Abstract
The epigenetic down-regulation of genes is emerging as a possible underlying mechanism of the GABAergic neuron dysfunction in schizophrenia. For example, evidence has been presented to show that the promoters associated with reelin and GAD67 are down-regulated as a consequence of DNA methyltransferase (DNMT)-mediated hypermethylation. Using neuronal progenitor cells to study this regulation, we have previously demonstrated that DNMT inhibitors coordinately increase reelin and GAD67 mRNAs. Here, we report that another group of epigenetic drugs, histone deacetylase (HDAC) inhibitors, activate these two genes with dose and time dependence comparable with that of DNMT inhibitors. In parallel, both groups of drugs decrease DNMT1, DNMT3A, and DNMT3B protein levels and reduce DNMT enzyme activity. Furthermore, induction of the reelin and GAD67 mRNAs is accompanied by the dissociation of repressor complexes containing all three DNMTs, MeCP2, and HDAC1 from the corresponding promoters and by increased local histone acetylation. Our data imply that drug-induced promoter demethylation is relevant for maximal activation of reelin and GAD67 transcription. The results suggest that HDAC and DNMT inhibitors activate reelin and GAD67 expression through similar mechanisms. Both classes of drugs attenuate, directly or indirectly, the enzymatic and transcriptional repressor activities of DNMTs and HDACs. These data provide a mechanistic rationale for the use of epigenetic drugs, individually or in combination, as a potential novel therapeutic strategy to alleviate deficits associated with schizophrenia.
The silencing of epigenetically regulated promoters is intimately associated with high levels of DNA methylation and decreased acetylation of core histones in these regions. DNA methylation, catalyzed by multiple DNA methyltransferases (DNMTs), may cause transcriptional repression by directly interfering with the binding of transcription factors to their recognition sites. Another more likely mechanism is that hypermethylation of CpG dinucleotides blocks transcription indirectly by facilitating the assembly of repressor complexes containing histone deacetylases (HDACs) in the vicinity of CpG island-containing promoters. This, in turn, leads to local histone deacetylation and the formation of a more condensed chromatin structure, which limits transcription factor accessibility and induces gene repression (Burgers et al., 2002; Klose and Bird, 2006).
Epigenetic gene regulation is critical for normal development and differentiation, including neurodevelopmental processes (Jaenisch and Bird, 2003). Accordingly, mutations in genes that encode epigenetic regulatory proteins, such as methyl CpG-binding protein 2 (MeCP2) or certain histone acetyltransferases, are associated with mental retardation in humans (Tsankova et al., 2007). In addition, increasing evidence suggests that adult neurons respond to various environmental signals via dynamic changes in DNA methylation and histone modifications. These processes are important to mechanisms of memory formation and cognition via modulation of genes involved in synaptic plasticity, such as BDNF and reelin (Levenson and Sweatt, 2005; Szyf et al., 2008). Epigenetic abnormalities, possibly introduced either during embryogenesis, puberty or adulthood, have also been noted in several psychiatric disorders, including schizophrenia, depression, and drug addiction (Tsankova et al., 2007).
Schizophrenia is associated with a dysfunction in GABAergic neurotransmission, which may be the consequence of reduced expression of GABAergic proteins, such as reelin and GAD67 (Guidotti et al., 2005; Costa et al., 2006). Reelin is an important neurodevelopmental protein that continues to be expressed in adult GABAergic neurons and has been implicated in synaptic plasticity and memory formation (Levenson et al., 2008). GAD67 is a key enzyme for GABA synthesis and is regulated by neuronal activity (Huang et al., 2007). Reduced reelin and GAD67 mRNA and protein levels have been among the more consistent findings reported in the post mortem examination of brains of patients with schizophrenia (Torrey Fuller et al., 2005; Costa et al., 2006). The down-regulation of reelin and GAD67 transcripts is accompanied by reelin promoter hypermethylation (Abdolmaleky et al., 2005; Grayson et al., 2005) and decreased amounts of trimethylation of lysine 4 on histone 3 at the GAD67 promoter (Huang et al., 2007), which has been associated with active transcription of GAD67. In addition, it has been reported that DNMT1 mRNA, which is selectively expressed in GABAergic interneurons of the adult brain, is significantly up-regulated in schizophrenia (Veldic et al., 2004; Ruzicka et al., 2007). Moreover, increased mRNA levels of the DNMT1-binding partner, HDAC1, have recently been found to correlate with GAD67 transcript down-regulation in a distinct hippocampal region of patients with schizophrenia (Benes et al., 2007). Taken together, these data are consistent with our hypothesis that alterations in reelin, GAD67, and probably other mRNAs and proteins that are expressed in GABAergic cortical neurons, are probably due to epigenetic changes triggered by DNMT1-induced promoter hypermethylation (Costa et al., 2006; Grayson et al., 2006). An epigenetic origin for the reduced expression of selected transcripts is further supported by a recent microarray-based genome-wide scan, which found significant psychosis-associated differences in DNA methylation associated with numerous genes involved in neuronal function (Mill et al., 2008).
Using NT-2 neuronal precursor cells, we have previously demonstrated that human reelin and GAD67 genes are coordinately regulated through DNA methylation-dependent mechanisms (Kundakovic et al., 2007). Here, we performed a detailed comparative analysis of the molecular events by which HDAC and DNMT inhibitors activate these two genes. We now provide evidence that DNMT1, DNMT3A, DNMT3B, MeCP2, and HDAC1 proteins form repressor complexes that negatively regulate expression of both the reelin and GAD67 promoters. Moreover, our data demonstrated that two classes of epigenetic drugs act via a similar mechanistic pathway. Both DNMT and HDAC inhibitors down-regulate all three DNMTs leading to the dissociation of a large repressor complex from the promoter. Moreover, each facilitates a reduction in DNA methylation and the reorganization of chromatin surrounding the reelin and GAD67 regulatory regions. These findings provide a better understanding of the molecular mechanisms that contribute to promoter up-regulation in neuronal progenitors. The studies are also relevant to an appreciation of promoter regulation in adult GABAergic neurons.
Materials and Methods
Cell Culture and Cell Nuclear Extracts. NT-2 cells (Stratagene, La Jolla, CA) were maintained as described previously (Kundakovic et al., 2007). Nuclear extracts from untreated and treated NT-2 cells were prepared using NE-PER Nuclear and Cytoplasmic Extraction kit as recommended by the manufacturer (Pierce Biotechnology, Rockford, IL). Protein concentrations were determined using protein assay reagent (Bio-Rad Laboratories, Hercules, CA) with bovine serum albumin as a standard.
Reagents and Antibodies. MS-275 and its inactive 3′-aminophenylbenzamide derivative (MS-IN) (Simonini et al., 2006) were dissolved in 100% dimethyl sulfoxide (DMSO) to a concentration of 10-2 M and stored at -20°C. Valproic acid (VPA), Trichostatin A (TSA), doxorubicin (DOXO), 5-aza-2′-deoxycytidine (AZA), and zebularine (ZEB) were obtained from Sigma-Aldrich (St. Louis, MO). Stock solutions of these drugs were prepared by dissolving the substances in either distilled water (VPA, DOXO), 50% acetic acid (AZA), or DMSO (TSA and ZEB), and stored at -20°C. Primary antibodies used for Western blot analysis were as follows: DNMT1 (1:1000 dilution; New England Biolabs, Ipswich, MA), DNMT3A (1:400 dilution; a generous gift from Dr. Shoji Tajima, Osaka University, Osaka, Japan), DNMT3B (1:100 dilution; Santa Cruz Biotechnology, Santa Cruz, CA), and MeCP2 antibody (1:500 dilution; Millipore, Billerica, MA). In addition, chromatin immunoprecipitation (ChIP) grade anti-DNMT1 monoclonal antibody (Imgenex, San Diego, CA), anti-DNMT3B and anti-MeCP2 polyclonal antibodies (Abcam, Cambridge, MA), anti-acetyl-histone H3 polyclonal antibody (Millipore), and DNMT3A (Santa Cruz Biotechnology) polyclonal antibody were used for ChIP assays. The same HDAC1 polyclonal antibody (Millipore) was used for both Western blot (1:2500 dilution) and ChIP analysis.
Quantitative RT-PCR Analysis. Total RNA was extracted after ultracentrifugation through CsCl (Chen et al., 2002). mRNA content was measured by competitive RT-PCR with internal standards. Specific conditions, primers and internal standards that were used to determine the levels of reelin, GAD67, and G3PDH transcripts were described previously (Kundakovic et al., 2007).
HDAC Enzymatic Assays. HDAC assays were performed using a fluorescent HDAC assay kit as recommended by the manufacturer (Millipore). A typical reaction (40 μl) included an appropriate volume of nuclear extract (containing 2 μg of protein) and 267 μM HDAC substrate in assay buffer (Millipore). To test HDAC inhibitory activity of the drugs, MS-275 (0.25, 1, 5, and 20 μM), MS-IN (5 μM), TSA (0.3 μM), or VPA (5 mM) was added to the reaction mixtures. All samples were incubated for 30 min at 30°C. Fluorescence intensity was measured (in triplicate) at an excitation wavelength of 360 nm and an emission wavelength of 460 nm, using a Spectra Max Gemini XS Microplate Spectrofluorometer (Molecular Devices, Sunnyvale, CA). Standard curves were generated using an HDAC Assay standard (Millipore) to verify assay linearity. In addition, the highest concentration of the standard was incubated with each drug to confirm that these compounds did not interfere with substrate activation.
Western Blot Analysis. Western blots were performed using the above-mentioned antibodies and according to Kundakovic et al. (2007).
DNMT Activity Assay. Nuclear extracts obtained from untreated or treated NT-2 cells were assayed for DNMT enzyme activity by the incorporation of S-adenosyl-l-[methyl-3H]methionine into [poly(dI-dC)·poly(dI-dC)] as described previously (Kundakovic et al., 2007).
RNA Interference Assay. DNMT1 and DNMT3B ShortCut siRNA Mixes (New England Biolabs) and DNMT3A ON-TARGET-plus SMARTpool siRNA (Dharmacon RNAi Technologies, Thermo Fisher Scientific, Waltham, MA) were used to down-regulate expression of the corresponding gene products. Equimolar concentrations of ON-TARGETplus Non-Targeting siRNA Pool (Dharmacon), which does not target any known transcript in human cells, were used as negative controls for all siRNA experiments. For transfections, 5 × 105 NT-2 cells were plated in a six-well plate, and 24 h later siRNAs were added with Lipofectamine 2000, according to the manufacturer's instructions (Invitrogen, Carlsbad, CA). Cells were harvested for nuclear extraction and RNA isolation 72 h after transfection. To minimize possible off-target effects of siRNA treatments, preliminary dose-response experiments were performed. For all three siRNA pools, the lowest concentrations (15 nM) that led to efficient protein knock-down were chosen for further experiments.
Chromatin Immunoprecipitation Assay. ChIP assays were performed using the ChIP assay kit protocol from Millipore as described previously (Kundakovic et al., 2007). In brief, NT-2 cells were treated with either 5 μM MS-IN or 5 μM MS-275 for 48 h. Crosslinked chromatin from 107 cells was sheared by sonication (Bioruptor UCD-200; Diagenode, Liege, Belgium) yielding 200- to 800-bp fragments. One percent of the sonicated lysate was used to quantitate the total amount of DNA present in different samples before immunoprecipitation (Inputs). Chromatin preparations were immunoprecipitated overnight using 10 μg of the following antibodies: anti-DNMT1, anti-DNMT3A, anti-DNMT3B, anti-MeCP2, anti-HDAC1, and anti-acetyl-histone H3 (Ac-H3). Chromatin eluted from samples incubated without antibody was used as negative (No Ab) control. DNA was recovered after phenol/chloroform extraction and ethanol precipitation and was analyzed by semiquantitative PCR. Conditions and primers that were used to amplify the reelin promoter region were described previously (Chen et al., 2002). The primers used to amplify the GAD67 promoter region were 5′-CTCGTGCGTGTCATCAACCTTCA-3′ (forward, -579 bp) and 5′-ACGGAGCTGGGATTGTTGTA-3′ (reverse, -320 bp). For the G3PDH control region, the forward primer (-122 bp) was 5′-CGGCTACTAGCGGTTTTACG-3′, whereas the reverse primer (+42 bp) was 5′-AGGAGGAGCAGAGAGCGAAG-3′. Both sets of primers are numbered relative to the RNA start site (GAD67, GenBank accession number NT_005403; G3PDH, GenBank accession number NM_002046). For both GAD67 and G3PDH promoters, PCR included an initial denaturation cycle (5 min, 94°C), 30 cycles of denaturation (1 min, 94°C), annealing (1 min, 62°C), and extension (1 min, 72°C), followed by the final extension (7 min, 72°C). PCR amplification products were separated on 1.6% agarose gels, and optical density readings were determined using a computer-assisted densitometry program (Kodak EDAS 290; Eastman Kodak Co., Rochester, NY). For all experiments, PCR products of input and immune-precipitated DNA samples were below saturation levels.
Luciferase Assay. An 842-bp region of the mouse reelin promoter (from -702 to +140 bp relative to RNA start site) was subcloned into the pCpGL luciferase reporter vector. The pCpGL is free of CpG dinucleotides, which makes this reporter appropriate for studying the effects of methylation on promoter activity (Klug and Rehli, 2006). The mouse reelin promoter region contains 108 CpGs. Cytosines in CpG sites were methylated in vitro by incubating 10 μg of promoter/reporter plasmid with 25 units of SssI DNA methylase in a reaction buffer (100 mM NaCl, 50 mM Tris-HCl, pH 7.5, 5 mM 2-mercaptoethanol, and 80 μM S-adenosylmethionine) for 16 h at 37°C. The extent of methylation was verified using the methylsensitive enzymes HpaII and MspI. The dual-luciferase reporter assay system (Promega, Madison, WI) was used to measure reporter activity. For each well of a six-well plate, a mixture of 2 to 4 μg of mouse reelin promoter/luciferase reporter vector and 5 ng of pRL-CMV (Renilla reniformis luciferase control) vector were cotransfected into NT-2 cells using Lipofectamine 2000, according to the manufacturer's instructions (Invitrogen). To examine the difference between the activities of the unmethylated and methylated promoters, cell lysates were prepared 4, 6, 8, 12, 24, 36, 48, 60, and 72 h after transfections. In addition, lysates were prepared from transfected cells exposed to different drug treatments 24 h after transfection. For dose-response experiments, NT-2 cells were incubated with vehicle (DMSO) or 250 nM, 1 μM, 2.5 μM, 5 μM, and 7.5 μM concentrations of either MS-275 or MS-IN for 48 h. For time-course experiments, transfections were followed by incubations with 5 μM MS-275 or 5 μM MS-IN for 6, 12, 24, and 48 h. In all cases, 20-μl aliquots of cell lysates were used for reporter activity determination in a luminometer (TD20/20; Turner Design, Sunnyvale, CA). For each individual transfection, firefly luciferase activity was normalized against R. reniformis luciferase activity to correct for variations in transfection efficiency.
Methylated DNA Immunoprecipitation Assay. To determine the methylation status of the reelin and GAD67 promoters, we used a modification of a previously published methylated DNA immunoprecipitation (MeDIP) method (Weber et al., 2005). In brief, DNA was extracted from cells that were either untreated (control for DOXO) or treated with 250 nM DOXO, 5 μM MS-IN, and 5 μM MS-275 for 48 h. Isolated DNA was sheared as indicated for the ChIP assay. For each MeDIP assay, 10 μg of DNA was diluted in Tris-EDTA buffer (10 mM Tris and 1 mM EDTA, pH 8.0) and denatured for 10 min in boiling water, and an aliquot was saved as input (control) DNA. The denaturated DNA was immunoprecipitated overnight using 10 μg of monoclonal antibody against 5-methylcytosine (Calbiochem, San Diego, CA) in a final volume of 500 μl of MeDIP buffer (10 mM sodium phosphate at pH 7.0, 140 mM NaCl, and 0.05% Triton X-100). Protein A/G-Sepharose beads (Biovision, Mountain View, CA), prewashed with phosphate-buffered saline/0.1% bovine serum albumin, were used to bind immunoprecipitated complexes. After a 2-h incubation with the reaction mixture, beads were washed three times with MeDIP buffer, resuspended in 500 μl of digestion buffer (50 mM Tris, pH 8.0, 10 mM EDTA, and 0.5% SDS) containing proteinase K (0.28 mg/ml), and incubated overnight with shaking at 55°C. DNA was recovered by centrifugation after phenol/chloroform extraction and ethanol precipitation. The resulting pellet was resuspended in 30 μl of nuclease-free water.
PCR and Real-Time PCR Analysis of MeDIP Samples. PCR and real-time PCR reactions were carried out with 40 ng of input DNA and 1/15 of the immunoprecipitated methylated DNA samples. For the GAD67 and G3PDH promoters, the primers were the same as those used for PCR analysis described for the ChIP assays. For the reelin promoter, the primers used after optimization for real-time PCR were 5′-CCGGGACACGTGTGGCGGCG-3′ (forward, -220 bp) and 5′-GGCGAGAAGAAGGCGGACGGG-3′ (reverse, +70 bp). These primers amplify a 310-bp promoter region, compared with the 482-bp region (from -220 to +262 relative to transcriptional start site) amplified after ChIP assay.
Real-time PCRs were performed using the Mx3000P real-time PCR system and data were analyzed with MxPro software (Stratagene, La Jolla, CA). PCR amplifications (in duplicates) were carried out in a 30-μl reaction volume, using Brilliant SYBR Green QPCR master mix and ROX as a reference dye (Stratagene, La Jolla, CA). In addition, Q-solution (QIAGEN, Valencia, CA) was added to reaction mixtures to increase the amplification efficiency of the GC-rich reelin promoter fragment. For all three promoter regions, PCR cycling conditions were as follows: initial denaturation at 95°C for 10 min, 40 cycles of denaturation (95°C, 30 s), annealing (65°C, 1 min), and extension (72°C, 30 s), followed by dissociation at 95°C for 1 min and 55°C for 30s. The dissociation curves of the PCR products confirmed that only the genes of interest were amplified. Before running the MeDIP samples, a standard curve for each gene was generated using serial dilutions (400-0.4 ng) of input DNA. In addition, the efficiencies of reelin versus G3PDH and GAD67 versus G3PDH PCR amplification were compared and calculated to be approximately equal. This validated the use of G3PDH as an endogenous reference for this assay.
To evaluate the relative enrichment of methylated reelin and GAD67 promoter fragments, the ratios of the signals in the immunoprecipitated DNA versus input DNA were calculated for each treatment. Those values were then standardized against the G3PDH control region. The final data are presented relative to normalized values obtained for control treatments (DOXO versus untreated, and MS-275 versus MS-275 treatment), which were given the value 1 (100%).
Results
HDAC Inhibitor MS-275 Coordinately Up-Regulate Reelin and GAD67 mRNA Expression. We have shown previously that the DNMT inhibitor DOXO (Yokochi and Robertson, 2004) induces the reelin and GAD67 mRNAs with similar dose and time dependence (Kundakovic et al., 2007). Here, we tested whether reelin and GAD67 mRNA expression might be similarly affected by the selective class I HDAC inhibitor MS-275. This benzamide derivative has been reported to inhibit class I enzymes at low micromolar concentrations, with a higher affinity for HDAC1 versus HDAC3 and no activity toward HDAC8 (Hess-Stumpp et al., 2007). To exclude drug effects that might be unrelated to the HDAC inhibitory activity, we used MS-IN, as a control, in this and other assays. MS-IN is a structural isomer of MS-275 which does not inhibit HDAC enzyme activity or affect histone acetylation status (Simonini et al., 2006).
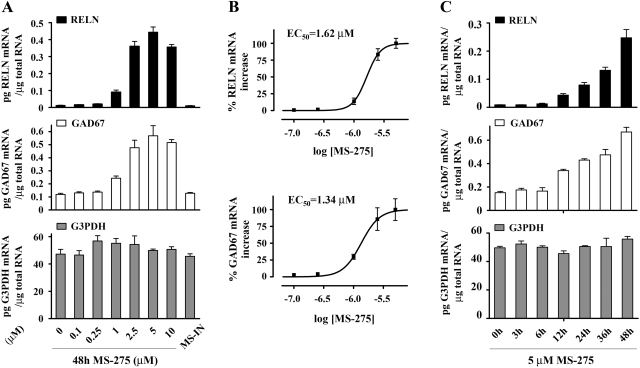
Using competitive RT-PCR with internal standards, we first determined the reelin and GAD67 mRNA levels in NT-2 cells 48 h after incubation with 0.1 to 10 μM MS-275. The data showed that MS-275 leads to a similar dose-dependent induction of these mRNAs (Fig. 1A). Barely detectable reelin mRNA expression (0.013 ± 0.0013 pg of reelin mRNA/μg of total RNA) was induced starting with a 1 μM dose of MS-275 (7.2-fold; 0.091 ± 0.012 pg of reelin mRNA/μg of total RNA). The maximal increase (35-fold) was observed with 5 μM MS-275 (0.44 ± 0.03 pg of reelin mRNA/μg of total RNA). Higher concentrations of the drug exhibited reduced activation of the reelin mRNA, as seen with the 10 μM concentration (28-fold increase; 0.36 ± 0.04 pg of reelin mRNA/μg of total RNA). Likewise, basal GAD67 mRNA levels (0.12 ± 0.01 pg of GAD67 mRNA/μg of total RNA) were induced by 1 μM MS-275 (2-fold; 0.24 ± 0.02 pg of GAD67 mRNA/μg of total RNA), and this peaked at the 5 μM dose (4.8-fold; 0.57 ± 0.08 pg of GAD67 mRNA/μg of total RNA). In this case, we also detected that higher concentrations (10 μM) of MS-275 were slightly less effective (4-fold induction; 0.52 ± 0.04 pg of GAD67 mRNA/μg of total RNA). The lower efficiency at higher drug concentrations may be due to cell toxicity, which was not observed with up to 5 μM MS-275 treatments (data not shown). In contrast to MS-275, a 48-h incubation with the inactive isomer, MS-IN (5 μM), did not have an effect on either reelin or GAD67 mRNA expression (Fig. 1A). The response to MS-275 was gene-specific, because there was no induction of G3PDH at any of the concentrations of drug used. Based on the competitive RT-PCR data, we constructed dose-response curves for the MS-275-mediated increase in reelin and GAD67 mRNAs (Fig. 1B). The calculated EC50 values for reelin (1.62 μM) and for GAD67 (1.34 μM) mRNA induction were not significantly different.
Fig. 1.
MS-275 induces reelin and GAD67 mRNA expression in similar dose-and time-dependent manners. A, results of quantitative analysis of reelin, GAD67, and G3PDH mRNA levels in NT-2 cells treated for 48 h with different concentrations of MS-275 or 5 μM MS-IN. Data are presented as amounts (picograms) of reelin, GAD67, and G3PDH mRNA per 1 μg of total RNA at the indicated doses (x-axis). B, dose-response curves for reelin and GAD67 mRNA induction after 48-h MS-275 treatment. To facilitate a comparison between the two dose-response curves, quantitative RT-PCR data are plotted as a percentage of maximal (reelin or GAD67) mRNA increase (y-axis) as a function of log drug concentration (x-axis). A baseline correction was performed: y-axis values are normalized so that baseline mRNA levels (before treatment) were defined as 0% response, whereas the highest mRNA values (which correspond to maximal gene induction) are defined as 100%. EC50 is the concentration of drug that leads to 50% of maximal reelin or GAD67 mRNA increase. Prism (ver. 5; GraphPad Software, San Diego, CA) was used to generate dose-response curves and to compare EC50 values by F test. C, graphs showing the results of quantitative analysis of reelin, GAD67, and G3PDH mRNA levels in NT-2 cells treated with 5 μM MS-275 for various times. Results are presented as amount (picogram) of reelin, GAD67, and G3PDH mRNA per 1 μg of total RNA at the indicated time points (x-axis). All data (A-C) are expressed as mean ± S.E.M. of three independent experiments (a minimum of two separate measurements were performed per treatment).
We next examined the temporal induction of these two genes by MS-275 (5 μM), performing time course experiments (up to 48 h). Quantification of mRNA levels showed that the reelin and GAD67 genes were induced in a comparable time frame. There were no associated changes in G3PDH mRNA levels (Fig. 1C). Significantly, the initial induction of both mRNAs coincided with the 12-h time point (5-fold increase for reelin and 2.2-fold increase for GAD67 mRNA levels). The similar dose- and time-dependent mRNA induction strongly suggested that MS-275 coordinately activates these two genes.
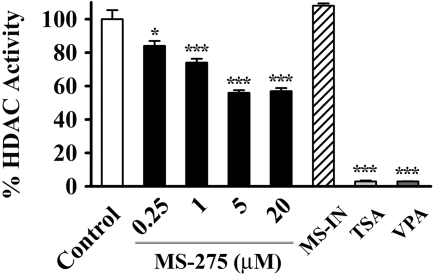
Reelin and GAD67 Promoter Induction Correlates with Inhibition of HDAC Activity. To confirm that MS-275 effectively inhibits HDAC activity in the same concentration range in which it induces reelin and GAD67 mRNAs, we assayed the HDAC enzymatic activity present in NT-2 cell nuclear extracts after their incubations with the drug (Fig. 2). We observed a dose-dependent decrease in HDAC activity by MS-275, whereas MS-IN had no effect. Comparable with its effect on reelin and GAD67 gene expression, MS-275 maximally inhibited HDAC enzymatic activity at the 5 μM concentration (46% decrease compared with control levels), whereas higher doses of the drug (20 μM) did not show additional inhibitory effects (43% decrease). Moreover, our data support the proposed selectivity of MS-275 for particular subclasses of HDACs, because TSA and VPA (two general HDAC inhibitors) led to almost complete inhibition of the enzyme activity under the same assay conditions (Fig. 2).
Fig. 2.
Inhibition of HDAC enzymatic activity correlates with reelin and GAD67 mRNA induction. Total HDAC enzymatic activity of untreated nuclear extracts (Control) and extracts treated with either MS-275 (0.25, 1, 5, and 20 μM), MS-IN (5 μM), TSA (0.3 μM), or VPA (5 mM). The fluorescence intensity was determined for each sample in triplicate. Data are expressed as a percentage of the HDAC activity detected in a control (untreated) sample. All results (from three independent experiments) represent mean ± S.E.M. *, p < 0.05; ***, p < 0.001 versus control group (one-way ANOVA followed by Bonferroni test).
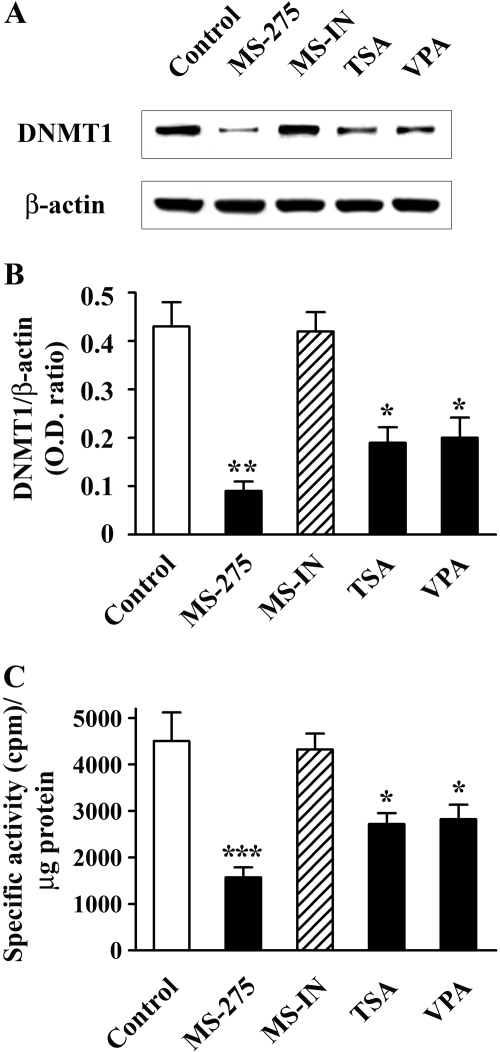
HDAC Inhibitors Down-Regulate DNMT1 Protein Expression and DNMT Enzyme Activity. Our previous study with DNMT inhibitors strongly supports a role for DNMT1 in the regulation of the reelin and GAD67 genes (Kundakovic et al., 2007). Therefore, we checked DNMT1 protein levels after incubation of NT-2 cells with either MS-275 (5 μM) for 48 h or TSA (0.3 μM) or VPA (5 mM) for 24 h. Shorter treatments with TSA and VPA were sufficient to significantly up-regulate reelin and GAD67 mRNAs while avoiding the excessive cell death seen after longer exposure to these drugs (data not shown). As depicted in Fig. 3, A and B, all three HDAC inhibitors tested led to a significant down-regulation of DNMT1 protein levels. DNMT1 protein was reduced by 79% after MS-275 treatment, and 46 and 43% after TSA and VPA treatments, respectively. In addition, down-regulation of DNMT1 protein content correlated with decreased methyltransferase activity, because MS-275, TSA, and VPA reduced nuclear DNMT enzymatic activity by 65, 40, and 37%, respectively (Fig. 3C). MS-IN (5 μM) had no effect on either DNMT1 protein levels or the methyltransferase activity of NT-2 cell nuclear extracts (Fig. 3, A-C).
Fig. 3.
HDAC inhibitors down-regulate DNMT1 expression and DNMT enzymatic activity. NT-2 cells were either not treated (Control) or treated with 5 μM MS-275 (48 h), 5 μM MS-IN (48 h), 0.3 μM TSA (24 h), or 5 mM VPA (24 h). After treatment, cells were harvested and nuclear extracts prepared. Representative DNMT1 and β-actin Western blots (A) and the ratio of the DNMT1 band (B) over the area of the β-actin band in nuclear extract protein samples from control and treated cells. For each experiment, two to three different protein concentrations per sample were run to ensure the linearity of the assay (in this and all other Western blot analyses). C, total DNA methyltransferase activity of nuclear extracts from untreated and treated cells. Data are expressed as specific radioactivity (total radioactivity - nonspecific radioactivity) normalized against the protein content present in the corresponding nuclear extracts. All results are expressed as mean ± S.E.M of three independent experiments. *, p < 0.05; **, p < 0.01; ***, p < 0.001 versus control group (one-way ANOVA followed by Bonferroni test).
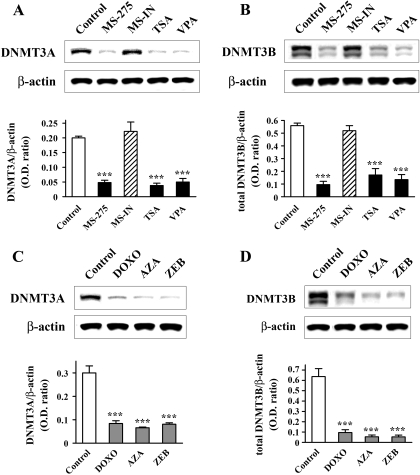
HDAC Inhibitors and DNMT Inhibitors Down-Regulate DNMT3A and DNMT3B Protein Levels. The decreased DNMT activity facilitated by HDAC inhibitors (shown above) and DNMT inhibitors (Kundakovic et al., 2007) can be explained, in part, by the down-regulation of DNMT1 protein. However, at least two additional proteins, DNMT3A and DNMT3B, contribute to total DNMT enzymatic activity. We tested whether the increased expression of the reelin and GAD67 mRNAs mediated by HDAC and DNMT inhibitors was also associated with changes in DNMT3A and DNMT3B protein levels. In nuclear extracts of NT-2 cells, we detected two different isoforms of DNMT3B protein (Figs. 4, B and D). The specificity of these two bands was confirmed using a peptide competition assay (data not shown). Western blot analysis showed that, similar to the effect on DNMT1, both groups of drugs induced a significant down-regulation of DNMT3A and both variants of DNMT3B protein (Fig. 4, A-D). MS-275 (5 μM), TSA (0.3 μM), and VPA (5 mM) treatments led to a 76, 81, and 75% reduction in DNMT3A (Fig. 4A), together with a 83, 69, and 76% reduction in total DNMT3B protein content (Fig. 4B), respectively. Likewise, DNMT3A protein was profoundly decreased after treatment with all three DNMT inhibitors [72% decrease for DOXO (250 nM), 78% for AZA (5 μM), and 73% for ZEB (500 μM); Fig. 4C]. These three drugs also led to nearly complete depletion of the two isoforms of DNMT3B protein (total reductions were as follows: 85% for DOXO, 91% for AZA, and 92% for ZEB; Fig. 4D). It is noteworthy that MS-IN (5 μM) did not have any affect on either DNMT3A or DNMT3B protein levels (Figs. 4, A or B).
Fig. 4.
HDAC and DNMT inhibitors decrease DNMT3A and DNMT3B protein levels. Western blot analyses of DNMT3A and DNMT3B proteins were performed using nuclear extracts prepared from control cells and cells treated with either HDAC inhibitors (A and B) or DNMT inhibitors (C and D). HDAC inhibitor treatments were performed as follows: 5 μM MS-275 (48 h), 5 μM MS-IN (48 h), 0.3 μM TSA (24 h), and 5 mM VPA (24 h). For assays with DNMT inhibitors, cells were treated with 250 nM DOXO (48 h), 5 μM AZA (48 h), or 500 μM ZEB (48 h followed by 48-h incubation with untreated medium). In all cases (A-D), representative Western blots are shown (top) together with the graphs depicting the ratio of the DNMT3A or DNMT3B (both isoforms) band over the area of the ß-actin band (bottom). Data represent mean ± S.E.M of three independent experiments. ***, p < 0.001 versus control group (one-way ANOVA followed by Bonferroni test).
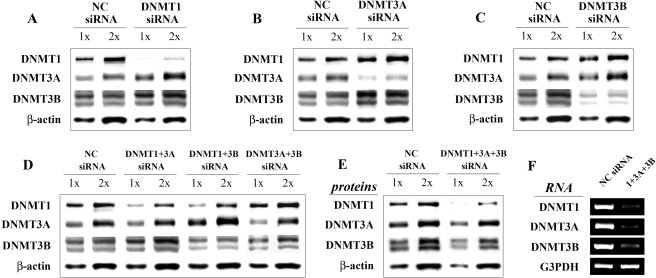
siRNA-Mediated Knock-Down of DNMT Proteins Leads to Overexpression of Nontargeted DNMTs. Because both classes of epigenetic drugs decrease DNMT protein levels, we further investigated whether down-regulation of individual DNMT proteins would be sufficient to activate reelin and GAD67 gene expression in NT-2 cells. We used specific siRNAs to target knockdown of DNMT1, DNMT3A, or DNMT3B proteins. As shown in Fig. 5, A to C, and Table 1, all single siRNA treatments efficiently down-regulated the corresponding protein levels (96, 70, and 88% decrease in DNMT1, DNMT3A, and DNMT3B proteins, respectively). However, none of these conditions led to a significant increase in either reelin or GAD67 mRNAs (see Table 1).
Fig. 5.
siRNA-mediated DNMT knock-down induces overexpression of the nontargeted DNMT proteins. siRNA pools (15 nM) targeting DNMT1, DNMT3A, and DNMT3B were transfected into NT-2 cells either individually or in different combinations. For all experiments, cells transfected with equimolar concentrations (15, 30, or 45 nM) of nontargeting siRNA pools were used as negative controls (NC siRNA). Seventy-two hours after transfection, cells were harvested for nuclear extraction and RNA isolation. Nuclear protein samples from single (A-C), double (D), and triple (E) DNMT siRNA treatments were separated on gels and transferred to nitrocellulose membranes. Blots were incubated with DNMT1, DNMT3A, and DNMT3B antibodies and were reprobed with a β-actin antibody to serve as a control for variations in sample loading. The representative Western blots (A-E) are depicted (1× corresponds to 5 μg of protein). DNMT protein levels were normalized to β-actin and expressed as a percentage of the same protein levels present in control siRNA samples. These data are given in Table 1. The corresponding reelin and GAD67 mRNA levels (normalized against G3PDH mRNA content) are shown in the same table. In all experiments, the targeted DNMT mRNAs were efficiently down-regulated. Validation RT-PCR experiment was shown for triple siRNA transfection only (F).
TABLE 1.
DNMT protein levels and corresponding reelin and GAD67 mRNA levels after treatments of NT-2 cells with siRNAs targeting indicated DNMT mRNAs
Values are means ± S.E.M. of two independent experiments (two separate measurements were performed per treatment).
| siRNA |
Protein Levels (Proportion of Control)
|
RNA Levels (Relative to Control)
|
|||
|---|---|---|---|---|---|
| DNMT1 | DNMT3A | DNMT3B | Reelin/G3PDH | GAD67/G3PDH | |
| % | % | % | |||
| Control siRNA | 100 | 100 | 100 | 1 | 1 |
| DNMT1 | 4 ± 0.9 | 230 ± 26 | 229 ± 20 | 0.94 ± 0.11 | 1.28 ± 0.08 |
| DNMT3A | 210 ± 16 | 30 ± 3.6 | 164 ± 19 | 1.30 ± 0.12 | 0.76 ± 0.08 |
| DNMT3B | 201 ± 23 | 185 ± 19 | 12 ± 2.8 | 1.1 ± 0.14 | 1.12 ± 0.07 |
| DNMT1 + 3A | 39 ± 5.2 | 81.5 ± 10 | 175 ± 15 | 0.89 ± 0.08 | 0.83 ± 0.1 |
| DNMT1 + 3B | 31 ± 5.7 | 204 ± 27 | 42 ± 6 | 0.95 ± 0.09 | 1.21 ± 0.14 |
| DNMT3A + 3B | 145 ± 16 | 95 ± 6 | 68 ± 11 | 0.74 ± 0.12 | 0.85 ± 0.09 |
| DNMT1 + 3A + 3B | 17 ± 3 | 93 ± 11 | 69.4 ± 7 | 0.93 ± 0.08 | 0.83 ± 0.06 |
It is noteworthy that, in all three cases, siRNA-mediated knock-down of one DNMT family member was associated with increased expression of the other two nontargeted DNMTs (approximate 2-fold induction; Table 1). These results implied that the DNMTs might functionally compensate one another. To explore this possibility, we also performed double and triple transfections of siRNAs targeting DNMT1, DNMT3A, and DNMT3B mRNAs. In all three double-siRNA treatments, we observed significantly reduced protein knock-down efficiency, compared with single siRNA transfections (Fig. 5D, Table 1). This was most pronounced with the DNMT3A siRNA, which barely showed any effect on DNMT3A protein levels when combined with siRNAs targeting the other two DNMTs. In addition, similar to the single knock-down experiments, the nontargeted DNMT protein level was increased in each case (Table 1). Finally, after the triple siRNA transfection, we observed a significant down-regulation of DNMT1 protein (83%), whereas DNMT3B protein was moderately reduced (30%), and DNMT3A protein was only slightly decreased if at all (Fig. 5E, Table 1). The reduced protein knock-down efficiency that occurs with the combined siRNA treatments does not seem to be caused by a technical limitation. We observed significant down-regulation of each targeted DNMT mRNA level after the same treatments, including the triple transfection (Fig. 5F). It remains a possibility that cells respond to the siRNA treatments by increasing the half-lives of the DNMT proteins to prevent depletion of all DNMT activity. Neither double- or triple-DNMT siRNA experiments led to significant changes in reelin and GAD67 mRNA levels (Table 1).
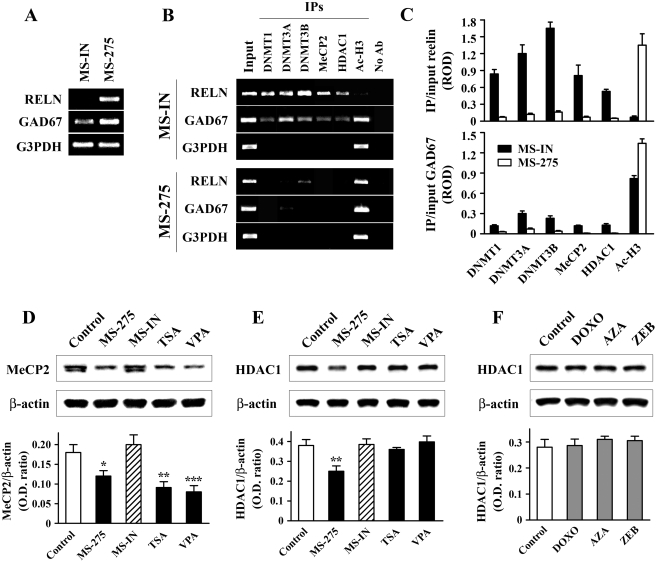
Activation of the reelin and GAD67 Genes by MS-275 Is Associated with Dissociation of DNMT1, DNMT3A, DNM3B, MeCP2, and HDAC1 from Both Promoters. We next examined whether DNMTs, together with MeCP2 and HDAC1, might form repressor complexes at the reelin and GAD67 promoters. To address this, we used antibodies against DNMT1, DNMT3A, DNMT3B, MeCP2, HDAC1, and Ac-H3 to immunoprecipitate chromatin preparations from cells treated for 48 h with either 5 μM MS-IN or 5 μM MS-275. In parallel, we performed RT-PCR analysis of RNA isolated from the same cell pools to confirm that reelin and GAD67 mRNAs were at significantly higher levels after MS-275 compared with MS-IN treatment. G3PDH mRNA levels were also monitored in the same samples (Fig. 6A). In addition, it was previously established that, compared with vehicle-treated cells, MS-IN does not have an effect on the binding of the examined proteins to the respective promoters (data not shown).
Fig. 6.
Reelin and GAD67 gene activation is accompanied by dissociation of repressor complexes from the corresponding promoters. NT-2 cells were treated with either 5 μM MS-IN or 5 μM MS-275 for 48h. A, RT-PCR analysis confirmed that reelin and GAD67 mRNAs were induced by MS-275. B, the corresponding chromatin preparations were immunoprecipitated with DNMT1, DNMT3A, DNMT3B, MeCP2, HDAC1, and Ac-H3 antibodies (IPs). Nonimmunoprecipitated samples were used as negative controls (No Ab). DNA isolated from inputs, IPs, and control samples were PCR-amplified using primers specific for the reelin, GAD67, and G3PDH promoter regions. Relative optical densities (RODs) of the bands derived from ethidium bromide-stained gels were quantified. C, graphs show the results (mean ± S.E.M of three independent experiments) of semiquantitative analysis of the occupancy of DNMT1, DNMT3A, DNMT3B, MeCP2, HDAC1, and Ac-H3 on the reelin (top) and GAD67 (bottom) promoters in MS-IN- and MS-275-treated cells, normalized to input DNA (each comparison MS-IN versus MS-275 showed statistical significance of at least p < 0.05, t test). In addition, Western blot analyses of MeCP2 protein after HDAC inhibitor treatment (D), and HDAC1 protein after treatments with HDAC inhibitors (E) and DNMT inhibitors (F) are shown. Drug treatments were as follows: 5 μM MS-275 (48 h), 5 μM MS-IN (48 h), 0.3 μM TSA (24 h), 5 mM VPA (24 h), 250 nM DOXO (48 h), 5 μM AZA (48 h), and 500 μM ZEB (48 h followed by 48-h incubation with untreated medium). In all three cases (D-F), top panels show the representative Western blots, whereas bottom panels show the graphs with the corresponding ratios of the MeCP2 or HDAC1 band to the area of the ß-actin band. All results (D-F, lower panels) are expressed as mean ± S.E.M of three independent experiments. *, p < 0.05; **, p < 0.01; ***, p < 0.001 versus control group (one-way ANOVA followed by Bonferroni test).
After chromatin immunoprecipitation, PCR amplification of the reelin and GAD67 regulatory regions showed that all three DNMTs, MeCP2, and HDAC1 bind to both promoters after MS-IN treatment (Fig. 6B). We also observed that, under these conditions, amplification of the immunoprecipitated reelin promoter gave a significantly higher signal relative to input compared with immunoprecipitated GAD67 promoter (Fig. 6C). NT-2 cells normally do not express reelin mRNA, whereas GAD67 is transcribed to some extent. The immunoprecipitation data suggest that the occupancy of individual repressor proteins is higher at the reelin than the GAD67 promoter. This is consistent with our model implying that the state of the reelin promoter is more compacted than the partially open state of the GAD67 promoter in NT-2 cells treated with MS-IN (Fig. 6A). We also showed that binding of DNMTs, MeCP2, and HDAC1 is gene-specific, because these proteins were not detected at the G3PDH gene promoter (Fig. 6B). Activation of the reelin and GAD67 mRNAs by MS-275 was accompanied by the dissociation of repressor proteins and hyperacetylation of H3 histones in the vicinity of the reelin and GAD67 regulatory regions (Fig. 6, B and C). As expected, histone H3 in the promoter of the housekeeping gene G3PDH was similarly acetylated under both treatment paradigms, which correlated with the robust expression of G3PDH mRNA seen under these conditions (Fig. 6, A and B).
MeCP2 Protein and HDAC1 Protein Levels Are Differently Affected after HDAC and DNMT Inhibitors Treatments. Dissociation of DNMT1, DNMT3A, and DNMT3B from the reelin and GAD67 promoters correlated with the drug-induced down-regulation of these proteins in nuclei of NT-2 cells. We further wanted to explore whether the decreased binding of HDAC1 and MeCP2 to these promoters might also have reflected changes in the corresponding protein levels after drug treatments. We have previously reported that DOXO treatments, which activate the reelin and GAD67 mRNAs, do not significant affect MeCP2 protein content (Kundakovic et al., 2007). However, the HDAC inhibitors MS-275, TSA, and VPA led to 33, 49, and 55% decreases in MeCP2 protein expression, respectively (Fig. 6D). In the case of HDAC1, only MS-275 showed moderate (34%) reduction in the nuclear protein levels (Fig. 6E), whereas other HDAC inhibitors (Fig. 6E) and DNMT inhibitors (Fig. 6F) were without an effect. MS-IN did not modulate expression of either MeCP2 or HDAC1 protein (Fig. 6, D and E).
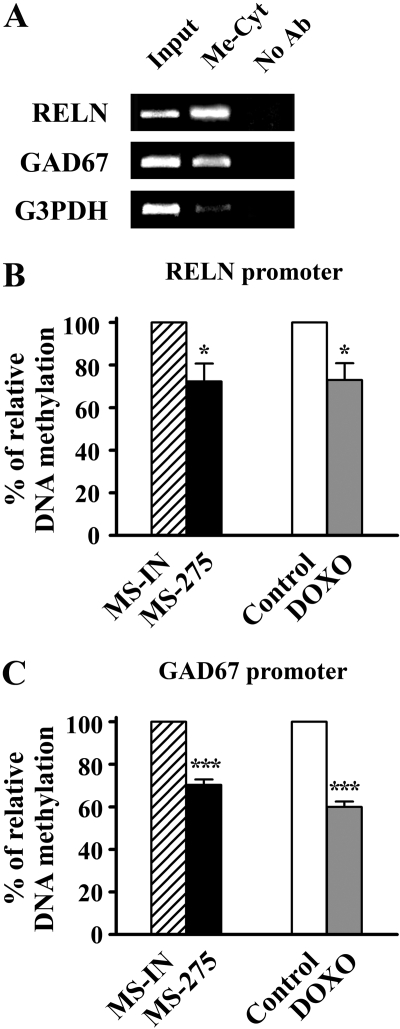
MS-275 and DOXO Decrease the Methylation Status of the Reelin and GAD67 Promoters. The drug-induced down-regulation of DNMT proteins was also accompanied by a reduced methyltransferase enzyme activity in nuclear extracts of NT-2 cells. Therefore, using a MeDIP assay, we tested whether the MS-275 and DOXO treatments were associated with changes in the methylation status of the endogenous promoters. To validate the method, we first checked the enrichment of the methylated reelin, GAD67, and G3PDH promoter sequences after incubation of the control DNA (from untreated NT-2 cells) with an antibody against 5-methylcytosine. As shown in Fig. 7A, in the untreated sample, we observed a robust signal corresponding to the methylated reelin promoter, implying that this region is highly methylated in NT-2 cells. This is consistent with data from our previous study obtained using bisulfite genomic sequencing (Chen et al., 2002). In the same sample, we also detected significant enrichment of the methylated GAD67 promoter, whereas the signal from a methylated G3PDH promoter was low, as expected for the promoter of a housekeeping gene.
Fig. 7.
MS-275 and DOXO decrease methylation of the endogenous reelin and GAD67 promoters. A, validation of the MeDIP assay. Genomic DNA extracted from untreated NT-2 cells was either immunoprecipitated with antibody against 5-methylcytosine (Me-Cyt) or incubated without an antibody (No Ab), and then amplified using PCR with primers specific for the reelin, GAD67, and G3PDH promoters. Graphs show relative enrichment of the methylated reelin (B) and methylated GAD67 (C) promoters after treatment of NT-2 cells with either 5 μM MS-275 or 250 nM DOXO for 48 h. Cells treated with 5 μM MS-IN for 48 h were used as control for MS-275 treatment, whereas untreated cells were used to control for DOXO treatment. Real-time PCR was used to quantify the amount of reelin and GAD67 promoter DNA in different samples after the MeDIP assays. Data were normalized to the G3PDH control region and expressed as a percentage of the amount of methylated promoter fragments detected in the control treatment (which were given the value 1 or 100%). Results are mean ± S.E.M. obtained from three independent experiments; *, p < 0.05 versus control group; ***, p < 0.001 versus control group (t test).
Using real-time PCR quantification, the relative enrichment of the methylated reelin and GAD67 promoter sequences was then compared between MS-IN and MS-275 treatment and between untreated sample (Control) and DOXO treatment. The results of this assay showed that MS-275 and DOXO led to moderate but significant decreases in the methylation status of both promoters (Fig. 7, B and C). MS-275 reduced reelin and GAD67 promoter methylation by 28 and 30%, respectively. Likewise, a 30% reduction in reelin and 40% reduction in GAD67 promoter methylation was seen after DOXO treatment.
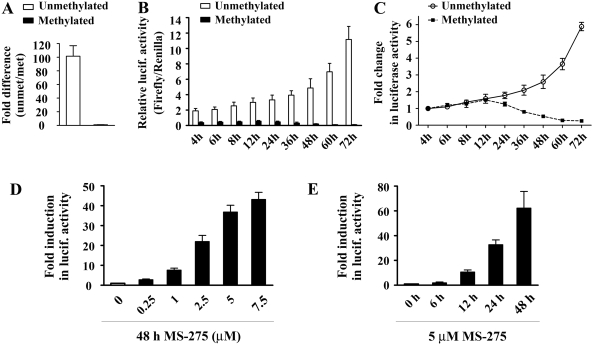
Cytosine Methylation Both Directly and Indirectly Inhibits the Reelin Promoter Activity. Previous data suggested that both DNA methylation and repressor protein assembly are important for silencing the reelin and GAD67 promoters. To further explore the contribution of these two processes, we performed in vitro reporter assays using a reelin promoter construct transfected into NT-2 cells. We have previously established that this episomal promoter efficiently models the behavior of the endogenous reelin gene (Chen et al., 2002, 2007). The unmethylated human reelin promoter construct is competent in driving reporter expression in NT-2 cells, whereas in vitro methylation of this construct before transfection significantly inhibits reporter activity. In the current study, we used a CG-rich mouse reelin promoter fragment (842 bp) subcloned into a CpG-free luciferase reporter vector and demonstrated that methylation represses its activity to a degree similar to that of its human counterpart (Fig. 8A).
Fig. 8.
Studies with the episomal reelin promoter. The mouse reelin promoter construct was either untreated (unmethylated) or treated overnight with SssI DNA methylase (methylated) before transfection into NT-2 cells. For each transfection, firefly luciferase activity was normalized against R. reniformis luciferase activity to control for plate to plate variations in transfection efficiency. A, graphs show luciferase signals from unmethylated versus methylated promoter, expressed as a fold difference in the activity. The reporter activity was measured at one time point, 72 h after transfections. Normalized (firefly/R. reniformis) values (B) and the corresponding time course trends (C) of the luciferase activity of the unmethylated and methylated reelin promoter at the indicated time points after transfection into NT-2 cells. To allow a direct comparison between the two time-course curves, the initial (4 h after transfection) luciferase signals for both promoters were given the value 1. Dose-response (D) and time course (E) effect of MS-275 on reporter activity arising from the methylated reelin promoter. Treatments were started 24 h after the plasmid/promoter constructs were transfected into NT-2 cells. Data (minimum of three transfections/construct) are expressed as a -fold induction over control transfection (cells transfected with methylated promoter and incubated with the corresponding MS-IN treatments).
We next explored whether DNA methylation per se could be sufficient to inhibit reelin promoter activity. Alternatively, to be silenced, a methylated template may require recruitment of repressor proteins and formation of repressive chromatin. Plasmid DNA adopts a nucleosomal structure after a latency period of at least 8 h after its introduction into cells (Buschhausen et al., 1987). Therefore, we compared the activity of the methylated versus unmethylated mouse reelin promoter construct at different times after transient transfection (Fig. 8, B and C). Our data showed that there was a significant difference in activity (around 5-fold) from the earliest time-point at 4 h after transfection (Fig. 8B). This difference was constant during the first 12 h after transfection (Fig. 8B), as in that period the luciferase signals from both promoters increased in a comparable fashion (Fig. 8C). However, in subsequent time points, the activity of the unmethylated construct continued to rise, whereas the signal corresponding to the methylated promoter progressively decreased and became barely detectable 60 h after transfection (Fig. 8C). Accordingly, the difference in activity between the methylated and unmethylated promoters dramatically increased over time (-fold differences in reporter activity for unmethylated versus methylated constructs were 7-fold after 24 h, 13-fold after 36 h, 24-fold after 48 h, 63-fold after 60 h, and 102-fold after 72 h). Therefore, our findings suggest that methylation can directly reduce, but is insufficient to block, the transcriptional activity of the reelin promoter. Complete repression of the promoter requires a 12- to 24-h latency period, during which, most likely, repressor complexes assemble on the DNA leading to the formation of a repressive chromatin domain.
The Transcriptional Activation Profile of the Methylated Episomal Reelin Promoter Mimics the Induction of the Endogenous Reelin and GAD67 Genes. Finally, we explored whether MS-275 treatment might reactivate expression of the methylated reelin promoter. The mouse reelin promoter/luciferase plasmid was first methylated in vitro with SssI methylase. After transfection (24 h), cells were treated with different concentrations of MS-IN and MS-275 for 48 h (Fig. 8D) or with a 5 μM concentration of these two drugs for various times (up to 48 h; Fig. 8E). The luciferase signal measured after various 48-h MS-IN treatments showed no significant variation (data not shown). Hence, the signals obtained after MS-275 treatments were normalized to the data obtained from the corresponding MS-IN treatments. It is striking that the results (Fig. 8, D and E) showed that MS-275 induced the methylated reelin promoter construct in a dose- and time-dependent manner that correlated with the induction of the endogenous reelin and GAD67 mRNAs (Figs. 1A and 2A). In dose-response studies, an induction was observed starting with 1 μM (7.6-fold), whereas luciferase activity peaked around 5 μM (37-fold increase for 5 μM and 43-fold increase for 7.5 μM MS-275) (Fig. 8D). The initial activation (11-fold) of the methylated reelin promoter was seen 12 h after MS-275 treatment (Fig. 8E). These results suggest that methylation-dependent mechanisms that induce repression of the episomal reelin promoter could be operative in silencing both the endogenous reelin and GAD67 genes.
Discussion
MS-275 is a selective class I HDAC inhibitor (Hess-Stumpp et al., 2007) that has been shown to hyperacetylate histone H3 at the reelin and GAD67 promoters in vivo (Simonini et al., 2006). Despite their different molecular targets, MS-275 and the DNMT inhibitor DOXO exhibited similar profiles for inducing reelin and GAD67 mRNAs in NT-2 neuronal progenitor cells. Like DOXO (Kundakovic et al., 2007), MS-275 increased both reelin and GAD67 mRNA expression in a comparable dose-dependent manner. Moreover, the initial induction of both mRNAs was detected 12 h after starting MS-275 treatment, mimicking the temporal pattern seen after treatment with DOXO. These data imply that there may be a similar mechanistic endpoint by which DNMT and HDAC inhibitors exert their action at the reelin and GAD67 promoters.
We previously presented evidence that DNMT inhibitors (DOXO, AZA, and ZEB) induce the reelin and GAD67 promoters, at least in part, by decreasing DNMT1 protein levels in NT-2 cells (Kundakovic et al., 2007). Other groups report that the HDAC inhibitors TSA and apicidin are able to down-regulate DNMT1 expression in Jurkat T and HeLa cells, respectively (Januchowski et al., 2007; You et al., 2008). In the current study, we demonstrated that three structurally unrelated HDAC inhibitors (MS-275, TSA, and VPA) also significantly reduce DNMT1 protein content in nuclei of NT-2 cells. In addition, some DNMT (AZA and ZEB) and HDAC inhibitors (depsipeptide and TSA) have been shown to down-regulate two additional DNMTs, DNMT3A and DNMT3B, that have important gene regulatory roles in a variety of cell types (Cheng et al., 2004; Xiong et al., 2005; Wu et al., 2008). As it turns out, all HDAC (MS-275, VPA and TSA) and DNMT inhibitor (DOXO, AZA and ZEB) treatments that induce reelin and GAD67 mRNA expression also significantly reduce DNMT3A and DNMT3B protein levels. Collectively, our data suggest that both classes of epigenetic drugs activate reelin and GAD67 expression through mechanisms that may include an action of all three DNMT proteins.
Moreover, siRNA-mediated knock-down of individual DNMT1, DNMT3A, or DNMT3B proteins had no effect on reelin and GAD67 mRNA levels. This suggests that the activation of both promoters in NT-2 cells might require simultaneous down-regulation of all three DNMTs. Furthermore, it is likely that there are additional effects of epigenetic drugs, besides their effect on DNMT proteins, that contribute to the activation of the reelin and GAD67 genes. It is noteworthy that in the same RNA interference assay, we also observed that cells responded to the partial loss of any one DNMT activity by increasing expression of the nontargeted DNMT proteins. These results imply that DNMTs cooperate in regulating gene expression and may functionally compensate for one another (Rhee et al., 2002; Leu et al., 2003). Functional cooperation of DNMTs is likely to be important for both the establishment and maintenance of promoter methylation patterns (Kim et al., 2002) and for their participation in large promoter/repressor complexes (Burgers et al., 2002). It is noteworthy that all three DNMTs, together with the Polycomb group protein EZH2, were found to be in the same repressor complex that keeps EZH2-target promoters in a silenced state (Viré et al., 2006). Our data suggest that similar DNMT-containing complexes exist at both the reelin and GAD67 promoters. It seems likely that siRNA-mediated knock-down of individual (or even two) DNMT proteins may not be sufficient to facilitate the dissociation of repressor complexes at the promoters because of functional compensation among DNMTs and other corepressors. It is also possible that the siRNAs are able to reduce unbound DNMT levels but that the treatment is insufficient to facilitate dissociation of DNMTs that are part of a bound repressor complex.
With this in mind, we hypothesized that DNMT1, DNMT3A, and DNMT3B might cooperatively silence expression of the reelin and GAD67 promoters by recruiting additional repressor proteins and bringing about a repressive local chromatin state. In support of this hypothesis, we demonstrated that all three DNMTs, together with MeCP2 and HDAC1, associate and form repressor complexes at both promoters. In addition, our data suggest that both classes of epigenetic drugs induce reelin and GAD67 expression by facilitating dissociation of repressor proteins and relaxing local chromatin structure. We have previously demonstrated that treatment with the DNMT inhibitor DOXO, resulted in release of DNMT1 and MeCP2 from the reelin promoter and an increase in local H3 histone acetylation (Kundakovic et al., 2007). Likewise, the activation of the reelin and GAD67 genes by MS-275 was accompanied by the dissociation of each of the repressor proteins (DNMT1, DNMT3A, DNMT3B, MeCP2, and HDAC1) from both promoters, and increased levels of acetyl histone H3 within the same regulatory domains.
DNMT and HDAC inhibitors activate the reelin and GAD67 promoters through similar mechanisms. It is very likely that dissociation of DNMT proteins from both promoters is a consequence of the down-regulation of nuclear DNMT1, DNMT3A, and DNMT3B protein levels, mediated by both DNMT inhibitors and HDAC inhibitors. However, we have previously reported that the DOXO-induced dissociation of MeCP2 from the reelin promoter was not accompanied by changes in MeCP2 protein levels in NT-2 cells (Kundakovic et al., 2007). MeCP2 specifically binds to methylated CpGs (Klose and Bird, 2006), and our data suggest that MeCP2 dissociates from the reelin promoter as a result of changes in methylation facilitated by DOXO treatment (Kundakovic et al., 2007). In contrast, results from the current study showed that each of the HDAC inhibitors tested significantly reduced MeCP2 protein levels that, together with promoter demethylation, probably contribute to decreased binding of this protein to the reelin and GAD67 promoters. In addition, with the exception of MS-275, the DNMT inhibitors and HDAC inhibitors tested had no effect on HDAC1 protein levels. HDAC1 is a corepressor (Jaenisch and Bird, 2003), and the most probable mechanism for decreased binding of this protein seems to be dissociation of other repressors that are needed for its recruitment to the promoters.
We further demonstrated that maximal activation of the endogenous reelin and GAD67 promoters by MS-275 and DOXO correlated with a significant decrease in methylation status of these regions. These data are consistent with our earlier findings, which showed that the HDAC inhibitors VPA and TSA, as well as the DNMT inhibitor AZA, led to demethylation of the reelin promoter in NT-2 cells (Mitchell et al., 2005). The drug-induced promoter demethylation could be an exclusively passive process caused by decreased expression of active DNMTs and the inability of cells to maintain DNA methylation patterns. Alternatively, the observed demethylation might involve an active process (Dong et al., 2008) triggered by the activation of MBD2b (Szyf et al., 2008), DNMT3A and/or DNMT3B (Ooi and Bestor, 2008), or another yet unknown protein(s) with demethylase activity. However, we would also like to leave open the possibility that slight or moderate transcriptional activation might be accomplished by solely facilitating the dissociation of repressor proteins without changing the promoter methylation status. Based on our in vitro methylated promoter/reporter studies, we propose that methylation per se is sufficient to reduce but not block transcription of the reelin and, most likely, the GAD67 promoters. Although we recognize that these data are open to alternative interpretations, it seems likely that DNA methylation primarily represents a prerequisite to silence promoters via the recruitment of repressor proteins which, in turn, induces the formation of repressive chromatin domains. Therefore, if the binding of repressors was relieved (for instance by drug-induced protein down-regulation), promoter methylation might still permit transcriptional initiation but interfere with maximal activation.
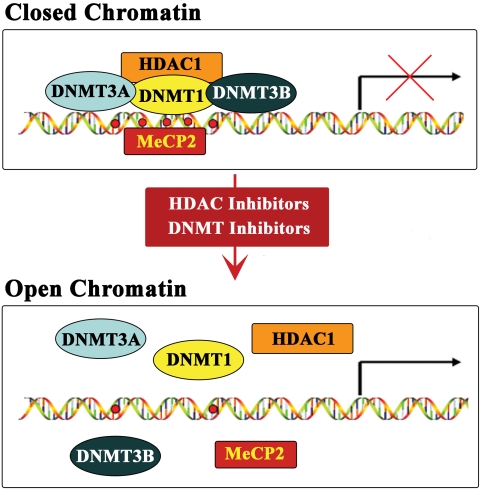
According to data described thus far, we propose the following model for activation of the reelin and GAD67 genes by DNMT and HDAC inhibitors (Fig. 9). The silenced state of the both promoters is characterized by cytosine methylation, the presence of repressor proteins (including DNMT1, DNMT3A, DNMT3B, MeCP2, and HDAC1, and most likely others), local histone deacetylation, and a compact or closed chromatin structure. Both groups of epigenetic drugs down-regulate all three DNMTs, which results in the dissociation of the repressor complexes, decreased cytosine methylation, and increased histone acetylation in the promoter regions. In the case of HDAC inhibitors, the above processes are facilitated by an inhibition of HDAC enzyme activity and reduced MeCP2 expression. The end result of the drug-induced epigenetic changes is a relaxation or opening of the chromatin structure that surrounds the reelin and GAD67 promoters, providing transcription factors access. We know that Sp1 and Pax6 bind to and trans-activate the reelin promoter (Chen et al., 2007), and functional Sp1 sites exist proximal to the GAD67 start site (Y. Chen, unpublished data).
Fig. 9.
Hypothetical model for promoter activation by HDAC and DNMT Inhibitors. For simplicity, the Fig. depicts a single stretch of promoter sequence that is applicable to both reelin and GAD67. Both promoters are represented schematically as either silent (closed) or fully active (open). The transcriptionally inactive chromatin structure (top) is the consequence of cytosine methylation (red-filled circles) and subsequent recruitment of repressor proteins, including DNMT1, DNMT3A, DNMT3B, MeCP2, and HDAC1 (most likely others, too). The down-regulation of DNMT proteins (by DNMT inhibitors and HDAC inhibitors), together with the inhibition of HDAC enzymatic activity and decrease in MeCP2 expression (in case of HDAC inhibitors), results in dissociation of these repressor complexes. This leads to demethylation of critical cytosines, histone acetylation and opening up of the chromatin surrounding the promoters (bottom). The more open chromatin configuration allows the recruitment of transcription factors, such as Sp1 (Chen et al., 2007), and the general transcriptional machinery to the promoters.
In conclusion, our findings suggest that the repressor and/or enzymatic activities of DNMT1, DNMT3A, DNMT3B, MeCP2, and HDAC1 proteins are associated with reelin and GAD67 gene silencing. Moreover, these negative regulators are targeted directly or indirectly by both classes of epigenetic drugs. Although our data are directly applicable to the epigenetic regulation of the reelin and GAD67 promoters in neuronal progenitor cells, there is increasing evidence that comparable regulatory mechanisms are operative in adult neurons (Levenson and Sweatt, 2005; Costa et al., 2006; Szyf et al., 2008). It is noteworthy that DNMT1 and HDAC1 overexpression correlates with the down-regulation of the reelin and GAD67 transcripts in postmortem brains of patients with schizophrenia (Veldic et al., 2004; Benes et al., 2007; Ruzicka et al., 2007). Therefore, these data provide a mechanistic rationale for our hypothesis that HDAC inhibitors and DNMT inhibitors used either individually or in combination may represent a novel therapeutic strategy for correcting the GABAergic deficits associated with schizophrenia (Guidotti et al., 2005; Levenson, 2007).
Acknowledgments
We thank Drs. Erminio Costa and Rajiv P. Sharma (Department of Psychiatry, University of Illinois at Chicago), for many helpful discussions and suggestions. We thank Dr. Shoji Tajima for his gift of the DNMT3A polyclonal antibody. We would also like to dedicate this work to the memory of Dr. Robert H. Costa (University of Illinois at Chicago), who died on September 1, 2006.
This work was supported in part by the National Institute of Mental Health [Grant MH062682].
ABBREVIATIONS: DNMT, DNA methyltransferase; HDAC, histone deacetylase; GAD67, glutamic acid decarboxylase 67; MeCP2, methyl CpG-binding protein 2; MS-275, N-(2-aminophenyl)-4-[N-(pyridin-3-yl-methoxycarbonyl) aminomethyl]benzamide; MS-IN, N-(3-aminophenyl)-4-[N-(pyridin-3-yl-methoxy carbonyl)aminomethyl]benzamide; NT-2, N-tera 2 neuronal progenitor cells; DMSO, dimethyl sulfoxide; VPA, valproic acid; TSA, Trichostatin A; DOXO, doxorubicin; AZA, 5-aza-2′-deoxycytidine; ZEB, zebularine; siRNA, small interfering RNA; ChIP, chromatin immunoprecipitation; RT-PCR, reverse transcription-polymerase chain reaction; bp, base pair(s); PCR, polymerase chain reaction; MeDIP, methylated DNA immunoprecipitation; G3PDH, glyceraldehyde 3-phosphate dehydrogenase; ANOVA, analysis of variance.
References
- Abdolmaleky HM, Cheng K, Russo A, Smith CL, Faraon SV, Wilcox M, Shafa R, Glatt SJ, Nguyen G, Ponte JF, Thiagalingam S, and Tsuang MT (2005) Hypermethylation of the reelin (RELN) promoter in the brain of schizophrenic patients: A preliminary report. Am J Med Genet B Neuropsychiatr Genet 134 60-66. [DOI] [PubMed] [Google Scholar]
- Benes FM, Lim B, Matzilevich D, Walsh JP, Subburaju S, and Minns M (2007) Regulation of the GABA cell phenotype in hippocampus of schizophrenics and bipolars. Proc Natl Acad Sci U S A 104 10164-10169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgers WA, Fuks F, and Kouzarides T (2002) DNA methyltransferases get connected to chromatin. Trends Genet 18 275-277. [DOI] [PubMed] [Google Scholar]
- Buschhausen G, Wittig B, Graessmann M, and Graessmann A (1987) Chromatin structure is required to block transcription of the methylated herpes simplex virus thymidine kinase gene. Proc Natl Acad Sci U S A 84 1177-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Sharma RP, Costa RH, Costa E, and Grayson DR (2002) On the epigenetic regulation of the human reelin promoter. Nucleic Acids Res 30 2930-2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Kundakovic M, Agis-Balboa RC, Pinna G, and Grayson DR (2007) Induction of the reelin promoter by retinoic acid is mediated by Sp1. J Neurochem 103 650-665. [DOI] [PubMed] [Google Scholar]
- Cheng JC, Weisenberger DJ, Gonzales FA, Liang G, Xu GL, Hu YG, Marquez VE, and Jones PA (2004) Continuous zebularine treatment effectively sustains demethylation in human bladder cancer cells. Mol Cell Biol 24 1270-1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa E, Dong E, Grayson DR, Ruzicka WB, Simonini MV, Veldic M, and Guidotti A (2006) Epigenetic targets in GABAergic neurons to treat schizophrenia. Adv Pharmacol 54 95-117. [DOI] [PubMed] [Google Scholar]
- Dong E, Nelson M, Grayson DR, Costa E, and Guidotti A (2008) Clozapine and sulpiride but not haloperidol or olanzapine activate brain DNA-demethylase. Proc Natl Acad Sci U S A 105 13614-13619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grayson DR, Jia X, Chen Y, Sharma RP, Mitchell CP, Guidotti A, and Costa E (2005) Reelin promoter hypermethylation in schizophrenia. Proc Natl Acad Sci U S A 102 9341-9346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grayson DR, Chen Y, Costa E, Dong E, Guidotti A, Kundakovic M, and Sharma RP (2006) The human reelin gene: transcription factors (+), repressors (-) and the methylation switch (+/-) in schizophrenia. Pharmacol Ther 111 272-286. [DOI] [PubMed] [Google Scholar]
- Guidotti A, Auta J, Davis JM, Dong E, Grayson DR, Veldic M, Zhang X, and Costa E (2005) GABAergic dysfunction in schizophrenia: new treatment strategies on the horizon. Psychopharmacology (Berl) 180 191-205. [DOI] [PubMed] [Google Scholar]
- Hess-Stumpp H, Bracker TU, Henderson D, and Politz O (2007) MS-275, a potent orally available inhibitor of histone deacetylases-the development of an anticancer agent. Int J Biochem Cell Biol 39 1388-1405. [DOI] [PubMed] [Google Scholar]
- Huang HS, Matevossian A, Whittle C, Kim SY, Schumacher A, Baker SP, and Akbarian S (2007) Prefrontal dysfunction in schizophrenia involves mixed-lineage leukemia 1-regulated histone methylation at GABAergic gene promoters. J Neurosci 27 11254-11262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenisch R and Bird A (2003) Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet 33 (Suppl): 245-254. [DOI] [PubMed] [Google Scholar]
- Januchowski R, Dabrowski M, Ofori H, and Jagodzinski PP (2007) Trichostatin A down-regulate DNA methyltransferase 1 in Jurkat T cells. Cancer Lett 246 313-317. [DOI] [PubMed] [Google Scholar]
- Kim GD, Ni J, Kelesoglu N, Roberts RJ, and Pradhan S (2002) Co-operation and communication between the human maintenance and de novo DNA (cytosine-5) methyltransferases. EMBO J 21 4183-4195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose RJ and Bird AP (2006) Genomic DNA methylation: the mark and its mediators. Trends Biochem Sci 31 89-97. [DOI] [PubMed] [Google Scholar]
- Klug M and Rehli M (2006) Functional analysis of promoter CpG methylation using a CpG-free luciferase reporter vector. Epigenetics 1 127-130. [DOI] [PubMed] [Google Scholar]
- Kundakovic M, Chen Y, Costa E, and Grayson DR (2007) DNA methyltransferase inhibitors coordinately induce expression of the human reelin and glutamic acid decarboxylase 67 genes. Mol Pharmacol 71 644-653. [DOI] [PubMed] [Google Scholar]
- Leu YW, Rahmatpanah F, Shi H, Wei SH, Liu JC, Yan PS, and Huang TH (2003) Double RNA interference of DNMT3b and DNMT1 enhances DNA demethylation and gene reactivation. Cancer Res 63 6110-6115. [PubMed] [Google Scholar]
- Levenson JM and Sweatt JD (2005) Epigenetic mechanisms in memory formation. Nat Rev Neurosci 6 108-118. [DOI] [PubMed] [Google Scholar]
- Levenson JM (2007) DNA (cytosine-5) methyltransferase inhibitors: a potential therapeutic agent for schizophrenia. Mol Pharmacol 71 635-637. [DOI] [PubMed] [Google Scholar]
- Levenson JM, Qiu S, and Weeber EJ (2008) The role of reelin in adult synaptic function and the genetic and epigenetic regulation of the reelin gene. Biochim Biophys Acta 1779 422-431. [DOI] [PubMed] [Google Scholar]
- Mill J, Tang T, Kaminsky Z, Khare T, Yazdanpanah S, Bouchard L, Jia P, Assadzadeh A, Flanagan J, Schumacher A, et al. (2008) Epigenomic profiling reveals DNA-methylation changes associated with major psychosis. Am J Hum Genet 82 696-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell CP, Chen Y, Kundakovic M, Costa E, and Grayson DR (2005) Histone deacetylase inhibitors decrease reelin promoter methylation in vitro. J Neurochemistry 93 483-492. [DOI] [PubMed] [Google Scholar]
- Ooi SK and Bestor TH (2008) The colorful history of active DNA demethylation. Cell 133 1145-1148. [DOI] [PubMed] [Google Scholar]
- Rhee I, Bachman KE, Park BH, Jair KW, Yen RW, Schuebel KE, Cui H, Feinberg AP, Lengauer C, Kinzler KW, et al. (2002) DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature 416 552-556. [DOI] [PubMed] [Google Scholar]
- Ruzicka WB, Zhubi A, Veldic M, Grayson DR, Costa E, and Guidotti A (2007) Selective epigenetic alteration of layer I GABAergic neurons isolated from prefrontal cortex of schizophrenia patients using laser-assisted microdissection. Mol Psychiatry 12 385-397. [DOI] [PubMed] [Google Scholar]
- Simonini MV, Camargo LM, Dong E, Maloku E, Veldic M, Costa E, and Guidotti A (2006) The benzamide MS-275 is a potent, long-lasting brain region-selective inhibitor of histone deacetylases. Proc Natl Acad Sci U S A 103 1587-1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szyf M, McGowan P, and Meaney MJ (2008) The social environment and the epigenome. Environ Mol Mutagen 49 46-60. [DOI] [PubMed] [Google Scholar]
- Torrey EF, Barci BM, Webster MJ, Bartko JJ, Meador-Woodruff JH, and Knable MB (2005) Neurochemical markers for schizophrenia, bipolar disorder, and major depression in postmortem brains. Biol Psychiatry 57 252-260. [DOI] [PubMed] [Google Scholar]
- Tsankova N, Renthal W, Kumar A, and Nestler EJ (2007) Epigenetic regulation in psychiatric disorders. Nat Rev Neurosci 8 355-367. [DOI] [PubMed] [Google Scholar]
- Veldic M, Caruncho HJ, Liu WS, Davis J, Satta R, Grayson DR, Guidotti A, and Costa E (2004) DNA-methyltransferase 1 mRNA is selectively overexpressed in telencephalic GABAergic interneurons of schizophrenia brains. Proc Natl Acad Sci U S A 101 348-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viré E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, Morey L, Van Eynde A, Bernard D, Vanderwinden JM, et al. (2006) The Polycomb group protein EZH2 directly controls DNA methylation. Nature 439 871-874. [DOI] [PubMed] [Google Scholar]
- Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, Lam WL, and Schübeler D (2005) Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet 37 853-862. [DOI] [PubMed] [Google Scholar]
- Wu LP, Wang X, Li L, Zhao Y, Lu S, Yu Y, Zhou W, Liu X, Yang J, Zheng Z, et al. (2008) Histone deacetylase inhibitor depsipeptide activates silenced genes through decreasing both CpG and H3K9 methylation on the promoter. Mol Cell Biol 28 3219-3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, Dowdy SC, Podratz KC, Jin F, Attewell JR, Eberhardt NL, and Jiang SW (2005) Histone deacetylase inhibitors decrease DNA methyltransferase-3B messenger RNA stability and down-regulate de novo DNA methyltransferase activity in human endometrial cells. Cancer Res 65 2684-2689. [DOI] [PubMed] [Google Scholar]
- Yokochi T and Robertson KD (2004) Doxorubicin inhibits DNMT1, resulting in conditional apoptosis. Mol Pharmacol 66 1415-1420. [DOI] [PubMed] [Google Scholar]
- You JS, Kang JK, Lee EK, Lee JC, Lee SH, Jeon YJ, Koh DH, Ahn SH, Seo DW, Lee HY, et al. (2008) Histone deacetylase inhibitor apicidin down-regulates DNA methyltransferase 1 expression and induces repressive histone modifications via recruitment of corepressor complex to promoter region in human cervix cancer cells. Oncogene 27 1376-1386. [DOI] [PubMed] [Google Scholar]