations, equivalent to 0.1, 2, and 10 times Kd are the means of five different experiments for each, and mutant ± S.E.M. pKd values for the NSAs are the mean ± S.E.M. of measurements using three different [3H]NMS concentrations (0.1, 2, and 10 times Kd); slope factors of binding curves were close to 1.0.
|
Helix & Mutant
|
Ligand pKd (−log M)
|
|||
|---|---|---|---|---|
| NMS | NDMC | AC42 | 77-LH-28-1 | |
| WT | 9.86 ± 0.03 | 7.41 ± 0.05 | 6.14 ± 0.12 | 6.56 ± 0.08 |
| TM2 | ||||
| M79A | 9.52 ± 0.12 | 7.62 ± 0.06 | 5.43 ± 0.04** | 5.78 ± 0.05** |
| Y82A | 9.46 ± 0.04 | 7.15 ± 0.10 | 6.25 ± 0.10 | 6.67 ± 0.20 |
| α = 0.12 | α = 0.046 | |||
| TM3 | ||||
| L100A | 9.72 ± 0.04 | 7.20 ± 0.03 | 5.81 ± 0.02 | 6.71 ± 0.02 |
| W101A | 8.90 ± 0.03 | 6.77 ± 0.06** | 8.00 ± 0.06*** | 9.00 ± 0.10*** |
| α = 0.018 | ||||
| L102A | 9.21 ± 0.01 | 6.58 ± 0.04** | 6.52 ± 0.04** | 6.61 ± 0.05 |
| α = 0.046 | α = 0.01 | |||
| D105E | 9.47 ± 0.02 | 7.04 ± 0.08* | 5.86 ± 0.05 | 6.7 ± 0.05 |
| TM7 | ||||
| Y404A | 8.11 ± 0.04 | 7.01 ± 0.03** | 6.03 ± 0.03 | 6.58 ± 0.03 |
| α = 0.046 | ||||
| Y404F | 8.80 ± 0.008 | 7.57 ± 0.04 | 5.89 ± 0.04 | 6.43 ± 0.05 |
| C407A | 8.31 ± 0.04 | 6.92 ± 0.03** | 5.74 ± 0.05* | 6.29 ± 0.03 |
| Y408A | 8.04 ± 0.03 | 7.85 ± 0.07** | 6.37 ± 0.03 | 6.37 ± 0.03 |
| Y408F | 8.98 ± 0.03 | 7.49 ± 0.05 | 5.41 ± 0.04** | 5.67 ± 0.03*** |
P < 0.05 significantly different from wild-type receptor.
P < 0.01 significantly different from wild-type receptor.
P < 0.001 significantly different from wild-type receptor.
The pKd values for binding of the NSAs (the structures are shown in Fig. 1a) to the wild-type M1 mAChR were 6.14 ± 0.12 (AC-42), 6.56 ± 0.08 (77-LH-28-1) and 7.41 ± 0.05 (NDMC; Table 1). The values for AC-42 and NDMC agree well with published estimates (Spalding et al., 2002, 2006; Sur et al., 2003; Langmead et al., 2006). All of the ligands behaved competitively with respect to [3H]NMS in equilibrium binding assays with the wild-type receptor, giving full inhibition of radioligand binding. A representative experiment is shown in Fig. 2a. This illustrates the good fit obtained with a competitive model of interaction between 77-LH-28-1 and [3H]NMS. Under our experimental conditions, this implies a cooperativity factor of α < 0.01.
Fig. 1.
a, the structures of agonists used in this study: AC-42, 77-LH-28-1, and NDMC. b, the locations of the residues mutated.
Fig. 2.
Inhibition by 77-LH-28-1 of specific binding of [3H]NMS to membranes prepared from COS-7 cells transfected with wild-type and mutant M1 mAChRs. Data are the mean ± S.E.M. of quadruplicate measurements obtained using three different [3H]NMS concentrations (0.1, 2, and 10 times Kd). Complete data sets were fitted globally to an allosteric ternary complex model of binding giving pKd values for [3H]NMS and 77-LH-28-1 and cooperativity factors (α) for mutants displaying noncompetitive allosteric binding behavior. Curves show the global fit to the allosteric ternary complex model. a, wild-type; [3H]NMS = 0.87 × 10-9 M (•), 1.7 × 10-10 M (▾), 1.8 × 10-11 (▪); pKd [3H]NMS = 9.74; pKd 77-LH-28-1 = 6.54. b, W101A: [3H]NMS = 3.17 × 10-9 M (•), 1 × 10-9 M (▾), 2.1 × 10-10 (▪); pKd [3H]NMS = 8.84; pKd 77-LH-28-1 = 9.00, α = 0.018; c, Y82A: [3H]NMS = 2.86 × 10-9 M (•), 5.7 × 10-10 M (▾), 5.8 × 10-11 (▪); pKd [3H]NMS = 9.45; pKd 77-LH-28-1 = 6.67, α = 0.046.
The affinities of the NSAs (Table 1) showed two different patterns of response to the mutations (the distribution of the mutants is shown in Fig. 1b). The changes in the pKd values are plotted in Fig. 3, where they are compared with the values for NMS. AC-42 and 77-LH-28-1 exhibited a similar pattern. They showed 5- to 8-fold reductions in affinity for the M79A and Y408F mutants. As described previously for AC-42 (Spalding et al., 2006), they displayed 70- and 275-fold increases in affinity for the W101A mutant (Table 1). Representative data are given in Fig. 2b. For 77-LH-28-1, the fit of the allosteric ternary complex model to the data was significantly improved (P < 0.01) by a value for the cooperativity factor slightly greater than 0 (α = 0.018), although the binding of AC-42 and [3H]NMS to the W101A mutant remained apparently competitive (α < 0.01). There were no affinity changes exceeding 2.5-fold in response to any of the other mutations, although an increase in affinity of AC-42 for the L102A mutant was statistically significant.
Fig. 3.
Mutation-induced changes in the affinities of the antagonist NMS (a) and novel selective agonists NDMC (b), AC-42 (c), and 77-LH-28-1 (d). The histogram represents variations of log-affinity constant ΔpKd ± S.E.M. for each mutant, compared with the wild-type, calculated from Table 1. NC designates noncompetitive behavior, defined by a value of the cooperativity factor α ≥0.01 (see Materials and Methods). *, P < 0.05; **, P < 0.01; ***, P < 0.001 with respect to wild type.
It is interesting that the alanine mutation of Tyr82 caused the emergence of clear allosteric (noncompetitive) binding behavior for both ligands with respect to [3H]NMS with cooperativity factors of 0.12 for AC-42 and 0.046 for 77-LH-28-1 (Table 1 and Fig. 2c). This implies that the removal of the Tyr82 side chain allows both of the interacting ligands to bind simultaneously to the mutant receptor to form a ternary complex. The cooperativity factor is the ratio of the affinity constants of the unlabeled ligand for the NMS-occupied and unoccupied receptor. Thus, we were also able to estimate the affinity of the ligand for the NMS-occupied receptor. Upper limits for the affinity constants at the wild-type NMS-occupied receptor were obtained by assuming that, in the case of competitive behavior, the cooperativity factor α must be ≤0.01. pKd values of 5.2 to 5.3 for AC-42 and 77-LH-28-1 were obtained at the NMS-occupied Y82A mutant, but these values must be ≤4.1 and 4.6, respectively, at the NMS-occupied WT receptor, representing increases in affinity of 5- to 10-fold. There was also evidence that the L102A mutation elicited allosteric binding behavior with AC-42 (Table 1) and possibly with 77-LH-28-1, although this was at the limit of detectability (α = 0.01).
NDMC showed a different pattern of behavior to AC-42 and 77-LH-28-1, with small reductions in affinity at W101A, L102A, D105E, Y404A, and C407A compared with the wild-type receptor, resembling, in this respect, an attenuated version of NMS or ACh (Table 1 and Fig. 3). However, unlike NMS and the other compounds, the affinity of NDMC was slightly (2.75-fold) increased by the Y408A mutation. The affinity of NDMC was not altered by the W101A mutation. There was evidence of allosteric interactions between NDMC and NMS in binding to the Y404A mutant (α = 0.046).
Phosphoinositide Assay. The functional responses of mutants to the binding of ACh and the NSAs have been studied by measuring total phosphoinositide breakdown using a scintillation proximity assay to quantitate the accumulation of labeled phosphoinositides. Analysis of the dose-response curve for each agonist at each mutant has yielded estimates of the potency, measured by the pEC50 value, and the maximum response (EMaxN) relative to that evoked by ACh at the same mutant. The results are summarized in Table 2.
TABLE 2.
Phosphoinositide responses evoked by the action of ACh and novel selective agonists on M1 mAChR mutants
Compound dilutions were added in triplicate in Krebs-Li solution. An equivalent concentration of DMSO (1%) was present in all incubations. In each experiment, atropine (10−6 M) was added to one triplicate set of assays to control for atropine-sensitive ACh-independent constitutive activity. This was maximally 8.5% for the wild-type receptor. Dose-response curves were fitted to a four-parameter logistic function, yielding EMax, EBasal, and pEC50 values for each NSA at the particular mutant under study. EMax values for the NSAs, corrected for basal signaling, were expressed relative to the corresponding values for ACh in the same experiment as follows: EMaxN = (EMaxNSA - EBasal)/(EMaxACh - EBasal). The value taken for EBasal was the measurement conducted in the presence of atropine, except in the case of L102A, D105E, and Y404A, where the values were not different. pEC50 and EMaxN values are tabulated. The values are mean ± S.E.M. of three to four separate measurements.
|
Helix & Mutant
|
ACh pEC50
|
NDMC
|
AC-42
|
77-LH-28-1
|
|||
|---|---|---|---|---|---|---|---|
| pEC50 | EMaxN | pEC50 | EMaxN | pEC50 | EMaxN | ||
| WT | 6.92 ± 0.04 | 6.42 ± 0.05 | 52.2 ± 7.0 | 5.79 ± 0.03 | 64.3 ± 2.7 | 6.68 ± 0.08 | 74.0 ± 0.4 |
| TM2 | |||||||
| M79A | 6.73 ± 0.18 | 6.00 ± 0.06 | 40.7 ± 6.5 | 5.29 ± 0.05* | 55.6 ± 8.0 | 6.14 ± 0.11* | 81.3 ± 8.3 |
| Y82A | 6.25 ± 0.07* | 6.16 ± 0.10 | 44.2 ± 5.0 | N.D. | N.D. | N.D. | N.D. |
| TM3 | |||||||
| L100A | 6.78 ± 0.14 | 6.04 ± 0.20 | 35.2 ± 2.7 | 5.52 ± 0.02 | 56.8 ± 3.9 | 6.58 ± 0.03 | 66.6 ± 4.1 |
| W101A | 5.38 ± 0.08*** | 6.17 ± 0.03 | 54.7 ± 3.6 | 7.57 ± 0.09*** | 70.8 ± 5.6 | 8.72 ± 0.02*** | 59.9 ± 10.1 |
| L102A | 4.81 ± 0.03*** | N.D. | N.D. | N.D. | N.D. | 5.41 ± 0.11*** | 30.0 ± 1.9** |
| D105E | 4.20 ± 0.03*** | N.D. | 0 | N.D. | 0 | N.D. | 0 |
| TM7 | |||||||
| Y404A | 4.58 ± 0.16*** | 6.42 ± 0.28 | 58.3 ± 3.9 | 6.07 ± 0.16 | 70.8 ± 13.9 | 7.07 ± 0.18 | 83.6 ± 11.3 |
| Y404F | 4.88 ± 0.25*** | 6.37 ± 0.07 | 21.5 ± 4.6** | 5.81 ± 0.17 | 68.4 ± 4.6 | 6.92 ± 0.23 | 75.4 ± 3.6 |
| C407A | 5.51 ± 0.07*** | 5.80 ± 0.04* | 29.7 ± 4.3** | 5.69 ± 0.05 | 80.1 ± 25.0 | 6.53 ± 0.05 | 104.1 ± 32 |
| Y408A | 5.11 ± 0.16*** | 6.98 ± 0.18* | 34.0 ± 4.4 | 4.61 ± 0.10*** | 21.4 ± 6.2* | 6.01 ± 0.10** | 34.0 ± 9.1 |
| Y408F | 5.34 ± 0.28*** | 6.74 ± 0.08 | 45.3 ± 5.1 | 4.93 ± 0.30** | 24.3 ± 2.2* | 5.26 ± 0.06*** | 44.6 ± 3.7 |
N.D., response too low to quantify.
P < 0.05 significantly different from wild-type receptor.
P < 0.01 significantly different from wild-type receptor.
P < 0.001 significantly different from wild-type receptor.
For ACh, the present study is in good agreement with previously published work (Page et al., 1995; Lu and Hulme, 1999; Lu et al., 2001; Bee and Hulme, 2007; Goodwin et al., 2007). ACh stimulated all of the mutants with EMax values between 65% (Y404A) and 94% (C407A) of the value obtained for the wild-type receptor (values not significantly different to wild-type receptor; P > 0.1). The ratio of the ACh-induced maximum signal to the basal ACh-independent signal ranged from 4.7- to 9-fold. The mean value for the wild-type receptor was 5.2. The pEC50 value of ACh at the wild-type receptor was 6.92 ± 0.04 (Table 2), which is in excellent agreement with previous values. The potency of ACh is reduced by almost 1000-fold by the D105E mutation (Page et al., 1995); this was reproduced (524-fold reduction) in the present study, which also confirmed large reductions in potency for the mutations of Trp101, Leu102, Tyr404, Cys407, and Tyr408 (Table 2).
The NSAs all stimulated the PhI response at the wild-type receptor (Fig. 4a and Table 2). AC-42 and 77-LH-28-1 acted as partial agonists with maximum responses at levels of 64 and 74% of the ACh-induced maximum response, respectively. NDMC (10-5 M) evoked 52% of the ACh-induced maximum PhI response of the wild-type receptor, but the highest concentration tested (10-4 M) showed a reduced signal, possibly indicating a nonspecific toxic effect on the cells (data not shown).
Fig. 4.
Representative phosphoinositide dose-response curves showing the action of ACh (•), NDMC (▿), AC-42 (▪), and 77-LH-28-1(⋄) on the wild-type M1 mAChR (a) and the following mutants: D105E (b), Y82A (c), L102A (d), Y408F (e), and W101A (f). The receptors were expressed in COS-7 cells labeled with [3H]inositol. Phosphoinositide response measurements were carried out using a scintillation proximity assay after 60 min of agonist stimulation. Points represent the mean ± S.E.M. of triplicate determinations. Curves show the fit to a four-parameter logistic function giving values for pEC50, EMax, EBasal, and a slope factor (usually approximately 0.9). EMax values were expressed as the percentage of the corresponding value for ACh (see Materials and Methods). Values are given as pEC50 (EMax) for ACh (•), NDMC (▿), AC-42 (▪), and 77-LH-28-1 (⋄), respectively. Values are designated N.D. if they could not be determined. Data shown are typical of three or more independent experiments. a, wild-type: 6.93 (100), 6.37 (42), 5.72 (67), 6.63 (73); b, D105E: 4.26 (100), N.D., N.D., N.D.; c, Y82A: 6.20 (100), 6.35 (36), N.D., N.D.; d, L102A: 4.75 (100), N.D., N.D., 5.40 (36); e, Y408F: 5.70(100), 6.59 (46), 4.64 (20), 5.33 (42); f, W101A: 5.49 (100), 6.12 (56), 7.46 (65), 8.70 (56).
The NSAs evoked maximum responses not significantly different from the wild-type values or the internal ACh controls (Table 2) from most of the mutant receptors. Four mutants showed strongly reduced signaling. These were D105E (Fig. 4b), for which receptor signaling by all three compounds was completely abolished; Y82A, at which only NDMC evoked a quantifiable response (Fig. 4c); L102A, in which the maximum response to all three of the NSAs was strongly reduced (Fig. 4d); and Y408A/F, in which the AC-42-induced response seemed to be more selectively reduced to less than 50% of the wild-type value (Fig. 4e). It is interesting that the AC-42 analogs evoked a maximum response comparable with that of ACh from the C407A mutant. In contrast, the maximum response to NDMC was diminished, as was also the case for the Y404F mutant (Table 2).
The relative potencies of AC-42 (pEC50 = 5.79 ± 0.03) and 77-LH-28-1 (pEC50 = 6.68 ± 0.08) at the wild-type M1 mAChR were consistent with published values, although the absolute values were 3- to 4-fold lower, perhaps reflecting our use of COS-7 rather than Chinese hamster ovary cells (Spalding et al., 2002; Langmead et al., 2008a). Particular mutations produced large changes in signaling potency (Table 2), best quantified with the more efficacious compound 77-LH-28-1. Consistent with previous reports (Spalding et al., 2006), there was a 100-fold increase in potency for W101A (Fig. 4f) and a 20-fold decrease for L102A (Fig. 4d). In contrast to ACh, the potencies of AC-42 and 77-LH-28-1 were diminished 3-fold by the mutation M79A. Mutations of Tyr408, which diminished the maximum signal, also decreased the potencies of these compounds by up to 25-fold (Fig. 4e), as seen for ACh. In contrast, mutations of Cys407 and Tyr404, although strongly inhibitory to ACh signaling (Lu et al., 2001), preserved or even slightly enhanced signaling by AC-42 and 77-LH-28-1.
For NDMC, the potency at the wild-type receptor (pEC50 = 6.42 ± 0.05) was in reasonable agreement with literature reports (Sur et al., 2003; Spalding et al., 2006). Nevertheless, the potency estimates at each mutant are less reliable because of the bell-shaped concentration-response curves produced by this compound. There was an apparent 3-fold increase in potency (P < 0.05) attributable to the Y408A mutation, as seen in the binding data (Fig. 3b). The C407A mutation showed 4-fold reduced potency (P < 0.05) for NDMC. With the exceptions of L102A and D105E, no strong effects of the remaining mutations on the potency and signaling efficacy of NDMC were observed.
Molecular Modeling and Energy Calculations. The W101A mutation strongly increased the affinity and potency of AC-42 and 77-LH-28-1. In contrast, the Y82A and L102A mutations decreased or abolished signaling by these compounds. These three residues are clustered, both in an optimized molecular model of the M1 mAChR based on the structure of rhodopsin (Goodwin et al., 2007; data not shown) and in an energy-minimized homology model of the M1 mAChR built on the structure of the β2-adrenergic receptor (Cherezov et al., 2007; see Materials and Methods). In the latter model, the indole ring of Trp101, which is in a trans conformation (χ1 = 167.7° after energy minimization), forms a stacking interaction with the side chain of Leu102 and an edge-to-face contact with the ring of Tyr82 (Fig. 5a). Energy deconvolution calculations suggested that both contacts involve favorable steric interactions in the range of 2.2 to 2.8 kcal/mol (Table 3). Preliminary calculations suggest that other neighboring residues, such as Trp91 (extracellular loop 1), make a smaller contribution (data not shown).
Fig. 5.
Model of the M1 mAChR based on the crystal structure of the β2-adrenergic receptor. The view is from outside of the TM helix bundle. Color-coding indicates the effects of the mutations of the residues studied on the potency and maximum signaling response of 77-LH-28-1: gray, little or no effect; green, reduced potency; red, reduced maximum signal; blue, increased potency. a, starting conformation showing the Trp101 side-chain in a trans conformation (χ1 = 167.7°). b, 77-LH-28-1 (magenta) docked into a binding pocket created by the isomerization of the side chain of Trp101 to a gauche+ conformation (χ1 = -66°).
TABLE 3.
Computed interaction energies of the indole side chain of W101 and of the ligand 77-LH-28-1 calculated using a structural model of the M1 mAChR
A homology model of the M1 mAChR was constructed on the structure of the β2-adrenergic receptor and optimized (see Materials and Methods). Values are the sum of the calculated interaction energies between the indicated moieties, given as kilocalories per mole. Interaction energies for the indole side chain of Trp101 with the side chains of Tyr82 and Leu102 were calculated by energy deconvolution using a trans conformation (ϰ1 = 167.7°) of the Trp101 side chain. After rotation of the side chain of Trp101 to give ϰ1 = −60° (gauche+ conformation), 77-LH-28-1 was docked into the resulting binding pocket, and the resulting structure was subjected to final energy minimisation resulting in a ϰ1 angle of −66° for Trp101. The interaction energies of the atoms of 77-LH-28-1 with Tyr82 and Leu102 were then calculated by energy deconvolution (see Materials and Methods).
|
Residue
|
Trp101 Indole Ring
|
77-LH-28-1
|
||||
|---|---|---|---|---|---|---|
| Electronic | Steric | Total | Electronic | Steric | Total | |
| kcal/mol | ||||||
| Tyr 82 | 0.064 | −2.246 | −2.181 | −0.131 | −5.839 | −5.969 |
| Leu 102 | −0.022 | −2.827 | −2.849 | −0.073 | −2.632 | −2.706 |
The large increases in the affinities of AC-42 (72-fold) and 77-LH-28-1 (275-fold) caused by the W101A mutation suggested that the indole ring of Trp101 may directly obstruct the binding of these ligands. We carried out energy calculations to explore the possibility that the side chain of 77-LH-28-1 might be capable of replacing the Trp101 indole ring, providing an anchor point for the side chain of the ligand. In the wild-type receptor, although not in the W101A mutant, this requires the preliminary conformational isomerization of the Trp101 side chain. Manual adjustment of the χ1 angle of Trp101 from a trans to a gauche+ conformation before the final minimization step (see Materials and Methods) increased the calculated overall free energy of the receptor structure by 3.4 to 3.5 kcal/mol. This unfavorable free-energy change is close to the 3.38 kcal/mol calculated from the increased affinity of 77-LH-28-1 binding resulting from the W101A mutation.
Figure 5b shows 77-LH-28-1 docked into the pocket created by the conformational isomerization of the Trp101 side chain, after energy minimization (see Materials and Methods). The view is from outside of the helix bundle. The final χ1 angle for Trp101 was -66°. The dihydroquinolinone ring of the selective agonist is complexed between the aromatic ring of Tyr82 and the side chain of Leu102, consistent with the positioning of the positively charged piperidine nitrogen close to the carboxylate group of Asp105 and the hydroxyl group of Tyr408. Met79 supports the ring of Tyr408, as proposed previously (Bee and Hulme, 2007). Packing interactions of the side chains of Tyr404 and Cys407 may restrict the binding of the bulky alkyl chain of 77-LH-28-1, modeled as extending into the “subsidiary” binding pocket between TM2, TM3, and TM7. A predicted contact between the alkyl side chain and Ile74 (2.53) is consistent with a significant reduction of the binding affinity of 77-LH-28-1 by the I74A mutation (G. Lebon, unpublished data). AC-42 can be docked in a similar manner (data not shown).
Table 3 summarizes the interaction energies, calculated by energy deconvolution, between the atoms of 77-LH-28-1 and the contact residues Tyr82 and Leu102. The bulk of the interaction energy is from the interactions of the dihydroquinolinone ring system, with a smaller contribution from interactions of the propyl linker with Tyr82; these components are not differentiated in Table 3. In the case of Leu102, the estimated interaction energy of 2.71 kcal/mol is similar to that of the indole ring of Trp101, so the substitution is broadly energetically neutral. For Tyr82, the computed interaction energy of 5.97 kcal/mol exceeds that computed for the Trp101 indole ring by 3.79 kcal/mol. This is of the right magnitude to overcome the unfavorable effect on the overall free energy of the receptor resulting from the trans-gauche+ isomerization of the Trp101 side chain. It should be noted that these calculations do not take into account factors such as the entropic costs of immobilization of the ligand side chain or desolvation of the ligand and must therefore be regarded as indicative in nature.
Discussion
The binding affinity and signaling activity of AC-42 and its more potent analog 77-LH-28-1 were unaffected or slightly potentiated by mutations of Tyr404 (7.39) and Cys407 (7.42), residues that contribute to the ACh binding pocket (Hulme et al., 2003). These results complement findings in TM3 and TM6 (Spalding et al., 2002, 2006) and further support a distinct mode of interaction for the AC-42 analogs. In contrast, the binding affinity or maximum signal evoked by NDMC was reduced, suggesting that its binding mode differs from that of the AC-42 analogs (Spalding et al., 2006).
Alanine substitution of Asp105 (3.32) abolishes signaling by AC-42 and NDMC (Spalding et al., 2006). However, this also occurs for ACh (Lu and Hulme, 1999). A less disruptive mutation may be D105E, which repositions the cationic amine counter-ion while preserving its charge. This may also perturb a hydrogen bond to the hydroxyl group of Tyr408 (7.43; Hulme et al., 1990) homologous to that in the carazololligated β2-adrenergic receptor (Cherezov et al., 2007). D105E reduces [3H]NMS affinity 2-fold and preserves 70% of the maximum signaling capability of ACh while reducing its potency by 1000-fold (Page et al., 1995).
D105E abolished signaling by AC-42, 77-LH-28-1, and NDMC without altering their binding affinities. Therefore, activation by these compounds depends on the precise position of the counter-ion within the receptor binding site. The AC-42 analogs also showed pronounced reductions in binding affinity, signaling potency, and efficacy in response to Y408F. In addition to abolishing direct interactions of the hydroxyl group with the ligand, this mutation may perturb Asp105 by disrupting the putative stabilizing hydrogen bond.
Signaling by the AC-42 analogs and NDMC is preserved by alanine mutation of Ser109 (3.36) but reduced for Asn110 (3.37) and Leu102 (3.29), which are also ACh “second shell” residues (Spalding et al., 2006). We confirmed the effect of L102A on signaling by all three NSAs. However, strong divergence between the AC-42 analogs on the one hand and ACh and NDMC on the other was uncovered in a cluster of residues at the top of TM2 and TM3.
The W101A (3.28) mutation decreases the affinity of ACh by 30-fold (Lu and Hulme, 1999). In contrast, W101A increased the affinities of AC-42 and 77-LH-28-1 by 100-fold or more, into the nanomolar range. NDMC imitated ACh, showing a 4-fold reduction in affinity. These changes were reflected in the signaling potencies rather than the maximum responses of the NSAs, implying that their signaling efficacies remained unchanged. In prior studies, we have assigned residues in which alanine substitution reduces the binding affinity but not the signaling efficacy of ACh to a ligand-anchor category, helping to ligate the agonist in both the ground and the activated states of the receptor (Hulme et al., 2003, 2007). From this viewpoint, Trp101 behaves as an antianchor or bump residue the indole side chain of which directly obstructs the binding of AC-42 and 77-LH-28-1 so that its removal substantially enhances their free energy of binding.
We have investigated two residues at the top of TM2 neighboring Trp101, namely Met79 (2.58) and Tyr82 (2.61). Confirming previous results (Lee et al., 1996; Baig et al., 2005; Bee and Hulme, 2007), ACh signaling potency was unaffected by M79A, whereas Y82A reduced the potency of ACh by 5-fold, suggesting that Met79 is not part of the ACh binding site, although Tyr82 may be a “second shell” residue. Again, NDMC responded to these mutations like ACh.
In contrast, M79A significantly reduced the affinity and potency of AC-42 and 77-LH-28-1, whereas Y82A virtually abolished signaling by both compounds. Y82A also changed the equilibrium interaction between the AC-42 analogs and [3H]NMS from apparently competitive (α ≤ 0.01) to clearly allosteric (α = 0.12 and 0.046 for AC-42 and 77-LH-28-1, respectively). The affinities of the selective ligands for the [3H]NMS-occupied receptor were increased at least 5- to 10-fold by the removal of the Tyr82 side chain, suggesting that it may help to transmit negatively cooperative interactions between the ligands.
It is interesting that Tyr82 and Leu102, mutation of which disrupted signaling by the AC-42 analogs, are in contact with the side chain of Trp101 in molecular models of the M1 mAChR based either on rhodopsin (Goodwin et al., 2007) or on the β2-adrenergic receptor (Fig. 5a). The carboxylate group of Asp105 is 3.6 Å from the indole NH, in the next turn of TM3. Met79 and Tyr408 form an extension of this cluster of residues. Leu100, the mutation of which is without functional effect, faces the lipid bilayer.
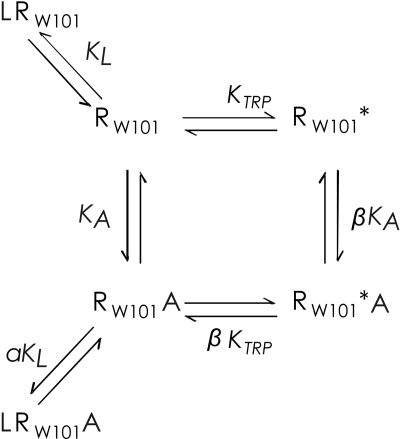
How does W101A increase the affinities of AC-42 and 77-LH-28-1? An attractive hypothesis is that deletion of the ring of Trp101 creates a novel binding pocket for a component of AC-42 and 77-LH-28-1. It is interesting that molecular modeling based on the structure of the β2-adrenergic receptor suggests that the indole ring of Trp101 can be rotated outward onto the receptor surface (Fig. 5b), potentially opening such a pocket in the wild-type receptor. In the case of 77-LH-28-1, the most likely candidate to occupy the cavity vacated by the tryptophan indole ring is the dihydroquinolinone side chain, which is similar in size and shape. Anchoring of this moiety may position the protonated nitrogen atom of the adjoining piperidine ring close to the Asp105-Tyr408 duplex, leading to receptor activation by the “back door.” Cys407, the mutation of which slightly potentiated the activity of AC-42 and 77-LH-28-1, may form a backstop to the resulting binding site. The proposed site lies within the subsidiary binding pocket between TM2, TM3, and TM7 and is distinct from the “common” allosteric site used by prototypical modulators such as gallamine and alcuronium (May et al., 2007). This model is formalized in the tryptophan-flip mechanistic scheme shown in Fig. 6, in which the equilibrium between the ground and conformationally isomerized states of the receptor is perturbed by formation of the agonist-receptor complex. The apparent affinity constant for the agonist is given by Kapp= KA · (1 + β · KTrp)/(1 + KTrp).
Fig. 6.
Trp-flip mechanism showing the equilibrium between the ground state of the receptor (RW101) and a state in which the side chain of Trp101 has undergone conformational isomerization (RW101*). A represents AC-42 or 77-LH-28-1, and L represents a nonselective orthosteric antagonist, such as NMS. The RW101*A state of the receptor is proposed to have signaling activity, whereas the RW101A state is inactive, in the case of AC-42 and 77-LH-28-1. LRW101A represents the formation of a (signaling-inactive) ternary complex between the orthosteric antagonist and the NSA.
KTrp, the isomerization constant, can be calculated from the overall free energy difference in the receptor resulting from the trans to gauche isomerization of the side chain of Trp101. This is computed to be ∼3.5 kcal/mol, yielding a value of 3 × 10-3 for KTrp. However, this can be offset by a large value of β, the cooperativity factor describing the ratio of the affinity of the agonist for the isomerized as opposed to the ground state of the receptor. By comparing the estimated free energy contributions of Leu102 and Tyr82 to the binding of the dihydroquinolinone side chain of 77-LH-28-1 and the indole ring of Trp101 (Table 3), the value of β is roughly estimated to be ∼430. The value is assumed to be comparably large for AC-42 but low for ACh and NDMC. The halogenated tricyclic ring system of NDMC may be too bulky to fit the cavity vacated by the indole side chain, whereas ligands with less bulky substituents, such as ACh, may be unable to make enough favorable contacts with the surrounding residues to stabilize the conformationally isomerized state. In the case of the AC-42 analogs, it is solely the complex with the conformationally isomerized RW101* state that is postulated to have signaling activity.
Generating a preformed cavity, as in the W101A mutant, means that the energetic cost (3.5 kcal/mol) of the trans-gauche isomerization of the side chain does not have to be paid from the free energy of ligand binding, increasing the effective value of KTrp by ∼340-fold. For the figures shown, which are derived from the indicative energy calculations performed on the β2-adrenergic receptor-based molecular model, a 340-fold increase in KTrp enhances Kapp by 96-fold, in good agreement with the experimentally observed effect of the W101A mutation on the affinities of the AC-42 analogs. In principle, the involvement of additional residues neighboring Trp101 could enhance this value further. In contrast, for the same values, a reduction in the value of β, reflecting the loss of stabilizing ligand interactions in the RW101* state in the Y82A and L102A mutants, reduces Kapp by a maximum of 2.3-fold but could decrease the proportion of RW101*A to an arbitrarily low value. In principle, the reduction in Kapp can be offset by an increase in KA, reflecting the opening up of alternative, but signaling-inactive, binding modes. This may be consistent with the observation of noncompetitive binding interactions between the AC-42 analogs and NMS in the Y82A and L102A mutants.
In summary, we suggest a novel conformational trapping mechanism for activation of the M1 mAChR by AC-42 analogs. The terminal moiety of the side chain may occupy and stabilize a cavity created by rotation of the side chain and indole ring of Trp101, positioning the positively charged ligand nitrogen close to Asp105 in TM3, following a vector distinct from that of ACh. The side chain rotation may be restricted by sequences elsewhere in the receptor, helping to explain the high functional selectivity of these compounds for the M1 subtype. It is interesting that tryptophan often occurs in protein binding sites that undergo large conformational changes on ligand binding (Gunasekaran and Nussinov, 2007). Tryptophan conformational isomerization may open up novel avenues in drug design. Finally NDMC, like ACh, does not exploit this mechanism.
Acknowledgments
We are grateful to Dr. Alex Goodwin for the construction of the Y82A mutant.
This work was supported by GlaxoSmithKline and by the Medical Research Council (United Kingdom) [Grant in Aid U.1175.03.003.00008.01].
ABBREVIATIONS: mAChR, muscarinic acetylcholine receptor; NSA, novel selective agonist; ACh, acetylcholine; TM, transmembrane domain; NMS, (-)-N-methyl scopolamine; PhI, phosphoinositide; NDMC, N-desmethylclozapine; AC-42, 4-n-butyl-1-[4-(2-methylphenyl)-4-oxo-1-butyl]-piperidine; 77-LH-28-1, 1-[3-(4-butyl-1-piperidinyl)propyl]-3,4-dihydro-2(1H)-quinolinone; PBS, phosphate-buffered saline; DMSO, dimethyl sulfoxide.
References
- Baig A, Leppik RA, and Birdsall NJ (2005) The Y82A mutant of the M1 receptor has increased affinity and cooperativity with acetylcholine for WIN 62,577 and WIN 51,708 Proceedings of the British Pharmacological Society at http://www.pA2online.org/abstracts/Vol3Issue2abst119P.pdf.
- Ballesteros JA and Weinstein H (1995) Integrated methods for the construction of three dimensional models and computational probing of structure-function relations in G-protein-coupled receptors, in Receptor Molecular Biology (Sealfon SC ed) pp 366-428, Methods in Neuroscience Vol. 25, Academic Press, San Diego. [Google Scholar]
- Bee MS and Hulme EC (2007) Functional Analysis of Transmembrane Domain 2 of the M1 Muscarinic Acetylcholine Receptor. J Biol Chem 282 32471-32479. [DOI] [PubMed] [Google Scholar]
- Blaney FE, Raveglia LF, Artico M, Cavagnera S, Dartois C, Farina C, Grugni M, Gagliardi S, Luttmann MA, Martinelli M, et al. (2001) Stepwise modulation of neurokinin-3 and neurokinin-2 receptor affinity and selectivity in quinoline tachykinin receptor antagonists. J Med Chem 44 1675-1689. [DOI] [PubMed] [Google Scholar]
- Brandish PE, Hill LA, Zheng W, and Scolnick EM (2003) Scintillation proximity assay of inositol phosphates in cell extracts: high-throughput measurement of G-protein-coupled receptor activation. Anal Biochem 313 311-318. [DOI] [PubMed] [Google Scholar]
- Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, and Karplus M (1983) CHARMM: a program for macromolecular energy minimization and dynamics calculations. J Comput Chem 4 187-217. [Google Scholar]
- Cherezov V, Rosenbaum DM, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, Choi HJ, Kuhn P, Weis WI, Kobilka BK, et al. (2007) High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor. Science 318 1258-1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies MA, Compton-Toth BA, Hufeisen SJ, Meltzer HY, and Roth BL (2005) The highly efficacious actions of N-desmethylclozapine at muscarinic receptors are unique and not a common property of either typical or atypical antipsychotic drugs: is M1 agonism a pre-requisite for mimicking clozapine's actions? Psychopharmacology (Berl) 178 451-460. [DOI] [PubMed] [Google Scholar]
- de Mendonça FL, da Fonseca PC, Phillips RM, Saldanha JW, Williams TJ, and Pease JE (2005) Site-directed mutagenesis of CC chemokine receptor 1 reveals the mechanism of action of UCB 35625, a small molecule chemokine receptor antagonist. J Biol Chem 280 4808-4816. [DOI] [PubMed] [Google Scholar]
- Dunbrack RL Jr and Karplus M (1993) Backbone-dependent rotamer library for proteins. Application to side-chain prediction. J Mol Biol 230 543-574. [DOI] [PubMed] [Google Scholar]
- Flanagan CA, Rodic V, Konvicka K, Yuen T, Chi L, Rivier JE, Millar RP, Weinstein H, and Sealfon SC (2000) Multiple interactions of the Asp(2.61(98)) side chain of the gonadotropin-releasing hormone receptor contribute differentially to ligand interaction. Biochemistry 39 8133-8141. [DOI] [PubMed] [Google Scholar]
- Goodwin JA, Hulme EC, Langmead CJ, and Tehan BG (2007) Roof and floor of the muscarinic binding pocket: variations in the binding modes of orthosteric ligands. Mol Pharmacol 72 1484-1496. [DOI] [PubMed] [Google Scholar]
- Govaerts C, Bondue A, Springael JY, Olivella M, Deupi X, Le Poul E, Wodak SJ, Parmentier M, Pardo L, and Blanpain C (2003) Activation of CCR5 by chemokines involves an aromatic cluster between transmembrane helices 2 and 3. J Biol Chem 278 1892-1903. [DOI] [PubMed] [Google Scholar]
- Gunasekaran K and Nussinov R (2007) How Different Are Structurally Flexible and Rigid Binding Sites? Sequence and Structural Features Discriminating Proteins That Do and Do Not Undergo Conformational Change Upon Ligand Binding. J Mol Biol 365 257-273. [DOI] [PubMed] [Google Scholar]
- Hulme EC, Bee MS, and Goodwin JA (2007) Phenotypic classification of mutants: a tool for understanding ligand binding and activation of muscarinic acetylcholine receptors. Biochem Soc Trans 35 742-745. [DOI] [PubMed] [Google Scholar]
- Hulme EC, Birdsall NJ, and Buckley NJ (1990) Muscarinic receptor subtypes. Annu Rev Pharmacol Toxicol 30 633-673. [DOI] [PubMed] [Google Scholar]
- Hulme EC, Lu ZL, and Bee MS (2003) Scanning mutagenesis studies of the M-1 muscarinic acetylcholine receptor. Receptors Channels 9 215-228. [PubMed] [Google Scholar]
- Jensen PC, Nygaard R, Thiele S, Elder A, Zhu G, Kolbeck R, Ghosh S, Schwartz TW, and Rosenkilde MM (2007) Molecular interaction of a potent nonpeptide agonist with the chemokine receptor CCR8. Mol Pharmacol 72 327-340. [DOI] [PubMed] [Google Scholar]
- Langmead CJ, Austin NE, Branch CL, Brown JT, Buchanan KA, Davies CH, Forbes IT, Fry VA, Hagan JJ, Herdon HJ, et al. (2008a) Characterization of a CNS penetrant, selective M1 muscarinic receptor agonist, 77-LH-28-1. Br J Pharmacol 154 1104-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead CJ, Fry VA, Forbes IT, Branch CL, Christopoulos A, Wood MD, and Herdon HJ (2006) Probing the molecular mechanism of interaction between 4-n-butyl-1-[4-(2-methylphenyl)-4-oxo-1-butyl]-piperidine (AC-42) and the muscarinic M1 receptor: direct pharmacological evidence that AC-42 is an allosteric agonist. Mol Pharmacol 69 236-246. [DOI] [PubMed] [Google Scholar]
- Langmead CJ, Watson J, and Reavill C (2008b) Muscarinic acetylcholine receptors as CNS drug targets. Pharmacol Ther 117 232-243. [DOI] [PubMed] [Google Scholar]
- Lee SY, Zhu SZ, and el-Fakahany EE (1996) Role of two highly conserved tyrosine residues in the M1 muscarinic receptor second transmembrane domain in ligand binding and receptor function. Recept Signal Transduct 6 43-52. [PubMed] [Google Scholar]
- Leonardi A, Barlocco D, Montesano F, Cignarella G, Motta G, Testa R, Poggesi E, Seeber M, De Benedetti PG, and Fanelli F (2004) Synthesis, screening, and molecular modeling of new potent and selective antagonists at the alpha 1d adrenergic receptor. J Med Chem 47 1900-1918. [DOI] [PubMed] [Google Scholar]
- Lu ZL and Hulme EC (1999) The functional topography of transmembrane domain 3 of the M1 muscarinic acetylcholine receptor, revealed by scanning mutagenesis. J Biol Chem 274 7309-7315. [DOI] [PubMed] [Google Scholar]
- Lu ZL, Saldanha JW, and Hulme EC (2001) Transmembrane domains 4 and 7 of the M1 muscarinic acetylcholine receptor are critical for ligand binding and the receptor activation switch. J Biol Chem 276 34098-34104. [DOI] [PubMed] [Google Scholar]
- May LT, Avlani VA, Langmead CJ, Herdon HJ, Wood MD, Sexton PM, and Christopoulos A (2007) Structure-function studies of allosteric agonism at M2 muscarinic acetylcholine receptors. Mol Pharmacol 72 463-476. [DOI] [PubMed] [Google Scholar]
- Page KM, Curtis CA, Jones PG, and Hulme EC (1995) The functional role of the binding site aspartate in muscarinic acetylcholine receptors probed by site-directed mutagenesis. Eur J Pharmacol 289 429-437. [DOI] [PubMed] [Google Scholar]
- Schetz JA, Benjamin PS, and Sibley DR (2000) Nonconserved residues in the second transmembrane-spanning domain of the D-4 dopamine receptor are molecular determinants of D-4-selective pharmacology. Mol Pharmacol 57 144-152. [PubMed] [Google Scholar]
- Simpson MM, Ballesteros JA, Chiappa V, Chen J, Suehiro M, Hartman DS, Godel T, Snyder LA, Sakmar TP, and Javitch JA (1999) Dopamine D4/D2 receptor selectivity is determined by a divergent aromatic microdomain contained within the second, third, and seventh membrane-spanning segments. Mol Pharmacol 56 1116-1126. [DOI] [PubMed] [Google Scholar]
- Spalding TA, Ma JN, Ott TR, Friberg M, Bajpai A, Bradley SR, Davis RE, Brann MR, and Burstein ES (2006) Structural requirements of transmembrane domain 3 for activation by the M1 muscarinic receptor agonists AC-42, AC-260584, clozapine and N-desmethylclozapine: evidence for three distinct modes of receptor activation. Mol Pharmacol 70 1974-1983. [DOI] [PubMed] [Google Scholar]
- Spalding TA, Trotter C, Skjaerbaek N, Messier TL, Currier EA, Burstein ES, Li D, Hacksell U, and Brann MR (2002) Discovery of an ectopic activation site on the M1 muscarinic receptor. Mol Pharmacol 61 1297-1302. [DOI] [PubMed] [Google Scholar]
- Stitham J, Stojanovic A, Merenick BL, O'Hara KA, and Hwa J (2003) The unique ligand-binding pocket for the human prostacyclin receptor. Site-directed mutagenesis and molecular modeling. J Biol Chem 278 4250-4257. [DOI] [PubMed] [Google Scholar]
- Sur C, Mallorga PJ, Wittmann M, Jacobson MA, Pascarella D, Williams JB, Brandish PE, Pettibone DJ, Scolnick EM, and Conn PJ (2003) N-Desmethylclozapine, an allosteric agonist at muscarinic 1 receptor, potentiates N-methyl-d-aspartate receptor activity. Proc Natl Acad Sci U S A 100 13674-13679. [DOI] [PMC free article] [PubMed] [Google Scholar]