Abstract
Biological membranes are two-dimensional mixtures of an enormous number of different components. Modeling cell membranes as simple bilayer mixtures reveals rich phase behavior, but how can we use the observed phase behavior to understand the real membranes?
A picture of a biological membrane that includes compositional and phase heterogeneity is consistent with many observations on cells. Simons and van Meer pointed this out to explain the sorting of proteins to their distinctive membrane locations1. Simons, in particular, advocated the idea that a special type of nonrandom mixing of the membrane lipids is involved in the sorting process—namely, that lipid domains in the plane of the membrane termed ‘rafts’ have high affinity for some proteins and low affinity for others, thereby contributing to sorting2.
The so-called membrane rafts might also come into play during signaling cascades, if the membrane phase behavior contributes to the activation and inactivation of membrane-bound enzymatic activities3. Other events at cell membranes might involve membrane lipid phase behaviors in a decidedly direct manner in viral entry and exit via preferred phases4, and bilayer fusions of exocytosis and fissions of endocytosis at particular phase domains or phase boundaries5.
Researchers want to know whether membrane composition directly influences how proteins cluster, whether certain proteins are excluded from regions of the membrane, how the membrane can influence enzymatic activities, and how membrane domains could be connected to the cell’s network of protein filaments. Yet, to a chemist’s way of thinking, it seems premature to try to think precisely about the great many components in the cell membrane in terms of clusters of lipids and proteins or even separated phases, without a firm grasp on the fundamental physical chemistry of lipid and protein mixing in a bilayer. We do not yet have this level of understanding. We do have phase data for some simple, relevant lipid mixtures. These data might imply a path toward a solution to the problem of this enormous complexity, as well as a molecular-level description of the nonrandom nature of the mixtures. Perhaps the thermodynamic and statistical ways of treating mixtures might be put to good use, but the issue of the cell membrane’s enormous complexity should be handled explicitly.
A problem with complex mixtures
Immediately, we run into a problem: biological membranes contain hundreds of chemically distinct lipid components that form the actual bilayer membrane. The most straightforward enumeration of each chemical species reveals several hundred amphiphilic lipid molecules, differing, for example, in hydrocarbon chain length and unsaturation and in the structure of the polar or charged headgroup6. As for the many proteins, their role in membrane phase behavior is even more difficult to assess. First, their numbers are enormous, with roughly a third of the human genome encoding proteins that are membrane associated—approximately 10,000 different molecules! Second, for perhaps 99% of these proteins we know very little about how they interact with their membrane surroundings. For most we do not even know what fraction of their amino acid residues are integral to the membrane as opposed to being quite external to the clustering and phase behaviors of the membrane components.
A treatment of the physical chemistry of mixing might involve as input data the phase boundaries of a mixture over all of composition space. Given the effort required to solve a three-component phase diagram, is there a practical alternative to measuring phase data for hundreds of multicomponent mixtures? One way might be to make use of the tricks that geological chemists have successfully used for many years to describe the phase behaviors of their complex mixtures, namely rocks such as basalts and granites: ignore some components and group together others7. In this way, geologists have effectively treated some of their favorite rocks as pseudo-three-component mixtures.
A problem that is as yet unsolved is how to sensibly group the lipid components. Perhaps the lipids that tend to form a fluid bilayer can be collected, and those that tend to form a solid bilayer can be collected, and the negatively charged lipids can be collected together as well. Experimental exploration of sensible grouping has not yet been done. Only a single major membrane component, cholesterol, is itself a distinct chemical species and so requires no grouping. However, even in this case, other membrane components might behave similarly to cholesterol and might profitably be grouped together with it.
A key problem in membrane research is simply the lack of reliable chemical analyses. The necessary lipid composition data have proven difficult to obtain for most real biological membranes. There are two main reasons for this. (i) With rare exceptions, animal cells contain a variety of different types of membranes: for example, the plasma membrane surrounding the cell, the nuclear membranes and the membranes around each of the organelles—lysosomes, peroxysomes, inner and outer mitochondrial membranes, endoplasmic reticulum, lipid bodies, Golgi. To perform lipid compositional analysis, one must physically separate the membrane of interest from all of the other membranes. This separation typically involves steps of centrifugation, chromatography or both, and invariably results in preparations that are contaminated with membranes other than the one of interest; most such membrane preparations are therefore more properly described as ‘enrichments’ rather than purifications. (ii) Each leaflet of the cell’s bilayer membranes has a different lipid composition and thus different phase behavior. Yet the separate determination of lipid compositions for each half of a bilayer is notoriously difficult. In addition, the methods used to purify biological membranes, as well as the methods used for their actual chemical analysis, cause some scrambling of the naturally separated inner- and outer-leaflet lipids. In part because of this difficulty, few research groups have undertaken to develop suitable analytical methods. The methodology has changed little over the past three decades and still involves the use of relatively non-penetrating reactive chemicals or phospholipases or lipid exchange proteins8.
Phase diagram for a model mixture
One possible starting place is to choose a biological membrane for which good compositional data are available, and attempt to model that membrane. The cholesterol-rich outer leaflet of animal cell plasma membranes meets this criterion, and in addition is especially influential in the life of a cell. The type of cell for which we have the very best data is the mature human red blood cell9. Even with a limitation to chemically simple models for the outer leaflet of plasma membranes, we find rich phase behavior. Veatch and co-workers have determined phase behavior for the three-component mixture 16:0,16:0-PC/18:1,18:1-PC/cholesterol (DPPC/DOPC/cholesterol), including examination of the temperature dependence10. We have examined a related mixture (Fig. 1), 18:0,18:0-PC/18:1,18:1-PC/cholesterol (DSPC/DOPC/cholesterol). This mixture was chosen, in part, to have a high degree of immiscibility of the three components, so as to clearly reveal the characteristics of the nonideal mixing. Mixtures of this sort—of cholesterol together with one phospholipid having a relatively high melting temperature and another phospholipid having a relatively low melting temperature—are widely viewed as useful models for the outer leaflet of animal-cell plasma membranes, as they contain a representative compound from each of the three major lipid groups.
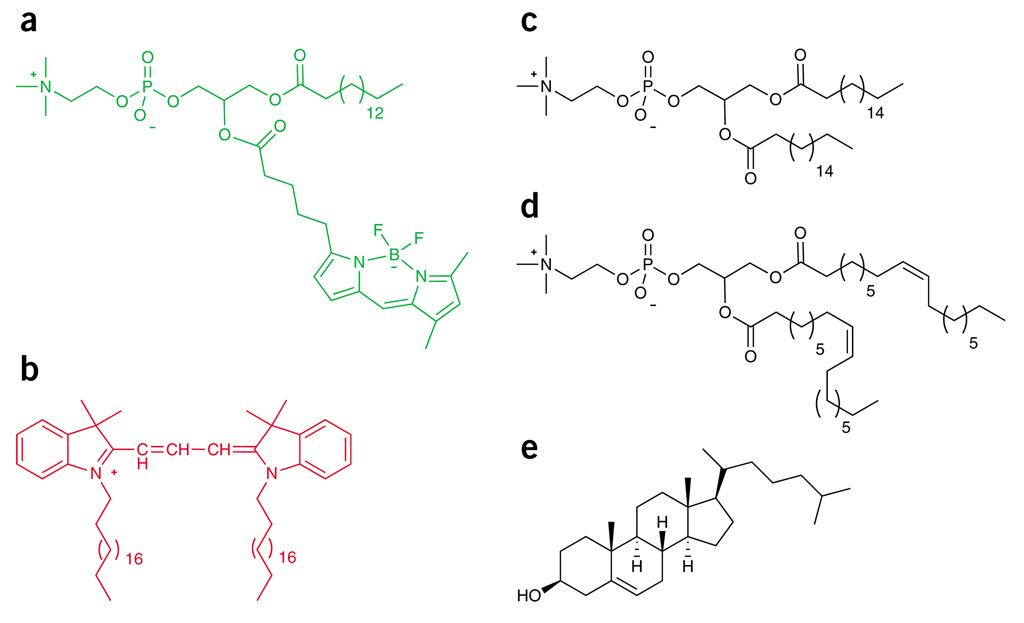
Figure 1.
Structures of probes and lipid components used for phase studies. (a) Bodipy-PC. (b) C20:0-DiI. (c) DSPC. (d) DOPC. (e) Cholesterol.
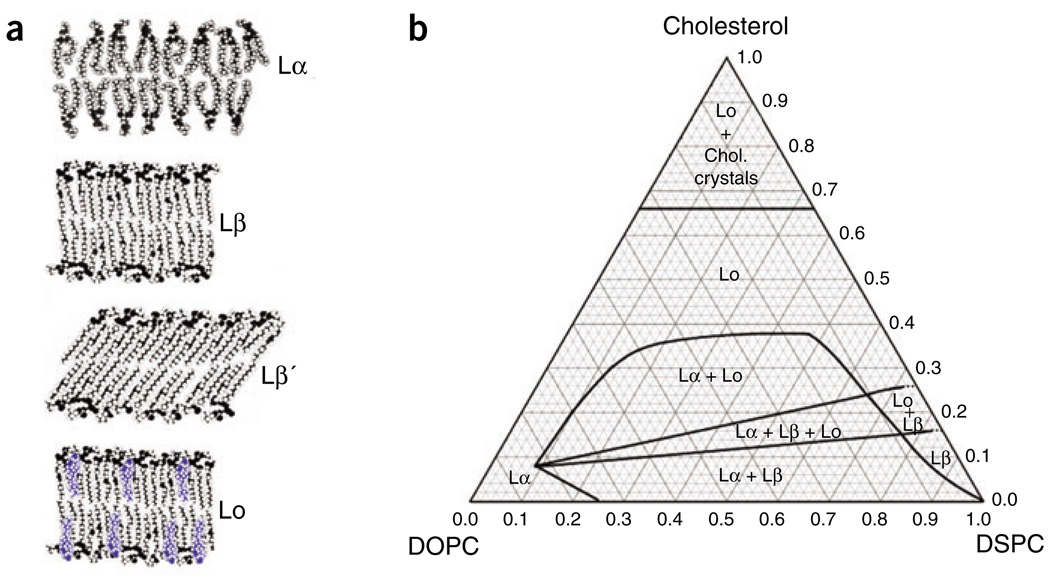
The phase behaviors of this simple model membrane are remarkable (Fig. 2). Near the bottom of the triangular phase diagram, at low cholesterol concentrations, Lβ′ domains of two-dimensional solid having tilted acyl chains are in equilibrium with Lα domains of two-dimensional disordered fluid. When cholesterol concentration increases, the tilted-chain solid domains first undergo a change to become the untilted but still solid Lβ phase. With a further increase in cholesterol concentration, this Lβ phase becomes saturated with cholesterol, and a new phase separates, the cholesterol-rich Lo phase. A region of three-phase coexistence results: Lα + Lβ + Lo. The Lo phase has an interesting combination of properties: a high degree of order, typical of a crystal, together with a high rate of translational diffusion, typical of a liquid. This unusual combination seems to arise less from the affinity of lipids for their neighbors and more from the requirement that cholesterol be shielded from water by neighboring phospholipid headgroups11.
Figure 2.
Bilayer mixtures of DSPC/DOPC/cholesterol at 23 °C show rich phase behavior. (a) Cartoons represent the four phase types found for this mixture. Lα is fluid and disordered; Lβ is rigid and ordered; Lβ is rigid and ordered, having tilted acyl chains; Lo is fluid and ordered. (b) The phase diagram shows four regions of two-phase coexistence, {Lα + Lβ}, {Lo + Lβ}, {Lα + Lo} and {Lo + crystals of cholesterol monohydrate}. There is one region of three-phase coexistence, {Lα + Lβ + Lo}.
When cholesterol concentration is further increased, the solid phase completely disappears, leaving the two coexisting phases Lα + Lo. The coexistence of these two different bilayer liquids is readily made visible by fluorescence microscopy (Fig. 3).
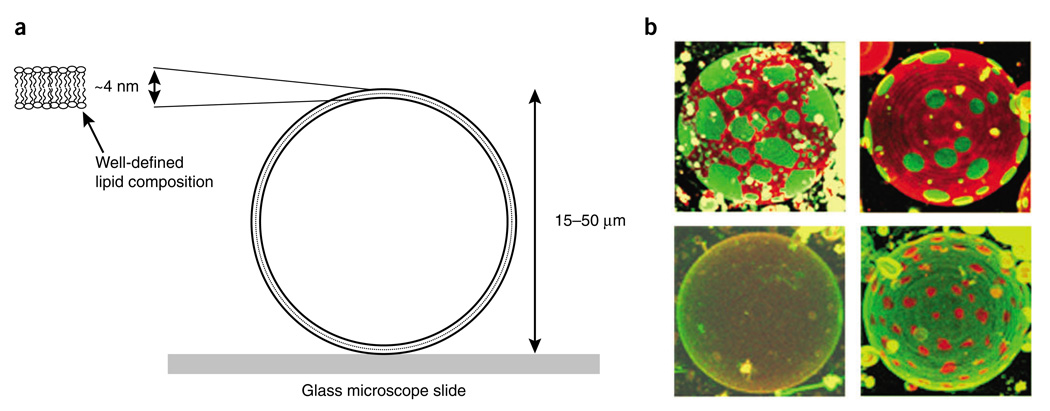
Figure 3.
Confocal fluorescence microscopy of giant unilamellar vesicles, labeled with fluorescent dyes (Fig. 1a), clearly reveals phase coexistence. (a) Schematic illustration of the GUV sample. The GUV is surrounded inside and out by an aqueous phase. (b) Images of phase behavior of GUVs of DSPC/DOPC/cholesterol of four mole ratios (clockwise starting from the top left) 50:36:14, 66:12:22, 20:52:28 and 42:19:39.
The construction of this phase diagram involved the use of some familiar methods: X-ray diffraction, fluorescence resonance energy transfer and simple direct fluorescence measurements of probes in the model membranes. One less familiar technique was indispensable for solving the phase behavior: fluorescence microscopy examination of bilayer spheres that are large enough (typically more than 20 µm in diameter) to allow study by optical microscopy. These bilayer spheres, referred to as ‘giant unilamellar vesicles’ or GUVs, are prepared to have a defined lipid composition and are labeled with fluorescent dyes that have selective partition behavior among the various lipid phases. The distinctive colors, the complementary partitioning of dyes and the shapes of the domains all help to identify the phases. Figure 3 illustrates this GUV-imaging method and shows some typical GUVs of different compositions.
Whereas researchers work hard to achieve near-equilibrium conditions for the measurements that go into a phase diagram, biological membranes in living cells are far from equilibrium. So, even when we have reliable lipid compositional data, and even if we had the data for each separate leaflet of a biological membrane, this composition could not be considered to lead inevitably to a particular phase behavior that we can map onto our experimental phase diagram. A more realistic view is that overall composition of a leaflet of a biological membrane is an ensemble average of different compositional regions having potentially different phase behaviors in different areas of the membrane. This is particularly relevant when considering (i) patches of selected lipid and protein composition fusing with, or pinching off from, a biological membrane during the normal life of the cell5 and (ii) protein activity in a membrane changing the local lipid composition, either catalytically to produce chemical changes in membrane lipids, or by aggregation or disaggregation to release or bind selected local membrane lipids. We can consider a model lipid bilayer phase diagram as indicating the various phase regions that could possibly occur—that is, different phase states in different patches of the membrane surface must depend upon the local compositional heterogeneities.
Even more complex mixtures
Does a three-component lipid model mixture behave similarly to a real biological membrane? When we look at the phase diagram (Fig. 2b), it is a small step to imagine experiments leading to the construction of a phase diagram for a four-component bilayer mixture. In that way, membrane proteins, reasonably grouped as a fourth component, could be examined in a systematic manner that is based on thermodynamic principles. But experimental considerations bring up a key issue: the information that lies in the patterns of these data, obtained at constant temperature, is the concentration dependence. To resolve features that change abruptly, such as straight-line phase boundaries, or that might involve special stoichiometries, a great many samples must be examined. For example, to achieve 2% compositional resolution for a three-component mixture requires about 1,000 samples. By use of methods such as simple fluorescence measurements, this many samples can be studied over a period of several months. However, determining a phase diagram with comparable resolution in composition space for a four-component mixture would take about 50 times longer. For this reason, study of a fourth component demands additional strategies, the simplest being to study only a small number of concentrations of the fourth component. This strategy might give us some clues for dealing with the great complexity of a cell’s membranes; that systematic study of many different lipids, added as fourth components at perhaps even a single concentration to a well-understood three-component mixture, might be sufficient to reveal the key interactions of that component.
As one example of an important additional component, cell biologists are intensely interested in membrane events that follow the activation of the enzyme sphingomyelinase, which catalyzes the hydrolysis of an abundant component of animal cell membranes, sphingomyelin, to produce a membrane-bound lipid product, ceramide12. Ceramide has potent effects in signaling—its influence is amplified to cause changes in cell metabolism and growth. We can ask: in what way, if at all, does ceramide change the phase behavior of the membrane area in which it appears?
Are phase domains small in biological membranes?
Even if we were to have a four-component phase diagram for a lipid mixture that includes ceramide or some other product of enzyme-catalyzed lipid hydrolysis, we might wonder, with good reason, whether there could possibly be any influence whatsoever on the phase behavior of the bilayer. Instead, the influence of lipid hydrolysis products might be connected to the direct binding of an individual hydrolysis product to a protein, thereby changing that protein’s behavior. The enzymes that catalyze lipid hydrolysis act relatively slowly, with turnover numbers of generally less than 10 s−1 according to the Brenda enzyme database (http://www.brenda.uni-koeln.de). Any lipid breakdown product could diffuse tens to hundreds of lipid diameters from the original site of enzymatic action before the next product appeared, and thus a hydrolysis product such as ceramide could never accumulate to an appreciable concentration.
Enzyme action that chemically changes lipids into new membrane components should hardly influence local lipid bilayer phase behavior—unless the new lipid components are restricted in diffusing out of a local area. The size scale of such a membrane area restriction should be not much more than the distance that a lipid hydrolysis product would diffuse before the next one is created, in order to increase the local concentration of that lipid, which would mean a few tens of nanometers. Such a small bilayer patch should possess the range of bilayer phase behaviors (Fig. 2) but also be subject to control of its composition by enzymes that have lipid substrates.
One means to confine a small number of lipid and protein molecules that are in a biological membrane is within an enclosure of membrane proteins that are bound to each other. Cell biologists have evidence for various types and sizes of such “confinement zones”13; the characteristics of these zones are expected to be important for cell function. For a lipid patch of this sort that is confined by proteins around its periphery: (i) this ‘phase-competent region’ could be uncoupled from the phase behavior of nearby areas of the bilayer; (ii) because the area of a lipid is a strong function of the phase state, ranging from >70 Å2 for unsaturated phospholipids in an Lα phase to <45 Å2 for lipids in an Lβ or perhaps Lo phase14, dramatic change in area, by ~50%, is possible when a phase change occurs, but could range from 0 to 50% depending upon the fractional phase change of that confined area. In turn, proteins that are bound to each other during a phase change of their entrapped bilayer patch would experience an alteration in protein-protein contacts to accommodate the phase change: phase behavior of a bilayer patch could be coupled to protein behavior. Likewise, a change in protein-protein contacts could initiate a phase change of the entrapped lipid, with higher lateral pressure on the lipid patch favoring a more condensed state. It remains to be seen how these changes might influence cell behavior or what insights will be gained from examining such systems.
The future: relating molecular interactions to lipid structure
What is an accessible goal for future studies of real and model biological membranes? Chemists place great value on having predictive power based on molecular structure—here, on knowing the phase description of any given mixture of membrane components. Because there are so many components, we must establish general principles that relate phase behavior to the structure of each lipid and to the composition of each mixture. In this era of screening large libraries of chemicals, a brute-force approach of directly examining many thousands of mixtures might possibly be devised.
Nonetheless, a physical chemist would be pleased by the discovery of the underlying nature of the molecular interactions. Once we have accurate and equilibrium phase boundaries, we then have thermodynamic parameters that can link theory with experiment. We aim to have a picture of the nonrandom mixing in all of the complexity of biological membranes that captures the most important features of compositional heterogeneity and size of any clusters, including—but by no means limited to—phase separations. The observation of phase separation in model bilayer mixtures, whether or not it reflects events in cell membranes, has the benefit that shifts of the phase boundary are thermodynamically well-defined properties of the mixture, which arise from the microscopic interactions. One strategy that might be up to the task of finding the energies of interaction among the components is Monte Carlo simulation15. We could start from a well-understood three-component mixture such as DSPC/DOPC/cholesterol and use Monte Carlo simulations of phase boundary shifts caused by a fourth component. Because the interaction energies obtained from Monte Carlo simulation are additive, each component of interest would have a ‘portable’ set of interaction energies that could be used in simulation of mixtures of that component with many others. In the simple bilayer mixtures having only two components, Monte Carlo simulation has worked well to describe how cholesterol mixes with various bilayer lipids and why cholesterol crystals precipitate at special stoichiometries11. But can Monte Carlo simulation be extended to deal with the enormous complexity of biological membranes? Active research to connect experiments and simulations should provide an answer to this question.
ACKNOWLEDGMENTS
This work was supported by US National Science Foundation grant MCB-0315330 to G.W.F. The author thanks J. Zhao and F. Heberle for helpful discussions.
References
- 1.Simons K, van Meer G. Biochemistry. 1988;27:6197–6202. doi: 10.1021/bi00417a001. [DOI] [PubMed] [Google Scholar]
- 2.Simons K, Ikonen E. Nature. 1997;387:569–572. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- 3.Holowka D, Baird B. Immunology. 2001;13:99–105. doi: 10.1006/smim.2000.0301. [DOI] [PubMed] [Google Scholar]
- 4.Scheiffele P, Rietveld A, Wilk T, Simons K. J. Biol. Chem. 1999;274:2038–2044. doi: 10.1074/jbc.274.4.2038. [DOI] [PubMed] [Google Scholar]
- 5.Mukherjee S, Maxfield FR. Annu. Rev. Cell Dev. Biol. 2004;20:839–866. doi: 10.1146/annurev.cellbio.20.010403.095451. [DOI] [PubMed] [Google Scholar]
- 6.van Meer G. EMBO J. 2005;24:3159–3165. doi: 10.1038/sj.emboj.7600798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morse SA. Basalts and Phase Diagrams. New York: Springer; 1980. [Google Scholar]
- 8.Op den Kamp JAF. Annu. Rev. Biochem. 1979;48:47–71. doi: 10.1146/annurev.bi.48.070179.000403. [DOI] [PubMed] [Google Scholar]
- 9.Roelofsen B, Op den Kamp JAF. Curr. Topics Membranes. 1994;40:7–46. [Google Scholar]
- 10.Veatch SL, Polozov IV, Gawrisch K, Keller SL. Biophys. J. 2004;86:2910–2922. doi: 10.1016/S0006-3495(04)74342-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang J, Feigenson GW. Biophys. J. 1999;76:2142–2157. doi: 10.1016/S0006-3495(99)77369-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bollinger CR, Teichgraber V, Gulbins E. Biochim. Biophys. Acta. 2005;1746:284–294. doi: 10.1016/j.bbamcr.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 13.Kusumi A, Murakoshi H, Murase K, Fujiwara T. Single-molecule imaging of diffusion, recruitment, and activation of signaling molecules in living cells Chapter 5. In: Damjanovich S, editor. Biophysical Aspects of Transmembrane Signaling. Heidelberg: Springer; 2005. pp. 23–152. [Google Scholar]
- 14.Nagle JF, Tristram-Nagle S. Biochim. Biophys. Acta. 2000;1469:159–195. doi: 10.1016/s0304-4157(00)00016-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang J. Biophys. J. 2002;83:1014–1025. doi: 10.1016/S0006-3495(02)75227-2. [DOI] [PMC free article] [PubMed] [Google Scholar]