Abstract
μ-Opioid receptor (MOR) mediates most of the pharmacological effects of opioid drugs. The expression of MOR is temporarily and spatially regulated at both the transcriptional and post-transcriptional levels. Long-term morphine treatment that induces tolerance does not alter MOR mRNA expression, suggesting no direct link between agonist treatment and MOR gene transcription. We previously identified the 3′-untranslated region (3′-UTR) of the major transcript of μ-opioid receptor (MOR1) and revealed a novel trans-acting factor, miRNA23b, that binds to the K box motif in the 3′-UTR. The interaction between miRNA23b with the MOR1 3′-UTR suppressed receptor translation by inhibiting polysome-mRNA association. In this report, we demonstrate that long-term morphine treatment increases miRNA23b expression in a dose- and time-dependent manner and represses the association of MOR1 mRNA with polysomes through the MOR1 3′-UTR. The translational luciferase reporter assay shows a suppression effect of morphine on reporter activity that requires the MOR1 3′-UTR. This suggests a potential link between MOR expression and morphine treatment at the post-transcriptional level in which a specific miRNA, miRNA23b, is involved.
Opioid drugs are widely used clinically to treat moderate to severe pain. Three major opioid receptors, μ, δ, and κ, belong to the G-protein-coupled receptor superfamily (Kieffer, 1995). μ-Opioid receptor (MOR) mediates most of the pharmacological effects of opiates; its regulation is of great importance to unravel the molecular mechanisms underlying the physical responses to opioid treatment, such as tolerance and dependence. In addition to the multiple cis-acting elements that regulate the transcription of MOR (Hwang et al., 2004; Kim et al., 2006; Choi et al., 2007; Song et al., 2007), a recent study on the 3′-UTR of the major μ-opioid receptor mRNA (MOR1) has started to address the regulation of MOR at the post-transcriptional level (Wu et al., 2008).
The phenomenon of MOR down-regulation has been observed in various cell lines and neurons caused by long-term agonist treatment (Yabaluri and Medzihradsky, 1997; Tao et al., 1998; Yamamoto et al., 2008). Down-regulation results primarily from the sequestration of membrane receptors to the cytosol via clathrin-coated pits and dynamin after receptor phosphorylation; the internalized endosomes merge with intracellular lysosomes, and the receptors are degraded proteolytically, resulting in a decrease of the total number of receptors in the cell (Binyaminy et al., 2008). In addition to this classic model, accumulating evidence shows the involvement of many other factors in the processes [e.g., protein kinase C (Kramer and Simon, 1999), mitogen-activated protein kinase (Schmidt et al., 2000) and Ca2+/calmodulin-dependent protein kinase (Koch et al., 1997)]. However, whether a decreased receptor biosynthesis is involved is still under debate. One study measured the receptor turnover rate in mouse neuronal N2A cells expressing a cloned μ-opioid receptor (N2A-MOR) and found that the down-regulation of MOR caused by agonist stimulation is the sum of both accelerated receptor degradation and decreased receptor biosynthesis (Afify, 2002).
The expression of MOR can be regulated at both the transcriptional and post-transcriptional levels. It is widely accepted that long-term morphine treatment does not alter MOR mRNA levels (Brodsky et al., 1995), indicating that morphine has no significant effect on the transcription of MOR gene. Nonetheless, it is not known whether morphine can regulate MOR mRNA at the post-transcriptional level, possibly through interactions between trans-acting factors and its 3′-UTR. Although morphine can induce discrete and fluctuating expression of important factors related to MOR function (Ammon-Treiber and Hollt, 2005), whether these factors are involved in the post-transcriptional regulation of MOR remains unknown. Our recent study identified miRNA23b as a trans-acting factor that represses MOR translation efficiency through interaction with the 3′-UTR of MOR1 (Wu et al., 2008).
Little is known about the mechanisms responsible for regulating miRNA expression. In some cells, the production of miRNAs seems to be actively regulated (Woods et al., 2007; Boyd, 2008). It would be interesting to investigate whether activating a membrane receptor could control the expression of a miRNA, thereby regulating expression of the receptor gene. In this report, we investigated whether morphine treatment could change miRNA23b expression and consequently regulate the translation of MOR mRNA.
miRNA23b interacts with the MOR1 3′-UTR through binding to a K Box motif (Wu et al., 2008). The MOR1 3′-UTR is absent from the plasmid DNA sequences in all cell lines that express cloned MOR, such as N2A-MOR and HEK-MOR (Chakrabarti et al., 1995; Wu and Wong, 2005). N2A cells are not known to express any opioid receptors (Im et al., 2001); N2A-MOR cells were established by stably transfecting MOR1 into N2A cells. MOR1, a 1.4-kb insert, was subcloned into the expression vector pRC/CMV; the cDNA sequence contains the entire coding region of the MOR, together with 200 base pairs of 5′ noncoding region and only 30 base pairs of 3′ noncoding region (i.e., the 3′-UTR). The cloned MOR (without the major part of MOR1 3′-UTR sequence) behaves like native receptor in terms of its desensitization and down-regulation effects (Chakrabarti et al., 1995). However, it has not yet been determined whether the absence of MOR1 3′-UTR influences the regulation of MOR gene. In this report, we investigated the effect of morphine on miRNA23b expression in N2A-MOR cells and the resulting changes in MOR1 mRNA translation efficiency. In addition, we revealed the critical roles of MOR receptor and MOR1 3′-UTR in this pathway.
Materials and Methods
Cell Culture, Transfection, and Luciferase Reporter Assay. Mouse neuronal cells N2A and N2A-MOR (Chakrabarti et al., 1995) and human neuronal cells NMB and SHSY-5Y were maintained in advanced Dulbecco's modified Eagle's medium or RPMI 1640 medium (for NMB) (Invitrogen, Carlsbad, CA) with 5% heat-inactivated fetal bovine serum in an atmosphere of 10% (for N2A and N2A-MOR) or 5% (for NMB and SHSY-5Y) CO2 at 37°C. The medium for N2A-MOR was supplemented with 0.2% G418 (Geneticin). Transfections of anti-23b or anti-miR negative control primer (Ambion, Austin, TX) were performed using Lipofectamine 2000 (Invitrogen) as described previously (Wu et al., 2008).
For the luciferase reporter assay, cells were plated at a density of 0.5 × 105 cells per well in 24-well plates 24 h before transfection; 2 ng of Renilla reniformis luciferase plasmid pCMV-Rluc (a gift from Dr. Yan Zeng, University of Minnesota, Minneapolis, MN) was included for normalization. Morphine was added 3 h before transfecting pSVUTR or pSVPA plasmids (Wu et al., 2008). Twenty-four hours after transfection, the firefly and R. reniformis luciferase activities were determined by a luminometer (Berthold, Oak Ridge, TN) using Dual-Luciferase Reporter Assay systems (Promega, Madison, WI) according to the manufacturer's protocol.
RT-PCR, Real-Time qPCR, and qRT-PCR. RNA was isolated from cells using TRI reagent (Molecular Research Center, Cincinnati, OH) and treated with Turbo DNase I (2 U/μg of RNA) (Ambion) before being reverse-transcribed. One-step RT-PCR was performed using the OneStep RT-PCR Kit (QIAGEN, Valencia, CA) and the following primers: mouse MOR1, 5′-CTGCTCGAATCCGTCAAAACA-3′ (sense) and 5′-AGCAACCTGATTCCAAGTAGA-3′ (antisense); HA-MOR1, 5′-CTGCTCGAATCCGTCAAAACA-3′ (sense) and 5′-GGCAACTAGAAGGCACAGTC-3′ (antisense); and mouse β-actin, 5′-TGGCCTTAGGGTGCAGGGGG-3′ (sense) and 5′-GTGGGCCGCTCTAGGCACCA-3′ (antisense). For MOR1 RNA, the product from the one-step RT-PCR was re-amplified for a second round using Taq polymerase (New England Biolabs, Ipswich, MA) and primers: 5′-CTGCTCGAATCCGTCAAAACA-3′ (sense) and 5′-GTAGATGGCAGCCTCTAA-3′ (antisense). PCR was performed on a GeneAmp PCR System 9600 (PerkinElmer Life and Analytical Sciences, Waltham, MA) using 30 cycles (for MOR1 and HA-MOR1) or 20 cycles (for β-actin) of 94°C for 1 min, 55°C for 1 min, and 72°C for 1 min followed by 72°C for 10 min. The linear range of PCR cycles for each gene had been predetermined using relative PCR, and cycle numbers for PCR and RT-PCR were optimized according to the results. PCR products were electrophoresed in 1 or 2% agarose gels, quantified by ImageQuant 5.2 (GE Healthcare, Chalfont St. Giles, Buckinghamshire, UK) and verified by DNA sequence analysis.
miRNA-enriched RNA was extracted and reverse transcribed followed by qPCR as described before (Wu et al., 2008). One-tenth of the reverse transcription mix was used for real-time qPCR. The miRNA primer sets hsa-miR23b and snoRNA234 (as an internal control) (Applied Biosystems) were used for reverse transcription, and qPCR was performed according to the manufacturer's protocol.
Real-time qPCR and qRT-PCR were performed on an iCycler (Bio-Rad Laboratories, Oakland, CA) using either an iQ Supermix Kit (Bio-Rad) for miRNA23b and snoRNA234 or a Quantitect SYBR Green RT-PCR kit (QIAGEN) for MOR1, HA-MOR1 and β-actin. The relative expression levels of miRNA23b were calculated using the Gene Expression Macro (Bio-Rad Laboratories, Hercules, CA) normalized to those of snoRNA234; and the levels of MOR1 and HA-MOR1 were calculated against those of β-actin.
Polysome mRNA Extraction. Polysome mRNA extraction was conducted as described previously (Wu et al., 2008). Polysomal mRNA was isolated from pellets using TRI reagent (Molecular Research Center) following the manufacturer's protocol.
Statistics. Data are presented as mean values ± S.D. Comparisons between groups were performed using the Student's t test. P < 0.05 was taken as significant.
Results
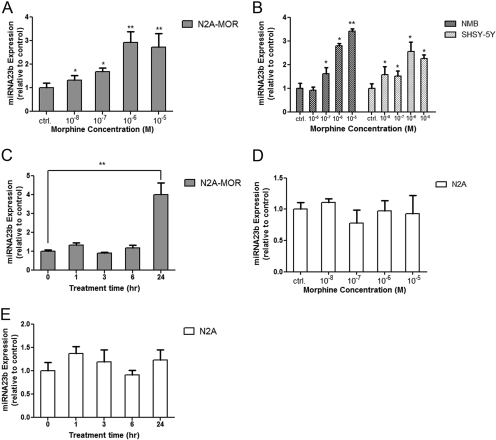
Long-Term Morphine Treatment Increases miRNA23b Levels. N2A-MOR cells expressing HA-tagged MOR1 receptor were treated with morphine (10-8 to 10-5 M) for 24 h. miRNA23b levels were determined by reverse transcription followed by real-time qPCR. There was a dose-dependent increase in miRNA23b levels, with the maximum effect reached under a treatment of 10-6 M morphine (Fig. 1A). Human and mouse miRNA23b sequences have 100% homology (http://microrna.sanger.ac.uk/). In human neuronal cell lines NMB and SHSY-5Y (both express MOR endogenously), morphine stimulated a similar dose-dependent increase of miRNA23b levels (Fig. 1B). N2A-MOR cells were treated with 10-6 M morphine for 1, 3, 6, or 24 h (Fig. 1C). The up-regulation of miRNA23b was only observed after treatments longer than 6 h, indicating that prolonged exposure to morphine was required to alter the expression of miRNA23b.
Fig. 1.
Morphine increases miRNA23b expression. A, miRNA-enriched RNA was extracted from N2A-MOR cells 24 h after morphine treatment. Twenty nanograms of RNA were used for reverse transcription using miRNA23b and snoRNA234 (control) primers, followed by real-time qPCR. The y-axis represents the levels of miRNA23b expression normalized relative to control (i.e., without treatment; miRNA23b/snoRNA234); the x-axis represents different concentrations of morphine (10-8 to 10-5 M). Student's t test was performed by comparing each sample to the control sample. The graph shows a single experiment performed in duplicate; the experiment was repeated three times with similar results. n = 3; *, p < 0.05; **, p < 0.01. B, miRNA was extracted from NMB and SHSY-5Y cells treated with morphine for 24 h. Student's t test was performed by comparing each sample to the control sample in each cell line. The legends are the same as in A. n = 3; *, p < 0.05; **, p < 0.01. C, miRNA was extracted from N2A-MOR cells treated with 10-6 M morphine for different lengths of time. The x-axis represents the different time points (1-24 h). Student's t test was performed by comparing each sample to the control (0 h treatment). n = 3; **, p < 0.01. D, miRNA was extracted from N2A cells 24 h after morphine treatment. The legends are the same as in A; n = 3. E, miRNA was extracted from N2A cells treated with 10-6 M morphine for different lengths of time. The legends are the same as in C. n = 3.
Wild-type N2A cells are devoid of opioid receptors (Im et al., 2001). To determine the role of MOR receptor, the dose-response and time course of miRNA23b in wild-type N2A cells was assessed. An increasing dose of morphine (10-8 to 10-5 M) did not change the miRNA23b levels in N2A cells (Fig. 1D). In addition, the same dose (10-6 M) that stimulated a 3-fold increase of miRNA23b expression in N2A-MOR cells did not induce any significant change in N2A cells over the course of a 24-h treatment period (Fig. 1E), confirming the role of MOR receptor in the morphine-induced up-regulation of miRNA23b.
Morphine Inhibits the Association of MOR1 mRNA with Polysomes through an Interaction between miRNA23b and the MOR1 3′-UTR. We reported previously that miRNA23b interacts with a K box motif in the MOR1 3′-UTR and suppresses the translation of MOR1 mRNA (Wu et al., 2008). Because morphine enhances miRNA23b expression in N2A-MOR cells, it is possible that these elevated levels of miRNA23b could repress the polysome association of MOR1 mRNA. In N2A-MOR cells, the pRC/CMV plasmid encoding the HA-tagged MOR1 protein lacks the MOR1 3′-UTR region. It is possible that RNA transcribed from the pRC/CMV plasmid (i.e., HA-MOR1) will not be affected by miRNA23b. In contrast, RNA transcribed from the native MOR DNA (i.e., MOR1) includes the 3′-UTR and should be regulated by miRNA23b via its interaction with the K Box.
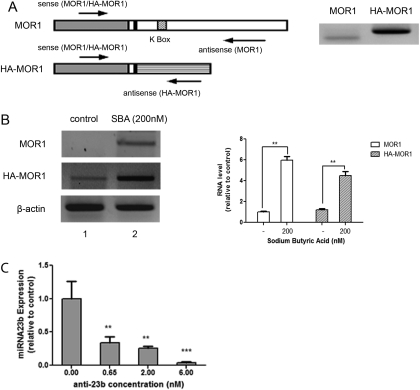
RT-PCR was used to distinguish the HA-MOR1 RNA from MOR1 RNA, by using two different antisense primers specific to pRC/CMV plasmid or the MOR1 3′-UTR, respectively (Fig. 2A). N2A-MOR cells express little native MOR1 RNA (Fig. 2B, lane 1). To detect changes in MOR1 transcript levels, the histone deacetylase inhibitor sodium butyric acid (SBA) was used to stimulate general transcription in N2A-MOR cells. Cells treated with 200 nM SBA for 24 h showed increased levels of both MOR1 and HA-MOR1 RNA by RT-PCR (Fig. 2B, lane 2). SBA treatment had no effect on the morphine-induced changes in miRNA23b expression (data not shown). Cells expressing these transcripts (i.e., HA-MOR1, without the MOR1 3′-UTR and MOR1, with the MOR1 3′-UTR) were used to examine the role of MOR1 3′-UTR in terms of their ability to associate with polysomes.
Fig. 2.
MOR1 and HA-MOR1 RNAs in N2A-MOR cells. A, structure of MOR1 and HA-MOR1 RNAs and the positions of RT-PCR primers. Left, MOR1 cDNA (gray box), MOR1 3′-UTR (large blank box), sequence in pRc/CMV plasmid downstream from the MOR1 cDNA (large striped box), K box (small striped box), and 30-base pair MOR1 3′-UTR that is included in both mRNAs (small blank box). The arrows represent the approximate positions for the sense and antisense RT-PCR primers. Right, N2A-MOR cells were treated with 200 nM SBA for 24 h, 1 μg of RNA was amplified by RT-PCR using MOR1 (left lane) or HA-MOR1 (right lane) primers. B, N2A-MOR cells were treated with 200 nM SBA for 24 h. RNA was extracted and 1 μg of RNA (or 500 ng for β-actin) was analyzed by one-step RT-PCR. Lane 1, control (mock treatment); lane 2, treated with 200 nM SBA. The graph shows the intensity of MOR1 or HA-MOR1 signals normalized to that of β-actin; data are expressed relative to the intensity of the control sample (lane 1). The experiments were repeated three times; the Student's t test was performed by comparing each sample to the control sample. n = 3; **, p < 0.01. C, Anti-23b primer (0, 0.65, 2, or 6 nM) was transfected into N2A-MOR cells and miRNA-enriched RNA was extracted 24 h later. Twenty nanograms of RNA were used for reverse transcription using miRNA23b and snoRNA234 primers, followed by real-time qPCR. The graph shows the miRNA23b levels, normalized to snoRNA234 from a single experiment performed in duplicate. The experiment was repeated three times with similar results. Student's t test was performed by comparing each sample to the control sample (0.00 nM anti-23b). n = 3; **, p < 0.01; ***, p < 0.001.
In NS20Y cells, when endogenous miRNA23b was knocked down using an anti-23b primer, the repression of miRNA23b on MOR1 translation was released, as shown by increased MOR protein levels (Wu et al., 2008). In N2A-MOR cells, anti-23b primer can also effectively knock down endogenous miRNA23b expression (Fig. 2C). miRNA23b inhibits the MOR1 expression mainly by repressing its association with polysomes rather than by inducing RNA degradation (Wu et al., 2008). N2A-MOR cells were treated with 200 nM SBA for 24 h before transfection with anti-23b primer. Total RNA was extracted and analyzed by one-step RT-PCR. Both MOR1 and HA-MOR1 RNA levels remained unchanged by anti-23b primer transfection alone, treatment with 10-6 M morphine alone, or by morphine treatment of anti-23b-transfected cells (Fig. 3A). This is consistent with the previous observation suggesting that miRNA23b does not induce significant RNA degradation (Wu et al., 2008).
Fig. 3.
Morphine inhibits the polysome association of MOR1 mRNA through the interaction between miRNA23b and MOR1 3′-UTR. A, N2A-MOR cells were pretreated with 200 nM SBA for 4 h before transfection with 2 nM anti-miR negative control primer (lanes 1 and 4) or 2 nM anti-23b primer (lanes 2 and 3). At the transfection, experimental cells were treated with 10-6 M morphine (lanes 3 and 4). Twenty-four hours after transfection, RNA was extracted and 1 μg (or 500 ng for β-actin) RNA was analyzed by one-step RT-PCR using primers specific for MOR1, HA-MOR1, and β-actin. The graph shows the intensity of the MOR1 and HA-MOR1 signals normalized to that of β-actin; data are expressed relative to the intensity of control sample (lane 1). Each experiment was repeated three times. Student's t test was performed by comparing each sample to the control sample; n = 3. (B) N2A-MOR cells were prepared as described in A. Twenty-four hours after transfection, polysomal mRNA was extracted and 2 μg of polysomal mRNA (or 500 ng for β-actin) was analyzed by one-step RT-PCR. The legends are the same as in B. Each experiment was repeated three times; n = 3; *, p < 0.05. C, one microgram of polysomal mRNA (or 500 ng for β-actin) was analyzed by real-time qRT-PCR. The graph shows the MOR1 and HA-MOR1 polysomal mRNA levels normalized to that of β-actin; data are expressed relative to the expression of control sample (i.e., w/o anti-23b transfection or morphine treatment). The experiment was repeated three times; Student's t test was performed by comparing each sample with the control; n = 3; *, p < 0.05.
Polysomal mRNA levels are a robust indicator for translation efficiency (del Prete et al., 2007). In N2A-MOR cells, the MOR1 polysomal mRNA levels increased significantly after transfection with anti-23b primer (Fig. 3B, lane 2). Treatment with 10-6 M morphine (i.e., a dose that increases miRNA23b expression) reversed the effect of anti-23b primer (Fig. 3B, lane 3). In contrast, when the 3′-UTR was truncated (e.g., in the HA-MOR1 transcript), neither anti-23b transfection nor morphine treatment affected the mRNA's ability to associate with polysomes (Fig. 3B), confirming that the MOR1 3′-UTR is required for the interaction with miRNA23b. This result was further confirmed by real-time qRT-PCR (Fig. 3C). In summary, long-term morphine treatment increases miRNA23b expression, thereby inhibiting the polysome-mRNA association of MOR1 via interactions with the MOR1 3′-UTR.
Morphine Inhibits the Translational Reporter Activity through MOR1 3′-UTR. pSVUTR and pSVPA plasmids were constructed by inserting the complete MOR1 3′-UTR or only MOR1 poly (A) [400-base pair sequence flanking the poly (A) signal] into a translational luciferase reporter construct, respectively (Wu et al., 2008). N2A-MOR cells were pretreated with morphine (10-8 to 10-5 M) before transfection. In cells transfected with pSVUTR (with the MOR1 3′-UTR), the reporter activity was repressed by morphine in a dose-dependent manner (Fig. 4A); however, no significant change was observed in cells transfected with pSVPA (without the MOR1 3′-UTR) (Fig. 4B). It shows that morphine treatment inhibits the translational reporter expression through the MOR1 3′-UTR.
Fig. 4.
Morphine suppresses reporter activity through MOR1 3′-UTR. A, pSVUTR reporter assay. Luciferase coding region (blank box); MOR1 poly (A) signal (gray box); MOR1 3′-UTR (broken line). N2A-MOR cells were treated with morphine (10-8 to 10-5 M) 3 h before transfected with 500 ng of pSVUTR plasmid; 2 ng of pCMV-Rluc was cotransfected for normalization. The morphine concentration is plotted against the normalized luciferase activity of reporter constructs (firefly luciferase/R. reniformis luciferase) relative to control. The graph shows a single experiment performed in duplicate. The experiment was repeated three times with similar results. Student's t test was performed by comparing each sample to the control. n = 3; *, p < 0.05. B, pSVPA reporter assay. Legends are the same as in A, except for 200 ng of pSVPA used in the transfection. The experiment was repeated three times with similar results. Student's t test was performed by comparing each sample to the control; n = 3.
Discussion
Morphine tolerance after long-term treatment includes a series of profound changes. Three phenomena have been studied extensively: desensitization, which happens when the activated receptor is phosphorylated by a G-protein-receptor-coupled kinase and associates with β-arrestin that uncouples the receptor-G-protein complex; internalization, which refers to the sequestration of the receptor from the membrane to the cytosol via clathrin-coated pits and dynamin; and down-regulation, which is shown by a general decrease in receptor numbers (Binyaminy et al., 2008).
For short-term treatment, it is generally agreed that morphine does not induce receptor down-regulation (Castelli et al., 1997). However, there are equivocal reports as to whether long-term morphine exposure can result in receptor down-regulation. Binding studies of MOR after long-term morphine treatment have shown significant μ-opioid receptor down-regulation in mouse (Yoburn et al., 1993), rat (Bhargava and Gulati, 1990), and cells in culture (Zadina et al., 1993). The decrease of μ-receptor protein quantity was also observed by Western blot in the mouse brainstem after long-term morphine administration (Bernstein and Welch, 1998). In contrast, some studies failed to show any change in MOR binding site numbers after long-term morphine treatment (Hitzemann et al., 1974). Several reports suggested that down-regulation of opioid receptors is readily observed after long-term exposure to high-intrinsic-efficacy agonists (e.g., etorphine), but not after low-intrinsic-efficacy agonists (e.g., morphine) (Duttaroy and Yoburn, 1995; Yabaluri and Medzihradsky, 1997; Whistler et al., 1999; Shen et al., 2000; Zaki et al., 2000).
The down-regulation of MOR can be seen as the sum of increased receptor degradation and decreased receptor synthesis (Afify, 2002). It is generally known that MOR mRNA levels do not change after morphine treatment (Brodsky et al., 1995), indicating no major alteration involved at the transcriptional level. However, whether morphine can affect MOR mRNA at the post-transcriptional level is still not clear.
In the past, we identified and cloned the MOR1 3′-UTR and demonstrated its ability to suppress the translation efficiency of receptor mRNA. A trans-acting factor, miRNA23b interacts with the cis-acting element K box in the MOR1 3′-UTR, inhibiting the association of MOR1 mRNA with polysomes, thereby arresting its translation. In this study, we employed the mouse neuronal N2A-MOR cell line to determine whether through miRNA23b morphine can affect the polysome-mRNA association of MOR1, a critical step in translation control. These cells stably express exogenous MOR encoded from a plasmid containing only MOR1 cDNA (i.e., excluding the major part of the MOR1 3′-UTR). The cell line produces a homogenous population of MOR proteins and imitates the native receptor in response to opioid agonists at the signal transduction level (Chakrabarti et al., 1995).
Morphine induced a dose-dependent increase of miRNA23b in N2A-MOR cells. A prolonged morphine treatment was required for the up-regulation of miRNA23b. This delayed response could result from the time it takes to alter the miRNA maturation pathway. In wild-type N2A cells, which do not express MOR protein, morphine did not change the expression level of miRNA23b, suggesting an indispensable role for MOR. In addition, the up-regulation of miRNA23b by morphine was confirmed in NMB and SHSY-5Y cells, which endogenously express MOR. We didn't detect significant change of miRNA23b in the mouse primary neuronal culture (hippocampus and cortex) (Supplementary Fig. 1). A reason for such discrepancy could be the high heterogeneity of primary cultures. Although these two regions are known to express MOR receptor (Arvidsson et al., 1995; Lin et al., 2004), the primary cultures are inevitably composed of MOR-positive and MOR-negative neurons and also glial cells. In contrast to N2A-MOR cell line that has homogenous population of MOR-expressing cells, the change of miRNA23b in individual neurons could be diluted in the heterogeneous primary cultures.
The expression of miRNA23b seems to be under complicated regulation. An opioid antagonist, naloxone also increases the miRNA23b levels but not in a dose-dependent manner (Supplemental Fig. 2A), and this effect was also seen in the MOR-negative cell line N2A (Supplemental Fig. 2B). The result suggests that naloxone regulates miRNA23b expression but probably through pathways other than that of the MOR receptor. In N2A-MOR cells pretreated with naloxone before adding morphine, the miRNA23b levels were not significantly different from those seen with morphine treatment alone (Supplementary Fig. 2C). This is probably the result of combined effect of naloxone's up-regulating miRNA23b and blocking the MOR receptor.
miRNAs are small noncoding RNAs that participate in the spatiotemporal regulation of mRNA and protein synthesis. Aberrant miRNA expression can lead to developmental abnormalities and diseases, but the stimuli and processes regulating miRNA biogenesis are largely unknown (Davis et al., 2008).
The maturation of miRNAs includes multiple-step processes from the transcription of pri-miRNAs to the association of mature miRNAs with the RISC complex targeting mRNAs (Boyd, 2008). miRNAs can be regulated exquisitely at each step through their maturation cascade by controlling the transcription of pri-miRNAs, altering the processing of pri- and pre-miRNAs, changing the miRNA turnover, or modulating the regulators involved in the biosysthesis of miRNAs, etc. (Ding et al., 2009).
miRNA23b was the first miRNA identified to regulate MOR. It suppresses the polysome-mRNA association of MOR1 through its interaction with a K box in the MOR1 3′-UTR (Wu et al., 2008). An important feature of this trans-acting element is that it responds to morphine treatment and can act as a “messenger” to regulate MOR gene expression at the post-transcriptional level. Morphine increases the miRNA23b level and leads to a decrease of the polysome association of MOR1 mRNA. This effect was observed only in native MOR1 mRNA (i.e., those with the MOR1 3′-UTR), not in HA-MOR1 mRNA (lacking the MOR1 3′-UTR). It confirms that the MOR1 3′-UTR with the K box motif is required for the repression by miRNA23b. In the reporter assay, morphine treatment induced a dose-dependent decrease of luciferase activity in the plasmid with the MOR1 3′-UTR (pSVUTR) but not in the one without MOR1 3′-UTR (pSVPA). Because of technical difficulties, we were unable to show the change of MOR protein level by Western blot. However, using the reporter assay, we confirmed the suppression of morphine on the reporter expression, which requires the MOR1 3′-UTR. Taken together, our data support a post-transcriptional pathway for MOR gene that is induced by morphine through increasing miRNA23b expression.
When studying the regulation of MOR gene, a long-existing dilemma is that the transcription level of MOR does not reflect the agonist activation. The current advance in studying the 3′-UTR of MOR has made it possible to inspect its post-transcriptional regulation, which has the potential to answer this critical question: how does the treatment by a MOR-agonist that induces tolerance regulate the gene expression of the receptor? Our study serves as a preliminary investigation in this purpose. miRNA23b not only inhibits the MOR translation efficiency but also is up-regulated by long-term morphine treatment. We note as well that miRNA23b seems to be under profound regulation; i.e., its level can be affected by agonists such as morphine as well as antagonists such as naloxone. The signaling pathways that regulate miRNAs are starting to unfold. It is of great interest to systemically examine the cellular events that transduce the morphine-activated signal to the synthesis or functioning of miRNAs. On the other hand, it must be noted that the 3′-UTR of MOR1 is absent from all plasmids that encode cloned MOR proteins in stably transfected cell lines. Caution should therefore be taken when studying the regulation of MOR1 that involves its 3′-UTR in these cell lines.
As the major molecular target of opiates, understanding the regulation of MOR, transcriptionally and post-transcriptionally, is important for unraveling the molecular mechanism of tolerance development. However, a direct link between agonist treatment and MOR gene transcription is lacking. From the aspect of post-transcriptional regulation, this report presents evidence for a new pathway that transduces membrane receptor signals to regulate the intracellular MOR expression in which a novel regulator miRNA is involved.
Supplementary Material
Acknowledgments
We would thank Dr. Dezhi Liao (University of Minnesota, Minneapolis, MN) for his technical assistance.
This work was supported by the National Institutes of Health National Institute on Drug Abuse [Grants DA00564, DA01583, DA11806, DA11190, K05-DA00153, K05-DA70554, K02-DA13926] and the F&A Stark Fund of the Minnesota Medical Foundation.
ABBREVIATIONS: MOR, μ-opioid receptor; 3′-UTR, 3′-untranslated region; MOR1, major μ-opioid receptor transcript; RT-PCR, reverse transcription-polymerase chain reaction; qPCR, quantitative polymerase chain reaction; qRT-PCR, quantitative real-time polymerase chain reaction; HA, hemagglutinin; SBA, sodium butyric acid; miRNA, microRNA.
The online version of this article (available at http://molpharm.aspetjournals.org) contains supplemental material.
References
- Afify EA (2002) Turnover of mu-opioid receptors in neuroblastoma cells. Brain Res Mol Brain Res 106 83-87. [DOI] [PubMed] [Google Scholar]
- Ammon-Treiber S and Höllt V (2005) Morphine-induced changes of gene expression in the brain. Addict Biol 10 81-89. [DOI] [PubMed] [Google Scholar]
- Arvidsson U, Riedl M, Chakrabarti S, Lee JH, Nakano AH, Dado RJ, Loh HH, Law PY, Wessendorf MW, and Elde R (1995) Distribution and targeting of a mu-opioid receptor (MOR1) in brain and spinal cord. J Neurosci 15 3328-3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein MA and Welch SP (1998) mu-Opioid receptor down-regulation and cAMP-dependent protein kinase phosphorylation in a mouse model of chronic morphine tolerance. Brain Res Mol Brain Res 55 237-242. [DOI] [PubMed] [Google Scholar]
- Bhargava HN and Gulati A (1990) Down-regulation of brain and spinal cord muopiate receptors in morphine tolerant-dependent rats. Eur J Pharmacol 190 305-311. [DOI] [PubMed] [Google Scholar]
- Binyaminy B, Gafni M, Shapira M, and Sarne Y (2008) Agonist-specific down regulation of mu-opioid receptors: Different cellular pathways are activated by different opioid agonists. Life Sci 82 831-839. [DOI] [PubMed] [Google Scholar]
- Boyd SD (2008) Everything you wanted to know about small RNA but were afraid to ask. Lab Invest 88 569-578. [DOI] [PubMed] [Google Scholar]
- Brodsky M, Elliott K, Hynansky A, and Inturrisi CE (1995) CNS levels of mu opioid receptor (MOR-1) mRNA during chronic treatment with morphine or naltrexone. Brain Res Bull 38 135-141. [DOI] [PubMed] [Google Scholar]
- Castelli MP, Melis M, Mameli M, Fadda P, Diaz G, and Gessa GL (1997) Chronic morphine and naltrexone fail to modify mu-opioid receptor mRNA levels in the rat brain. Brain Res Mol Brain Res 45 149-153. [DOI] [PubMed] [Google Scholar]
- Chakrabarti S, Law PY, and Loh HH (1995) Neuroblastoma Neuro2A cells stably expressing a cloned mu-opioid receptor: a specific cellular model to study acute and chronic effects of morphine. Brain Res Mol Brain Res 30 269-278. [DOI] [PubMed] [Google Scholar]
- Choi HS, Kim CS, Hwang CK, Song KY, Law PY, Wei LN, and Loh HH (2007) Novel function of the poly(C)-binding protein alpha CP3 as a transcriptional repressor of the mu opioid receptor gene. FASEB J 21 3963-3973. [DOI] [PubMed] [Google Scholar]
- Davis BN, Hilyard AC, Lagna G, and Hata A (2008) SMAD proteins control DRO-SHA-mediated microRNA maturation. Nature 454 56-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Prete MJ, Vernal R, Dolznig H, Müllner EW, and Garcia-Sanz JA (2007) Isolation of polysome-bound mRNA from solid tissues amenable for RT-PCR and profiling experiments. RNA 13 414-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding XC, Weiler J, and Großhans H (2009) Regulating the regulators: mechanisms controlling the maturation of microRNAs. Trends Biotechnol 27 27-36. [DOI] [PubMed] [Google Scholar]
- Duttaroy A and Yoburn BC (1995) The effect of intrinsic efficacy on opioid tolerance. Anesthesiology 82 1226-1236. [DOI] [PubMed] [Google Scholar]
- Hitzemann RJ, Hitzemann BA, and Loh HH (1974) Binding of 3H-naloxone in the mouse brain: effect of ions and tolerance development. Life Sci 14 2393-2404. [DOI] [PubMed] [Google Scholar]
- Hwang CK, Kim CS, Choi HS, McKercher SR, and Loh HH (2004) Transcriptional regulation of mouse μ opioid receptor gene by PU. 1. J Biol Chem 279 19764-19774. [DOI] [PubMed] [Google Scholar]
- Im HJ, Smirnov D, Yuhi T, Raghavan S, Olsson JE, Muscat GE, Koopman P, and Loh HH (2001) Transcriptional modulation of mouse mu-opioid receptor distal promoter activity by Sox18. Mol Pharmacol 59 1486-1496. [DOI] [PubMed] [Google Scholar]
- Kieffer BL (1995) Recent advances in molecular recognition and signal transduction of active peptides: receptors for opioid peptides. Cell Mol Neurobiol 15 615-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CS, Choi HS, Hwang CK, Song KY, Lee BK, Law PY, Wei LN, and Loh HH (2006) Evidence of the neuron-restrictive silencer factor (NRSF) interaction with Sp3 and its synergic repression to the mu opioid receptor (MOR) gene. Nucleic Acids Res 34 6392-6403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch T, Kroslak T, Mayer P, Raulf E, and Höllt V (1997) Site mutation in the rat mu-opioid receptor demonstrates the involvement of calcium/calmodulin-dependent protein kinase II in agonist-mediated desensitization. J Neurochem 69 1767-1770. [DOI] [PubMed] [Google Scholar]
- Kramer HK and Simon EJ (1999) Role of protein kinase C (PKC) in agonist-induced mu-opioid receptor down-regulation: II. Activation and involvement of the alpha, epsilon, and zeta isoforms of PKC. J Neurochem 72 594-604. [DOI] [PubMed] [Google Scholar]
- Lin H, Huganir R, and Liao D (2004) Temporal dynamics of NMDA receptor-induced changes in spine morphology and AMPA receptor recruitment to spines. Biochem Biophys Res Commun 316 501-511. [DOI] [PubMed] [Google Scholar]
- Schmidt H, Schulz S, Klutzny M, Koch T, Händel M, and Höllt V (2000) Involvement of mitogen-activated protein kinase in agonist-induced phosphorylation of the mu-opioid receptor in HEK 293 cells. J Neurochem 74 414-422. [DOI] [PubMed] [Google Scholar]
- Shen J, Benedict Gomes A, Gallagher A, Stafford K, and Yoburn BC (2000) Role of cAMP-dependent protein kinase (PKA) in opioid agonist-induced mu-opioid receptor down-regulation and tolerance in mice. Synapse 38 322-327. [DOI] [PubMed] [Google Scholar]
- Song KY, Hwang CK, Kim CS, Choi HS, Law PY, Wei LN, and Loh HH (2007) Translational repression of mouse mu opioid receptor expression via leaky scanning. Nucleic Acids Res 35 1501-1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao PL, Han KF, Wang SD, Lue WM, Elde R, Law PY, and Loh HH (1998) Immunohistochemical evidence of down-regulation of mu-opioid receptor after chronic PL-017 in rats. Eur J Pharmacol 344 137-142. [DOI] [PubMed] [Google Scholar]
- Whistler JL, Chuang HH, Chu P, Jan LY, and von Zastrow M (1999) Functional dissociation of mu opioid receptor signaling and endocytosis: implications for the biology of opiate tolerance and addiction. Neuron 23 737-746. [DOI] [PubMed] [Google Scholar]
- Woods K, Thomson JM, and Hammond SM (2007) Direct regulation of an oncogenic micro-RNA cluster by E2F transcription factors. J Biol Chem 282 2130-2134. [DOI] [PubMed] [Google Scholar]
- Wu EH and Wong YH (2005) Activation of delta-, kappa-, and mu-opioid receptors induces phosphorylation of tuberin in transfected HEK 293 cells and native cells. Biochem Biophys Res Commun 334 838-844. [DOI] [PubMed] [Google Scholar]
- Wu Q, Law PY, Wei LN, and Loh HH (2008) Post-transcriptional regulation of mouse μ-opioid receptor (MOR1) via its 3′ untranslated region: a role for microRNA23b. FASEB J 22 4085-4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabaluri N and Medzihradsky F (1997) Down-regulation of mu-opioid receptor by full but not partial agonists is independent of G protein coupling. Mol Pharmacol 52 896-902. [DOI] [PubMed] [Google Scholar]
- Yamamoto J, Kawamata T, Niiyama Y, Omote K, and Namiki A (2008) Down-regulation of mu opioid receptor expression within distinct subpopulations of dorsal root ganglion neurons in a murine model of bone cancer pain. Neuroscience 151 843-853. [DOI] [PubMed] [Google Scholar]
- Yoburn BC, Billings B, and Duttaroy A (1993) Opioid receptor regulation in mice. J Pharmacol Exp Ther 265 314-320. [PubMed] [Google Scholar]
- Zadina JE, Chang SL, Ge LJ, and Kastin AJ (1993) Mu opiate receptor down-regulation by morphine and up-regulation by naloxone in SH-SY5Y human neuroblastoma cells. J Pharmacol Exp Ther 265 254-262. [PubMed] [Google Scholar]
- Zaki PA, Keith DE Jr, Brine GA, Carroll FI, and Evans CJ (2000) Ligand-induced changes in surface mu-opioid receptor number: relationship to G protein activation? J Pharmacol Exp Ther 292 1127-1134. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.