Abstract
Allostery is essential for controlled catalysis, signal transmission, receptor trafficking, turning genes on and off, and apoptosis. It governs the organism’s response to environmental and metabolic cues, dictating transient partner interactions in the cellular network. Textbooks taught us that allostery is a change of shape at one site on the protein surface brought about by ligand binding to another. For already several years it has been broadly accepted that the change of shape is not induced; rather, it is observed simply because a larger protein population presents it. Current data indicate that while side-chains can reorient and rewire, allostery may not even involve a change of (backbone) shape. Assuming that the enthalpy change does not reverse the free energy change due to the change in entropy, entropy is mainly responsible for binding.
Keywords: residue communication, thermodynamics, entropy, network, rewiring
Introduction
Biological systems are networks. To optimally address functional requirements, avoiding waste yet making available the right components in the needed quantities at any given time necessitates orchestration with appropriate switches. Efficiency mandates regulation which dictates the response of the system. The response is triggered by the presence or absence of certain interactions with other molecules. Intermolecular interactions are physical binding events: between proteins and proteins, proteins and DNA (or, RNA), proteins and small molecules and drugs; they relate to genetic relationships which govern how genes combine leading to the observed phenotypes. Physical interactions control the switches of cellular machines, sensitive to their quantitative yield versus the dynamically changing needs. Allostery is the vehicle translating and transmitting the effects of these physical interactions.
Under given environmental conditions, allostery regulates the increase or decrease in catalytic activities; it controls the transport of proteins and ligands; and it coordinates enzymatic and signaling pathways. The hallmark of allostery has long been that binding at one site affects the conformation of the other1–5. This occurs through an allosteric effector, which may be another protein molecule or any other ligand. The effector interacts with the target protein and via successive making and breaking of (non-covalent or covalent) bonds, the effector eventually leads to a conformational change at the second site. Yet, crucial to the understanding of allostery is that such events do not create new populations of conformations with altered binding site shapes. Instead, allosteric regulation takes place via the re-distribution of the existing protein conformational ensembles. This implies that native protein structures do not consist of a single conformation species; rather, currently there is ample evidence that the native state is a certain distribution of pre-existing conformational substates6; 7 some of which already have altered binding site shapes8–10. The allosteric re-distribution increases the relative population of these substates11. The binding of the allosteric effector can then be viewed as changing the environment of the target protein; and this change is transmitted, leading to a shift in the distributions of the conformational substates. The two binding sites - that of the allosteric ligand and of the substrate, may be nearby or far away on the protein surface.
Wyman’s thermodynamic linkage theory and the allosteric models
Introduced in 1948 by Wyman12; 13, the theory of linked function establishes the fundamental linkage equation for a given macromolecule with multiple binding sites. Derived from thermodynamic principles, the Linkage theory provides the mathematical relationships among measurable data. The linkage theory has shown quantitative predictive power, such as in the well known oxygen Bohr effect where it has correctly predicted the variation in the affinity of oxygen to hemoglobin as a function of the pH value. Coupled with the concept of the allosteric binding potential14, the linkage theory has been applied to express the allosteric transition. However, the linkage equation itself does not explain the mechanism of the allosteric effect; rather, it merely presents the outcome of the inherent inter-dependence between the binding sites.
On the other hand, the two classical allosteric models (MWC15 and KNF16), provide the conceptual mechanism to explain the allosteric effect. Both models describe the allosteric effect as a binding event at one site affecting the activity at another site via a conformational change. While the MWC model emphasizes that the conformational transition is a concerted action between two co-existing, discrete states (R and T), the KNF model formulates it as a sequential, induced conformational change by the binding at the first site which is responsible for the allosteric effect.
The recent landscape shift model indicates that if there is an accompanied conformational change in the allosteric process, it is not an induced conformational change; rather, it is a population shift from one conformation to the other. As in the MWC model, this model states that different allosteric states co-exist; however, they are not necessary under a concerted transition.
Currently, there is increasing evidence illustrating that the allosteric effect - in terms of the binding cooperativity - may not need a conformational change at all. Below we propose that these observations can be understood via the free energy in terms of the thermodynamic enthalpy and entropy, which is totally different from the basic consideration of Wyman’s linkage theory.
Allostery: the dogma, the concept and the expanded view
To date, the prevailing view of allostery tends to focus on structure. Yet, since allostery is fundamentally thermodynamic in nature, communication across the protein could be mediated not only by changes in the mean conformation but also by changes in the dynamic fluctuations about the mean conformation. That is, allosteric communication could involve not solely the enthalpic component, which is the key factor responsible for the observed alteration in the binding site shape, but also has an entropic contribution17–20. Currently, there is mounting evidence indicating the necessity of an update of the prevailing view of allostery: there are now clear data illustrating that allostery need not involve a backbone conformational change. Allosteric signals initiating at one site need not culminate in a change in a target site shape. In particular, on the backbone level, there are striking examples where it has been convincingly demonstrated that allostery may involve mainly an entropic component. This appears to dismiss the central dogma of allostery, stipulating that the effector binds at one site and induces a conformational change in a second site. This dogma had two components: first, that there are two distinct conformations, the R and T states; in the absence of a ligand they exist in a ratio governed by the equilibrium constant L15; and second, that allostery involves a change of shape. Actually, the term allostery comes from allos, “other”, and stereos, which according to Webster’s New Collegiate Dictionary means “solid” or “an object in three dimensions”. Yet, allostery was interpreted as a “different shape”, simply since visually in early allostery-regulated cases a change of shape was observed. In a way, this is reminiscent of the expanded view of enzymes to include RNA, and of “amyloids”, a term derived from the Greek amylon (or amylum in Latin), meaning “starch”. Nonetheless, in this case, despite the well-recognized complete early misconception, the misnomer term amyloid still stuck.
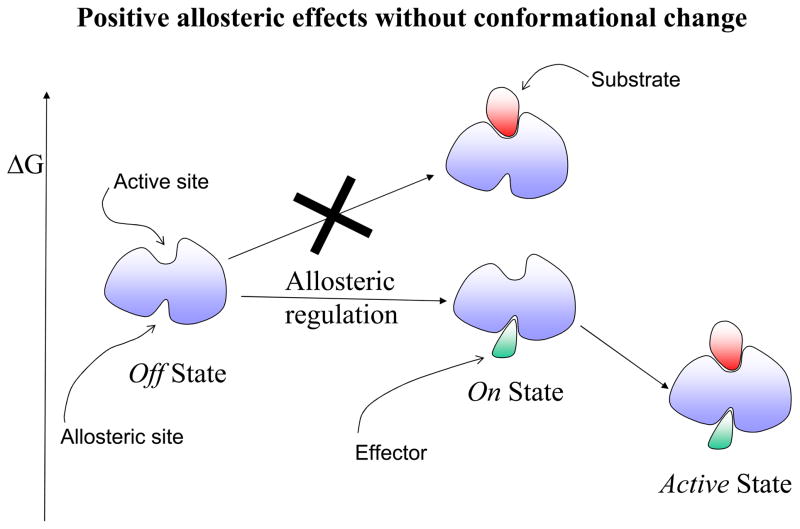
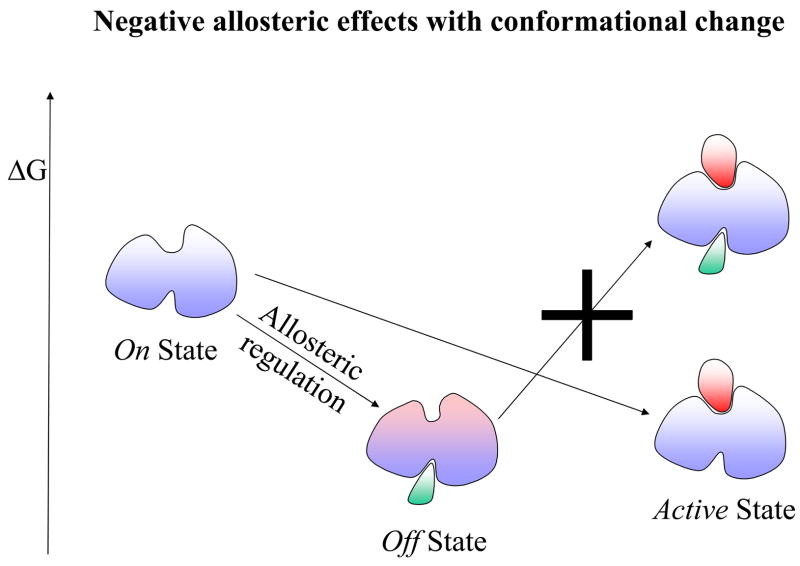
The first step toward a new expanded view of allostery derived from viewing the native state as consisting of an ensemble, and hence allostery as involving a conformational shift of pre-existing conformations. Yet, the accepted outcome was still a visible change in the binding site shape. Now, in terms of the protein backbone, current evidence clearly indicates that there may not even be a conformational change. This emphasizes the pre-existence of conformational substates and leads to a new definition of allostery as purely thermodynamic phenomena. A definition in these terms underlines the fact that visual inspection of the effector-free and effector-bound states may not show any differences; and in particular, that the absence of marked shape changes does not imply that allosteric regulation is not involved. The latter has vast implications in recognizing new allosteric switches and drug targets. Hence, allostery is much broader than envisioned by the Monod, Wyman and Changeux “MWC” model. Figure 1 provides a schematic description of allosteric effects. Here, an allosteric effect is referred to as a change at one site (the allosteric site) affecting the activity at another site (the active site) without (Figure1A) or with (Figure 1B) a backbone conformational change.
Figure 1.
A schematic drawing illustrating the allosteric effect. Here the allosteric effect is referred to as a change at one site (allosteric site) affecting the activity of another site (active site). In this drawing, the activity at both sites is depicted as a ligand binding event: an effector binding to the allosteric site and a substrate binding to the active site. The location of the two binding sites could be adjacent or distal (as shown in the drawing) to each other. The allosteric effect is said to show positive cooperativity if the effector binding increases the affinity for the substrate (favorable binding free energy). Conversely, if the effector binding lowers the substrate affinity, it exhibits negative cooperativity. The change at the allosteric site (an effector binding in the drawing) might, or might not, alter the conformation at the active site. Therefore, there are four combinations of allosteric effects in terms of positive/negative cooperativity versus with/without conformational changes. Here we illustrate only two of these cases in terms of the relative free energy change: positive allosteric regulation without conformational change in Figure 1A; and negative allosteric regulation with conformational change in Figure 1B. In positive allosteric regulation (Figure 1A), the event of the effector binding at the allosteric site (indicated as Allosteric regulation) switches it from an Off regulation state to an On regulation state. Note that the discrete On or Off regulation state in the Figure is highlighted in order to reflect the outcome of the cellular regulation functionality. For a general definition of the allosteric effect, the degree of change is always continuous either increasing or decreasing the affinity at the substrate site. The allosteric binding clearly shows that the previous unfavorable substrate binding for the Off state (indicated by a big cross) becomes favorable for the On state. In negative allosteric regulation (Figure 1B), an effector binding at the allosteric site (also indicated as an Allosteric regulation) switches it from an On regulation state to an Off regulation state. Here, the allosteric binding indicates that the previous favorable substrate binding for the On state becomes unfavorable (indicated by a big cross) for the Off state. The conformational change due to the effector binding is highlighted in pink color around the active site.
A new definition is useful only if it leads to deeper comprehension and in particular has predictive power. The definition of allostery in thermodynamic terms permits dividing allosteric proteins into three types of cases. Type I is governed by entropy; Type II by enthalpy and entropy; and Type III is dominantly governed by enthalpy. Accordingly, in Type I there is no or a subtle backbone structural change; in Type II minor conformational changes are coupled with entropic effects; and in Type III there is relatively large domain or local (the classification “order-disorder” here could lead to confusion by suggesting entropic changes) conformational change opening/closing the active site. Assessment of the backbone conformational change, which may permit such classification, is provided below. The statistics of the side-chain reorientation has been provided by Daily and Gray1. The emergence of accurate and sensitive tools4; 21–23 should lead to an increasing number of proteins illustrating this spectrum. Below, we provide examples and discuss allostery and allosteric pathways in this light. Eventually, a complete physical description of the energy transmission in the protein structure accounting for this behavior to elicit dynamic and conformational changes would assist in a better comprehension of how proteins have evolved to perform their biological functions. Understanding how energy is transduced through a protein to communicate a signal is a major challenge in structural biology24. In enthalpy-driven allostery (Type III) the landscape changes, shifting the locations of the minima corresponding to populations with altered shapes. In contrast, in entropy-driven allostery the free energy landscape also changes; however rather than a shift in the population and thus in the minima, what we observe is a change in the depth of the corresponding well, due to the entropy contribution (Type I). In this paper we largely focus on entropy-dominated allostery. We emphasize that here we address solely backbone level conformational change. We do not treat side-chain conformational changes which could be substantial even in the absence of a backbone conformational change, leading to residue rewiring25; 26.
Allosteric effects are cooperative
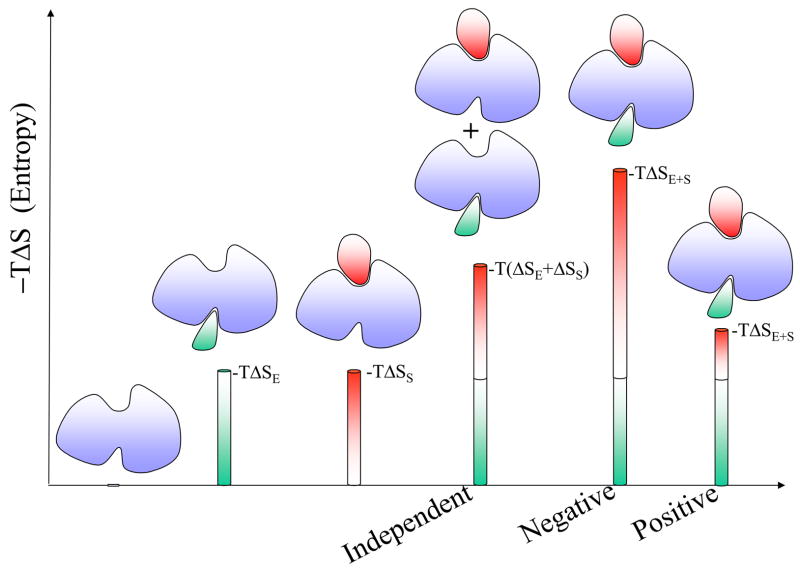
Cooperativity is non-independence. Proteins are widely believed to fold cooperatively. Non-cooperative folding events would lead to exhaustive search of the conformational space to reach the global minimum. Cooperativity, which largely derives from the hydrophobic effect, leads to preferred protein folding pathways. Cooperativity is similarly observed in intermolecular interactions27. To understand cooperativity, we need to think of the system as a cohesive unit, where the behavior of the parts may depend on each other. That is, the overall behavior is the outcome of the properties of the entire system; it is not the sum of the properties of its components. Two terms contribute to the behavior of the system: entropy and enthalpy. In intermolecular binding the entropy term reflects the loss of freedom of motions of the interacting partners including their internal motions; consequently it is unfavorable. The enthalpy term reflects the added interactions; consequently it is favorable. The contributions of the two terms during the interaction process are not independent: as an increasing number of interactions get tighter (i.e. an increasingly favorable enthalpy term) there is an increased loss of degrees of freedom, that is, lower mobility (a more unfavorable entropy term), and vice-versa, loosening of the interactions leads to higher mobility27. A given binding process may present a positive (compensation) or a negative (anti-compensation) correlation between enthalpy and entropy. Allostery involves (at least) two binding processes (Figure 1) presenting positive or negative cooperativity. Positive cooperativity is observed when the binding of an effector at one site increases the affinity for the ligand at another. On the other hand, if the binding of an effector decreases the affinity for the ligand, the protein exhibits negative cooperativity (see Figure 1 legend). The origin of positive cooperativity can be entropic or enthalpic. Positive entropic effects can occur when the combined entropic cost of both binding events is lower than the summation of the two independent events. Such a case was observed in glycopeptide antibiotics28. Hence, positive cooperativity can occur even in the absence of a conformational change (Figure 2). On the other hand, enthalpy can lead to positive cooperativity through a favorable tightening of the interactions, also called structural tightening, partially compensated by less favorable entropy27; 29. For negative cooperativity, the main factor is the loss of configurational entropy. This is the case for the dimeric enzyme CTP:glycerol-3-phosphate cytidylyl transferase, which displays strong negative cooperativity between the binding of the first and second CTP ligands30. Analysis of the binding of agonists and antagonists to G protein-coupled receptors and to ligand gated ion channels receptors coupled with hydrogen/deuterium exchange experiments31 has shown that tightening of the receptor leads to positive cooperativity while loosening leads to negative cooperativity. Interestingly, the agonist and antagonist binding can be driven by different factors. A positive cooperative effect is observed in the binding of Ca2+ ions to calbindin. Ca2+ binding leads to a loss of flexibility in the C-terminal EF-hand, but with a favorable significant conformational reorganization32. Figure 2 explains both positive and negative allosteric effects via a non-additive entropic contribution for the entropy-governed Type I case. Table 1 presents collected thermodynamic data for cooperative allosteric binding. Unfortunately, currently available data are mainly available for Type I. In the cytochrome P450eryF there is a single large binding pocket accommodating the two ligands33. Three of the studied ligands present positive cooperativity (see Table 1).
Figure 2.
A schematic drawing to illustrate both positive and negative allosteric effects via a non-additive entropic contribution. Since entropy involving solvent is excluded in this drawing, a binding event here is assumed to be accompanied by an entropy loss. The higher the bar with an indicated quantity, -TΔS (entropy in terms of free energy), the more unfavorable the relative entropy loss. The green bar -TΔSE reflects the entropy loss when an effector molecule binds to the allosteric site; the red bar -TΔSS represents the entropy loss when a substrate binds to the allosteric protein active site. If there is no allosteric effect at play, each individual binding is considered independent of the other. Hence, the entropy loss is additive as -T(ΔSE+ΔSS). In positive cooperative binding, the entropy loss of the first (effector) binding prepays most of the entropy loss that the second (substrate) binding has to pay. Hence the entropy loss -TΔSE+S is much less than the sum of the entropy loss of the individual binding events. While in negative cooperative binding, the first binding does not pay for the entropy loss due to the second binding and together they incur an extra substantial entropy loss. The resulting -TΔSE+S is much higher than -T(ΔSE+ΔSS).
Table 1.
Thermodynamic data of allosteric effect with cooperative binding
| Allostery Type | Complex | ΔG (kcal/mol) |
ΔH (kcal/mol) |
TΔS (kcal/mol) |
Dominant Thermodynamic Factor | Cooperativity |
|---|---|---|---|---|---|---|
| Cooperative Binding | CAP dimer: cAMP1 (or cAMP2) | −10.3 | −1.8 | 8.5 | Entropic | Negative |
| CAP dimer: cAMP1 + cAMP2 | −7.5 | −2.9 | 4.6 | Entropic | ||
| Cooperative Binding | MetJ: DNA | −10.0 | −2.4 | 7.6 | Entropic | Positive |
| MetJ: SAM + DNA | −11.4 | −23.9 | −12.3 | Enthalpic | ||
| Cooperative Binding | P450eryF: ANF1 | −6.7 | −16.6 | −9.9 | Enthalpic | Positive |
| P450eryF: ANF1 + ANF2 | −12.9 | −9.6 | 3.3 | Entropic | ||
| Cooperative Binding | P450eryF: 1-PB1 | −6.8 | 3.9 | 10.7 | Entropic | Positive |
| P450eryF: 1-PB1 + 1-PB2 | −13.1 | 8.2 | 22.3 | Entropic | ||
| Cooperative Binding | P450eryF: 9-AP1 | −7.2 | −5.3 | 1.9 | Enthalpic | Positive |
| P450eryF: 9-AP1 + 9-AP2 | −13.9 | −11.5 | 2.4 | Enthalpic |
CAP: Catabolite activator protein
cAMP: Cyclic adenosine monophosphate
P450eryF: Bacterial Cytochrome P450
MetJ: Methionine repressor
SAM: S-adenosyl methionine
9-Ap: 9-aminophenanthrene
1-PB: 1-pyrenebutanol
ANF: α-naphthoflavone
Subscript number 1 refers the ligand is at the first binding site and 2 at the second site.
Assessment of the conformational change
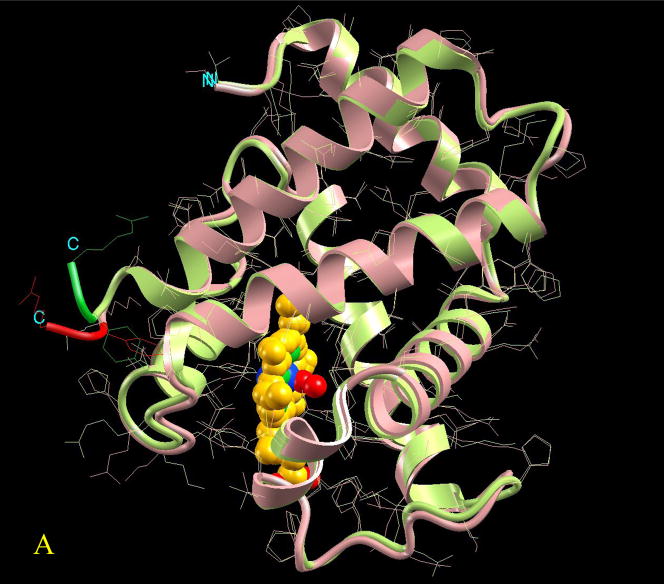
Since allostery is a thermodynamic phenomenon, which does not mandate a change in the (backbone) shape, a definition in thermodynamics terms (entropy and enthalpy) is appropriate. The Type I, II, III definition above provides one such possibility. To obtain some threshold which allows assessment and classification of Type I (no or subtle conformational change; governed largely by entropy) versus Types II and III (involvement of enthalpy to different extents), the allosteric protein benchmark1 is useful. The benchmark provides a systematic analysis of the conformational changes for 51 pairs of known inactive (off state) and active (on state) allosteric protein structures. In this collected data set, all effector molecules are either small molecules or short peptides and most of the substrate molecules are not present at the active sites (see Figure 1A for the definition of off state, on state, effector, and substrate molecules), thus not allowing better measurements of the changes in the second site. Daily and Gray assess the local motions for each residue in the allosteric pairs based on six merits. In spite of the presence of only small effector molecules, the authors conclude that statistically 20% of the residues exhibit substantial conformational changes. Since there is no classification of the overall conformational change for these allosteric pairs, we recalculate the backbone Cα atoms superposition of the two states. The decade old geometric hashing method34 has been utilized to superimpose the allosteric protein pairs with the cut-off distance between two to-be-matched Cα atoms set at 2.0 Å. The final match converges via an iterative procedure similar to that of Daily and Gray. While the conformational change is a continuum, visual classification based on either the absolute number or the percentage of unmatched residues suggests that allosteric pairs can be roughly classified into five categories: domain-movement, minor, disorder→order, subtle, and no-change. At one end of the spectrum, for the domain-movement category, we expect more than 35% or 60 unmatched residues. At the other end, pairs with less than 1% or 3 unmatched residues are classified as no-change. An allosteric pair belongs to the subtle category if the match can not meet the criteria of no-change but has less than 10% or 10 unmatched residues. The rest are classified as minor category with more than 10% but under 35% unmatched residues. The disorder→order category represents a special case. If any allosteric pair lacks a significant part of its PDB coordinate file, which implies possible existence of a disordered fragment(s), this allosteric pair is classified as a disorder→order class. Figure 3 provides one example for each category except the disorder→order class. Type I cases are those classified as subtle, and no-change. The rest are Type II/III. The conformational change for the benchmark pairs (whose structures are available in the PDB) is given at http://protein3d.ncifcrf.gov/tsai/allostery. Here crystal structures have been compared to evaluate the conformational changes. Yet, it behooves us to remember that a single structure may not represent the major population at equilibrium. Crystallization conditions and crystal effects may influence the observed conformation.
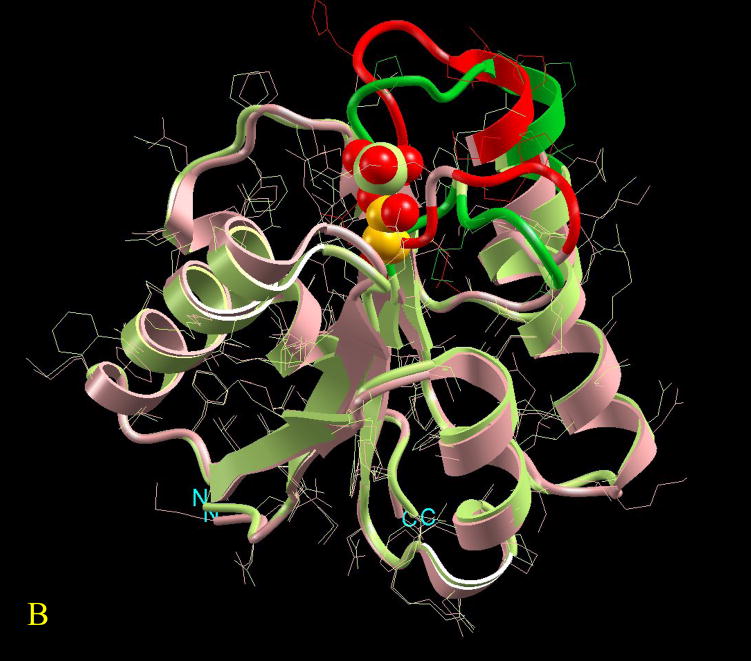
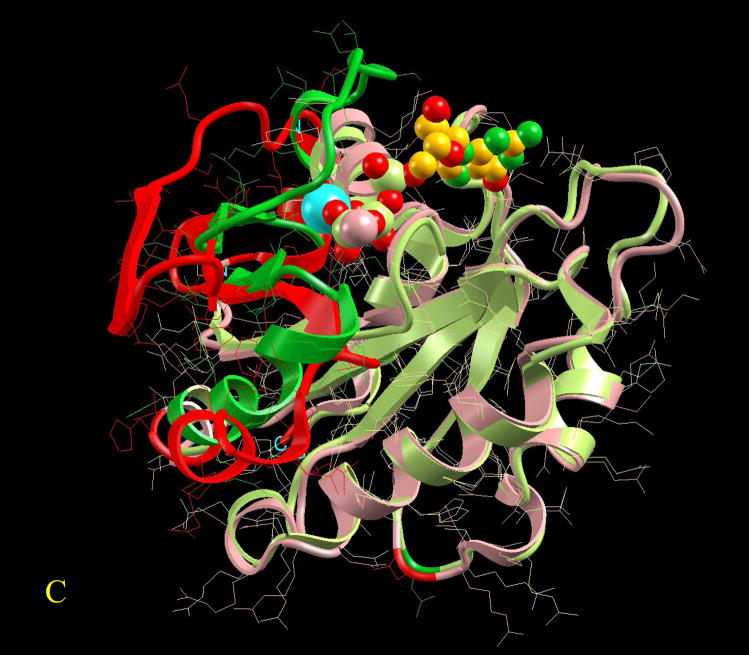
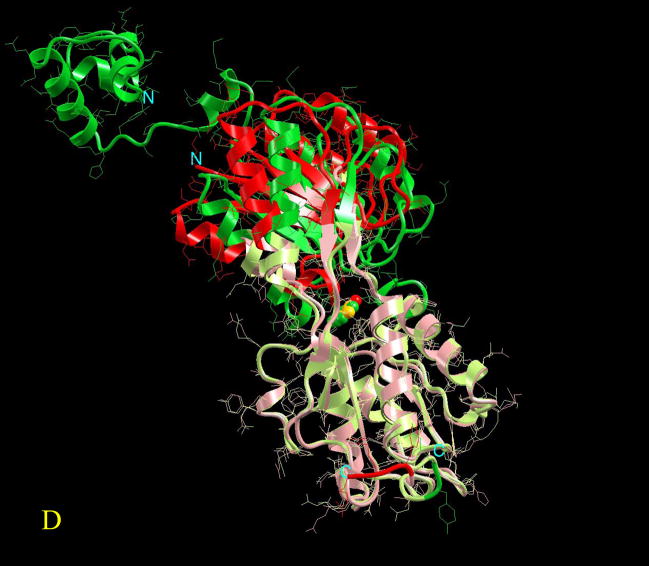
Figure 3.
Visualization of the overall conformational changes in allosteric proteins. Four pairs of known inactive and active allosteric protein structures from the Protein Data Bank are illustrated. In each pair, the protein backbone trace is represented by solid ribbon and the side-chains are drawn as thin lines. The effector molecule binding (or covalent modification) to the active allosteric protein is shown in space-fill (atom color codes are Carbon yellow, Nitrogen green, Oxygen red, Phosphorus light green, Sulfur pink, Magnesium cyan, and Iron blue). The ligand is located at the allosteric site. However, even if present in the PDB file of the inactive protein, it is not shown here for clarity. The superposition is based on matched residues with the distance between superimposed Cα atoms <= 2.0 Å. The scaffold of the matched residues is colored pink and light green, respectively for the inactive and active allosteric proteins. The conformational changes (unmatched residues) are highlighted in red and green, respectively for the inactive and active allosteric proteins. Here four pictures are illustrated as examples of conformational changes in allostery: (A) hemoglobin (PDB codes: 2hhb A and 1hho A); this case has been classified in the no-change category, (B) fixJ (1dbw A; 1d5w A): classified as subtle (C) arf6 (1e0s A; 1hfv A): classified minor, (D) purR (1dbq A; 1wet A): classified domain-movement.
Signals are transmitted through the network when there are, or there aren’t conformational changes
How the signals are transmitted through the network in response to internal and external events is still not completely understood and is the subject of intense research. Nonetheless, it is clear that there is no single chain of events in the ensemble and that multiple paths are involved.
To date, transmission of signals has largely been considered in terms of a conformational change. For such a (Type II or III) enthalpy-driven case, the “classical mechanical” view stipulates that allostery is communicated through a series of discrete conformational changes propagating the energy through the interaction network. Since it is a property of the ensemble, the response to the conformational perturbations can be expected to be transmitted through multiple pathways across the population. The mean structure may not reflect the extent of the structural deviations in distinct conformers of the ensemble. Yet, the energetic coupling between residues (positive or negative) observed for a growing number of cases35–38 (though not all) suggests that as in cellular networks, there are major communication routes. A further reflection of highly populated routes is the occurrence of fold-central residues through which the shortest pathways proceed, corresponding to the experimentally observed functional residues39. Similar to cellular networks, these attest to the robustness of the system, yet to sensitivity of some central (hub-like) residues20; 40–42. Allosteric communications leading to conformational changes are governed by the topology of the protein, the location of the binding sites and the chemical nature of the interacting residues. Large structural changes are more likely to take place if the two binding sites, allosteric and target are far from each other and the signals transmitted over large distances. Structural changes are largely enthalpy-driven. In contrast, entropy (Types I and II) is communicated by changes in the dynamic fluctuations about the mean conformation, mediated solely by changes in protein dynamics. Allostery which is entropy-driven (Type I) (with no backbone conformational change though with side-chain re-orientation, see the reported statistics1) is likely to occur between binding sites which are relatively nearby. However, it was also proposed that it may travel over long distances through “channels” created by coiled coils43 via an altered pattern of the internal dynamics of the protein induced by the binding of a ligand. The case presented by Hawkins and McLeish relates to dynein.
While currently there are examples of one extreme type (Type I), where allostery is dominated by entropy, there are no available data on the other extreme, where there are only structural changes with no entropy involvement. While the literature abounds with allostery-mediated long-range structural changes data, the existence of possible changes in protein dynamics in many of these systems has not been probed.
A case in point where allosteric transition was shown to be driven by enthalpy is the E. coli biotin repressor. This biotin repressor is an allosteric DNA binding protein which is activated by the small molecule bio-5′-AMP. The binding of this small molecule promotes the assembly of the repressor and the biotin biosynthetic operator through the repressor dimerization. Upon adenylate binding the free energy of the dimerization becomes more favorable (by ~ −4 kcal/mol). Activation of dimerization was measured using analytical ultracentrifugation, while isothermal titration calorimetry was used to measure binding of the weak (modest coupling free energy) and strong (large coupling free energy) effectors to the repressor monomer44–46. These results suggested that the activation of repressor dimerization is associated with an enthalpic penalty47.
The role of dynamics in protein allostery
Below, we describe a few examples of Type II. The two-domain protein calbindin D9k mentioned above, a member of the EF-hand family of Ca2+-binding proteins32 was shown to possess an allosteric dynamic entropy effect, accompanying a conformational change48. The dynamic network of communication observed even in the presumably nonallosteric small globular protein eglin C by NMR spin relaxation, residual dipolar couplings, and scalar couplings illustrated local perturbations transmitted as dynamic and structural changes to distal sites as far as 16 Å away24. Both contiguous pathways of enhanced dynamics with no conformational change and noncontingous changes in methyl rotation rates appear to result from backbone deformation. Energy transmission was unidirectional. These observations in a small rigid protein believed to be nonallosteric, lend experimental weight to the proposition that all proteins inherently possess allosteric features5 and illustrate that dynamics may provide mechanistic insight into communication pathways48. In a third example, mutagenesis and NMR relaxation methods were used to investigate the communication networks in the PDZ (second post-synaptic density-95/discs large/zonula occludens-1) domain of the human tyrosine phosphatase 1E protein (hPTP1E). Three mutants with significant changes in binding also displayed dynamic effects. Two of these, one with a mutation at a partially exposed site and another at a buried core position led to only limited side-chain 2H-based dynamic response. On the other hand, a change at another core position (I20F), previously shown to belong to the energetic and dynamic network, resulted in extensive changes in side-chain dynamics reminiscent of those observed upon peptide ligand binding suggesting that position 20 is critical for transmitting changes in dynamics throughout the PDZ domain49. Here too, the dynamic changes occur in the absence of significant conformational changes4. A yet additional example is that of barnase50. Although the 3D structures of the free and bound barnase are very similar, 2H relaxation measurements and 1H, 15N and 13C chemical shifts indicated changes in the dynamics of 11 residues in an extended β-sheet located far from the binding site. Side-chain dynamics and chemical shifts are sensitive to binding-induced variations in the ensemble populations with structural changes which are small enough to escape detection. Simulations revealed relatively rigid domains separated by the extended β-sheet, further suggesting propagation of ligand binding effects across the interfaces between rigid modules via changes in the dynamics. Frederick et al51 have recently used changes in conformational dynamics as representing changes in conformational entropy upon binding illustrating that the apparent change in the conformational entropy is linearly related to the change in the overall binding entropy.
Experimentally, X-ray structure B-factors at the backbone level have been used as indices of fluctuation. However, these parameters can only provide a rough estimate of the fluctuations. NMR relaxation experiments are quite sensitive for detecting motions in the picosecond-nanosecond and microsecond-milliseconds time scales. On the other hand, computationally, molecular dynamics simulations can provide theoretical atomic-level detail on the dynamical behavior of proteins; however, simulations encounter difficulties when dealing with large systems.
Thermodynamic and conformational data indicate that allosteric communications can be mediated solely by changes in motions
E. coli methionine repressor has two intertwined monomers. Conformational data illustrate a Type I case with only a subtle change between the repressor with and without bound co-repressor52; concomitantly, crystal structure B-factors indicate stiffening upon co-repressor binding and NMR suggests a significant decrease in dynamics upon SAM binding. Thermodynamic (Table 1), calorimetric data indicate large compensatory entropic and enthalpic allosteric energies53. Hawkins and McLeish illustrate that the entropic term is too large to be accounted for by the slow modes alone and propose a mechanism (“dynamic enthalpic allostery”) in which high frequency modes may contribute to the allosteric signaling by coupling to the global modes54. Further, experiments carried out by Popovytch et al. presented direct evidence indicating that allostery can be mediated solely by changes in protein dynamics without any conformational change20. This work substantiated the current paradigm in allostery5: there is no “induced fit” and it is not necessary to invoke creation of new conformational states to explain allostery. Rather, there is a population shift toward lesser populated (allosteric) states. In the Popovytch et al Type I case there is no backbone conformational change; nonetheless, allosteric effects are observed. Allosteric interactions - mediated exclusively by changes in protein motions - still cause a free energy landscape change, yet without any “induced fit” mechanism. Popovych et al characterized the negatively cooperative binding of cAMP (Table 1) to the dimeric catabolite activator protein (CAP). They observed that binding of the first cAMP to one subunit of a CAP dimer did not alter the conformation of the other subunit as evidenced in NMR chemical shift data. However, the system dynamics were modulated by the sequential binding process: the first cAMP partially enhanced and the second cAMP completely quenched protein motions. Consequently, the second cAMP binding incurred a pronounced conformational entropy penalty leading to the observed negative cooperative binding of cAMP to CAP.
Entropy is the count of the density of states. A binding event always gains in enthalpy but loses in entropy. In allostery, binding at site A could boost the binding at site B by reducing the entropy loss (by rigidifying binding site B) or promoting enthalpy gain (by a conformational change at binding site B). Alternatively, binding at site A could repress site B binding by partially unfolding site B, leading to a larger entropy loss (see also Figure 2). In the case of cAMP binding to CAP, the first binding is favorable since the enthalpy gain can compensate for the entropy loss. However, the second binding is unfavorable since the entropy loss is much larger than the enthalpy gain (Table 1).
These examples highlight the proposition of Cooper and Dryden over 20 years ago48; 55. By separating the motions into vibrational (normal mode) and conformational effects, they illustrated the feasibility of an extreme case of no enthalpic term in the allosteric free energy: they showed that even in the case of total absence of a conformational change, ligand-induced changes in protein dynamics could produce free energies sufficient for an allosteric communication between distinct binding sites. The effect derives from the possible changes in frequencies and amplitudes of macromolecular thermal fluctuations in response to ligand binding, and can involve dynamic behavior ranging from highly correlated low-frequency normal mode vibrations to random local anharmonic motions of individual atoms or groups. Dynamic allostery of this form is primarily an entropy effect.
Conclusions
There is a growing interest in allostery. By now it is well established that allostery is a natural property of all (non-fibrous) proteins; that even if the protein is not known to be allosteric, under given conditions such as the presence of appropriate allosteric effectors, or the presence of mutations, the proteins will be observed to be allosteric. Further, an increasing number of proteins not known to be allosteric have been shown to be allosteric. Allosteric effects can be the outcome of changes in ligand concentration, pH, ionic strength, crystallization buffer, mutations or the binding of another protein, peptide, or small molecule ligand, DNA or RNA, and post-translational modifications, such as phosphorylation or glycosylation. Evolution has made use of this property: allostery plays an indispensable role in all processes in the living cell.
So what is allostery? The central dogma of allostery stipulated that the effector binds at one site and thereby induces a conformational change in a second site. Current view recognizes that the effector does not induce a conformational change, and that allostery involves a conformational shift of pre-existing conformations. Nonetheless, the broadly accepted outcome was still a visible change in the binding site shape. However, accumulating solid evidence clearly indicates that on the backbone level, there may not even be a conformational change. Thus, the hallmark of allostery that an observable visual change in the overall shape provides proof that an allosteric mechanism is at play holds; yet, allostery can be at play even in the absence of a change in shape. Allostery is a thermodynamic phenomenon: it may involve enthalpic, enthalpic and entropic, or solely entropic effects. This validates the view that allostery does not dictate the creation of new conformational species; rather, it leads to a change in their relative concentrations. So what is allostery? It is not a change of shape; we refer to it as a change at one site (allosteric site) affecting the activity at another. Further, if the enthalpy change does not reverse the free energy change due to the change in entropy, entropy may be the factor responsible for the ligand binding. This may particularly hold if the binding sites are close to each other.
Acknowledgments
This project has been funded in whole or in part with Federal funds from the National Cancer Institute, National Institutes of Health, under contract number NO1-CO-12400. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. This research was supported (in part) by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Daily MD, Gray JJ. Local motions in a benchmark of allosteric proteins. Proteins Struct Funct Genet. 2007;67:385–399. doi: 10.1002/prot.21300. [DOI] [PubMed] [Google Scholar]
- 2.Wilson CJ, Zhan H, Swint-Kruse L, Matthews KS. The lactose repressor system: paradigms for regulation, allosteric behavior and protein folding. Cell Mol Life Sci. 2007;64:3–16. doi: 10.1007/s00018-006-6296-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lindsley JE, Rutter J. Whence cometh the allosterome? Proc Natl Acad Sci USA. 2006;103:10533–10535. doi: 10.1073/pnas.0604452103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Swain JF, Gierasch LM. The changing landscape of protein allostery. Curr Opin Struct Biol. 2006;16:102–108. doi: 10.1016/j.sbi.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 5.Gunasekaran K, Ma BY, Nussinov R. Is allostery an intrinsic property of all dynamic proteins? Proteins Struct Funct Genet. 2004;57:433–443. doi: 10.1002/prot.20232. [DOI] [PubMed] [Google Scholar]
- 6.Miyashita O, Onuchic JN, Wolynes PG. Nonlinear elasticity, proteinquakes, and the energy landscapes of functional transitions in proteins. Proc Natl Acad Sci USA. 2003;100:12570–12575. doi: 10.1073/pnas.2135471100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miyashita O, Wolynes PG, Onuchic JN. Simple energy landscape model for the kinetics of functional transitions in proteins. J Phys Chem B. 2005;109:1959–1969. doi: 10.1021/jp046736q. [DOI] [PubMed] [Google Scholar]
- 8.Ma BY, Shatsky M, Wolfson HJ, Nussinov R. Multiple diverse ligands binding at a single protein site: A matter of pre-existing populations. Protein Sci. 2002;11:184–197. doi: 10.1110/ps.21302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma BY, Kumar S, Tsai CJ, Nussinov R. Folding funnels and binding mechanisms. Protein Eng. 1999;12:713–720. doi: 10.1093/protein/12.9.713. [DOI] [PubMed] [Google Scholar]
- 10.Kumar S, Ma BY, Tsai CJ, Sinha N, Nussinov R. Folding and binding cascades: Dynamic landscapes and population shifts. Protein Sci. 2000;9:10–19. doi: 10.1110/ps.9.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fetler L, Kantrowitz ER, Vachette P. Direct observation in solution of a preexisting structural equilibrium for a mutant of the allosteric aspartate transcarbamoylase. Proc Natl Acad Sci USA. 2007;104:495–500. doi: 10.1073/pnas.0607641104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wyman J. HEME PROTEINS. Adv Protein Chem. 1948;4:407–531. doi: 10.1016/s0065-3233(08)60011-x. [DOI] [PubMed] [Google Scholar]
- 13.Wyman J. LINKED FUNCTIONS AND RECIPROCAL EFFECTS IN HEMOGLOBIN - A 2ND LOOK. Adv Protein Chem. 1964;19:223–286. doi: 10.1016/s0065-3233(08)60190-4. [DOI] [PubMed] [Google Scholar]
- 14.Wyman J. ALLOSTERIC LINKAGE. J Am Chem Soc. 1967;89:2202. [Google Scholar]
- 15.Monod J, Wyman J, Changeux JP. On Nature of Allosteric Transitions - a Plausible Model. J Mol Biol. 1965;2:88. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- 16.Koshland DE, Nemethy G, Filmer D. Comparison of Experimental Binding Data and Theoretical Models in Proteins Containing Subunits. Biochemistry. 1966;5:365. doi: 10.1021/bi00865a047. [DOI] [PubMed] [Google Scholar]
- 17.Wand AJ. Dynamic activation of protein function: A view emerging from NMR spectroscopy. Nat Struct Biol. 2001;8:926–931. doi: 10.1038/nsb1101-926. [DOI] [PubMed] [Google Scholar]
- 18.Hawkins RJ, McLeish TCB. Coarse-grained model of entropic allostery. Phys Rev Lett. 2004;93 doi: 10.1103/PhysRevLett.93.098104. [DOI] [PubMed] [Google Scholar]
- 19.Homans SW. Probing the binding entropy of ligand-protein interactions by NMR. Chembiochem. 2005;6:1585. doi: 10.1002/cbic.200500010. [DOI] [PubMed] [Google Scholar]
- 20.Popovych N, Sun SJ, Ebright RH, Kalodimos CG. Dynamically driven protein allostery. Nat Struct Mol Biol. 2006;13:831–838. doi: 10.1038/nsmb1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Das R, Abu-Abed M, Melacini G. Mapping allostery through equilibrium perturbation NMR spectroscopy. J Am Chem Soc. 2006;128:8406–8407. doi: 10.1021/ja060046d. [DOI] [PubMed] [Google Scholar]
- 22.Korzhnev DM, Orekhov VY, Kay LE. Off-resonance R1(p) NMR studies of exchange dynamics in proteins with low spin-lock fields: An application to a fyn SH3 domain. J Am Chem Soc. 2005;127:713–721. doi: 10.1021/ja0446855. [DOI] [PubMed] [Google Scholar]
- 23.Korzhnev DM, Salvatella X, Vendruscolo M, Di Nardo AA, Davidson AR, Dobson CM, Kay LE. Low-populated folding intermediates of Fyn SH3 characterized by relaxation dispersion NMR. Nature. 2004;430:586–590. doi: 10.1038/nature02655. [DOI] [PubMed] [Google Scholar]
- 24.Clarkson MW, Gilmore SA, Edgell MH, Lee AL. Dynamic coupling and allosteric behavior in a nonallosteric protein. Biochemistry. 2006;45:7693–7699. doi: 10.1021/bi060652l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sathyapriya R, Vishveshwara S. Structure networks of E-coli glutaminyl-tRNA synthetase: Effects of ligand binding. Proteins Struct Funct Genet. 2007;68:541–550. doi: 10.1002/prot.21401. [DOI] [PubMed] [Google Scholar]
- 26.Daily MD, Upadhyaya TJ, Gray JJ. Contact map rearrangements reveal allosteric pathways through protein structure. Biophys J. 2007;19A [Google Scholar]
- 27.Camara-Campos A, Hunter CA, Tomas S. Cooperativity in the self-assembly of porphyrin ladders. Proc Natl Acad Sci USA. 2006;103:3034–3038. doi: 10.1073/pnas.0508071103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jusuf S, Loll PJ, Axelsen PH. Configurational entropy and cooperativity between ligand binding and dimerization in glycopeptide antibiotics. J Am Chem Soc. 2003;125:3988–3994. doi: 10.1021/ja027780r. [DOI] [PubMed] [Google Scholar]
- 29.Calderone CT, Williams DH. An enthalpic component in cooperativity: The relationship between enthalpy, entropy, and noncovalent structure in weak associations. J Am Chem Soc. 2001;123:6262–6267. doi: 10.1021/ja003016y. [DOI] [PubMed] [Google Scholar]
- 30.Stevens SY, Sanker S, Kent C, Zuiderweg ERP. Delineation of the allosteric mechanism of a cytidylyltransferase exhibiting negative cooperativity. Nat Struct Biol. 2001;8:947–952. doi: 10.1038/nsb1101-947. [DOI] [PubMed] [Google Scholar]
- 31.Williams DH, O’Brien DP, Sandercock AM, Stephens E. Order changes within receptor systems upon ligand binding: Receptor tightening/oligomerisation and the interpretation of binding parameters. J Mol Biol. 2004;340:373–383. doi: 10.1016/j.jmb.2004.04.056. [DOI] [PubMed] [Google Scholar]
- 32.Mäler L, Blankenship J, Rance M, Chazin WJ. Site-site communication in the EF-hand Ca2+-binding protein calbindin D9k. Nat Struct Biol. 2000;7:245–250. doi: 10.1038/73369. [DOI] [PubMed] [Google Scholar]
- 33.Muralidhara BK, Negi SS, Halpert JR. Dissecting the thermodynamics and cooperativity of ligand binding in cytochrome P450eryF. J Am Chem Soc. 2007;129:2015–2024. doi: 10.1021/ja066303w. [DOI] [PubMed] [Google Scholar]
- 34.Tsai CJ, Lin SL, Wolfson HJ, Nussinov R. A dataset of protein-protein interfaces generated with a sequence-order-independent comparison technique. J Mol Biol. 1996;260:604–620. doi: 10.1006/jmbi.1996.0424. [DOI] [PubMed] [Google Scholar]
- 35.Rod TH, Radkiewicz JL, Brooks CL. Correlated motion and the effect of distal mutations in dihydrofolate reductase. Proc Natl Acad Sci USA. 2003;100:6980–6985. doi: 10.1073/pnas.1230801100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rajagopalan PTR, Lutz S, Benkovic SJ. Coupling interactions of distal residues enhance dihydrofolate reductase catalysis: Mutational effects on hydride transfer rates. Biochemistry. 2002;41:12618–12628. doi: 10.1021/bi026369d. [DOI] [PubMed] [Google Scholar]
- 37.Agarwal PK, Billeter SR, Rajagopalan PTR, Benkovic SJ, Hammes-Schiffer S. Network of coupled promoting motions in enzyme catalysis. Proc Natl Acad Sci USA. 2002;99:2794–2799. doi: 10.1073/pnas.052005999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sawaya MR, Kraut J. Loop and subdomain movements in the mechanism of Escherichia coli dihydrofolate reductase: Crystallographic evidence. Biochemistry. 1997;36:586–603. doi: 10.1021/bi962337c. [DOI] [PubMed] [Google Scholar]
- 39.del Sol A, Fujihashi H, Amoros D, Nussinov R. Residues crucial for maintaining short paths in network communication mediate signaling in proteins. Mol Syst Biol. 2006 doi: 10.1038/msb4100063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Changeux JP, Edelstein SJ. Allosteric mechanisms of signal transduction. Science. 2005;308:1424–1428. doi: 10.1126/science.1108595. [DOI] [PubMed] [Google Scholar]
- 41.Bray D, Duke T. Conformational spread: The propagation of allosteric states in large multiprotein complexes. Annu Rev Biophys Biomol Struct. 2004;33:53–73. doi: 10.1146/annurev.biophys.33.110502.132703. [DOI] [PubMed] [Google Scholar]
- 42.Koshland DE. Conformational changes: How small is big enough? Nat Med. 1998;4:1112–1114. doi: 10.1038/2605. [DOI] [PubMed] [Google Scholar]
- 43.Hawkins RJ, McLeish TCB. Dynamic allostery of protein alpha helical coiled-coils. J R Soc Interf. 2006;3:125–138. doi: 10.1098/rsif.2005.0068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wilson KP, Shewchuk LM, Brennan RG, Otsuka AJ, Matthews BW. Escherichia-Coli Biotin Holoenzyme Synthetase Biorepressor Crystal-Structure Delineates the Biotin-Binding and DNA-Binding Domains. Proc Natl Acad Sci USA. 1992;89:9257–9261. doi: 10.1073/pnas.89.19.9257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weaver LH, Kwon K, Beckett D, Matthews BW. Corepressor-induced organization and assembly of the biotin repressor: A model for allosteric activation of a transcriptional regulator. Proc Natl Acad Sci USA. 2001;98:6045–6050. doi: 10.1073/pnas.111128198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wood ZA, Weaverl LH, Brown PH, Beckett D, Matthews BW. Co-repressor induced order and biotin repressor dimerization: A case for divergent followed by convergent evolution. J Mol Biol. 2006;357:509–523. doi: 10.1016/j.jmb.2005.12.066. [DOI] [PubMed] [Google Scholar]
- 47.Brown PH, Beckett D. Use of binding enthalpy to drive an allosteric transition. Biochemistry. 2005;44:3112–3121. doi: 10.1021/bi047792k. [DOI] [PubMed] [Google Scholar]
- 48.Kern D, Zuiderweg ERP. The role of dynamics in allosteric regulation. Curr Opin Struct Biol. 2003;13:748–757. doi: 10.1016/j.sbi.2003.10.008. [DOI] [PubMed] [Google Scholar]
- 49.Fuentes EJ, Gilmore SA, Mauldin RV, Lee AL. Evaluation of energetic and dynamic coupling networks in a PDZ domain protein. J Mol Biol. 2006;364:337–351. doi: 10.1016/j.jmb.2006.08.076. [DOI] [PubMed] [Google Scholar]
- 50.Zhuravleva A, Korzhnev DM, Nolde SB, Kay LE, Arseniev AS, Billeter M, Orekhov VY. Propagation of dynamic changes in barnase upon binding of barstar: An NMR and computational study. J Mol Biol. 2007;367:1079–1092. doi: 10.1016/j.jmb.2007.01.051. [DOI] [PubMed] [Google Scholar]
- 51.Frederick KK, Marlow MS, Valentine KG, Wand AJ. Conformational entropy in molecular recognition by proteins. Nature. 2007;448:325–U3. doi: 10.1038/nature05959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rafferty JB, Somers WS, Stgirons I, Phillips SEV. 3-Dimensional Crystal-Structures of Escherichia-Coli Met Repressor with and without Corepressor. Nature. 1989;341:705–710. doi: 10.1038/341705a0. [DOI] [PubMed] [Google Scholar]
- 53.Cooper A, McAlpine A, Stockley PG. Calorimetric Studies of the Energetics of Protein-DNA Interactions in the Escherichia-Coli Methionine Repressor (Metj) System. FEBS Lett. 1994;348:41–45. doi: 10.1016/0014-5793(94)00579-6. [DOI] [PubMed] [Google Scholar]
- 54.Hawkins RJ, McLeish TCB. Coupling of global and local vibrational modes in dynamic allostery of proteins. Biophys J. 2006;91:2055–2062. doi: 10.1529/biophysj.106.082180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cooper A, Dryden DTF. Allostery without Conformational Change - a Plausible Model. Eur Biophys J Biophys Lett. 1984;11:103–109. doi: 10.1007/BF00276625. [DOI] [PubMed] [Google Scholar]