Abstract
Many eubacterial DNA polymerases are bifunctional molecules having both polymerization (P) and 5′ nuclease (N) activities, which are contained in separable domains. We previously showed that the DNA polymerase I of Thermus aquaticus (TaqNP) endonucleolytically cleaves DNA substrates, releasing unpaired 5′ arms of bifurcated duplexes. Here, we compare the substrate specificities of TaqNP and the isolated 5′ nuclease domain of this enzyme, TaqN. Both enzymes are significantly activated by primer oligonucleotides that are hybridized to the 3′ arm of the bifurcation; optimal stimulation requires overlap of the 3′ terminal nucleotide of the primer with the terminal base pair of the duplex, but the terminal nucleotide need not hybridize to the complementary strand in the substrate. In the presence of Mn2+ ions, TaqN can cleave both RNA and circular DNA at structural bifurcations. Certain anti-TaqNP mAbs block cleavage by one or both enzymes, whereas others can stimulate cleavage of nonoptimal substrates.
Structure-sensing nucleases are ubiquitous in biology, being essential for both the synthesis and the repair of DNA (1–12). Several of these enzymes cleave bifurcated duplex DNAs endonucleolytically, releasing the single-stranded 5′ arm (13). The 5′ nuclease activity accounts for the ability of these enzymes to remove RNA primers or damaged DNA nucleotides (for review, see ref. 15). In eubacteria, 5′ nucleases are discrete domains in the DNA polymerases, but in Eukarya and Archaea, they are separate from DNA polymerases and have been called DNA endonuclease IV (1) or, more recently, FEN1 nuclease (16). We refer to the DNA polymerase of T. aquaticus as TaqNP, because it contains both the nuclease and polymerase domains in a single polypeptide; likewise, we refer to the isolated nuclease domain of this enzyme as TaqN.
We previously showed that the 5′ nuclease activity of TaqNP is increased by several orders of magnitude if an oligonucleotide (the primer) is hybridized to the 3′ arm of the bifurcation (13). The role of the primer in the activation and location of cleavage was unclear. We also showed that cleavage required a free 5′ end of the single-stranded arm, indicating that the enzyme moved to the site of cleavage by threading the single strand through a hole or a narrow groove in the enzyme, and a requirement for a free 5′ end was subsequently observed for the calf FEN1 nuclease (14). Recent crystal structures have demonstrated the existence of helical arches or holes in a similar nuclease, T5 exonuclease, as well as in TaqNP and FEN nucleases (15–21).
The influence of the polymerase domain on the activity of the 5′ nuclease has not been determined. Structure–function probing of the nuclease and polymerase domains of TaqNP by using mAbs generated against the intact enzyme (22) demonstrated some functional overlap between the nuclease and polymerase domains of the enzyme (23). Here, we compare the activities and substrate requirements of TaqN, the isolated 5′ nuclease of TaqNP, and the same functional domain when it is part of the intact TaqNP holoenzyme. We find that the site of cleavage is fixed not only by the point of duplex bifurcation but also by the end of the oligonucleotide that is hybridized to the 3′ arm; this end is recognized directly by the isolated nuclease domain and is most effective if it overlaps at least one base pair at the end of duplex. The polymerase domain can influence the choice of cleavage site and, in most cases, appears to impose additional stringency on substrate processing.
MATERIALS AND METHODS
Materials.
Escherichia coli Klenow fragment, polynucleotide kinase, T4 DNA polymerase, and terminal deoxynucleotidyltransferase (TdT) were from Promega. Anti-TaqNP mAbs were the gift of J. Daiss (Eastman Kodak, now Ortho Diagnostics). Oligonucleotides were from Integrated DNA Technologies (Coralville, IA); unless noted, A, G, C, and T refer to deoxyribonucleotides.
Cloning of TaqNP Gene and Its Deletion Mutants.
Cloning of the TaqNP gene was done as described by Engelke et al. (24). The TaqNP gene was PCR-amplified with 5′-GTGAGATCTATCACTCCTTGGCGGAGAGCCAGTC-3′ and 5′-CACGAATTCGGGGATGCTGCCCCTCTTTGAGCCCAAG-3′ as primers, by using as a template DNA from T. aquaticus strain YT-1. The amplified product was cloned into the pTTQ18 expression plasmid (25) at EcoRI and BamHI sites, and the construct pTaq1 carrying the full-size TaqNP gene was identified by restriction analysis and the presence of 5′ nuclease activity in crude cell extracts. A blunt-ended EcoRI/SalI fragment containing the TaqNP I gene was ligated into a blunt-ended BamHI site of the pET3c expression vector (26).
The TaqN template containing the N-terminal part of TaqNP gene was amplified from pTaq1 by using 5′-TAATACGACTCACTATAGG-3′ and 5′-CGGCCAGGGCCAGAAGATCG-3′ as PCR primers. The amplified DNA was ligated into pET21b vector (Novagen) by using XbaI and blunt-ended BstXI sites of the PCR fragment and XbaI and blunt-ended NotI of the vector. The resulting construct had the N-terminal part of the TaqNP gene linked at Ala-293 to the His tag of the pET21b expression vector. Sequence analysis of the amplified genes for TaqN and TaqNP showed that they both encoded the same protein up to position 293. Other templates were constructed similarly.
Expression and Purification of TaqNP I and Its Deletion Mutants.
TaqNP was expressed and purified as described by Engelke et al. (24), and TaqN was expressed and purified by His tag affinity chromatography from E. coli strain BL21(DE3) (Novagen). Crude extracts containing soluble proteins, TaqNP, and TaqN were prepared by lysis of pelleted cells in 50 ml of 10 mM Tris⋅HCl, pH 8.3/1 mM EDTA/0.5 mg/ml lysozyme during incubation at room temperature for 15 min; the lysate was mixed with 50 ml of 10 mM Tris⋅HCl, pH 7.8/50 mM KCl/1 mM EDTA/0.5% Tween 20/0.5% Nonidet P-40, heated at 72°C for 1 hr, and cell debris was removed by centrifugation at 12,000 × g for 5 min (24). Enzyme concentrations were determined by measuring absorption at 279 nm (27); when TaqNP from Promega was used, quantities are noted in units. All enzymes were dialyzed and stored in 50% (vol/vol) glycerol/20 mM Tris⋅HCl, pH 8/50 mM KCl/0.5% Tween 20/0.5% Nonidet P-40/100 μg/ml BSA.
DNA Preparation for 5′ Nuclease Assays.
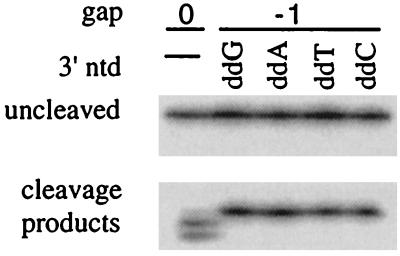
To construct D16 DNA, the PvuII–AvaI fragment of pGEM1 plasmid DNA (Promega) was ligated into pGEM2 DNA digested with the same enzymes, creating a pGEM2 plasmid with a 32-nt inverted repeat of the polylinker region. D16 DNA was generated by PCR amplification across this repeat, with the Stoffel fragment of TaqNP (Perkin–Elmer) with 32P-labeled 5′-TAATACGACTCACTATAGGG-3′ and 5′-GAATTCGATTTAGGTGACACTATAGAA-3′ as primers. The shorter D9 and D6 DNA fragments were derived from D16 by digesting it with SacI or TaqI, respectively, to remove internal portions of the 36-nt inverted repeat, and the flanking fragments were joined by ligation before use as templates for PCR amplification as described above. D16-ΔA DNA was constructed essentially as described above for D16 DNA except that one of the primers was 5′-TCACTATAGGGAGCCGGAATTCG-3′, which deleted one A from the D16 sequence and shortened the 5′ arm by 9 nt. The 32P-labeled 206-bp fragment and oligonucleotides 30-12 and 30-0 were prepared as described (13). On occasion (Fig. 2), primer oligonucleotides were elongated by one dideoxynucleotide, by using TdT. U1 RNA was made by SP-6 transcription as described (28), by using as template a gene encoding a variant of U1 RNA (29).
Figure 2.
Efficient cleavage of gap −1 substrates with unpaired 3′ ends. The digestion products were generated by TaqNP on incubation with the gap 0 substrate shown in Fig. 1B or with the same substrate but with the 3′ end of the primer elongated with terminal deoxytransferase and the appropriate dideoxynucleoside triphosphate to produce a the 3′ terminal nucleotides indicated. Digestion conditions were 1 nM D16 hairpin substrate, 0.2 μM gap primers, and 0.5 unit of TaqNP (Promega) in 10 μl of 10 mM Tris⋅HCl, pH 8/50 mM KCl/1 mM MgCl2/10 μg/ml tRNA incubated at 55°C for 2 min.
5′ Nuclease Assays.
PCR products D16, D16-ΔA, D9, and D6 labeled at their 5′ ends with P32 were gel-purified, diffusion-eluted, ethanol-precipitated, and resuspended in TE buffer (10 mM Tris⋅HCl, pH 8/1 mM EDTA) to make a 10 nM solution. To prepare substrates for 5′ nuclease assays, the DNA stock solutions were heated at 95°C for 15 sec to denature the DNA and cooled on ice to form hairpin substrates. Cleavage reactions were carried out in 10 μl of 10 mM Tris⋅HCl, pH 8.5/50 mM KCl/1 mM MgCl2/10 μg/ml tRNA (Sigma) containing 1 nM hairpin substrate and primer oligonucleotide as appropriate. The gap 0 primer was 5′-ATTTAGGTGACACTATAGAATACA-3′ and the gap 4 primer was 5′-GAATTCGATTTAGGTGACACTATAGAA-3′. Primers with intermediate gaps were progressively shorter at their 3′ ends and contained at least 24 nt complementary to the 3′ arm of the template strand. Reactions were started by addition of enzyme at 55°C, as described in the figure legends, and stopped by addition of 4 μl of buffer containing 90% formamide/20 mM EDTA. Unless otherwise indicated, the products were separated by electrophoresis in 12% (19:1) polyacrylamide gels.
RESULTS
Several eubacterial DNA polymerases can both synthesize and degrade DNA. Although the enzymes are single polypeptides, they have several functionally distinct domains. The 5′ nuclease cleaves bifurcated DNAs in a structure-specific manner during removal of primers or damaged DNA, so its activity is likely to be coordinated with the polymerizing activity of the enzyme. We asked whether the specificity and cleavage activities of the 5′ nuclease of the eubacterial enzyme TaqNP is affected by separation of the polymerase and nuclease activities.
Functional Domains of TaqNP and the Cleavage Substrate.
An N-terminal fragment of TaqNP containing the first 293 aa of TaqNP had 5′ nuclease activity; however, fragments with only the first 249 or 207 aa did not (data not shown). Conversely, a fragment containing only the carboxyl two thirds of TaqNP is capable of polymerization but not cleavage (30). Moreover, mutants of TaqNP with amino acid changes in the C-terminal domain (E465G and Q754R, as predicted from the gene sequence) had nuclease activity but lacked significant polymerase activity; in E. coli DNA polymerase I, Q849, which is comparable to Q754, is important in making contact with the DNA substrate (31). Thus, like E. coli DNA polymerase I, the 5′ nuclease domain of TaqNP is in the N-terminal third of the protein (32). It is likely that the nuclease and polymerase domains are joined in a hinge region at amino acids 290–293 (17).
Individual features of cleavage substrates are structurally similar to regions that function in DNA synthesis (13), and we refer to them by an analogous nomenclature. The template strand is paired with two other strands, the substrate and the primer strands, and aligns them relative to each other, as illustrated in Fig. 1A. For convenience and duplex stability, in many of the experiments described here, the substrate and template strands were linked in a hairpin structure such as that shown in Fig. 1.
Figure 1.
Cleavage of bifurcated DNA helices by TaqNP and TaqN in the presence of various upstream primers. (A) Proposed secondary structure of the D16 substrate indicating the nomenclature used to describe particular regions. (B) Products of cleavage of substrates containing gaps of various lengths. Standard cleavage reactions contained 1 nM hairpin substrate labeled at the 5′ end with 32P and 10 nM primer incubated at 55°C for 30 min with 0.5 units of TaqNP (Promega) or 20 ng of TaqN in 10 μl of 20 mM Tris⋅HCl, pH 8/10 mM KCl/1 mM MgCl2/10 μg/ml tRNA. The products were separated by electrophoresis in 10% polyacrylamide gel. −E, no enzyme; −P, no primer. Locations of the 3′ ends of the cleavage products are indicated to the right. (C) Proposed secondary structures of the DNA substrates formed in the presence of primers that leave 0, 2, or 4 unpaired nucleotides in the template strand making gap 0, gap 2, and gap 4 substrates, respectively. The gap −1 substrate has an overlap of 1 nt at the 3′ end of the primer and the bifurcation of the substrate duplex. The gap2ΔA substrate has a deletion in the 5′ arm to alter its ability to form the alternative structures shown. Arrows indicate the major sites of cleavage by TaqN.
Effects of the Primer on Cleavage.
Previously, we reported that efficient cleavage by TaqNP at high ionic strength required a primer (13), and the same appears to be true for TaqN nuclease (Fig. 1B, lanes -P). In the absence of primer, cleavage by either enzyme, although inefficient, occurred between the last two base-paired nucleotides of the substrate duplex (ref.13 and data not shown).
Surprisingly, primers both increased the efficiency of cleavage by TaqN and influenced the site of cleavage; these effects were unexpected because TaqN lacks a polymerization domain, which would contribute a primer binding site. The structure with no gapped nucleotide (gap 0) was a relatively poor substrate for TaqN, whereas the structure with a gap of −1 (a single nucleotide of overlap between the 3′ end of the primer and the end of the substrate duplex) was cleaved very efficiently. The sites of cleavage, determined from mapping of the ends of the products, are indicated on the right side of Fig. 1B. The presence of 2′, 3′-dideoxy- or 2′-deoxynucleotides at the 3′ end of the primer did not affect the site of cleavage (data not shown).
The ability of TaqN to cleave the substrate with a gap of −1 specifically and efficiently led us to ask whether a single mechanism could account for all of the products generated from the various substrates. Structures with single nucleotide overlaps could be drawn for the other complexes that had gaps of various lengths, if small bulges in the substrate duplex were allowed (Fig. 1C). In all of these cases, the site of cleavage by TaqN nuclease was between the first two base-paired nucleotides of the redrawn structure. Fig. 1C shows how substrates with gaps of +2 and +4 could be folded to generate overlapping structures, and how cleavage in the 5′ arm of such structures could be described as occurring at the alternative −1 cleavage site.
If most or all cleavage by TaqN occurs at a site corresponding to the 3′ end of a gap −1 primer, mutations that would alter the alternative structures should affect the site of cleavage in a predictable way. We generated variants of the gapped substrates (called ΔA), in which an A residue next to the bifurcation junction was deleted from the 5′ arm; this deletion would destroy the ability of the oligonucleotides to form several of the bulged structures proposed in Fig. 1C but would allow formation of a set of alternative structures. Digestion of the set of ΔA gapped substrates produced a different set of products that could be explained by the new set of alternative structures (data not shown); for example, a possible structure using the gap +2 primer is shown at the bottom of Fig. 1C, in which the observed site of cleavage by TaqN is indicated.
Analysis of the products generated by digestion of the gapped substrates with TaqNP (Fig. 1B, Center) revealed a more complex pattern of products, which included those produced by TaqN. In contrast to TaqN, TaqNP cleaved the gap 0 substrate efficiently, at the bifurcation and 1 nt into the single stranded arm (Fig. 1B). Substrates with gaps of between 1 and 4 nt were also cleaved in the 5′ arm at sites 1 and 3 nt from the bifurcation. These other cleavage sites may result from additional alternative structures on the substrate, stabilized by interactions with the primer-binding site of the polymerase domain.
Lack of Base Pairing at the 3′ End of the Primer.
The increased cleavage efficiency when the substrate has a gap of −1 indicates that in the preferred substrate, the primer and 5′ arm overlap at the site of cleavage. Assuming that both nucleotides do not hybridize to the template strand simultaneously, one of the overlapping nucleotides must be unpaired. The inefficient cleavage by TaqN of a substrate with a completely paired primer (gap 0) indicates that an unpaired nucleotide at the 3′ end of the primer promotes cleavage. To test whether the 3′ end of the primer is unpaired in overlapping structures, we generated a series of gap −1 primers that were identical except for their 3′ bases, only one of which had the ability to pair with the nucleotide opposite it in the template. Structures with all four primers were cleaved at the same point, and with equal efficiency, either by TaqNP (Fig. 2) or by TaqN nuclease (data not shown), showing that the substrate strand, but not the 3′ end of the primer, is base-paired to the template at the site of cleavage. We propose that the 3′ end of the primer interacts with the nuclease domain to position it at the optimal point for cleavage, between the last two base pairs of the substrate and template strands.
Other Substrate Requirements.
The length of the duplex region affects cleavage by TaqNP (Fig. 3A). In the presence of primers, TaqNP efficiently cleaved a hairpin substrate with 9 (D9) but not 6 (D6) bp of template duplex. The hairpin end stabilized the short duplexes at the assay temperature (data not shown). In the presence of a gap 4 primer, a low amount of cleavage product was produced from D6, presumably as a result of extended duplex formation by using nucleotides in the two single-stranded arms (as illustrated in Figs. 1C and 3A). Similar results were obtained with the TaqN nuclease (data not shown).
Figure 3.
Structural requirements of substrates for cleavage by TaqN and TaqNP. (A) Effect of the substrate duplex length. Upper shows suggested secondary structures of representative substrates complexed with various primers, in which the substrate duplexes contain either 9 (D9) or 6 (D6) base pairs. Incubation of these substrates TaqNP was as in Fig. 2 (but for 4 min), and the products are shown Lower. Arrows in the structures indicate deduced cleavage sites. (B) Requirement for a 3′ arm of the substrate duplex. The top of the figure shows suggested secondary structures of substrates made from a 5′ 32P-labeled 206-nt fragment and oligonucleotides 30 or 42 nt long, which differ by the presence of a 12-nt self-complementary 3′ end. These substrates, formed by annealing of 2 nM 206-nt fragment and 100 nM 30-0 or 30-12 oligonucleotides, were incubated for 30 min at 55°C with either 50 ng of TaqNP or 300 ng of TaqN in 10 μl of 10 mM Tris⋅HCl, pH 8/100 mM KCl/1 mM MgCl2/10 μg/ml tRNA. −, no enzyme. Arrows in the structures indicate deduced cleavage sites. (C) Requirement for a 5′ end of the substrate strand. Circular ssM13 mp19 DNA (50 ng) was incubated with 2.5 ng of TaqNP or 1 ng of TaqN in 10 mM Tris⋅HCl, pH 8/50 mM KCl buffer in the presence of 1 mM MgCl2 (lanes 2 and 4) or 1 mM MnCl2 (lanes 1, 3, and 5) at 55°C for 4 hours. The products were separated by electrophoresis in 1% agarose gel in 45 mM Tris⋅borate, pH 8.3/1 mM EDTA buffer.−, no enzyme.
The need for a 3′ arm was studied using two substrates generated by combining a 5′-labeled substrate strand with partially complementary template strands, one of which lacked a 3′ arm and primer (Fig. 3B). TaqN nuclease cleaved both DNAs efficiently, releasing a fragment whose size indicated cleavage at the bifurcation of the substrate duplex. In contrast, TaqNP did not efficiently cleave the substrate lacking a 3′ extension; this low efficiency of cleavage probably results from binding of the polymerase domain of this enzyme to the end of the duplex, which resembles a template–primer complex.
To test whether the 5′ arm must be free, we incubated circular M13 mp19 DNA with TaqN or TaqNP in the presence of MnCl2 or MgCl2. In MnCl2 TaqN, but not TaqNP, converted the circular DNA into a linear form, but in MgCl2, neither enzyme cleaved the circular DNA (Fig. 3C). The major site of cleavage was mapped by primer extension to the bifurcation of a potential 13-bp hairpin formed in the lac operator region (position 6175 of M13 sequence) (data not shown). On linearization, M13 mp19 DNA was cleaved by both enzymes at this position, as has been reported by Tombline et al. (33). Cleavage of circular DNA by TaqN required an internal hairpin and did not occur in unstructured regions or at sites created by addition of oligonucleotides complementary to other regions of the DNA (data not shown). Therefore, a free 5′ end is required for almost all conditions, the only exception being TaqN-catalyzed cleavage at the base of a hairpin, in the presence of MnCl2.
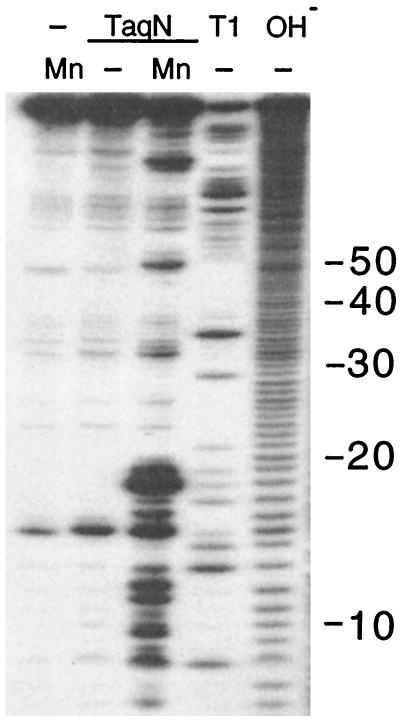
TaqNP can cleave RNA that is hybridized to a DNA template strand (13), but is unable to cleave duplexes composed entirely of RNA, in the presence of either MgCl2 or MnCl2. However, TaqN cleaved the structured U1 small nuclear RNA in a specific and reproducible way, in the presence of MnCl2 (Fig. 4). The sizes of the 5′ labeled fragments generated indicated that most cleavages were between the first 2 bp of duplexed regions of the proposed secondary structure of wild-type U1RNA (ref. 29 and data not shown), although additional cleavages occurred at sites that could reflect alternative structures of the RNA.
Figure 4.
Endonucleolytic cleavage of U1 snRNA by TaqN. A variant of U1 RNA (29) labeled in its 5′ end cap with 32P was incubated for 15 min at 55°C with 50 ng of TaqN in 10 μl of 10 mM Mes, pH 6/100 mM NaCl/50 mg/ml tRNA buffer/1 mM MnCl2 (lanes 1 and 3). Size markers were generated from the RNA by digestion with 0.1 unit of RNase T1 in 10 mM Tris⋅HCl, pH 8/1 mM EDTA buffer at 37°C for 10 min (T1, lane 4) or 25 mM Tris⋅base, pH 9.4/9.5 for 3 min (OH−, lane 5).
Effects of mAbs on Cleavage by TaqNP and TaqN.
We probed interactions between the polymerase and nuclease domains and the cleavage structure by using anti-TaqNP mAbs (22) that had previously been characterized with respect to their epitope specificities and their abilities to inhibit polymerization and primer–template binding (23). Examples of assays to test the effects on cleavage activities are shown in Fig. 5 and summarized in Table 1. Five of the antibodies (≈1/4) had no effect on nuclease activity, regardless of the domain to which they bound.
Figure 5.
Inhibition and stimulation of cleavage activity by mAbs generated against TaqNP. (Top) D16 substrate (5 nM) and gap 4 primer (0.1 μM) were incubated at 55°C for 15 min with 0.8 ng of TaqNP in 10 μl of 10 mM Tris⋅HCl, pH 7.8/50 mM KCl/1 mM MgCl2/0.2% Tween 20/100 μg/ml BSA in the presence of 150 ng of TaqNP-specific mAb. The products, separated from uncleaved substrate by PAGE, are shown. (Middle) The D16 substrate, gap 4 primers, and antibodies were incubated with 10 ng of TaqN nuclease as described above at 55°C for 40 min. (Bottom) The D6 substrate, gap 4 primers, and antibodies were incubated at 55°C for 15 min with 0.8 ng of TaqNP, as above. In all of these experiments, reaction samples containing all components except MgCl2 were preincubated at room temperature for 10 min, heated to 55°C, and reactions were started by addition of MgCl2. A total of 22 different mAbs were assayed and representative products of cleavage are shown; the results are summarized in Table 1.
Table 1.
Inhibition of TaqNP and TaqN nucleases by anti-TaqNP antibodies
| Antibody | Inhibition
|
Subunit | |
|---|---|---|---|
| DNA polymerization | DNA binding | ||
| No effect | |||
| 4-4 | − | − | N |
| 4-1 | − | ++ | N |
| 4-5 | − | − | P |
| 4-11 | − | − | N |
| 4-14 | − | − | P |
| Inhibit TaqNP | |||
| 4-9 | + | ++ | P |
| 4-3 | + | ± | P |
| TP-2 | + | ± | P |
| TP-7 | + | ++ | P |
| TP-9 | + | ± | P |
| Inhibit TaqNP and TaqN | |||
| TP-8 | + | − | N |
| 4-15 | − | ++ | N |
| 4-17 | − | ++ | N |
| Inhibit TaqN | |||
| 3-1 | + | ± | N |
| TP-5 | + | − | N |
| Stimulate TaqNP* | |||
| 4-8 | − | − | P |
| 4-13 | − | − | P |
| TP-3 | + | ± | P |
| TP-4 | + | − | P |
| TP-6 | ± | − | P |
| 3-5 | − | ± | P |
| 3-7 | − | − | P |
Five mAbs, which recognized epitopes in the polymerase domain (23), inhibited cleavage by TaqNP but not by TaqN. Among mAbs specific for epitopes in the nuclease domain, three inhibited the nuclease activities of both TaqNP and TaqN, whereas two mAbs, which have been reported to block polymerization activity of TaqNP, inhibited cleavage by TaqN but not TaqNP. It is unclear how these latter two mAbs can inhibit cleavage by TaqN but only polymerization by TaqNP.
Unexpectedly, seven of the mAbs stimulated cleavage by TaqNP of D6, the substrate that has a duplex only 6 bp long. Such a substrate is normally cleaved very inefficiently (Fig. 3A), but addition of these seven mAbs stimulated cleavage significantly. Perhaps these antibodies, all of which recognize epitopes in the polymerase domain, prevent binding of this domain to the 3′ end of the primer, thereby allowing formation of an extended substrate duplex, such as is shown in Fig. 3A. This extended substrate duplex would have the requisite 10 bp needed for binding to the nuclease, and the size of the cleavage product is consistent with this structure. The small amount of product seen in Fig. 3A, which is not apparent in Fig. 5, could result from the longer incubation and greater amount of enzyme use in the first experiment.
DISCUSSION
The experiments described here demonstrate that the nuclease domain of TaqNP retains many characteristics but also is changed in several significant ways, when separated from the polymerase domain. In both cases, TaqN is strongly influenced by the structure of the substrate DNA, cleaving the substrate at the end of a bifurcated duplex and releasing a free 5′ arm; generally, the site of cleavage is 1 nt into the duplex. As we observed previously for TaqNP, the efficiency of cleavage by the isolated nuclease, TaqN, is increased significantly by a strand of DNA (the primer) that is base-paired to the 3′ arm of the bifurcation.
Here we mapped the site of cleavage by TaqN to the phosphodiester bond between the first 2 bp of the bifurcated duplex. This site is adjacent to the 3′ end of the primer, which need not be base paired with the template strand of the bifurcated duplex (Fig. 2). We propose that the preferred cleavage substrate is composed of the last 2 bp of a duplex and the 3′ end of an overlapping primer.
When the primer is too short to overlap with the substrate duplex, we propose that the nuclease can stabilize alternative structures containing bulged nucleotides, which allow for extension of the duplex up to the site of primer overlap. Although we have no direct evidence for the existence of the structures illustrated in Fig. 1C, the change in cleavage pattern on deletion of on A residue in the substrate arm argues in favor of such structures. Similar alternative structures could be stabilized by the polymerase domain, accounting for the ability of TaqNP to cleave at additional sites; for example, bulging of one C in the center of the substrate strand would allow for cleavage of the gap 0 substrate by TaqNP. Other structures that could account for additional sites for cleavage by TaqNP seem likely because a direct correspondence between gap size and position of cleavage in the 5′ arm is lacking.
When the hairpin substrate duplex was 6 bp (D6), cleavage was very inefficient, setting a lower limit for the length of duplex that could be recognized. However, cleavage of this substrate was observed when a gap downstream of the primer supported formation of an extended substrate duplex (as illustrated in Fig. 3A); in this case, cleavage was in the 5′ arm, reflecting the preferences for cleavage site choice described above. Thus, the apparent ability of TaqN and TaqNP to stabilize structures that promote cleavage indicates that care must be exercised in making predictions about the resistance of particular structures to cleavage by these enzymes and could account for apparently contradictory results on the role of primers in fixing the site of cleavage.
It was somewhat surprising that TaqN was able to cleave a circular DNA molecule in the presence of MnCl2. We and others have proposed that a free end of the 5′ arm must feed through a hole or channel in the nuclease of TaqNP and FEN-1 (13, 14), and such an arch is near the active site in bacteriophage T5 5′ exo/endonuclease (18). We propose that in the presence of MnCl2, the circular DNA may access the active site of TaqN by feeding a hairpin through a comparable arch toward the active site, in contrast to threading the 5′ arm through the arch, away from the active site. The Mn2+ may allow for passage of the duplex through an arch that normally accommodates only a single strand of DNA. T5 exonuclease can cleave single stranded circular M13 DNA even in the presence of Mg2+ (34) and can digest double-stranded circluar plasmid DNA in the presence of Mn2+ (S. J. Garforth and J. R. Sayers, personal communication).
TaqN, TaqNP, and probably other 5′ nucleases, use at least two points of reference in orienting themselves on the substrate for optimal cleavage; these are the bifurcation of the duplex and the 3′ end of the primer. Two features of the end of the primer were unexpected; its overlap with the end of the substrate duplex and the apparent lack of base pairing with the template. During DNA synthesis, the newly incorporated 3′ nucleotide is paired with the template, when the structure is in the polymerase domain of TaqNP. Recent crystal structures of various forms of TaqNP (17, 35) indicate the possibility of conformational changes and reorientation of the two domains relative to each other, on binding substrates. These movements could facilitate displacement of the end of the primer strand by the nucleotide at the 5′ end of the bifurcated substrate duplex. Cleavage of the 5′ arm occurs immediately opposite the unpaired nucleotide at the 3′ end of the primer. Thus, this 3′ nucleotide, through interactions with the nuclease, serves as a point of reference for placement of the scissile bond in the active site of the enzyme. The resulting cleavage creates a nick rather than a gap in the cleaved strand (36, 37).
Our results show that the nuclease of TaqNP, although able to function independently of the polymerase domain, is influenced by it. Under some conditions, the polymerase domain increases the range of substrates that can be cleaved, such as when alternative structures must be stabilized, while in others, such as in cleavage of RNA, circular DNA, or a substrate lacking a 3′ arm, the polymerase domain interferes with cleavage. Although some of these effects can be explained, in many cases the explanation requires a better understanding of the interactions of the nuclease and its substrates, as could be obtained from structures of TaqNP and TaqN cocrystalized with DNA substrates in the active sites of their nuclease domains.
Acknowledgments
We thank D. Scalice and J. Daiss for TaqNP-specific mAbs, E. Lund and J. R. Sayers for valuble comments on the manuscript, and G. Q. Pennabble for comments. This work was supported by National Institutes of Health Grant GM30230 to J.E.D. and Biotechnology Training Grant GM08349 to V.E.V.
ABBREVIATIONS
- TaqNP
T. aquaticus DNA polymerase I
- TdT
terminal deoxynucleotidyltransferase
References
- 1.Lindahl T, Gally J A, Edelman G M. Proc Natl Acad Sci USA. 1969;62:597–603. doi: 10.1073/pnas.62.2.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deutscher M P, Kornberg A. J Biol Chem. 1969;244:3029–3037. [PubMed] [Google Scholar]
- 3.Lundquist R C, Olivera B M. Cell. 1982;31:53–60. doi: 10.1016/0092-8674(82)90404-4. [DOI] [PubMed] [Google Scholar]
- 4.Hollingsworth H C, Nossal N G. J Biol Chem. 1991;266:1888–1897. [PubMed] [Google Scholar]
- 5.Siegal G, Turchi J J, Myers T W, Bambara R A. Proc Natl Acad Sci USA. 1992;89:9377–9381. doi: 10.1073/pnas.89.20.9377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carr A M, Sheldrick K S, Murray J M, al-Harithy R, Watts F Z, Lehmann A R. Nucleic Acids Res. 1993;21:1345–1349. doi: 10.1093/nar/21.6.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.MacInnes M A, Dickson J A, Hernandez R R, Learmonth D, Lin G Y, Mudgett J S, Park M S, Schauer S, Reynolds R J, Strniste G F, et al. Mol Cell Biol. 1993;13:6393–6402. doi: 10.1128/mcb.13.10.6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Habraken Y, Sung P, Prakash L, Prakash S. Nature (London) 1993;366:365–368. doi: 10.1038/366365a0. [DOI] [PubMed] [Google Scholar]
- 9.Scherly D, Nouspikel T, Corlet J, Ucla C, Bairoch A, Clarkson S G. Nature (London) 1993;363:182–185. doi: 10.1038/363182a0. [DOI] [PubMed] [Google Scholar]
- 10.Harrington J J, Lieber M R. EMBO J. 1994;13:1235–1246. doi: 10.1002/j.1460-2075.1994.tb06373.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murray J M, Tavassoli M, al-Harithy R, Sheldrick K S, Lehmann A R, Carr A M, Watts F Z. Mol Cell Biol. 1994;14:4878–4888. doi: 10.1128/mcb.14.7.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O’Donovan A, Davies A A, Moggs J G, West S C, Wood R D. Nature (London) 1994;371:432–435. doi: 10.1038/371432a0. [DOI] [PubMed] [Google Scholar]
- 13.Lyamichev V, Brow M A, Dahlberg J E. Science. 1993;260:778–783. doi: 10.1126/science.7683443. [DOI] [PubMed] [Google Scholar]
- 14.Murante R S, Rust L, Bambara R A. J Biol Chem. 1995;270:30377–30783. doi: 10.1074/jbc.270.51.30377. [DOI] [PubMed] [Google Scholar]
- 15.Ceska T A, Sayers J R. Trends Biochem Sci. 1998;23:331–336. doi: 10.1016/s0968-0004(98)01259-6. [DOI] [PubMed] [Google Scholar]
- 16.Lieber M R. BioEssays. 1997;19:233–240. doi: 10.1002/bies.950190309. [DOI] [PubMed] [Google Scholar]
- 17.Kim Y, Eom S H, Wang J, Lee D S, Suh S W, Steitz T A. Nature (London) 1995;376:612–616. doi: 10.1038/376612a0. [DOI] [PubMed] [Google Scholar]
- 18.Ceska T A, Sayers J R, Stier G, Suck D. Nature (London) 1996;382:90–93. doi: 10.1038/382090a0. [DOI] [PubMed] [Google Scholar]
- 19.Mueser T C, Nossal N G, Hyde C C. Cell. 1996;85:1101–1112. doi: 10.1016/s0092-8674(00)81310-0. [DOI] [PubMed] [Google Scholar]
- 20.Hwang K Y, Baek K, Kim H Y, Cho Y. Nat Struct Biol. 1998;5:707–713. doi: 10.1038/1406. [DOI] [PubMed] [Google Scholar]
- 21.Hosfield D J, Mol C D, Shen B, Tainer J A. Cell. 1998;95:135–146. doi: 10.1016/s0092-8674(00)81789-4. [DOI] [PubMed] [Google Scholar]
- 22.Scalice E R, Sharkey D J, Daiss J L. J Immunol Methods. 1994;172:147–163. doi: 10.1016/0022-1759(94)90102-3. [DOI] [PubMed] [Google Scholar]
- 23.Daiss J L, Scalice E R, Sharkey D J. J Immunol Methods. 1995;183:15–26. doi: 10.1016/0022-1759(95)00019-7. [DOI] [PubMed] [Google Scholar]
- 24.Engelke D R, Krikos A, Bruck M E, Ginsburg D. Anal Biochem. 1990;191:396–400. doi: 10.1016/0003-2697(90)90238-5. [DOI] [PubMed] [Google Scholar]
- 25.Stark M J. Gene. 1987;51:255–267. doi: 10.1016/0378-1119(87)90314-3. [DOI] [PubMed] [Google Scholar]
- 26.Studier F W, Rosenberg A H, Dunn J J, Dubendorff J W. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 27.Gill S C, von Hippel P H. Anal Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 28.Grimm C, Lund E, Dahlberg J E. EMBO J. 1997;16:793–806. doi: 10.1093/emboj/16.4.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Branlant C, Krol A, Ebel J P, Gallinaro H, Lazar E, Jacob M. Nucleic Acids Res. 1981;9:841–858. doi: 10.1093/nar/9.4.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lawyer F C, Stoffel S, Saiki R K, Myambo K, Drummond R, Gelfand D H. J Biol Chem. 1989;264:6427–6437. [PubMed] [Google Scholar]
- 31.Polesky A H, Steitz T A, Grindley N D, Joyce C M. J Biol Chem. 1990;265:14579–14591. [PubMed] [Google Scholar]
- 32.Longley M J, Bennett S E, Mosbaugh D W. Nucleic Acids Res. 1990;18:7317–7322. doi: 10.1093/nar/18.24.7317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tombline G, Bellizzi D, Sgaramella V. Proc Natl Acad Sci USA. 1996;93:2724–2728. doi: 10.1073/pnas.93.7.2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sayers J R, Eckstein F. Nucleic Acids Res. 1991;19:4127–4132. doi: 10.1093/nar/19.15.4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murali R, Sharkey D J, Daiss J L, Murthy H M. Proc Natl Acad Sci USA. 1998;95:12562–12567. doi: 10.1073/pnas.95.21.12562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaiser, M. W., Lyamicheva, N., Ma, W., Miller, C., Neri, B. & Lyamichev, V. I. (1999) J. Biol. Chem., in press. [DOI] [PubMed]
- 37.Lyamichev V, Mast A L, Hall J G, Prudent J R, Kaiser M W, Takova T, Kwiatkowski R, Sander T, de Arruda M, Arco D, et al. Nat Biotech. 1999;17:292–296. doi: 10.1038/7044. [DOI] [PubMed] [Google Scholar]