Abstract
Bacillus anthracis causes anthrax disease and exerts its deleterious effects by the release of three exotoxins: lethal factor, protective antigen, and edema factor (EF), a highly active calmodulin-dependent adenylyl cyclase (AC). However, conventional antibiotic treatment is ineffective against either toxemia or antibiotic-resistant strains. Thus, more effective drugs for anthrax treatment are needed. Previous studies from our laboratory showed that mammalian membranous AC (mAC) exhibits broad specificity for purine and pyrimidine nucleotides (Mol Pharmacol 70 878-886,2006). Here, we investigated structural requirements for EF inhibition by natural purine and pyrimidine nucleotides and nucleotides modified with N-methylanthraniloyl (MANT)- or anthraniloyl groups at the 2′(3′)-O-ribosyl position. MANT-CTP was the most potent EF inhibitor (Ki, 100 nM) among 16 compounds studied. MANT-nucleotides inhibited EF competitively. Activation of EF by calmodulin resulted in effective fluorescence resonance energy transfer (FRET) from tryptophan and tyrosine residues located in the vicinity of the catalytic site to MANT-ATP, but FRET to MANT-CTP was only small. Mutagenesis studies revealed that Phe586 is crucial for FRET to MANT-ATP and MANT-CTP and that the mutations N583Q, K353A, and K353R differentially alter the inhibitory potencies of MANT-ATP and MANT-CTP. Docking approaches relying on crystal structures of EF indicate similar binding modes of the MANT nucleotides with subtle differences in the region of the nucleobases. In conclusion, like mAC, EF accommodates both purine and pyrimidine nucleotides. The unique preference of EF for the base cytosine offers an excellent starting point for the development of potent and selective EF inhibitors.
The spore-forming bacterium Bacillus anthracis exerts its deleterious effects by production of three major exotoxins: EF, protective antigen, and lethal factor (Jedrzejas, 2002). EF and lethal factor enter host cells via a complex with membrane-associated protective antigen, which acts as a pH-dependent protein transporter. Lethal factor, a specific zinc-metalloprotease, inactivates mitogen-activated protein kinase (Hong et al., 2005). EF possesses ∼800 amino acid residues and an apparent molecular mass of ∼89 kDa and is a CaM-dependent AC (Drum et al., 2002). After entering host cells, EF forms a complex with CaM, a mammalian regulatory protein that mediates many aspects of calcium-regulated signaling (Shen et al., 2002). The binding of CaM induces a major conformational change in the catalytic domain of EF (Drum et al., 2002). This rearrangement renders EF highly efficient at catalyzing the conversion of ATP into cAMP, disrupting intracellular signaling pathways through excessive activation of cAMP-dependent signaling pathways (Shen et al., 2005).
We resolved several crystal structures of nucleotide-EF-CaM complexes and characterized the amino acids that are important for binding of the substrate ATP and catalysis (Drum et al., 2002; Shen et al., 2002, 2005). In addition, we showed that mAC and bacterial AC toxins are potently inhibited by MANT-substituted nucleoside 5′-triphosphates (Gille et al., 2004; Mou et al., 2005, 2006; Göttle et al., 2007). MANT-nucleotides are fluorescent, and we exploited this property to suggest conformational changes associated with activation in purified catalytic subunits of mAC (Mou et al., 2005, 2006) and the Bordetella pertussis AC toxin CyaA (Göttle et al., 2007). In addition, by combining crystallographic and molecular modeling approaches, we developed a three-site pharmacophore model for mAC and CyaA, with binding domains for the base, the MANT group, and the polyphosphate chain (Gille et al., 2005; Mou et al., 2006; Göttle et al., 2007; Wang et al., 2007). Those studies revealed that the catalytic sites of mAC and CyaA exhibit substantial conformational flexibility, accommodating both purine and pyrimidine nucleotides. Despite this flexibility, the structure/activity relationships of MANT-nucleotides at mAC and CyaA are quite different, offering the opportunity to design potent and isoform-selective AC inhibitors.
In contrast to mAC (Gille et al., 2004, 2005; Mou et al., 2005, 2006) and CyaA (Göttle et al., 2007), a detailed analysis of MANT-nucleotide/EF interactions has not yet been presented. Therefore, in the present study, we systematically examined the interactions of natural purine and pyrimidine nucleotides and several (M)ANT-substituted analogs with EF in terms of catalysis, fluorescence changes and molecular modeling.
Materials and Methods
Materials. MANT-GTP, ANT-GTP, MANT-ATP, and MANT-ADP were purchased from Jena Bioscience, Germany. PMEApp was supplied by Gilead Sciences (Foster City, CA). GTP, UTP, and CTP were purchased from Roche (Mannheim, Germany). ITP, ampicillin, kanamycin, lysozyme enzyme, β-mercaptoethanol, MES buffer (low moisture content), and dithiothreitol (for molecular biology) were purchased from Sigma-Aldrich (Steinheim, Germany). Tryptone and yeast were purchased from BD Biosciences (Franklin Lakes, NJ). [α-32P]ATP (800 Ci/mmol) was purchased from PerkinElmer Life and Analytical Sciences (Rodgau Jügesheim, Germany). Aluminum oxide 90 active (neutral, activity 1; particle size, 0.06-0.2 mm) was purchased from Biomedicals (Eschwege, Germany). Bovine serum albumin (fraction V, highest quality) was bought from Sigma-Aldrich. Imidazole (highest quality), CaCl2, MnCl2 tetrahydrate. and MgCl2 hexahydrate (highest quality) were purchased from Merck Biosciences (Darmstadt, Germany). For all experiments double-distilled water was used.
(M)ANT-Nucleotide Synthesis: General Procedure. MANT-CTP, MANT-CDP, MANT-UTP, MANT-UDP, MANT-ITP, MANT-IDP, ANT-ATP, and ANT-ADP were synthesized according to Hiratsuka (1983) with modifications. The nucleotide (0.33 mmol, 1 eq) was propounded in a small two-neck round flask and dissolved in a minimum amount of water (3 ml). Under continuous stirring, a crystalline preparation of (methyl)isatoic anhydride (0.5 mmol, 1.5 eq) was added. After heating to 38°C, the pH value was adjusted to 8.6 and maintained by titration of 1 N NaOH solution for 2 h. The reaction mixture was extracted three times with 20 ml of chloroform (only for MANT-nucleotides, not for ANT-nucleotides). The aqueous phase was dry-frozen. The received foam showed white to brown color and was applied to a Sephadex LH-20 column (85 × 2 cm) and subsequently eluted with double-distilled water. The desired product could be detected directly by its blue fluorescence in the collection tubes at λex of 366 nm and by thin-layer chromatography. For further purification, reversed-phase preparative HPLC was used to separate (M)ANT-NTP from (M)ANT-NDP. After final dry-freezing, white solid compounds (purity >99%) were obtained.
HPLC Analysis of (M)ANT-Nucleotides. The samples were filtered using a PTFE filter (Chromafil, O-20/15, organic, pore size 0.2 mm; Machery-Nagel, Düren, Germany). A 10-μl sample was analyzed using an HPLC (model 1100; Agilent Technologies, Waldbronn, Germany) fitted with a C18 analytical column (particle size, 3 μm; 150 × 4.60 mm; Luna; Phenomenex, Aschaffenburg, Germany) and diode array detection. Data were analyzed using an HPLC-3D ChemStation (Rev. A.10.01 [1635]). Gradient elution was performed with 0.05 M ammonium acetate (solvent A) and acetonitrile (solvent B) at a constant flow rate of 1.0 ml/min. A gradient profile with the following proportions of solvent B was applied: [0 min, 5% B], [10 min, 5% B], [30 min, 45% B], and [40 min, 80% B]. The chromatograms were monitored at UV absorption at 220 and 254 nm. In addition, a fluorescence detector was used for the analysis of the fluorescent anthraniloylic compounds at λex of 350 nm and λem of 450 nm.
LC/MS Online Coupling. All samples were filtered using a PTFE filter and injected into an HPLC (model 1100; Agilent Technologies). The compound to be analyzed was separated by a C18 column (particle size, 3 μm; 150 × 2 mm; Luna; Phenomenex). A binary eluent mixture consisting of water (10 mM ammonium acetate) (eluent A) and acetonitrile (eluent B) was pumped with a constant flow of 0.3 ml/min. The following gradient profile was used: [0 min, 5% B], [10 min, 5% B], [30 min, 45% B], and [40 min, 80% B]. The injected volume was 3 μl. The mass of the respective compound was determined with the use of a triple-stage mass spectrometer (Finnigan TSQ 7000; Thermo Fisher Scientific, Waltham, MA).
Preparative HPLC. Compound mixtures were dissolved in water (concentration, 30-50 mg/ml) and filtered using a PTFE filter. Compounds were separated by an HPLC (model 1100; Agilent Technologies) fitted with a C18 preparative column (particle size, 10 μm; 250 × 21.2 mm; Luna; Phenomenex). Gradient elution was performed with 0.05 M ammonium acetate (solvent A) and acetonitrile (solvent B) at a constant flow rate of 21 ml/min. The chromatograms were monitored by UV absorption at 220 and 254 nm.
MANT-UTP (N-Methyl-2′(3′)-O-anthraniloyl-uridine-5′-triphosphate) or [(2R,3S,4R,5R)-5-(2,4-Dioxopyrimidin-1-yl)-4(3)-hydroxy-2-[[hydroxy-(hydroxy-phosphonooxyphosphoryl) oxyphosphoryl]oxymethyl]oxolan-3(4)-yl]2-methylaminobenzoate. One hundred eighty-two milligrams of introduced disodium salt of UTP yielded over all purification steps 32 mg (52 μmol, 15%) of pure product. Rf = 0.21 (1-propanol:H2O:NH3 (32%) = 2:1:1). HPLC (analytic): Rt = 18.78 min, 19.15 min; k = 12.49, 12.75; LC/MS (ESI, H2O/CH3CN): m/z = 635.2 [M+NH4+] (Rt = 19.69 min, 19.88 min, 100%), 652.2 [M-H + 2NH4+] (Rt = 20.15 min, 100%); (-ESI, H2O/CH3CN): m/z = 616.2 [M-H-] (Rt: 19.53 min, 20.01 min, 100%); HPLC (preparative), gradient (t [min], % B: [0, 14], [20, 14], [30, 80]): Rt = 6.07 min, 6.79 min; empirical formula: C17H22N3O16P3; MW = 617.29.
MANT-UDP (N-Methyl-2′(3′)-O-anthraniloyl-uridine-5′-diphosphate) or [(2R,3S,4R,5R)-5-(2,4-dioxopyrimidin-1-yl)-4(3)-hydroxy-2-[(hydroxy-phosphonooxyphosphoryl)oxymethyl] oxolan-3(4)-yl]2-methylaminobenzoate. One hundred eighty-two milligrams of introduced disodium salt of UDP yielded over all purification steps 8 mg (15 μmol, 3.8%) of pure product. Rf = 0.25 (1-propanol:H2O:NH3 (32%) = 2:1:1). HPLC (analytic): Rt = 19.75 min, 19.93 min; k = 13.19, 13.32; LC/MS (ESI, H2O/CH3CN): m/z = 555.2 [M+NH4+] (Rt = 20.64 min, 20.94 min, 100%), 572.2 [M-H + 2NH4+] (Rt = 20.64 min, 20.94 min, 40%); (-ESI, H2O/CH3CN): m/z = 536.2 [M-H-] (Rt: 20.50 min, 20.78 min, 100%); HPLC (preparative) gradient: [0 min, 14% B], [20 min, 14% B], [30 min, 80% B]. Rt = 8.79 min, 9.49 min; empirical formula: C17H21N3O13P2; MW = 537.31.
MANT-CTP (N-Methyl-2′(3′)-O-anthraniloyl-cytosine-5′-triphosphate) or [(2R,3S,4R,5R)-5-(4-Amino-2-oxopyrimidin-1-yl)-4(3)-hydroxy-2-[[hydroxy-(hydroxy-phosphonooxyphosphoryl) oxyphosphoryl]oxymethyl]oxolan-3(4)-yl]2-methylaminobenzoate. Two hundred milligrams of introduced trisodium salt of CTP yielded over all purification steps 30 mg (48 μmol, 14%) of pure product. Rf = 0.24 (1-propanol:H2O:NH3 (32%) = 2:1:1). HPLC (analytic): Rt = 16.86 min, 17.60 min; k = 11.17, 11.70; LC/MS (ESI, H2O/CH3CN): m/z = 634.2 [M+NH4+] (Rt = 12.98 min, 16.98 min, 100%); (-ESI, H2O/CH3CN): m/z = 615.2[M-H-] (Rt: 13.75 min, 17.24 min, 100%); HPLC (preparative) gradient: [0 min, 14% B], [20 min, 14% B], [30 min, 80% B]. Rt = 4.08 min, 4.61 min; empirical formula: C17H23N4O15P3; MW = 616.30.
MANT-CDP (N-Methyl-2′(3′)-O-anthraniloyl-cytosine-5′-diphosphate) or [(2R,3S,4R,5R)-5-(4-Amino-2-oxopyrimidin-1-yl)-4(3)-hydroxy-2-[(hydroxy-phosphonooxyphosphoryl)oxymethyl] oxolan-3(4)-yl]2-methylaminobenzoate. Two hundred milligrams of introduced disodium salt of CDP yielded over all purification steps 2 mg (3.7 μmol, 1%) of pure product. Rf = 0.28 (1-propanol: H2O:NH3 (32%) = 2:1:1). HPLC (analytic): Rt = 18.38 min; k = 12.27; LC/MS (ESI, H2O/CH3CN): m/z = 554.2 [M+NH4+](Rt = 18.55 min, 100%); (-ESI, H2O/CH3CN): m/z = 535.2 [M-H-] (Rt: 18.66 min, 100%); HPLC (preparative) gradient: [0 min, 14% B], [20 min, 14% B], [30 min, 80% B]. Rt = 5.80 min; empirical formula: C17H22N4O12P2; MW = 536.32.
MANT-ITP (N-Methyl-2′(3′)-O-anthraniloyl-inosine-5′-triphosphate) or [(2R,3S,4R,5R)-5-(6-Oxo-1H-purin-9-yl)-4(3)-hydroxy-2-[[hydroxy-(hydroxy-phosphonooxyphosphoryl)oxyphosphoryl]oxymethyl]oxolan-3(4)-yl]2-methylaminobenzoate. One hundred eighty-nine milligrams of introduced trisodium salt of ITP yielded overall purification steps 39 mg (61 μmol, 18%) of pure product. Rf = 0.31 (1-propanol:H2O:NH3 (32%) = 2:1:1). HPLC (analytic): Rt = 19.74 min, 19.86 min; k = 12.65, 12.73; LC/MS (ESI, H2O/CH3CN): m/z = 676.2 [M-H + 2NH4+](Rt = 20.92 min, 100%), 659.2 [M+NH4+] (Rt = 20.92 min, 80%); (-ESI, H2O/CH3CN): m/z = 640.2 [M-H-] (Rt: 20.92 min, 100%); HPLC (preparative) gradient: [0 min, 14% B], [20 min, 14% B], [30 min, 80% B]. Rt = 8.03 min, 8.23 min; empirical formula: C18H22N5O15P3; MW = 641.31.
MANT-IDP (N-Methyl-2′(3′)-O-anthraniloyl-inosine-5′-diphosphate) or [(2R,3S,4R,5R)-5-(6-Oxo-1H-purin-9-yl)-4(3)-hydroxy-2-[(hydroxy-phosphonooxyphosphoryl)oxymethyl]oxolan-3(4)-yl] 2-methylaminobenzoate. One hundred eighty-nine milligrams of introduced disodium salt of IDP yielded over all purification steps 15 mg (27 μmol, 7%) of pure product. Rf = 0.35 (1-propanol:H2O:NH3 (32%) = 2:1:1). HPLC (analytic): Rt = 20.34 min, 20.58 min; k = 13.07, 13.23; LC/MS (ESI, H2O/CH3CN): m/z = 596.3 [M-H + 2NH4+] (Rt = 21.38 min, 100%), 579.3 [M+NH4+] (Rt = 21.38 min, 70%); (-ESI, H2O/CH3CN): m/z = 560.2[M-H-] (Rt: 21.39 min, 21.55 min, 100%); HPLC (preparative) gradient: [0 min, 14% B], [20 min, 14% B], [30 min, 80% B]. Rt = 10.62 min, 11.36 min; empirical formula: C18H21N5O12P2; MW = 561.33.
ANT-ATP (2′(3′)-O-Anthraniloyl-adenosine-5′-triphosphate) or [(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-4(3)-hydroxy-2-[[hydroxy-(hydroxy-phosphonooxyphosphoryl)oxyphosphoryl] oxymethyl]oxolan-3(4)-yl]2-aminobenzoate. One hundred eighty-nine milligrams of introduced disodium salt of ATP yielded over all purification steps 59 mg (94 μmol, 26%) of pure product. Rf = 0.24 (1-propanol:H2O:NH3 (32%) = 2:1:1). HPLC (analytic): Rt = 17.44 min; k = 10.07; LC/MS (ESI, H2O/CH3CN): m/z = 661.3 [M-H + 2NH4+] (Rt = 18.12 min, 18.32 min, 100%), 644.2 [M+NH4+] (Rt = 18.12 min, 18.32 min, 80%); (-ESI, H2O/CH3CN): m/z = 625.2 [M-H-] (Rt = 18.12 min, 18.33 min, 100%); HPLC (preparative) gradient: [0 min, 11% B], [9 min, 11% B], [19 min, 80% B]. Rt = 7.07 min, 7.51 min; empirical formula: C17H21N6O14P3; MW = 626.30.
ANT-ADP (2′(3′)-O-Anthraniloyl-adenosine-5′-diphosphate) or [(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-4(3)-hydroxy-2-[(hydroxy-phosphonooxyphosphoryl)oxymethyl]oxolan-3(4)-yl]2-aminobenzoate. Two hundred milligrams of introduced disodium salt of ADP yielded over all purification steps 8 mg (15 μmol, 4%) of pure product. Rf = 0.27 (1-propanol:H2O:NH3 (32%) = 2:1:1). HPLC (analytic): Rt = 18.09 min; k = 10.73; LC/MS (ESI, H2O/CH3CN): m/z = 564.3 [M+NH4+](Rt = 18.81 min, 19.04 min, 100%); (-ESI, H2O/CH3CN): m/z = 545.2 [M-H-] (Rt = 18.81 min, 19.04 min, 100%); HPLC (preparative) gradient: [0 min, 11% B], [9 min, 11% B], [19 min, 80% B]. Rt = 8.99 min; empirical formula: C17H20N6O11P2; MW = 546.32.
Expression and Purification of EF and EF3(F586A). The plasmids pProExH6-EF and pProExH6-EF3F586A were prepared as described and amplified in Escherichia coli BL21 (DE3)/pUBS520 cells (Drum et al., 2002; Shen et al., 2002, 2005; Guo et al., 2004). The EF3 mutants (H577A, N583A, N583Q, N583H, K353A, and K353R) were purified as described previously (Drum et al., 2002; Shen et al., 2002, 2005; Guo et al., 2004).
Expression and purification of EF was essentially performed as described previously (Shen et al., 2002) with minor modifications. Specifically, the imidazole concentration in the elution buffer for the HisTrap fast-flow-rate Ni2+ column was increased to 200 mM. Moreover, imidazole (20 mM) was added into the sample, thus yielding the same imidazole concentration as in the second wash buffer of the Ni2+ column. The following columns were used for EF protein purification; HisTrap fast-flow-rate affinity Ni2+ column (5 ml) and resource Q (quaternary ammonium salt) strong anionic exchange column (6 ml) (GE Healthcare, Freiburg, Germany). EF3(F586A) mutant was expressed and purified essentially as described previously (Shen et al., 2002) with minor modifications. Specifically, the imidazole concentration in the elution buffer for the Ni2+ column was 200 mM, and imidazole (20 mM) was included in the sample. The HisTrap fast-flow-rate affinity Ni2+ column (5 ml) was used for immobilized matrix affinity chromatography and the HiPrep 16/10 SP XL column (GE Healthcare) was used in cation exchange chromatography. CaM was extracted and purified from bovine brain as described previously (Gopalakrishna and Anderson, 1982). The HiPrep 16/10phenyl FF (high sub) column (GE Healthcare) was used in hydrophobic chromatography purification of CaM.
AC Activity Assay. For the determination of the potency of AC toxin inhibitors, assay tubes contained 10 μl of MANT-nucleotides at final concentrations from 10 nM to 100 μM as appropriate to obtain saturated inhibition curves and 20 μl of EF, EF3, or EF3(F586A) (final concentration, 10 pM) in 75 mM Tris/HCl, pH 7.4, containing 0.1% (m/v) bovine serum albumin. Tubes were preincubated for 2 min at 25°C, and reactions were initiated by the addition of 20 μl of reaction mixture consisting of the following components to yield the given final concentrations; 100 mM KCl, 100 μM free Ca2+, 5 mM free Mn2+ or Mg2+, 100 μM EGTA, 100 μM cAMP, and 100 nM CaM. ATP was added as nonlabeled substrate at a final concentration of 40 μM and as radioactive tracer [α-32P]ATP (0.2 μCi/tube). For the determination of Km and Vmax values, 10 μM to 1 mM ATP/Mn2+ or ATP/Mg2+ were added, plus a 5 mM concentration of free Mn2+ or Mg2+, respectively. Tubes were incubated for 10 min at 25°C, and reactions were stopped by the addition of 20 μl of 2.2 N HCl. Denaturated protein was sedimented by a 1-min centrifugation at 13,000g. [32P]cAMP was separated from [α-32P]ATP by transferring the samples to columns containing 1.4 g of neutral alumina. [32P]cAMP was eluted by the addition of 4 ml of 0.1 M ammonium acetate solution, pH 7.0. Blank values were approximately 0.02% of the total amount of [α-32P]ATP added; substrate turnover was <3% of the total amount of [α-32P]ATP added. Samples collected in scintillation vials were filled with 10 ml of double-distilled water and Čerenkov radiation was measured in a liquid scintillation counter (Tricarb 2800TR; PerkinElmer Life and Analytical Sciences). Free concentrations of divalent cations were calculated with WinMaxC (http://www.stanford.edu/~cpatton/maxc.html). Vmax and Km values reported in Table 1 and Ki values reported in Tables 2 and 3 were calculated using the Prism 4.02 software (Graphpad, San Diego, CA).
TABLE 1.
Km and Vmax values of EF, EF3, and various EF3 mutants
AC toxin activities were determined as described under Materials and Methods. Km and Vmax values were obtained by nonlinear regression analysis of substrate-saturation experiments and are the means ± S.D. of at least three independent experiments performed in triplicates. For the determination of the Vmax and Km of EF, EF3, and EF3(F586A), reaction mixtures contained 100 μM free Ca2+, 5 mM free Mn2+, 100 μM EGTA, 0.2-1.0 μCi/tube [α-32P]ATP, 100 μM cAMP, 100 nM calmodulin, and 10 pM enzyme in 75 mM Tris/HCl, pH 7.4. In the case of the other EF3 mutants, reaction mixtures contained 0.4 μCi/tube [α-32P]ATP and 2 nM enzyme. For determination of Vmax and Km without CaM and Ca2+, the enzyme concentration was 5 nM. The unlabeled ATP/Mn2+ concentrations ranged from 100 nM to 4 mM as appropriate to obtain saturated curves.
| Toxin | Ca2+/CaM | Vmax | Km |
|---|---|---|---|
| s−1 | μM | ||
| EF | + | 232 ± 34 | 84.5 ± 8.3 |
| EF3 | + | 223 ± 12 | 82.6 ± 8.2 |
| EF3(F586A) | + | 128 ± 5.0 | 92.0 ± 14.7 |
| EF3(H577A) | + | 0.013 ± 0.003 | 34.5 ± 9.7 |
| EF3(N583A) | + | 15.4 ± 0.13 | 224 ± 7.3 |
| EF3(N583Q) | + | 18.4 ± 0.08 | 855 ± 39 |
| EF3(N583H) | + | 59.7 ± 10.4 | 3890 ± 1,220 |
| EF3(K353A) | + | 3.93 ± 0.02 | 804 ± 137 |
| EF3(K353R) | + | 32.3 ± 0.06 | 75.4 ± 1.35 |
| EF3 | − | 0.11 ± 0.007 | 60.0 ± 0.89 |
TABLE 2.
Inhibitory potencies of MANT-ATP and MANT-CTP at EF, EF3 and EF3 mutants in the presence of Mn2+
Inhibitory potencies of MANT-ATP and MANT-CTP at EF, EF3, and various EF3 mutants were determined as described under Materials and Methods. Ki values are given as micromolar values and are the means ± S.D. of three experiments performed in triplicate. The relative potencies (Rel. Pot.) of MANT-ATP and MANT-CTP are given, too, EF3 being the reference. For determination of the inhibitory potencies of MANT-ATP and MANT-CTP at EF, EF3, and EF3(F586A), reaction mixtures contained 100 μM free Ca2+, 5 mM free Mn2+, 100 μM EGTA, 40 μM ATP, 0.2 to 1.0 μCi/tube [α-32P]ATP, 100 μM cAMP, 100 nM CaM, and 10 pM enzyme in 75 mM Tris/HCl, pH 7.4. For other EF3 mutants, reaction mixtures contained 1.0 μCi of [α-32P]ATP per tube. The enzyme concentration was 2 nM. For the determination of Ki values of MANT-ATP and MANT-CTP without CaM and Ca2+, the ATP/Mn2+ concentration was 20 μM and the enzyme concentration was 5 nM. Nucleotides were added at different concentrations as appropriate to obtain saturated concentration-response curves. Inhibition curves were analyzed by nonlinear regression using the Prism 4.02 software.
| Toxin | Ca2+/CaM | MANT-ATP Ki | MANT-ATP Rel. Pot. | MANT-CTP Ki | MANT-CTP Rel. Pot. |
|---|---|---|---|---|---|
| μM | μM | ||||
| EF | + | 0.58 ± 0.13 | 169 | 0.11 ± 0.04 | 91 |
| EF3 | + | 0.98 ± 0.08 | 100 | 0.10 ± 0.008 | 100 |
| E3(F586A) | + | 4.54 ± 1.15 | 22 | 0.61 ± 0.11 | 16 |
| EF3(H577A) | + | 0.52 ± 0.05 | 189 | 0.11 ± 0.017 | 91 |
| EF3(N583A) | + | 91.7 ± 2.5 | 1.1 | 10.4 ± 0.02 | 1.0 |
| EF3(N583Q) | + | 67.4 ± 4.8 | 1.5 | 248 ± 23 | 0.04 |
| EF3(N583H) | + | 308 ± 89 | 0.3 | 22 ± 5.4 | 0.5 |
| EF3(K353A) | + | 17.4 ± 4.0 | 5.6 | 54.8 ± 0.83 | 0.2 |
| EF3(K353R) | + | 3.27 ± 0.08 | 30 | 6.53 ± 0.26 | 1.5 |
| EF3 | − | 1.07 ± 0.04 | 92 | 0.16 ± 0.02 | 63 |
TABLE 3.
Inhibitory potencies of NTPs and (M)ANT-nucleotides at EF in the presence of Mn2+ and Mg2+
Inhibitory potencies of various purine and pyrimidine nucleotides at EF were determined as described under Materials and Methods. Ki values are given in μM and are the means ± S.D. of three to four independent experiments performed in triplicate. The Km value of EF for ATP in the presence of Mg2+ was 120 ± 6.5 μM. Reaction mixtures contained 5 mM MnCl2 or 5 mM MgCl2, 100 mM KCl, 100 μM CaCl2, 40 μM ATP, [α-32P]ATP (0.2 μCi/tube), 100 μM cAMP, and 100 nM CaM. Nucleotides were added at different concentrations as appropriate to construct saturated concentration-response curves. Inhibition curves were analyzed by nonlinear regression using the Prism 4.02 software.
|
Nucleotide |
Ki | |
|---|---|---|
| Mn2+ | Mg2+ | |
| μM | ||
| GTP | 9.2 ± 0.79 | 73.6 ± 6.02 |
| ITP | 45.4 ± 3.60 | 233 ± 15.8 |
| UTP | 63.9 ± 9.21 | 138 ± 2.53 |
| CTP | 5.10 ± 0.38 | 45.2 ± 9.18 |
| MANT-ATP | 0.58 ± 0.09 | 1.36 ± 0.57 |
| MANT-ADP | 3.26 ± 0.50 | 34.7 ± 3.34 |
| ANT-ATP | 0.44 ± 0.09 | 5.15 ± 2.12 |
| ANT-ADP | 3.85 ± 1.25 | 57.9 ± 25.1 |
| MANT-GTP | 2.49 ± 0.08 | 4.70 ± 0.32 |
| ANT-GTP | 4.10 ± 1.09 | 13.0 ± 1.86 |
| MANT-ITP | 4.06 ± 0.06 | 10.6 ± 3.29 |
| MANT-IDP | 11.0 ± 1.89 | 48.5 ± 3.16 |
| MANT-UTP | 3.67 ± 0.08 | 32.2 ± 0.82 |
| MANT-UDP | 38.7 ± 0.48 | 107 ± 20.6 |
| MANT-CTP | 0.10 ± 0.01 | 1.26 ± 0.09 |
| MANT-CDP | 1.30 ± 0.12 | 13.8 ± 0.06 |
For determination of the potency of AC toxin inhibitors at various EF3 mutants (H577A, N583A, N583Q, N583H, K353A, and K353R), the experiments were essentially performed as described for EF and EF3 with some modifications. Specifically, the final enzyme concentrations were increased up to 2 nM to account for the lower catalytic activity of the mutants. Moreover, the reaction time was prolonged to 20 min at 30°C. For the determination of Vmax and Km, the ATP/Mn2+ concentration ranged from 100 μM to 4 mM. The higher substrate concentrations compared with EF were essential to obtain saturated enzyme kinetics.
For determination of the potency of AC toxin inhibitors at EF3 in absence of both CaM and Ca2+, experiments were performed as described for standard experiments with some modifications to increase assay sensitivity. Specifically, the final enzyme concentration was increased up to 5 nM, and the reaction was stopped after 30 min incubation at 30°C. Moreover, in competition experiments, the concentration of unlabeled ATP was decreased to 20 μM, and [α-32P]ATP was used at a high amount (1.0 μCi/tube). For the determination of Vmax and Km, the same modifications were performed, except that the ATP/Mn2+ concentration was varied.
For studying of the inhibition mechanism of the EF3 by MANT-nucleotides, enzyme saturation experiments were performed in the presence of various inhibitor concentrations (Fig. 1). Assay tubes contained MANT-nucleotides at final concentrations from 0.5 to 20 μM as appropriate according to the potency of the inhibitor. For the basal saturation curve, double-distilled water was added instead of the inhibitor. Assay tubes contained 50 to 600 μM ATP/Mn2+, 10 pM EF3 in 75 mM Tris/HCl, pH 7.4, and 0.1% (m/v) bovine serum albumin. Tubes were preincubated for 2 min at 25°C, and reactions were initiated by the addition of 20 μl of reaction mixture consisting of the following components to yield the given final concentrations: 100 mM KCl, 100 μM free Ca2+, 5 mM free Mn2+, 100 μM EGTA, 100 μM cAMP, 100 nM CaM, and [α-32P]ATP (0.2 μCi/tube).
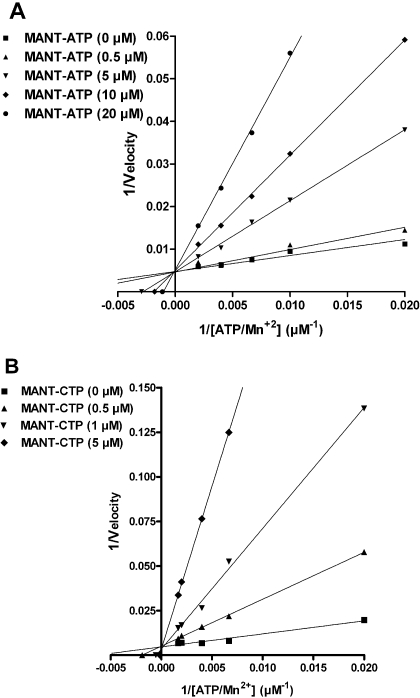
Fig. 1.
Lineweaver-Burk analysis of the inhibition of EF3 AC activity by MANT-ATP and MANT-CTP. AC activities were determined as described under Materials and Methods with the indicated concentrations of MANT-ATP (0, 0.5, 5, 10, and 20 μM) (A) and MANT-CTP (0, 0.5, 1, and 5 μM) (B). Reaction mixtures contained 10 pM EF3, 100 mM KCl, 100 μM free Ca2+, 5 mM free Mn2+, 100 μM EGTA, 100 μM cAMP, 100 nM calmodulin, 0.2 μCi/tube [α-32P]ATP, and unlabeled ATP/Mn2+ concentrations indicated in the graph. Data were plotted reciprocally and analyzed by linear regression according to Lineweaver-Burk. Shown are the results of a representative experiment performed in triplicates. Similar results were obtained in two independent experiments.
Fluorescence Resonance Energy Transfer Experiments for Monitoring Inhibitor Binding to EF. Fluorescence experiments were performed using quartz UV ultra-microcuvettes, types 105.251-QS (light path length, 3 × 3 mm; center, 15 mm; total volume, 70 μl) and 105.250-QS (light path length, 10 × 2 mm; center, 15 mm; total volume, 150 μl; Hellma, Müllheim, Germany) in a multicell holder equipped with a thermostat at 25°C in a Cary Eclipse fluorescence spectrometer (Varian, Darmstadt, Germany). In the case of 150-μl cuvettes, 140 μl of buffer consisting of 100 mM KCl, 100 μM CaCl2, 10 mM MnCl2, and 25 mM HEPES/NaOH, pH 7.4, was added into the cuvette. Five microliters of 10 μM full-length EF (final concentration, 300 nM), 5 μl of 10 μM CaM (final concentration, 300 nM), and MANT-ATP or MANT-CTP (300 nM each) were added. In the case of experiments with 70-μl cuvettes, volumes were adjusted stoichiometrically. The results obtained with 70- and 150-μl cuvettes were identical, with the 70-μl cuvettes offering an opportunity to save EF/EF3 mutant protein.
Steady-state fluorescence emission spectra of nucleotides were recorded at low speed in the scan mode from λem 300 to 550 nm with λex at 280 nm. Fluorescence recordings were analyzed with the spectrum package of the Varian Cary Eclipse software version 1.1. Baseline fluorescence (buffer alone) and the baseline-corrected nucleotide-dependent emission of each concentration of the ligand (buffer + nucleotide) were subtracted from the spectra shown in Fig. 2. In the competition experiments shown in Fig. 3, MANT-nucleotides were displaced from the EF catalytic site using PMEApp. In some experiments, we also directly excited MANT-nucleotides at λex 350 nm and monitored fluorescence emission from 400 to 500 nm.
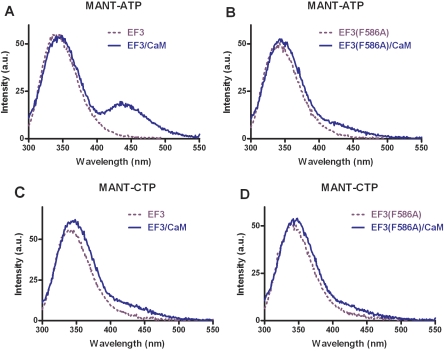
Fig. 2.
Analysis of MANT-nucleotide binding to the catalytic site of EF3 and EF3(F586A) using FRET. FRET experiments were performed as described under Materials and Methods. The assay buffer consisted of 75 mM HEPES/NaOH, 100 μM CaCl2, 100 mM KCl, and 5 mM MnCl2, pH 7.4. Nucleotides were added to the buffer to yield 300 nM final concentrations. EF3/EF3(F586A) (300 nM final concentration) was added followed by the addition of CaM (final concentration, 1 μM). Emission was scanned at an excitation wavelength of 280 nm after each addition. The buffer and the MANT-nucleotide basal fluorescence was subtracted from the fluorescence after addition of EF3/EF3(F586A) (dotted purple line) and CaM (solid blue line). Shown are superimposed recordings of a representative experiment. Similar data were obtained in five independent experiments. a.u., arbitrary unit.
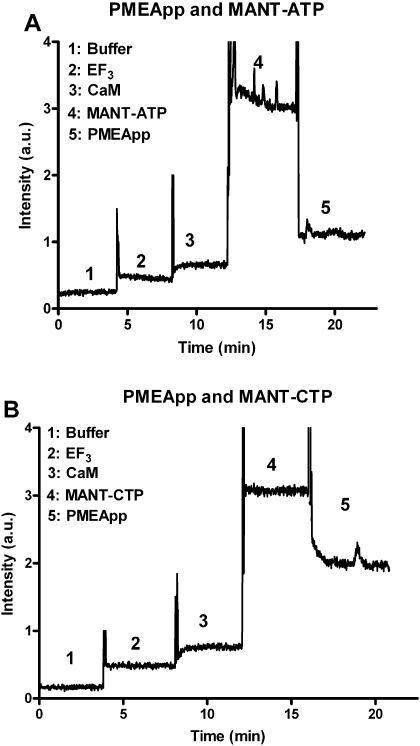
Fig. 3.
Kinetic analysis of the interaction of EF3 with MANT-nucleotides and CaM in FRET experiments. Kinetic experiments were performed as described under Materials and Methods. The excitation wavelength was 280 nm and emission was detected at 430 nm over time. Buffer (1), 300 nM EF3 (2), 1 μM CaM (3), nucleotide (A, MANT-ATP; B, MANT-CTP; 300 nM each) (4), and 1 μM PMEApp (5) were added in sequence. A recording of a representative experiment is shown. Similar data were obtained in four independent experiments. a.u., arbitrary unit.
Modeling of the Nucleotide Binding Mode to EF-CaM. Docking studies were performed with the molecular modeling package SYBYL 7.3 (Tripos International, St. Louis, MO) on an Octane workstation (SGI, Mountain View, CA). An initial computer model was generated from the crystal structure of EF-CaM in complex with 2′-deoxy-3′ANT-ATP, PDB 1lvc, chain C (Shen et al., 2002). Yb2+ was replaced by Mg2+. Water molecules were added from the crystal structure of EF-CaM in complex with 3′-deoxy-ATP, PDB 1k90 (Drum et al., 2002). Hydrogens were added and AMBER_FF99 charges assigned to the protein and the water. 2′-Deoxy-3′ANT-ATP was provided with Gasteiger-Hueckel-charges. The model was then preoptimized with the AMBER_FF99 force field (Cornell et al., 1995) (ligand and Mg2+ fixed, distant dependent dielectric constant ε = 4, 25 cycles steepest descent, followed by Powell conjugate gradient, end gradient of 0.1 kcal · mol-1 · Å-1) were performed.
Initial docking positions of 2′-MANT-CTP, 3′-MANT-CTP, and 3′-MANT-ATP were based on the localization and conformation of 2′-deoxy-3′-ANT-ATP in this model, allowing the modification of rotatable bonds. Each complex was refined in a stepwise approach: 1) ∼25 minimization cycles with fixed ligand (AMBER_FF99 force field, steepest descent, ε = 4); 2) minimization of the ligand and the surrounding protein residues (distance up to 6 Å) with the Tripos force field (Clark et al., 1989) (Powell conjugate gradient, ε = 1, end gradient of 0.05 kcal · mol-1 · Å-1); and 3) final refinement with fixed ligand (AMBER_FF99 force field, Powell conjugate gradient, ε = 4, end gradient of 0.01 kcal · mol-1 · Å-1). The root-mean-square deviation among the three minimized models amounts to 0.2 to 0.25 Å if all atoms of the protein are considered.
Results
Kinetic Analysis of the Catalytic Activities of EF, EF3, and Various EF3 Mutants. In Table 1, the Vmax and Km values of EF, EF3, and various EF3 mutants in the presence of Ca2+/CaM and Mn2+ are summarized. The difference between EF and EF3 is that EF3 does not contain the binding domain for protective antigen (Drum et al., 2002; Shen et al., 2005). The Vmax and Km values were very similar for EF and EF3. In EF3(F586A), Vmax decreased by approximately 50% compared with EF3 with little change in Km. The H577A mutation dramatically reduced Vmax and moderately decreased Km. Substitution of Asn583 with other hydrogen-bond-forming residues, such as Gln583 or His583, significantly reduced Vmax together with a 10- to 50-fold increase in Km. Mutation of Lys353 into Ala353 reduced Vmax 55-fold and increased Km 10-fold. EF(K353R) exhibited approximately 5-fold reduced Vmax and similar Km compared with the EF3. There are some differences in the kinetic parameters of EF3 and EF3 mutants in the presence of Mg2+ (Drum et al., 2002) and Mn2+ (Table 1), supporting the view that these divalent cations interact differentially with ACs (Gille et al., 2004; Mou et al., 2005, 2006; Göttle et al., 2007). Because the FRET signals presented below were much smaller in the presence of Mg2+ than Mn2+ (data not shown), we decided to conduct all enzymatic studies in the presence of Mn2+ as well.
We also determined the kinetic parameters of EF3 in the presence of Mn2+ but in the absence of Ca2+/CaM. By removing both CaM and Ca2+ from the reaction mixture, Vmax was reduced approximately 2000-fold, but there was no effect on Km.
Inhibition of the Catalytic Activity of EF, EF3, and Various EF3 Mutants by (M)ANT-Nucleotides. Table 2 summarizes the Ki values of MANT-ATP and MANT-CTP at EF, EF3, and EF3 mutants in the presence of Mn2+. At EF and EF3, MANT-CTP was a 5- to 10-fold more potent inhibitor than MANT-ATP. The F586A mutation reduced the inhibitory potencies of MANT-ATP and MANT-CTP by 5- to 6-fold, whereas the H577A mutation did not decrease inhibitor potency. The N583A mutation decreased nucleotide potency by 90- to 100-fold. The N583Q substitution reduced the potency of MANT-ATP by 70-fold and the potency of MANT-CTP by 2500-fold. For the N383H mutant, a 200- to 300-fold decrease in potency of MANT-ATP and MANT-CTP was observed. The K353A substitution reduced the potency of MANT-ATP by less than 20-fold, whereas the potency of MANT-CTP was reduced by more than 500-fold. The Ki value of MANT-ATP at EF3(K353R) increased just 3-fold, whereas the Ki value of MANT-CTP increased by 65-fold. The omission of Ca2+/CaM had only little effect on the Ki values of both MANT-ATP and MANT-CTP at EF3.
Figure 1 shows the double-reciprocal analysis of EF3 inhibition kinetics by MANT-ATP and MANT-CTP according to Lineweaver-Burk. The linear regression lines intersected at the y-axis; i.e., Vmax remained constant, whereas Km increased with increasing inhibitor concentration.
Table 3 summarizes the Ki values of various natural purine and pyrimidine nucleotides and various (M)ANT-NDPs and -NTPs at EF in the presence of Mn2+ and Mg2+. In general, inhibitor potencies were higher with Mn2+ than with Mg2+, but the impact of the cation varied with the nucleotide studied. For example, ANT-ATP, MANT-CTP, and MANT-CDP were approximately 10-fold more potent in the presence of Mn2+ than in the presence of Mg2+, whereas for MANT-GTP, the affinity difference was less than 2-fold. A differential impact of Mg2+ and Mn2+ on MANT-nucleotide affinity was also noted for mAC and CyaA (Gille et al., 2004; Mou et al., 2005, 2006; Göttle et al., 2007).
All natural nucleoside 5′-triphosphates studied inhibited the catalytic activity of EF with CTP being the most potent inhibitor. Substitution of the 2′(3′)-O-ribosyl group with a (M)ANT group substantially increased inhibitor potency. For example, in the presence of Mn2+, MANT-CTP showed approximately 60-fold higher potency at EF than the unmodified nucleotide. The potency-increasing effect of the (M)ANT-group depended on the specific nucleotide examined. For example, in the presence of Mn2+, the potency of MANT-GTP was just 3.5-fold higher than the potency of GTP. (M)ANT-NDPs were less potent EF inhibitors than the corresponding (M)ANT-NTPs. In the presence of Mn2+, MANT-ATP and ANT-ATP were similarly potent EF inhibitors, whereas in the presence of Mg2+, MANT-ATP was approximately 4-fold more potent than ANT-ATP. MANT-GTP was up to 2.5-fold more potent than ANT-GTP.
Analysis of the Interaction of MANT-Nucleotides with EF3 in FRET Experiments. At an excitation wave-length of 280 nm, tryptophan and tyrosine residues in proteins are excited, emitting light at 350 nm (Lakowicz 1999), which can then excite the (M)ANT group of nucleotides (Hiratsuka 1983), provided sufficient proximity between donor and acceptor. Such energy transfer results in increased fluorescence of the MANT-group at 420 to 450 nm in mAC and CyaA, reflecting the fact that the MANT group is in a hydrophobic environment (Mou et al., 2005, 2006; Göttle et al., 2007). Previous studies with mAC showed that in the presence of Mn2+, FRET signals were much larger than in the presence of Mg2+ (Mou et al., 2005). Preliminary studies with EF3 showed that the Mn2+-preference also applies to the toxin (data not shown). Therefore, all subsequent FRET studies with EF3 were conducted in the presence of Mn2+ (see Materials and Methods).
In the absence of CaM, EF3 exhibits a strong emission peak at 350 nm when excited at 280 nm under steady-state conditions (Fig. 2, A and C). Upon addition of the EF activator CaM, an additional prominent fluorescence peak with an emission maximum at 440 nm appeared with MANT-ATP (Fig. 2A), reflecting FRET from tryptophan and tyrosine residues to the MANT group. In EF(F586A), the endogenous tryptophan and tyrosine fluorescence was similar as in EF3, but the FRET signal (difference in fluorescence at an emission wavelength of 440 nm in the presence and absence of calmodulin) was reduced by 55% (Fig. 2B). With MANT-CTP, only a small FRET signal was apparent with EF3 upon addition of CaM (Fig. 2C). Even an increase of the final CaM concentration to 3 μM, yielding a 10-fold molar excess of CaM relative to EF3, did not increase FRET with MANT-CTP (data not shown). The small FRET signal with MANT-CTP was reduced by 43% in EF(F586A) (Fig. 2D). Analysis of the EF mutants H577A, N583A, N583Q, N583H, and K353A with MANT-ATP and MANT-CTP revealed no FRET at all (data not shown). With CaM and MANT-nucleotides alone; i.e., in the absence of EF3, no FRET signal was observed (data not shown).
In FRET experiments, the appearance of the new fluorescence peak at λem 440 nm would have been expected to be accompanied by a decrease in fluorescence at λem 350 nm as was the case for mAC (Mou et al., 2005, 2006) and CyaA (Göttle et al., 2007). Rather, a small increase in EF3 fluorescence upon CaM addition was observed (Fig. 2). An explanation for these findings could be that the endogenous tyrosine and tryptophan fluorescence of EF3 is quenched by surrounding polar amino acids such as aspartate, glutamate, and histidine (Lakowicz, 1999). Upon addition of CaM, a large conformational change occurs in EF3 (Drum et al., 2002; Shen et al., 2002), abrogating the quenching effects of polar amino acids and thereby masking the expected decrease in fluorescence at λem 350 nm.
Control experiments with dimethyl sulfoxide ranging from 0 to 100% (v/v) (Hiratsuka, 1983) revealed that MANT-ATP and MANT-CTP possess similar relative increases in fluorescence upon exposure to a hydrophobic environment (λex 350 nm; λem 400-500 nm) (data not shown). Thus, differences in biophysical properties of nucleotides do not account for the different FRET responses observed with MANT-ATP and MANT-CTP upon interaction with EF. We also studied the interaction of EF with MANT-nucleotides by exciting nucleotides directly at λem 350 nm. As was true in the FRET experiments (Fig. 2), the response observed with MANT-ATP at λem 450 nm was greater than with MANT-CTP (data not shown).
Figure 3 shows the kinetics of FRET experiments with MANT-ATP and MANT-CTP at a fixed emission wavelength of 440 nm. Sequential addition of EF3 and CaM resulted only in small fluorescence increases, reflecting the far end of the tryptophan/tyrosine emission spectrum (see Fig. 2). Addition of MANT-nucleotides to cuvettes instantaneously resulted in substantial fluorescence increases, reflecting FRET. Addition of the high-affinity EF inhibitor and nonfluorescent nucleotide analog PMEApp (1 μM) (Shen et al., 2004) to cuvettes reduced the fluorescence signals with both MANT-nucleotides (300 nM each), but the inhibitory effect of PMEApp was more pronounced and more rapid in onset with MANT-ATP than with MANT-CTP.
Modeling of the Binding Modes of MANT-Nucleotides to EF-CaM. The crystal structure of EF in complex with CaM and 2′-deoxy-3′-ANT-ATP (PDB 1lvc; Shen et al., 2002) served as basis for the docking of MANT-CTP and MANT-ATP. Figure 4A shows that the nucleotide binding site of EF is a spacious cavity located at the interface of two structural domains, CA (Asp294-Asn349, Ala490-Lys622) and CB (Val350-Thr489). Three switches, A (Gln507-Leu549), B (Gly578-Asn591), and C (Arg630-Thr659), which strongly change their conformation and position on the transition from EF alone to EF-CaM (Drum et al., 2002, Shen et al., 2002), cover the catalytic site in the EF-CaM state. A metal cation (Yb2+ in the template, replaced by Mg2+ in the models) is involved in ionic interactions with Asp491, Asp493, and the α-phosphate of the nucleotides. The MANT group is aligned in parallel with the phenyl ring of Phe586. Thus, hydrophobic interactions account for the generally higher potency of the ANT and MANT derivatives compared with their natural parent nucleotides (Table 3).
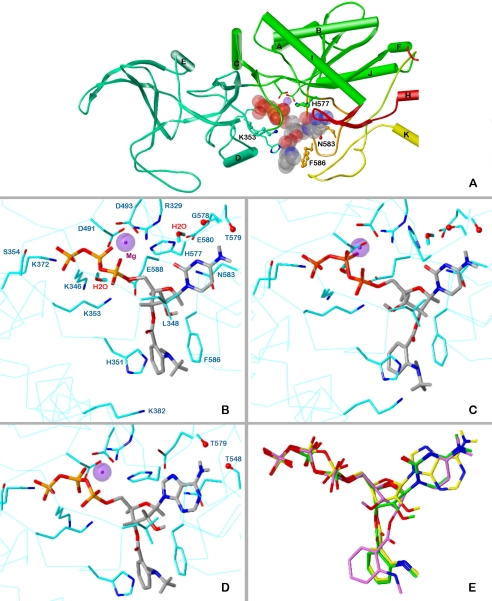
Fig. 4.
Docking of MANT-CTP and MANT-ATP on EF-CaM. The minimized models are based on the crystal structure of EF-CaM-3′ANT-2′deoxy-ATP, PDB 1lvt (Shen et al., 2002). Colors of atoms, unless otherwise indicated: P, orange; O, red; N, blue; C, H, gray; Mg2+, purple spheres. A, topology diagram of the EF domains surrounding the binding site with docked 3′-MANT-CTP (represented as transparent Corey/Pauling/Koltun model). Domains CA, green; CB, green-blue; switch A, red; switch B, orange; switch C, yellow. Amino acids subjected to mutation, ball-and-stick models. B to D, detailed representation of interactions of MANT-nucleotides with EF-CaM. The side chains of amino acids within a sphere of 3 Å around the ligand and Mg2+ are drawn as sticks (C atoms, cyan) and labeled. For clarity, repeating labels are omitted in C and D (see B). Cα-trace, cyan line. B, docking of 3′-MANT-CTP. C, docking of 2′-MANT-CTP. D, docking of 3′-MANT-ATP. E, superposition of the docked MANT nucleotides based on alignment of the three EF models (all heavy protein atoms considered). Colors of C and essential H atoms: 3′-MANT-CTP, green; 2′-MANT-CTP, purple; 3′-MANT-ATP, yellow.
The docking approaches were performed to suggest possible reasons why MANT-CTP inhibits EF-CaM more potently than MANT-ATP. Figure 4, B and D, compares the putative binding mode of 3′-MANT-CTP and 3′-MANT-ATP in more detail. The common structures of both molecules interact with EF-CaM in the same manner. In the minimized models, the ribosyl rings adopt a 3′-exo conformation like the ribosyl moiety of 3′-MANT-ATP in complex with mAC (Mou et al., 2006). The triphosphate moieties fit into a deep polar pocket forming ionic interactions with the lysines 346, 353, and 372 as well as an H bond with Ser354 (γ-phosphate). These interactions account for the higher inhibitory potencies of the triphosphates compared with the diphosphate analogs in Table 3. The ring oxygens of the ribosyl moieties are hydrogen-bonded to the amide NH2 of Asn583. The 2′-OH groups approach the side chain of Leu348. In addition to the parallel fit to Phe586, the 3′-MANT substituents interact via their amino function with His351.
The nucleobases show both shared and different interactions. Most striking is the flat alignment with the side chain of Asn583. In the case of 3′-MANT-CTP, such interaction is more favorable because the oxygen in the 2′-position of the base may align with the positive pole of the amide dipole. However, this difference cannot be the only reason for the higher potency compared with MANT-ATP, because, also on the N583A mutant, MANT-CTP is more potent than its ATP analog (Table 3). Considering the more distant environment of the cytosine oxygen, additional reasons can be suggested. In the minimized model, it is 3.3 Å from the guanidino group of Arg329. Moreover, a water molecule can be placed in an ideal position into the EF-MANT-CTP model, where it forms three H bonds, bridging the cytosine oxygen with the side chains of Arg329 and Glu580. The NH2 groups of the cytosine and adenine moieties interact similarly with EF-CaM via H bonds to backbone oxygens (3′-MANT-CTP to Gly578 and/or Thr579, 3′MANT-ATP to Thr548 and Thr579). The significance of these interactions is confirmed by the lower potency of MANT-UTP and MANT-GTP (Table 3). It is noteworthy that the N583H mutant disproportionately reduces the potency of MANT-ATP, whereas on the N583Q species, MANT-CTP is even less potent than its ATP analog. In the former case, an alignment of the heterocycles should be still possible. However, the larger glutamine side chain in N583Q restricts the binding cavity for the nucleobases so that the mode of interaction changes. As suggested for MANT-CTP, the affinity of the other nucleotides to EF3 and EF mutants may also be affected by a specific arrangement of water molecules.
The MANT nucleotides used in the assays are mixtures of 2′-MANT and 3′-MANT isomers, and there is spontaneous isomerization between the two species under physiological conditions (Hiratsuka, 1983, 2003; Jameson and Eccleston, 1997). So far, only the putative binding mode of 3′-MANT isomers has been considered. The question arises whether the MANT group may fit to the same EF-CaM site in both isomers as shown for the binding of MANT-nucleotides to CyaA (Göttle et al., 2007). Figure 4C indicates that 2′-MANT-CTP may indeed interact with EF-CaM in a very similar position to that of its 3′-MANT analog. The main difference is a 3′-endo conformation of the ribosyl moiety, present also in the complex of 3′-deoxy-ATP with EF-CaM (Drum et al., 2002). This geometry implies an axial position of the 2′-MANT group, which may then project to the space between His351 and Phe586. In addition, an intramolecular H bond between the 3′-hydroxy group and the proximal α-phosphate oxygen is possible. Figure 4E, showing the alignment of 2′-MANT-CTP, 3′-MANT-CTP, and 3′-MANT-ATP in the docked poses, indicates a close fit of the 2′-MANT and 3′-MANT isomers. Only the positions of the MANT groups themselves are slightly different (variability of the triphosphate conformation is due to the individual minimization courses).
Discussion
The major goal of the present study was to characterize the interaction of the catalytic site of EF with (M)ANT-nucleotides possessing various purine and pyrimidine bases to better understand the molecular mechanisms of EF inhibition and to provide the basis for the rational development of potent and selective EF inhibitors. Such EF inhibitors could be useful compounds to treat EF toxinemia and antibiotic-resistant Bacillus anthracis strains (Jedrzejas, 2002).
In previous studies, we developed a three-site pharmacophore model for mAC and CyaA toxin with binding regions for the base, the MANT group, and the polyphosphate chain (Gille et al., 2005; Mou et al., 2006; Göttle et al., 2007; Wang et al., 2007). Those studies revealed that the MANT group and the polyphosphate chain are the major determinants of inhibitor potency, whereas the base plays a relatively small role in this respect. This is reflected by the fact that the catalytic sites of mAC and CyaA are conformationally flexible and accommodate both purine and pyrimidine nucleotides.
Likewise, in EF, the (M)ANT-group and the length of the polyphosphate chain have a substantial impact on inhibitor potency, and EF accommodates various purine and pyrimidine bases (Table 3). These data indicate that the three-site binding model developed for mAC and CyaA can also be extended to EF. The structure/activity relationships of (MANT)-nucleotides at EF, CyaA and mAC are different, indicating that in principle, the development of potent and AC isoform-specific inhibitors is feasible. Most strikingly, CTP inhibited EF more than 400-fold more potently than mAC (Table 3) (Gille et al., 2005). Substitution of the 2′(3′)-O-ribosyl position of CTP with a MANT group decreased the Ki value from 5 μM to 100 nM, yielding an EF inhibitor that is even 5- to 10-fold more potent than MANT-ATP in the presence of Mn2+ (Tables 2 and 3).
The highly unexpected preference of EF for the base cytosine prompted us to analyze EF inhibition by MANT-CTP and MANT-ATP in more detail. The analysis of enzyme inhibition kinetics revealed that both MANT-CTP and MANT-ATP are competitive EF inhibitors; i.e., they bind to the same site as, and freely compete with, the substrate ATP (Fig. 1). These data rule out the existence of a hitherto unidentified cytosine base-preferring nucleotide-binding site in the structurally very complex EF protein (Fig. 4A) (Drum et al., 2002; Shen et al., 2002). Kinetic FRET competition experiments with the nonfluorescent ATP analog PMEApp revealed that both MANT-ATP and MANT-CTP reversibly bind to the catalytic site (Fig. 3), corroborating the competitive inhibition mode and the existence of a single nucleotide-binding site in EF. The faster displacement of MANT-ATP from EF by PME-App compared with the displacement of MANT-CTP is explained by the higher affinity of EF for MANT-CTP.
To dissect possible differences in the binding modes of MANT-ATP and MANT-CTP to EF, we studied their interaction with several EF mutants in terms of enzyme inhibition and fluorescence spectroscopy. A previous study had shown that Phe586 mediates π-stacking interactions with 2′-deoxy-3′-ANT-ATP, resulting in a fluorescence increase upon excitation of the ANT-group (Shen et al., 2002). In agreement with those data, mutation of Phe586 reduced the potency of MANT-ATP and largely reduced the CaM-dependent FRET of MANT-ATP (Table 2 and Fig. 2). Phe586 is also important for the interaction with MANT-CTP, as revealed by the 6-fold reduction in potency. However, compared with MANT-ATP, the FRET signal with MANT-CTP in EF was much smaller, and the F586A mutation had a smaller inhibitory effect on the FRET. This difference in FRET cannot be explained by a lower CaM affinity of EF bound to MANT-CTP compared with the protein complex bound to MANT-ATP, because a 10-fold molar excess of CaM relative to EF did not yield a larger FRET with MANT-CTP. The models shown in Fig. 4 suggest a similar binding mode of MANT-CTP and its ATP analog. However, subtle differences caused by the nucleobases occur and may account for the higher potency and the small FRET signal of MANT-CTP. In particular, the cytosine moiety may form water-mediated hydrogen bonds with Arg329 and Glu580 and favorably fit to the amide dipole of Asn583. In addition, the flexibility of the bound cytosine should be greater than in the case of the bulkier adenine ring. Together with the specific charge distribution in the vicinity of the nucleobase, this may lead to absorption and thus attenuation of the FRET energy, which is due mainly to tyrosine and tryptophan residues in switch C.
His577 plays a crucial role in catalysis, as reflected by the very low catalytic activity of the H577A mutant (Table 1) (Drum et al., 2002; Guo et al., 2004). Nonetheless, the catalytic activity of the H577A mutant was sufficiently large to determine substrate and MANT-nucleotide affinity. Indeed, this mutation does not exert detrimental effect on substrate and inhibitor binding per se (Tables 1 and 2).
Asn583 forms a crucial hydrogen bond with the ribosyl moiety of nucleotides bound to the catalytic site of EF (Drum et al., 2002). Accordingly, replacement of Asn583 by a non-hydrogen-bond-forming amino acid (N583A) or hydrogen-bond-forming amino acid with a different spatial arrangement of the bonding partners (N583Q and N383H) substantially decreases catalytic activity of the resulting EF mutants and also increases Km (Table 1) (Drum et al., 2002). Thus, it was also not surprising that Asn583 mutants substantially reduced the potencies of MANT-ATP and MANT-CTP (Table 2). However, whereas the N583A mutation and the N583H mutation affected inhibitor potencies to a similar extent, the potency of MANT-CTP was much more strongly reduced by the N583Q mutation than the potency of MANT-ATP. These findings suggest that the binding of MANT-CTP is severely impaired by the longer side chain of Q compared with N, indicative of a substantial spatial constraint in this part of the binding pocket.
The carboxyl group of Glu588 and the amino group of Lys353 form an ionic bond that locks the base into the catalytic site (Drum et al., 2002). Disruption of this ionic bond by the K353A mutation largely reduces catalytic activity and lowers substrate affinity (Drum et al., 2002) (Table 1). The K353R mutation that alters the spatial arrangement of the catalytic site but still allows ionic bridge formation displays less severe impairment of catalysis and no change in Km (Table 1). Intriguingly, binding of MANT-CTP to the catalytic site is much more sensitive to disruption of ionic bond formation between Glu588 and Lys353 and reorientation of the ionic bond than binding of MANT-ATP (Table 2), further corroborating the notion that there are subtle differences in the binding modes of MANT-ATP and MANT-CTP.
The high sensitivity of our AC assay, which was largely due to very low blank values (see Materials and Methods), allowed us to precisely determine the kinetic parameters not only of EF mutants but also of wild-type EF in the absence of CaM. In fact, the catalytic activity of EF is not absolutely CaM-dependent. Even in the absence of CaM, we could accurately determine kinetic parameters using appropriate experimental conditions (i.e., higher protein and [α-32P]ATP amounts, higher incubation temperature, and longer incubation time). It was rather surprising that in the absence of CaM, EF displays unaltered substrate- and inhibitor affinity despite the largely reduced catalytic activity. Even the MANT-CTP/MANT-ATP potency ratio is conserved. Comparing the crystal structures of EF-CaM (PDB 1lvc; Shen et al., 2002) and EF alone (PDB 1k8t; Drum et al., 2002), 13 mutually resolved amino acids of the MANT-nucleotide binding sites (Fig. 4B) fit very well (RMSD of the backbone atoms 0.99 Å; only Gly578 and Thr579 at the N terminus of switch B are outliers with distances of greater than 1.5 Å). However, switch B stabilized by switch C in EF-CaM is disordered in the structure of EF alone and contains amino acids involved in substrate- and MANT-nucleotide binding to EF-CaM (Thr579, Glu580, Asn583, Phe586, Glu588) or in the stabilization of residues that participate in catalysis (e.g., Asp590 forming a salt bridge with Arg329). To explain the unaltered potency of the MANT inhibitors at EF alone, we postulate that the nucleotides stabilize switch B in a conformation like that in EF-CaM. The substrate may act similarly in terms of affinity, but the resulting switch B conformation is insufficient for high catalytic activity. Experiments with membrane-permeable CaM inhibitors (Wolberg and Zimmerman, 1984) will have to answer the question of whether the CaM-independent catalytic activity of EF is of pathophysiological relevance.
In conclusion, through a combination of enzymological, fluorescence spectroscopic, mutagenesis, and molecular modeling approaches, we have shown that there are subtle differences in the binding modes of MANT-ATP and MANT-CTP to EF. EF, unlike all other ACs studied so far, including mAC and CyaA toxin from Bordetella pertussis, exhibits a unique preference for the base cytosine, offering an excellent starting point for the development of EF inhibitors with specificity for the toxin relative to mAC. Finally, our studies also raise the intriguing question whether CTP, exhibiting an unusually high affinity for EF, is not only an inhibitor of cAMP synthesis but also, perhaps, a substrate itself.
Acknowledgments
We thank Astrid Seefeld for expert technical assistance. Thanks are also due to the reviewers of the manuscript for helpful critique and suggestions.
This work was supported by the Deutsche Forschungsgemeinschaft [Grant Graduiertenkolleg 760 “Medicinal Chemistry: Molecular Recognition-Ligand-Receptor Interactions” and Research Grant Se 529/5-1], by a predoctoral fellowship from the Arabic Republic of Egypt, and by the “International Study and Training Partnerships (ISAP) Program” of the German Academic Exchange Service (DAAD).
ABBREVIATIONS: EF, full-length edema factor adenylyl cyclase toxin; AC, adenylyl cyclase; ANT, anthraniloyl-; CaM, calmodulin; CyaA, Bordetella pertussis adenylyl cyclase toxin; ESI, electrospray ionization; FRET, fluorescence resonance energy transfer; HPLC, high pressure liquid chromatography; k, capacity factor; mAC, mammalian membranous adenylyl cyclase; MANT, methylanthraniloyl-; MES, 2-(N-morpholino)ethanesulfonic acid; LC, liquid chromatography; MS, mass spectroscopy; MW, molecular weight; NDP, nucleoside 5′-diphosphate; NTP, nucleoside 5′-triphosphate; PDB, Protein Data Bank; PMEApp, 9-[2-(phosphonomethoxy)ethyl]adenine diphosphate; PTFE, polytetrafluoroethylene; EF3, catalytic domain of edema factor adenylyl cyclase toxin; Rf, retention factor; Rt, retention time.
References
- Clark M, Cramer RD 3rd, and Van Opdenbosch N (1989) Validation of the general purpose Tripos 5.2 force field. J Comput Chem 10982 -1012. [Google Scholar]
- Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KMJ, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, and Kollman PA (1995) A second generation force field for the simulation of proteins and nucleic acids. J Am Chem 1175179 -5197. [Google Scholar]
- Drum CL, Yan SZ, Bard J, Shen YQ, Lu D, Soelaiman S, Grabarek Z, Bohm A, and Tang WJ (2002) Structural basis for the activation of anthrax adenylyl cyclase exotoxin by calmodulin. Nature 415396 -402. [DOI] [PubMed] [Google Scholar]
- Gille A, Lushington GH, Mou TC, Doughty MB, Johnson RA, and Seifert R (2004) Differential inhibition of adenylyl cyclase isoforms and soluble guanylyl cyclase by purine and pyrimidine nucleotides. J Biol Chem 27919955 -19969. [DOI] [PubMed] [Google Scholar]
- Gille A, Guo J, Mou TC, Doughty MB, Lushington GH, and Seifert R (2005) Differential interactions of G-proteins and adenylyl cyclase with nucleoside 5′-triphosphates, nucleoside 5′-[γ-thio]phosphates and nucleoside 5′-[β,γ-imido]triphosphates. Biochem Pharmacol 7189 -97. [DOI] [PubMed] [Google Scholar]
- Gopalakrishna R and Anderson WB (1982) Ca2+-induced hydrophobic site on calmodulin: application for purification of calmodulin by phenyl-Sepharose affinity chromatography. Biochem Biophys Res Commun 104830 -836. [DOI] [PubMed] [Google Scholar]
- Göttle M, Dove S, Steindel P, Shen Y, Tang WJ, Geduhn J, König B, and Seifert R (2007) Molecular analysis of the interaction of Bordetella pertussis adenylyl cyclase with fluorescent nucleotides. Mol Pharmacol 72526 -535. [DOI] [PubMed] [Google Scholar]
- Guo Q, Shen Y, Zhukovskaya NL, Florián J, and Tang WJ (2004) Structural and kinetic analyses of the interaction of anthrax adenylyl cyclase toxin with reaction products cAMP and pyrophosphate. J Biol Chem 27929427 -29435. [DOI] [PubMed] [Google Scholar]
- Hiratsuka T (1983) New ribose-modified fluorescent analogs of adenine and guanine nucleotides available as substrates for various enzymes. Biochim Biophys Acta 742496 -508. [DOI] [PubMed] [Google Scholar]
- Hiratsuka T (2003) Fluorescent and colored trinitrophenylated analogs of ATP and GTP. Eur J Biochem 2703479 -3485. [DOI] [PubMed] [Google Scholar]
- Hong J, Beeler J, Zhukovskaya NL, He W, Tang WJ, and Rosner MR (2005) Anthrax edema factor potency depends on mode of cell entry. Biochem Biophys Res Commun 335850 -857. [DOI] [PubMed] [Google Scholar]
- Jameson DM and Eccleston JF (1997) Fluorescent nucleotide analogs: synthesis and applications. Methods Enzymol 278363 -390. [DOI] [PubMed] [Google Scholar]
- Jedrzejas MJ (2002) The structure and function of novel proteins of Bacillus anthracis and other spore-forming bacteria: development of novel prophylactic and therapeutic agents. Crit Rev Biochem Mol Biol 37339 -373. [DOI] [PubMed] [Google Scholar]
- Lakowicz JR (1999) Principles of Fluorescence Spectroscopy. Kluwer Academic/Plenum, New York.
- Mou TC, Gille A, Fancy DA, Seifert R, and Sprang SR (2005) Structural basis for the inhibition of mammalian membrane adenylyl cyclase by 2′(3′)-O-(N-methylanthraniloyl)-guanosine 5′-triphosphate. J Biol Chem 2807253 -7261. [DOI] [PubMed] [Google Scholar]
- Mou TC, Gille A, Suryanarayana S, Richter M, Seifert R, and Sprang SR (2006) Broad specificity of mammalian adenylyl cyclase for interaction with 2′,3′-substituted purine- and pyrimidine nucleotide inhibitors. Mol Pharmacol 70878 -886. [DOI] [PubMed] [Google Scholar]
- Shen Y, Lee YS, Soelaiman S, Bergson P, Lu D, Chen A, Beckingham K, Grabarek Z, Mrksich M, and Tang WJ (2002) Physiological calcium concentrations regulate calmodulin binding and catalysis of adenylyl cyclase exotoxins. EMBO J 216721 -6732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Zhukovskaya NL, Zimmer MI, Soelaiman S, Bergson P, Wang CR, Gibbs CS, and Tang WJ (2004) Selective inhibition of anthrax edema factor by adefovir, a drug for chronic hepatitis B virus infection. Proc Natl Acad Sci U S A 1013242 -3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Zhukovskaya NL, Guo Q, Florián J, and Tang WJ (2005) Calcuim-independent calmodulin binding and two-metal-ion catalytic mechanism of anthrax edema factor. EMBO J 24929 -941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JL, Guo JX, Zhang QY, Wu JJ, Seifert R, and Lushington GH (2007) A conformational transition in the adenylyl cyclase catalytic site yields different binding modes for ribosyl-modified and unmodified nucleotide inhibitors. Bioorg Med Chem 152993 -3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolberg G and Zimmerman TP (1984) Effects of calmodulin antagonists on immune mouse lymphocytes. Mol Pharmacol 26286 -292. [PubMed] [Google Scholar]