Abstract
A DNA lesion created by oxidative stress is 7,8-dihydro-8-oxo-guanine (8-oxoG). Because 8-oxoG can mispair with adenine during DNA synthesis, it is of interest to understand the efficiency and fidelity of 8-oxoG bypass by DNA polymerases. We quantify bypass parameters for two DNA polymerases implicated in 8-oxoG bypass, Pols δ and η. Yeast Pol δ and yeast Pol η both bypass 8-oxoG and misincorporate adenine during bypass. However, yeast Pol η is 10-fold more efficient than Pol δ, and following bypass Pol η switches to less processive synthesis, similar to that observed during bypass of a cis-syn thymine-thymine dimer. Moreover, yeast Pol η is at least 10-fold more accurate than yeast Pol δ during 8-oxoG bypass. These differences are maintained in the presence of the accessory proteins RFC, PCNA and RPA and are consistent with the established role of Pol η in suppressing ogg1-dependent mutagenesis in yeast. Surprisingly different results are obtained with human and mouse Pol η. Both mammalian enzymes bypass 8-oxoG efficiently, but they do so less processively, without a switch point and with much lower fidelity than yeast Pol η. The fact that yeast and mammalian Pol η have intrinsically different catalytic properties has potential biological implications.
INTRODUCTION
One of the most common lesions resulting from oxidative stress is 7,8-dihydro-8-oxo-guanine (8-oxoG) [(1) and references therein]. Compared to guanine, 8-oxoG contains only one extra oxygen atom, yet presents a major problem for a cell because during DNA synthesis it can pair with either correct deoxycytidine (dC) or incorrect deoxyadenine (dA). NMR and X-ray crystallographic studies with either a dC·8-oxoG or dA·8-oxoG basepair shows that neither lesion causes a serious distortion of the overall helical structure of the DNA (2–5). This is because the dC·8-oxoG pair exists in the standard anti·anti conformation, while the dA·8-oxoG mispair exists in an anti·syn conformation, leading to base pairing along the Hoogsteen edge of the incoming dA. The dA·8-oxoG mispair can result in a G·C → T·A transversion, a substitution linked to somatic cancers (6). To avoid the adverse consequences of 8-oxoG·dA mispairs, cells devote a number of enzymes to processing 8-oxoG (7). The base excision repair (BER) pathway recognizes and removes 8-oxoG from the dC·8-oxoG pair using the MutM/OGG1 glycosylase in E. coli and human cells, respectively. The dA base from the dA·8-oxoG mispair is removed by the MutY/MYH glycosylase (E. coli/human). In addition, MutT/MTH1 (E. coli/human) can hydrolyze the oxidized precursor 8-oxoGTP to its monophosphate form (8-oxoGMP), thereby preventing incorporation opposite either dA or dC in the template. Lastly, the DNA mismatch repair system has been shown to recognize 8-oxoG mispairs (8).
DNA repair systems are not perfect, such that template 8-oxoG is sometimes encountered by DNA polymerases during replication and gap-filling synthesis associated with DNA repair. Given the tendency for mispairing by DNA polymerases with associated increase in mutations, many studies have investigated the activities of specific DNA polymerases when they encounter template 8-oxoG. In an early report, human Pols α and β, calf thymus Pol δ and E. coli Pol I were all able to incorporate both dC and dA opposite 8-oxoG, in ratios varying from 7:1 to 1:200 (9). All these polymerases more efficiently extended the 8-oxoG·dA mispair than the 8-oxoG·dC pair. More recent studies have reported ratios of dC:dA insertion opposite 8-oxoG of 2:1 for Pol β (10) and T7 DNA polymerase (11,12), 3:1 for E. coli Pol I (13), 1:1 for E. coli Pol II (13), 1:14 for HIV RT (11), 1:9 for Bacillus stearothermophilus Pol I (14), 20:1 for RB69 DNA polymerase (15), 3:1 for calf thymus Pol δ (16), 20:1 for Saccharomyces cerevisiae Pol η (17), between 70 and 90:1 for Sulfolobus solfataricus DNA polymerase 4 (Dpo4) (18,19) and between 1:1 and 1:3 for human Pol κ (20,21). Most of these studies involved kinetic analysis of insertion of a single nucleotide opposite 8-oxoG, and in some cases, the ability to add a correct nucleotide onto primer termini containing either dA·8-oxoG or dC·8-oxoG pairs. These studies clearly demonstrate that relative to unmodified template guanine, 8-oxoG reduces the nucleotide selectivity of DNA polymerases, but to varying degrees. The structural basis for this reduced selectivity, and for polymerase-specific variations in the ratio of dC:dA insertion opposite 8-oxoG, has been investigated in several studies showing that both base pairs can in fact exist within the active site of a polymerase, but in different conformations and with different polymerase-specific interactions (12,14,18,19,22,23). Interestingly, one such study shows that the dA·8-oxoG anti·syn mispair can mimic the geometry of a correct base pair and thereby escape efficient proofreading by the 3′ exonuclease activity of the replicative T7 DNA polymerase (9,12).
The present study examines the consequences of attempts by Pols δ and η to bypass a template 8-oxoG. The choice of these two particular polymerases with this lesion was motivated by biological evidence clearly indicating that Pol η has an important role in suppressing mutagenesis induced by sunlight (24–30), which not only generates photodimers but also lesions due to oxidative stress (31–34). Moreover, S. cerevisiae Pol η also has a role in suppressing mutagenesis in cells defective in removing 8-oxoG due to a defect in the Ogg1 glycosylase (17,35,36). These data imply that in a manner akin to the prevailing model for Pol η-dependent bypass of UV photoproducts (37,38), when a major replicative polymerase like Pol δ encounters 8-oxoG, it pauses, thereby initiating a switch to allow Pol η to bypass this lesion. We have been investigating this model based on the specific hypothesis that polymerase switching during translesion synthesis (TLS) occurs during transitions from preferred to disfavored use of damaged primer-templates and that the polymerase used for each successive nucleotide incorporated is the one whose properties result in the highest efficiency. Here, we test this hypothesis by investigating the properties of 8-oxoG bypass by Pols δ and η using assays designed to quantify the efficiency and the fidelity of TLS (39–42).
MATERIALS AND METHODS
Materials and reagents
All bacterial strains, plasmids, bacteriophage and other materials for the assays performed were from previously described sources (42,43). DNA modification and restriction enzymes were purchased from New England Biolabs (Ipswich, MA), and oligonucleotides were purchased from Oligos Etc., Inc. (Wilsonville, OR). Streptavidin was purchased from Roche Applied Science (Indianapolis, IN), and dNTPs were purchased from Amersham Biosciences (Piscataway, NJ).
DNA substrates
Bypass assays used substrates with 70-mer templates (70Bio-G: 5′-biotin-ATGACCATGATTACGAATTCCA GCTCGGTACCGGGTTAGCCTTTGGAGTCGACCT GCAGAAATTCACTGG; 70Bio-Go: 5′-biotin-ATGAC CATGATTACGAATTCCAGCTCGGTACCGGGTTA xCCTTTGGAGTCGACCTGCAGAAATTCACTGG; 70Bio-Go2 5′-biotin-ATGACCATGATTACGAATTCC AGCTCGGTACCGGxTTAGCCTTTGGAGTCGACC TGCAGAAATTCACTGG) containing a 5′ biotin moiety. The position of the 8-oxoG residue is indicated by ‘x’. Primer strand oligonucleotides were BP14 (5′-biotin-CCAGTGAATTTCTG) with a 5′-biotin moiety, LP16 (5′-CAGGTCGACTCCAAAG), LP20 (5′-CAGGTCGACTCCAAAGGCTA) and 30Fid (5′-biotin-CCAGTGAATTTCTGCAGGTCGACTCCAAAG) with a 5′-biotin moiety. Bypass efficiency assay substrates used the ‘blocking’ primer BP14 and the ‘labeled’ primer LP16 that was labeled at the 5′ end using 32P-γ-ATP and T4 polynucleotide kinase. Substrates were prepared by mixing template oligonucleotide (70Bio-G/70Bio-Go/70Bio-Go2) with 1.2 M equivalents each of BP14 and LP16 in 50 mM Tris–Cl (pH 7.5) and 1X SSC, followed by incubation at 75°C for 5 min, then cooling to 25°C over 2 h, protected from light. Substrates used in the bypass fidelity assay were prepared similarly using only the unlabeled 30Fid primer oligonucleotide. For apurinic/apyrimidinic (AP) site bypass assays, a 102-mer template (V9/V9AP1: 5′-biotin-CCTTTGCGAATTCT25GCGxCTCCCTTCTTCTCCTCCCTCTCCCTTCCCT T29-biotin) with both 5′ and 3′ biotin moieties was used, with the position indicated by ‘x’ containing either an undamaged G or a synthetic AP site (tetrahydrofuran) residue. The primer used (C12-4: 5′-AGGGAAGGGAGAGGGAGGAGAAGAAG) pairs with the region in italics. AP site bypass substrates were prepared as described above using a 1.2× molar excess of template to 5′ 32P end-labeled primer. All bypass efficiency experiments with human and mouse Pol η used a 45-mer template (45TTAG/45TTAGo: 5′-CCAGCTCGGTACCGGGTTAxCCTTTGGAGTCGACCTGCAGAAATT) that contained either an undamaged G or 8-oxoG at the position indicated by ‘x’ with 5′ 32P-labeled primer (LBP-24: 5′-AATTTCTGCAGGTCGACTCCAAAG), prepared as described earlier. Bypass fidelity reactions using the 45-mer template were prepared with unlabeled LBP-24 primer. Templates with different sequence contexts indicated in the text, tables and figure legends are limited to the region in italics in the above 45-mer sequence.
Protein isolation and purification
S. cerevisiae polymerase η was purified as described previously (44) using a plasmid kindly provided by Dr. Zhigang Wang (University of Kentucky). Two separate preparations gave similar results. S. cerevisiae PCNA, RFC, RPA and three subunit polymerase δ, both exonuclease proficient (δexo+) and deficient forms (δexo−) were purified as previously described (45–48). Mouse and human Pol η were expressed and purified as described previously (49).
Bypass efficiency assay
Bypass efficiency reactions using the 70-mer template substrate and S. cerevisiae proteins were performed as previously described (39,41,50). All reactions contained 40 mM Tris–Cl (pH 7.8), 75 mM NaCl, 5 mM MgCl2, 1 mM ATP, 2 mM DTT and 100 µg/ml BSA. Reactions (30 μl) contained 1 pmol of DNA substrate that was first incubated with 10 pmol of streptavidin. When present, PCNA trimer, RFC complex and RPA were added in 1.2-fold excess compared to DNA substrate. Reactions with Pol δ contained 25 µM of each of the four dNTP, while reactions with Pol η contained 100 µM each. All components except polymerase were mixed on ice and then incubated at 30°C for 2 min. Polymerase (5 fmol) was added to initiate the reaction, and 6 µl samples were removed at the indicated times and mixed with 12 µl formamide loading buffer (95% deionized formamide, 25 mM EDTA, 0.1% bromophenol blue, 0.1% xylene cyanol). Products were heated to 95°C for 5 min and separated through a 12% denaturing polyacrylamide gel. Dried gels were visualized and quantified using a Molecular Dynamics Typhoon 9400 imager and ImageQuant software. Calculation of termination probability, bypass amount and relative bypass efficiency were performed as previously described (39,41). Reactions with human and mouse Pol η using 45-mer template substrates were performed as previously described (41,51), using 4 pmol of substrate and either 10 fmol human Pol η or 16 fmol mouse Pol η. In all reactions used to calculate bypass efficiency parameters, short incubation times and high DNA to polymerase ratios were used. The conditions used were chosen to assure that termination probabilities remained with time, thereby empirically demonstrating that the majority of product chains result from a single cycle of synthesis (39).
Bypass fidelity assay
The assay was performed as previously described (39,42). Seventy-mer template substrates containing a single primer (30Fid) were blocked with streptavidin, while 45-mer template substrates did not have biotin–streptavidin complexes blocking the ends. Reaction conditions were the same as described earlier with the following exceptions: S. cerevisiae Pol δ experiments contained 8 pmol of substrate and 4 pmol of polymerase; S. cerevisiae Pol η experiments contained 2 pmol of substrate and 1 pmol of enzyme. Reactions with S. cerevisiae proteins were incubated at 30°C for 30 min. Reactions with mouse and human Pol η contained 4 pmol substrate and 0.8 pmol enzyme and were incubated at 37°C for 20 min. Recovery of the synthesized strand, annealing to gapped M13 DNA molecules, hybridization efficiency controls, transfection into E. coli and determination of error rates and spectra were performed as previously described (39,42).
RESULTS
The efficiency of 8-oxoG bypass by yeast Pol δ
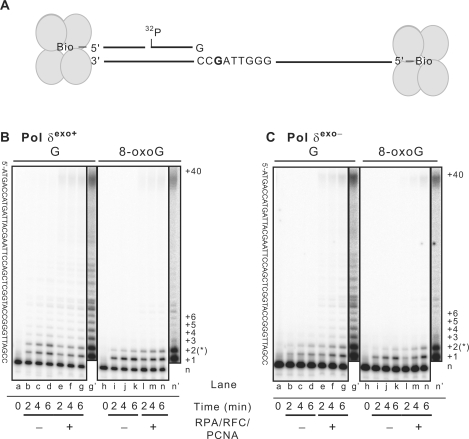
First, we examined the ability of three-subunit yeast Pol δ to bypass 8-oxoG (Figure 1). In comparison to bypass of undamaged guanine in the same sequence context (lanes b–d, in Figure 1B and C), 8-oxoG inhibited polymerization by wild type (i.e., exo+) Pol δ (Figure 1B, lanes i–k) and exo− Pol δ (Figure 1C, lanes i–k). When band intensities were quantified and used to calculate bypass efficiencies (Table 1), wild type and exo– Pol δ were found to bypass 8-oxoG with 15% and 31% efficiency, respectively, compared to bypass of undamaged G in the same sequence context. This reduced bypass efficiency is due mainly to reduced extension following incorporation opposite 8-oxoG (Figure 1B, compare band intensities at n + 3 in lanes d and k). We also performed reactions with a template of the same sequence but with the 8-oxoG located four bases downstream, i.e., in a different sequence context. In this case, Pol δexo+ bypassed 8-oxoG with an efficiency that was 25% of that observed with undamaged G in the same context (Table 1). Thus in two different contexts, 8-oxoG substantially reduces processive bypass synthesis by Pol δ. The product chains generated during 8-oxoG bypass by Pol δexo+ (Figure 1B) and Pol δexo− (Figure 1C) were similar, and the bypass efficiency of Pol δexo– was only slightly higher than the bypass efficiency of Pol δexo+, indicating that the intrinsic 3′ exonuclease activity of Pol δ has limited influence on bypass of 8-oxoG.
Figure 1.
Inhibition of S. cerevisiae Pol δ DNA synthesis by 8-oxoG. (A) Schematic diagram of the substrate used in TLS assays. The template is a 70-mer, the biotinylated primer is a 14-mer and the radio-labeled primer is a 16-mer. (B) Twelve percent dPAGE image of reactions containing a 200:1 substrate:enzyme (S:E) ratio of exonuclease proficient Pol δ (Pol δexo+) and either undamaged (G) or damaged (8-oxoG) templates with blocked ends (see ‘Materials and Methods’ section), without and with the accessory proteins RPA, RFC and PCNA. The template sequence is given to the left of the image and the number of nucleotide incorporation (+1, +2, etc.) is given on the right. The +40 incorporation represents synthesis to the end of the template. The position of the 8-oxoG residue is indicated with a star (*). Lanes g′ and n′ are overexposures of lanes g and n, respectively. (C) Gel image of reactions containing a 200:1 S:E ratio of exonuclease deficient Pol δ (Pol δexo−). Details are the same as in (B).
Table 1.
Bypass efficiency of 8-oxoG by pols δ and η
| Yeasta |
Humanb,c | Mouseb | ||||||
|---|---|---|---|---|---|---|---|---|
| δexo+ |
δexo− |
η |
η | η | ||||
| RPA/RFC/PCNA | − | + | − | + | − | + | − | − |
| Bypass (%)d | ||||||||
| 3′-CCGATT | 8.9 | 25 | 4.8 | 24 | 47 | 54 | 24 | 17 |
| 3′-CCGoATT | 1.3 | 4.1 | 1.5 | 5.9 | 62 | 69 | 36 | 31 |
| Relative bypass (%) | 15 | 16 | 31 | 25 | 130 | 130 | 150 | 180 |
| Switch | No | No | No | No | Yes | Yes | No | No |
| Bypass (%) | ||||||||
| 3′-TTGGG | 71 | 75 | 34 | 23 | ||||
| 3′-TTGoGG | 18 | 23 | 86 | 57 | ||||
| Relative bypass (%) | 25 | 31 | 250 | 250 | ||||
| Switch | No | No | Yes | Yes | ||||
aEfficiency measurements made using 70-mer template with streptavidin blocked ends (see ‘Materials and Methods’ section).
bEfficiency measurements made using 45-mer template (see ‘Materials and Methods’ section).
cBypass efficiency measured in three other template sequence contexts was 3′ … CGoATC … = 220%; 3′- … CAGoTC … = 180%; 3′- … CAGoTT … = 180%.
dExperiments depicted in Figures 1 and 2 were quantified and the bypass amount on each template calculated as previously described (39,41). Relative bypass is the amount of 8-oxoG bypass compared to undamaged DNA. Switch is defined as a change from lower to higher termination probability when comparing values at the 8-oxoG and next undamaged base.
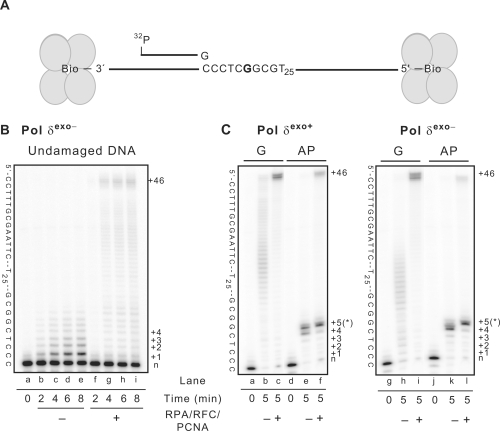
Addition of RFC, PCNA and RPA stimulated the processivity of both forms of Pol δ (e.g. Figure 1B and C, see full-length 70-mer products in lanes g and n). In addition, the accessory proteins increased the percentage of product chains reflecting bypass of G and 8-oxoG by several-fold (compare values in first four columns and top two lines of Table 1). However, the degree of stimulation by accessory proteins was similar for both the undamaged G and 8-oxoG templates, such that the accessory proteins did not selectively increase the relative 8-oxoG bypass efficiency for either exo+ or exo– Pol δ (third line in Table 1). To further confirm that the accessory proteins are active under the reaction conditions used here, we performed reactions using a second undamaged template, in this case one that yields lower termination probabilities after the first two incorporations (Figure 2). The processivity of Pol δ is somewhat higher on this template (compare Figure 1B and C, lanes b–d with Figure 2B, lanes b–e), and the ability of RPA, RFC and PCNA to increase processivity is clearly seen (Figure 2B, lanes f–i). We also performed experiments with 2-fold excess of Pol δ over primer-template, in this case with both an undamaged template and a template containing a synthetic abasic site (tetrahydrofuran). These experiments more closely approximate the many published TLS studies that use high-enzyme concentrations and permit multiple cycles of polymerization, evidenced here by extension of nearly all the starting DNA substrate (Figure 2C). Under these reaction conditions, the accessory proteins clearly stimulate formation of full-length products, both with undamaged template (Figure 2C, lanes compare lanes b to c and lanes h to i) and with the lesion-containing template (Figure 2C, compare lanes e to f and lanes k to l). The stimulation of bypass of a synthetic abasic site is consistent with earlier observations with yeast and calf thymus Pol δ (52,53).
Figure 2.
Stimulation of Pol δ activity by accessory proteins. (A) Schematic diagram of the substrate used in TLS assays. The template is a 102-mer with biotin at both the five and three ends. The radio-labeled primer is a 26-mer. (B) Time course of reactions of Pol δexo− using undamaged template that does not have sequence specific pause sites. Reactions are confirmed to be under single-interaction conditions (see ‘Materials and methods’ section). The template sequence is given to the left of the image and the number of nucleotide incorporation (+1, +2, etc.) is given on the right. The +46 incorporation represents synthesis to the end of the template. (C) Gel images of reaction products created using a S:E ratio of 1:2 (polymerase excess) with 102-mer templates containing either undamaged G or tetrahydrofuran (an AP site mimic) at the fifth incorporation position (*). These images show that in the presence of the accessory proteins, stimulation of AP site bypass occurs, similar to a previous report (53). Details are the same as in (B). Panels (B) and (C) confirm that the accessory proteins are active under the conditions used.
Efficiency of 8-oxoG bypass by yeast, mouse and human Pol η
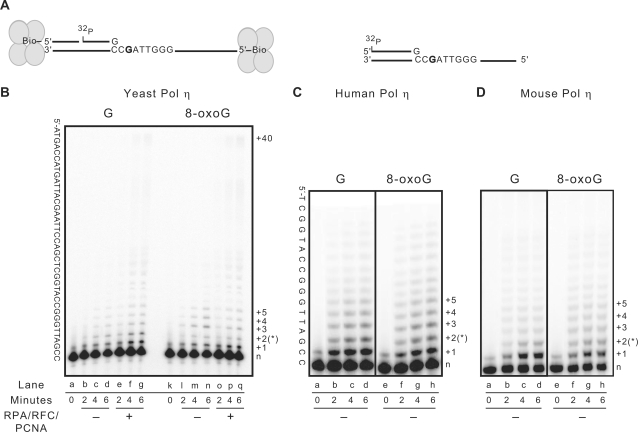
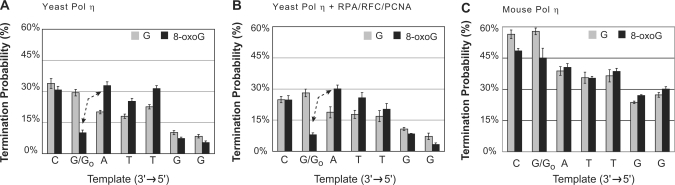
While bypass of 8-oxoG is somewhat problematic for yeast Pol δ, yeast Pol η bypasses the lesion with ease (Figure 3B, lanes l–n). In two sequence contexts examined (one not shown), bypass values are higher for the lesion than for the corresponding undamaged G, giving relative bypass efficiencies of 130–250% (Table 1). Also, as previously reported for bypass of a cis-syn TT dimer by yeast (40,50) and human Pol η (41), yeast Pol η has a lower probability of terminating processive synthesis after insertion opposite 8-oxoG (12%, Figure 4A, black bar) as compared to undamaged G (29%, Figure 4A, gray bar). This lower termination opposite 8-oxoG corresponds to a higher probability of adding the next base, i.e. after insertion opposite 8-oxoG, yeast Pol η extends the 8-oxoG containing terminus even more efficiently than it extends the undamaged terminus. Yeast Pol η then inserts nucleotides opposite the next three undamaged bases less efficiently compared to the equivalent positions in the undamaged template. In other words, after incorporation opposite 8-oxoG and the next undamaged template nucleotide (i.e. after bypass), synthesis by yeast Pol η becomes disfavored.
Figure 3.
High-efficiency bypass of 8-oxoG by Pol η. (A) Schematic diagram of the substrates used in TLS assays. For yeast Pol η the substrate is as described in Figure 1. For human and mouse Pol η, the template is a 45-mer and the radio-labeled primer is a 24-mer. (B) Reactions were performed as described in Figure 1 legend using a 200:1 S:E ratio of S. cerevisiae Pol η, either in the absence of any accessory proteins (lanes b–d and l–n), or with RPA, RFC and PCNA (lanes e–g and o–q). The template sequence is given to the left of the image and the number of nucleotide incorporation is given on the right. The position of the 8-oxoG residue is indicated with a star (*). (C and D) Bypass efficiency reactions were performed using a 400:1 S:E ratio of human Pol η and a 250:1 S:E ratio of mouse Pol η.
Figure 4.
Termination probability analysis during 8-oxoG bypass by Pol η. Gel images of reaction products seen in Figure 3 were quantified using ImageQuant software, as described previously (39,41). (A) Graph of termination probability (vertical axis, 0–60%) at each of the first seven incorporations (horizontal axis) for S. cerevisiae Pol η without accessory proteins. Undamaged template and 8-oxoG template values are indicated by light gray and black bars, respectively. Error bars are the standard deviation for six measurements (three time points each for two separate experiments). The switch from low to high termination probability after 8-oxoG has been bypassed is indicated with dashed-line arrows. (B) Graph of termination probability for S. cerevisiae Pol η with RPA, RFC and PCNA. Details are the same as in (A). (C) Graph of termination probability for mouse Pol η without accessory proteins on a 45-mer template (see ‘Materials and methods’ section). Note that mouse and human Pol η display nearly identical properties (41,49,51).
Neither the high efficiency of 8-oxoG bypass by yeast Pol η (Table 1) nor the switch to less processive synthesis (Figure 4A and B) is strongly influenced by the presence of the accessory proteins. As reported earlier at a higher enzyme to substrate ratio (53), there is clear stimulation of Pol η by RPA, PCNA and RFC (Figure 3B, compare amount of product with 6 or more incorporations in lanes b–d with e–g and in lanes l–n with o–q). The stimulation is similar with the undamaged and damaged templates. The results were somewhat different with human and mouse Pol η. Both polymerases are less processive than yeast Pol η [compare termination probabilities in Figure 4A and C and reference (50)]. Despite this lower processivity, both mammalian polymerases readily bypass 8-oxoG, with no detectable block to synthesis (Figure 3C and D). In fact, they do so with relative bypass efficiencies consistently higher than for the undamaged template (150–220%, Table 1 and its legend). These values in excess of 100% are largely due to a slightly higher efficiency of insertion opposite the lesion compared to the equivalent undamaged bases. However, after the 8-oxoG was copied, the mammalian enzymes failed to switch from low-to-high termination probabilities (Figure 3C and D, Figure 4C and data not shown). This differs from yeast Pol η and from the behavior of human and mouse Pol η when copying a TT dimer (41,51).
Fidelity of 8-oxoG bypass
Next, we measured the error rates of Pol δexo+, Pol δexo− from yeast and Pol η from yeast, human and mouse when bypassing an 8-oxoG in the presence of all four dNTPs. Enough enzyme and time were provided to allow complete synthesis to the end of the template. The newly synthesized strand was recovered and hybridized to gapped M13mp2 DNA, which was introduced into E. coli cells that were then plated to score M13 plaque color phenotype (39,42). The undamaged G or 8-oxoG is within a TAG codon in the lacZα gene in M13mp2 DNA. Correct incorporation of dC results in M13 plaques that are faint blue (due to slight read through of the nonsense codon), whereas stable misincorporation of dA or dG opposite the G or 8-oxoG results in dark blue plaques. Sequencing M13 DNA from dark blue plaques identifies the substitution and allows calculation of error rates.
In this assay, Pol δexo+ and Pol δexo− have much lower fidelity when bypassing 8-oxoG than when copying the undamaged template (Table 2). This is reflected in the dramatically higher dark blue plaque frequencies when copying the damaged template (23–30%) as compared to the undamaged template (0.05–0.15%). Sequence analysis followed by error rate calculations was performed for Pol δexo+ and Pol δexo−, in each case in the absence and in the presence of RFC, PCNA and RPA (Table 3). The results indicate that 40–50% of bypass of 8-oxoG by Pol δ results in stable misincorporation of dA and/or dG opposite the lesion. Rates are similar with Pol δexo+ and Pol δexo−, indicating that these damaged mismatches are not proofread by the 3′ exonuclease activity of yeast Pol δ. The accessory proteins have little effect on error rates for dA·8-oxoG mismatches generated by either Pol δexo+ or Pol δexo−. In the presence of the accessory factors, both forms of Pol δ showed a 6-fold decrease in the rate of 8-oxoG·dG mismatches. This difference is statistically significant (Fisher's exact test, P = 0.012 and 0.011 for Pol δexo+ and Pol δexo−, respectively).
Table 2.
Dark blue plaque frequency for G and 8-oxoG bypass reactions
| −RPA/RFC/PCNA |
+RPA/RFC/PCNA |
|||
|---|---|---|---|---|
| G | 8-oxoG | G | 8-oxoG | |
| Yeast | ||||
| δexo+ | ||||
| Totala | 54 728 | 2441 | 29 786 | 446 |
| Dark blue | 26 | 705 | 14 | 132 |
| MF (%) | 0.05 | 29 | 0.05 | 30 |
| δexo− | ||||
| Total | 24 049 | 866 | 12 710 | 3528 |
| Dark blue | 37 | 210 | 17 | 800 |
| MF (%) | 0.15 | 24 | 0.13 | 23 |
| η | ||||
| Total | 6869b | 5506 | 5342 | 7590 |
| Dark blue | 121b | 165 | 92 | 242 |
| MF (%) | 1.8b | 3.0 | 1.7 | 3.2 |
| Human ηc | ||||
| Total | 4083 | 3702 | ||
| Dark blue | 134 | 1081 | ||
| MF (%) | 3.3 | 29 | ||
| Mouse η | ||||
| Total | 4345 | 3420 | ||
| Dark blue | 166 | 1031 | ||
| MF (%) | 3.8 | 30 | ||
aValues given are the numbers of plaques of each type counted and the resulting mutation frequencies for G/8-oxoG bypass reactions. Data shown is from experiments using a 70-mer template with streptavidin-blocked ends (see ‘Materials and Methods’ section).
bThis data has been previously published (50).
cAdditional experiments using a 45-mer template (see ‘Materials and Methods’ section) gave dark blue plaque frequencies of 32% (3′-CCGoATC), 29% (3′-CCAGoTC) and 32% (3′-CCGoAT T).
Table 3.
Error specificity during G and 8-oxoG bypass
| RPA/RFC/PCNA | Yeast |
Human | Mouse | |||||
|---|---|---|---|---|---|---|---|---|
| δexo+ |
δexo− |
η |
ηa | η | ||||
| − | + | − | + | − | + | − | – | |
| 3′-CGATT | ||||||||
| Sequencedb | 23 | 12 | 24 | 23 | 52c | 72 | 32 | 28 |
| G → C | 1.4 | <0.7 | 8.5 | 4.8 | 17 | 12 | 34 | <23 |
| G → T | 1.4 | 0.7 | 2.1 | 2.9 | <5.6 | 4.0 | 52 | 68 |
| 3′-CGoATT | ||||||||
| Sequenced | 23 | 22 | 24 | 24 | 49 | 70 | 32 | 31 |
| G → C | 1300 | <220 | 1000 | <160 | 41 | 76 | 300 | <20 |
| G → T | 3100 | 4900 | 2900 | 3800 | 260 | 150 | 4400 | 4500 |
aAdditional experiments using a 45-mer template (see ‘Materials and Methods’ section) with slight sequence difference gave error rates of: (3′- … CGoATC …) G → C = 610 × 10−4, G → T = 4600 × 10−4; (3′- … CAGoTC …) G → C = <230 × 10−4, G → T = 4500 × 10−4; (3′- … CAGoT T …) G → C = 350 × 10−4 = 4000 × 10−4.
bDNA from dark blue plaques described in Table 2 was sequenced and error rates (×10−4) were calculated as previously described (42). Only changes at the G/8-oxoG site are given (the TAG stop codon is in italics in the left-most column). Note that G → A changes are not detectable by color screening as the amber stop codon (TAG) changes instead to an ochre stop codon (TAA) in the lacZα sequence.
cThese sequenced samples are the same as those used in a published report (50). The specific error rates shown here have not previously been reported.
Pol η lacks intrinsic proofreading activity and, when copying undamaged DNA, it is much less accurate [Tables 2 and 3, and refs (49,50,54)] than Pol δ [Tables 2 and 3, and ref. (55)]. However, when the fidelity of Pol η was measured for bypass of 8-oxoG, yeast Pol η was more accurate than yeast Pol δ, as reflected in lower dark blue plaque frequencies (Table 2) and in much lower error rates for dA·8-oxoG and dG·8-oxoG mismatches calculated after sequence analysis (Table 3). The accessory proteins had little if any affect on the fidelity of 8-oxoG bypass by yeast Pol η. Error rates for 8-oxoG bypass by human and mouse Pol η are much higher than for yeast Pol η. Dark blue plaque frequencies of ∼30% were observed (Table 2), and the error rates for dA misincorporation (44% for mouse, 45% for human) are 17-fold higher than for yeast Pol η. Similar and high error rates for 8-oxoG bypass by human Pol η were observed in three other sequence contexts (Table 3, note c).
DISCUSSION
This study tests the hypothesis that enzymatic switching during TLS occurs during transitions from preferential to disfavored use of damaged primer-templates and that the polymerase or 3′-exonuclease used for each successive nucleotide incorporated is the one whose properties result in the highest efficiency and the highest fidelity of bypass. We previously examined this for two lesion-polymerase combinations that are highly relevant to mutagenesis and cancer susceptibility, i.e. Pols η and δ bypass of a cis-syn TT dimer resulting from sunlight exposure (40,41). Here, we extend the effort to bypass of template 8-oxoG. This lesion results from exposure to sunlight and other types of environmental stress, both exogenous and endogenous, and convincing evidence shows that yeast Pol η suppresses mutagenesis in yeast strains deficient the Ogg1 glycosylase that normally removes 8-oxoG from DNA (17,35,36). Those genetic data are strongly supported by the biochemical properties of yeast Pols δ and η reported here. Template 8-oxoG impedes synthesis by yeast pol δ (Figure 1), while yeast pol η efficiently incorporates opposite 8-oxoG and the next undamaged template nucleotide (Figure 3A) to achieve very efficient bypass (Table 1) and then switches to less processive synthesis (Figure 4A and B). Yeast pol η stably incorporates C rather than A opposite 8-oxoG by a factor of 67-fold (Table 3, error rate of 150 × 10−4 for yeast Pol η with accessory proteins). This result is consistent with kinetic studies of yeast Pol η (17,56). By comparison, bypass of 8-oxoG by wild type yeast pol δ is 33-fold less accurate (Table 3, error rate of 4900 × 10−4 with accessory proteins), and neither its proofreading activity nor accessory proteins improve fidelity (Table 3). All these properties strongly support the proposal (17) that yeast Pol η, although clearly not error-free, contributes to bypass of spontaneously generated 8-oxoG in a manner that suppresses mutagenesis.
It is well known that 8-oxoG not only arises spontaneously but is also generated by exposure to UV radiation. This and the fact that XPV patients lacking functional Pol η have greatly increased susceptibility to sunlight-induced skin cancer raises the interesting issue of whether mammalian Pol η also bypasses 8-oxoG in a manner that suppresses mutagenesis. This possibility is supported by a report (17) that, like yeast Pol η, human Pol η also bypasses 8-oxoG accurately. This led to the suggestion that, in addition to suppressing sunlight-induced skin cancer, human Pol η may also suppress internal cancers that would otherwise result from mutagenic bypass of 8-oxoG in DNA (17). Although there is no compelling evidence that XPV patients have increased susceptibility to internal cancers, a recent study of human cell lines has concluded that human Pol η suppresses 8-oxoG-dependent mutations when plasmid DNA that was damaged in vitro by methylene blue plus light is replicated in vivo (57).
On the other hand, we find that the human and mouse Pol η error rate for bypass of 8-oxoG approaches 50% (Table 3). These high error rates are observed in multiple sequence contexts and are consistent with single nucleotide insertion studies (58,59) showing that human Pol η inserts dA and dC opposite 8-oxoG with similar efficiencies and then extends the resulting termini with similar efficiencies. Importantly, the high error rates we observe for human and mouse Pol η differ by only about 2-fold from the error rates for bypass of 8-oxoG by bovine Pol δ (83%, 5:1 ratio of A to C incorporation) and human Pol α (99.5%, 200:1 A to C incorporation) (9). These comparisons predict that switching from mammalian Pol δ or Pol α to mammalian Pol η in order to bypass 8-oxoG would have at most a 2-fold affect on 8-oxoG-dependent mutagenesis. Consistent with this is a recent demonstration that the frequency of 8-oxoG-dependent mutagenesis in a gapped plasmid is high, and, more importantly, it is similar in Pol η-deficient and Pol η-proficient human cells (60). Also relevant here may be the observation that, unlike for bypass of a TT dimer (41), we see no evidence for a switch to less processive synthesis after bypass of 8-oxoG by mouse and human Pol η (Figure 2B, Table 1). This represents a second difference between the yeast and mammalian Pol η. Given structure–function studies of other polymerases (10,12,14,15,18,19,23), these differences may depend on amino acids variations at the active sites and/or the little finger domains of yeast and mammalian pol η.
It is of course possible that the intrinsically low 8-oxoG bypass fidelity of the catalytic subunit of mammalian Pol η that we observe here can be improved. Our data (Table 1) indicate that RFC, PCNA and RPA may slightly improve bypass efficiency, but do not improve the 8-oxoG bypass fidelity of either yeast Pol δ or yeast Pol η (Table 3). This is consistent with earlier studies showing that these accessory proteins have at most a 1.5-fold effect on the fidelity of yeast Pol η when bypassing a TT dimer (50) or on the base substitution or indel error rates of yeast Pol δ and yeast Pol η when copying undamaged DNA (50,61). Theoretically, the situation could be different for the human proteins, based on the observation that PCNA and RPA allowed correct insertion of dC opposite 8-oxoG to proceed 68-fold more efficiently than incorporation of dA opposite 8-oxoG in single nucleotide insertion experiments (59). Thus, it will be interesting to measure the error rate for a complete 8-oxoG bypass reaction by mammalian Pol η in the presence of the accessory proteins. Although accessory proteins do not strongly influence single base error rates by many polymerases, including yeast Pol η during 8-oxoG bypass (Table 3), it remains possible that mammalian accessory proteins might improve Pol η fidelity. Other possibilities for improving the fidelity of 8-oxoG-dependent bypass by Pol η include (but are not limited to) correction of 8-oxoG-containing mismatches by mismatch repair (8,62) or by proofreading. Either correction process could suppress mutagenesis during TLS, even for TLS by a highly inaccurate polymerase, especially if multiple cycles of polymerization-error correction are permitted. Relevant here is the finding that 8-oxoG containing mismatches made by Pol δ are not proofread by its intrinsic exonuclease activity (Table 3), which is consistent with studies showing that the large fragment of E. coli DNA polymerase I (9), T7 DNA Pol (12) and RB69 Pol (15,63) also do not efficiently proofread dA·8-oxoG mismatches. This has been rationalized by the observation that an dA·8-oxoG mismatch mimics the geometry of a correct base pair, especially regarding interactions of atoms in the DNA minor groove with side chains at the polymerase active site (12,14). Nonetheless, even though Pol η lacks an intrinsic exonuclease activity, it is theoretically possibly that mismatches made during Pol η bypass of some lesions, e.g. 8-oxoG that retains base coding potential, may be proofread by a separate exonuclease. Genetic evidence exists for extrinsic proofreading of Pol α replication errors by Pol δ (64) and biochemical evidence has been obtained for extrinsic proofreading of human Pol η errors during SV40 origin-dependent replication of undamaged DNA (65) and for Pols δ and ε-dependent proofreading of Pol η misincorporations opposite the 3′ T of a TT dimer (40). For extrinsic proofreading to get around the geometric mimicry mentioned earlier, Pol δ and/or Pol ε may need to bind to the mismatched terminus directly via the exonuclease active site, as already reported for two other replicative DNA polymerases, T7 DNA pol (66) and RB69 pol (67).
The capacity of Pol η to suppress 8-oxoG-dependent mutagenesis may also depend on when the lesion is encountered, whether by a replication fork or during post-replication gap-filling (17). Also relevant may be the physiological state of the cell when the 8-oxoG is generated. For example, the error rates for bypass of 8-oxoG by Pol δ (9) and Pol η (Table 3) were all measured for reactions containing 100 µM dNTPs. This is approximately the dNTP concentration induced when budding yeast are exposed to the UV mimetic 4-NQO (68). Thus comparatively high-dNTP concentrations may be relevant to bypass of 8-oxoG or other lesions induced by sunlight or other exogenous environmental stress. On the other hand, the strongest evidence that Pol η suppresses 8-oxoG-dependent mutagenesis is for 8-oxoG arising spontaneously (17). Interestingly, dNTP concentrations in yeast are normally several-fold lower than in the induced state (69). Given that the efficiency of bypass of 8-oxoG by yeast Pol ε depends on the dNTP concentration (69), the successful competition among the many polymerases present in a cell for a lesion may depend not only on lesion identity, but also on whether the lesion arose spontaneously or was induced by exogenous environmental stress. Because the four dNTPs are not present in equimolar amounts in vivo [ref. (69) and earlier studies reviewed in refs (70,71)], the nature of the polymerase competition, as well as the fidelity of TLS, may vary depending on the lesion, the 5′ flanking template sequence and the amount of each dNTP available for insertion opposite the lesion and subsequent extension.
FUNDING
NIH Grant GM032431 [to P.M.B.]; Intramural Research Program of the NIH, National Institute of Environmental Health Sciences [to T.A.K.]. Funding for open access charge: Intramural Research Program, National Institutes of Health, National Institute of Environmental Health Sciences.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Katarzyna Bebenek and Zachary Pursell for thoughtful comments on the manuscript.
REFERENCES
- 1.Marnett LJ. Oxyradicals and DNA damage. Carcinogenesis. 2000;21:361–370. doi: 10.1093/carcin/21.3.361. [DOI] [PubMed] [Google Scholar]
- 2.Kouchakdjian M, Bodepudi V, Shibutani S, Eisenberg M, Johnson F, Grollman AP, Patel DJ. NMR structural studies of the ionizing radiation adduct 7-hydro-8-oxodeoxyguanosine (8-oxo-7H-dG) opposite deoxyadenosine in a DNA duplex. 8-Oxo-7H-dG(syn).dA(anti) alignment at lesion site. Biochemistry. 1991;30:1403–1412. doi: 10.1021/bi00219a034. [DOI] [PubMed] [Google Scholar]
- 3.Oda Y, Uesugi S, Ikehara M, Nishimura S, Kawase Y, Ishikawa H, Inoue H, Ohtsuka E. NMR studies of a DNA containing 8-hydroxydeoxyguanosine. Nucleic Acids Res. 1991;19:1407–1412. doi: 10.1093/nar/19.7.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lipscomb LA, Peek ME, Morningstar ML, Verghis SM, Miller EM, Rich A, Essigmann JM, Williams LD. X-ray structure of a DNA decamer containing 7,8-dihydro-8-oxoguanine. Proc. Natl Acad. Sci. USA. 1995;92:719–723. doi: 10.1073/pnas.92.3.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McAuley-Hecht KE, Leonard GA, Gibson NJ, Thomson JB, Watson WP, Hunter WN, Brown T. Crystal structure of a DNA duplex containing 8-hydroxydeoxyguanine-adenine base pairs. Biochemistry. 1994;33:10266–10270. doi: 10.1021/bi00200a006. [DOI] [PubMed] [Google Scholar]
- 6.Hainaut P, Hernandez T, Robinson A, Rodriguez-Tome P, Flores T, Hollstein M, Harris CC, Montesano R. IARC Database of p53 gene mutations in human tumors and cell lines: updated compilation, revised formats and new visualisation tools. Nucleic Acids Res. 1998;26:205–213. doi: 10.1093/nar/26.1.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Friedberg EC, Lehmann AR, Fuchs RP. Trading places: how do DNA polymerases switch during translesion DNA synthesis? Mol. Cell. 2005;18:499–505. doi: 10.1016/j.molcel.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 8.Mazurek A, Berardini M, Fishel R. Activation of human MutS homologs by 8-oxo-guanine DNA damage. J. Biol. Chem. 2002;277:8260–8266. doi: 10.1074/jbc.M111269200. [DOI] [PubMed] [Google Scholar]
- 9.Shibutani S, Takeshita M, Grollman AP. Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature. 1991;349:431–434. doi: 10.1038/349431a0. [DOI] [PubMed] [Google Scholar]
- 10.Miller H, Prasad R, Wilson SH, Johnson F, Grollman AP. 8-OxodGTP incorporation by DNA polymerase beta is modified by active-site residue Asn279. Biochemistry. 2000;39:1029–1033. doi: 10.1021/bi991789x. [DOI] [PubMed] [Google Scholar]
- 11.Furge LL, Guengerich FP. Analysis of nucleotide insertion and extension at 8-oxo-7,8-dihydroguanine by replicative T7 polymerase exo− and human immunodeficiency virus-1 reverse transcriptase using steady-state and pre-steady-state kinetics. Biochemistry. 1997;36:6475–6487. doi: 10.1021/bi9627267. [DOI] [PubMed] [Google Scholar]
- 12.Brieba LG, Eichman BF, Kokoska RJ, Doublie S, Kunkel TA, Ellenberger T. Structural basis for the dual coding potential of 8-oxoguanosine by a high-fidelity DNA polymerase. EMBO J. 2004;23:3452–3461. doi: 10.1038/sj.emboj.7600354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lowe LG, Guengerich FP. Steady-state and pre-steady-state kinetic analysis of dNTP insertion opposite 8-oxo-7,8-dihydroguanine by Escherichia coli polymerases I exo− and II exo. Biochemistry. 1996;35:9840–9849. doi: 10.1021/bi960485x. [DOI] [PubMed] [Google Scholar]
- 14.Hsu GW, Ober M, Carell T, Beese LS. Error-prone replication of oxidatively damaged DNA by a high-fidelity DNA polymerase. Nature. 2004;431:217–221. doi: 10.1038/nature02908. [DOI] [PubMed] [Google Scholar]
- 15.Freisinger E, Grollman AP, Miller H, Kisker C. Lesion (in)tolerance reveals insights into DNA replication fidelity. EMBO J. 2004;23:1494–1505. doi: 10.1038/sj.emboj.7600158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Einolf HJ, Guengerich FP. Fidelity of nucleotide insertion at 8-oxo-7,8-dihydroguanine by mammalian DNA polymerase delta. Steady-state and pre-steady-state kinetic analysis. J. Biol. Chem. 2001;276:3764–3771. doi: 10.1074/jbc.M006696200. [DOI] [PubMed] [Google Scholar]
- 17.Haracska L, Yu SL, Johnson RE, Prakash L, Prakash S. Efficient and accurate replication in the presence of 7,8-dihydro-8-oxoguanine by DNA polymerase η. Nat. Genet. 2000;25:458–461. doi: 10.1038/78169. [DOI] [PubMed] [Google Scholar]
- 18.Rechkoblit O, Malinina L, Cheng Y, Kuryavyi V, Broyde S, Geacintov NE, Patel DJ. Stepwise translocation of Dpo4 polymerase during error-free bypass of an oxoG lesion. PLoS Biol. 2006;4:e11. doi: 10.1371/journal.pbio.0040011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zang H, Irimia A, Choi JY, Angel KC, Loukachevitch LV, Egli M, Guengerich FP. Efficient and high fidelity incorporation of dCTP opposite 7,8-dihydro-8-oxodeoxyguanosine by Sulfolobus solfataricus DNA polymerase Dpo4. J. Biol. Chem. 2006;281:2358–2372. doi: 10.1074/jbc.M510889200. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Y, Yuan F, Wu X, Wang M, Rechkoblit O, Taylor JS, Geacintov NE, Wang Z. Error-free and error-prone lesion bypass by human DNA polymerase κ in vitro. Nucleic Acids Res. 2000;28:4138–4146. doi: 10.1093/nar/28.21.4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jaloszynski P, Ohashi E, Ohmori H, Nishimura S. Error-prone and inefficient replication across 8-hydroxyguanine (8-oxoguanine) in human and mouse ras gene fragments by DNA polymerase κ. Genes Cells. 2005;10:543–550. doi: 10.1111/j.1365-2443.2005.00858.x. [DOI] [PubMed] [Google Scholar]
- 22.Duarte V, Muller JG, Burrows CJAG. Insertion of dGMP and dAMP during in vitro DNA synthesis opposite an oxidized form of 7,8-dihydro-8-oxoguanine. Nucleic Acids Res. 1999;27:496–502. doi: 10.1093/nar/27.2.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krahn JM, Beard WA, Miller H, Grollman AP, Wilson SH. Structure of DNA polymerase beta with the mutagenic DNA lesion 8-oxodeoxyguanine reveals structural insights into its coding potential. Structure. 2003;11:121–127. doi: 10.1016/s0969-2126(02)00930-9. [DOI] [PubMed] [Google Scholar]
- 24.Maher VM, Ouellette LM, Curren RD, McCormick JJ. Caffeine enhancement of the cytotoxic and mutagenic effect of ultraviolet irradiation in a xeroderma pigmentosum variant strain of human cells. Biochem. Biophys. Res. Commun. 1976;71:228–234. doi: 10.1016/0006-291x(76)90272-2. [DOI] [PubMed] [Google Scholar]
- 25.Maher VM, Ouellette LM, Curren RD, McCormick JJ. Frequency of ultraviolet light-induced mutations is higher in xeroderma pigmentosum variant cells than in normal human cells. Nature. 1976;261:593–595. doi: 10.1038/261593a0. [DOI] [PubMed] [Google Scholar]
- 26.Roush AA, Suarez M, Friedberg EC, Radman M, Siede W. Deletion of the Saccharomyces cerevisiae gene RAD30 encoding an Escherichia coli DinB homolog confers UV radiation sensitivity and altered mutability. Mol. Gen. Genet. 1998;257:686–692. doi: 10.1007/s004380050698. [DOI] [PubMed] [Google Scholar]
- 27.McDonald JP, Levine AS, Woodgate R. The Saccharomyces cerevisiae RAD30 gene, a homologue of Escherichia coli dinB and umuC, is DNA damage inducible and functions in a novel error-free postreplication repair mechanism. Genetics. 1997;147:1557–1568. doi: 10.1093/genetics/147.4.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gibbs PE, McDonald J, Woodgate R, Lawrence CW. The relative roles in vivo of Saccharomyces cerevisiae Pol eta, Pol zeta, Rev1 protein and Pol32 in the bypass and mutation induction of an abasic site,T-T (6-4) photoadduct and T-T cis-syn cyclobutane dimer. Genetics. 2005;169:575–582. doi: 10.1534/genetics.104.034611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu SL, Johnson RE, Prakash S, Prakash L. Requirement of DNA polymerase η for error-free bypass of UV-induced CC and TC photoproducts. Mol. Cell Biol. 2001;21:185–188. doi: 10.1128/MCB.21.1.185-188.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang H, Siede W. UV-induced T → C transition at a TT photoproduct site is dependent on Saccharomyces cerevisiae polymerase eta in vivo. Nucleic Acids Res. 2002;30:1262–1267. doi: 10.1093/nar/30.5.1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kozmin S, Slezak G, Reynaud-Angelin A, Elie C, de Rycke Y, Boiteux S, Sage E. UVA radiation is highly mutagenic in cells that are unable to repair 7,8-dihydro-8-oxoguanine in Saccharomyces cerevisiae. Proc. Natl Acad Sci. 2005;102:13538–13543. doi: 10.1073/pnas.0504497102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang X, Rosenstein BS, Wang Y, Lebwohl M, Mitchell DM, Wei H. Induction of 8-oxo-7,8-dihydro-2′-deoxyguanosine by ultraviolet radiation in calf thymus DNA and HeLa cells. Photochem. Photobiol. 1997;65:119–124. doi: 10.1111/j.1751-1097.1997.tb01886.x. [DOI] [PubMed] [Google Scholar]
- 33.Kvam E, Tyrrell RM. Induction of oxidative DNA base damage in human skin cells by UV and near visible radiation. Carcinogenesis. 1997;18:2379–2384. doi: 10.1093/carcin/18.12.2379. [DOI] [PubMed] [Google Scholar]
- 34.Kielbassa C, Roza L, Epe B. Wavelength dependence of oxidative DNA damage induced by UV and visible light. Carcinogenesis. 1997;18:811–816. doi: 10.1093/carcin/18.4.811. [DOI] [PubMed] [Google Scholar]
- 35.de Padula M, Slezak G, Auffret van Der Kemp P, Boiteux S. The post-replication repair RAD18 and RAD6 genes are involved in the prevention of spontaneous mutations caused by 7,8-dihydro-8-oxoguanine in Saccharomyces cerevisiae. Nucleic Acids Res. 2004;32:5003–5010. doi: 10.1093/nar/gkh831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sakamoto AN, Stone JE, Kissling GE, McCulloch SD, Pavlov YI, Kunkel TA. Mutator alleles of yeast DNA polymerase zeta. DNA Repair. 2007;6:1829–1838. doi: 10.1016/j.dnarep.2007.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Masutani C, Araki M, Yamada A, Kusumoto R, Nogimori T, Maekawa T, Iwai S, Hanaoka F. Xeroderma pigmentosum variant (XP-V) correcting protein from HeLa cells has a thymine dimer bypass DNA polymerase activity. EMBO J. 1999;18:3491–3501. doi: 10.1093/emboj/18.12.3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johnson RE, Prakash S, Prakash L. Requirement of DNA polymerase activity of yeast Rad30 protein for its biological function. J. Biol. Chem. 1999;274:15975–15977. doi: 10.1074/jbc.274.23.15975. [DOI] [PubMed] [Google Scholar]
- 39.Kokoska RJ, McCulloch SD, Kunkel TA. The efficiency and specificity of apurinic/apyrimidinic site bypass by human DNA polymerase η and Sulfolobus solfataricus Dpo4. J. Biol. Chem. 2003;278:50537–50545. doi: 10.1074/jbc.M308515200. [DOI] [PubMed] [Google Scholar]
- 40.McCulloch SD, Kokoska RJ, Chilkova O, Welch CM, Johansson E, Burgers PM, Kunkel TA. Enzymatic switching for efficient and accurate translesion DNA replication. Nucleic Acids Res. 2004;32:4665–4675. doi: 10.1093/nar/gkh777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McCulloch SD, Kokoska RJ, Masutani C, Iwai S, Hanaoka F, Kunkel TA. Preferential cis-syn thymine dimer bypass by DNA polymerase η occurs with biased fidelity. Nature. 2004;428:97–100. doi: 10.1038/nature02352. [DOI] [PubMed] [Google Scholar]
- 42.McCulloch SD, Kunkel TA. Measuring the fidelity of translesion DNA synthesis. Methods Enzymol. 2006;408:341–355. doi: 10.1016/S0076-6879(06)08021-9. [DOI] [PubMed] [Google Scholar]
- 43.Bebenek K, Kunkel TA. Analyzing fidelity of DNA polymerases. Methods Enzymol. 1995;262:217–232. doi: 10.1016/0076-6879(95)62020-6. [DOI] [PubMed] [Google Scholar]
- 44.Yuan F, Zhang Y, Rajpal DK, Wu X, Guo D, Wang M, Taylor JS, Wang Z. Specificity of DNA lesion bypass by the yeast DNA polymerase η. J. Biol. Chem. 2000;275:8233–8239. doi: 10.1074/jbc.275.11.8233. [DOI] [PubMed] [Google Scholar]
- 45.Ayyagari R, Gomes XV, Gordenin DA, Burgers PM. Okazaki fragment maturation in yeast. I. Distribution of functions between FEN1 and DNA2. J. Biol. Chem. 2003;278:1618–1625. doi: 10.1074/jbc.M209801200. [DOI] [PubMed] [Google Scholar]
- 46.Fien K, Stillman B. Identification of replication factor C from Saccharomyces cerevisiae: a component of the leading-strand DNA replication complex. Mol. Cell Biol. 1992;12:155–163. doi: 10.1128/mcb.12.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Henricksen LA, Umbricht CB, Wold MS. Recombinant replication protein A: expression, complex formation, and functional characterization. J. Biol. Chem. 1994;269:11121–11132. [PubMed] [Google Scholar]
- 48.Burgers PM, Gerik KJ. Structure and processivity of two forms of Saccharomyces cerevisiae DNA polymerase δ. J. Biol. Chem. 1998;273:19756–19762. doi: 10.1074/jbc.273.31.19756. [DOI] [PubMed] [Google Scholar]
- 49.Matsuda T, Bebenek K, Masutani C, Rogozin IB, Hanaoka F, Kunkel TA. Error rate and specificity of human and murine DNA polymerase η. J. Mol. Biol. 2001;312:335–346. doi: 10.1006/jmbi.2001.4937. [DOI] [PubMed] [Google Scholar]
- 50.McCulloch SD, Wood A, Garg P, Burgers PM, Kunkel TA. Effects of accessory proteins on the bypass of a cis-syn thymine-thymine dimer by Saccharomyces cerevisiae DNA polymerase eta. Biochemistry. 2007;46:8888–8896. doi: 10.1021/bi700234t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lin Q, Clark AB, McCulloch SD, Yuan T, Bronson RT, Kunkel TA, Kucherlapati R. Increased susceptibility to UV-induced skin carcinogenesis in polymerase η-deficient mice. Cancer Res. 2006;66:87–94. doi: 10.1158/0008-5472.CAN-05-1862. [DOI] [PubMed] [Google Scholar]
- 52.Mozzherin DJ, Shibutani S, Tan CK, Downey KM, Fisher PA. Proliferating cell nuclear antigen promotes DNA synthesis past template lesions by mammalian DNA polymerase δ. Proc. Natl Acad. Sci. USA. 1997;94:6126–6131. doi: 10.1073/pnas.94.12.6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Garg P, Burgers PM. Ubiquitinated proliferating cell nuclear antigen activates translesion DNA polymerases η and REV1. Proc. Natl Acad. Sci. USA. 2005;102:18361–18366. doi: 10.1073/pnas.0505949102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Matsuda T, Bebenek K, Masutani C, Hanaoka F, Kunkel TA. Low fidelity DNA synthesis by human DNA polymerase-η. Nature. 2000;404:1011–1013. doi: 10.1038/35010014. [DOI] [PubMed] [Google Scholar]
- 55.Fortune JM, Pavlov YI, Welch CM, Johansson E, Burgers PM, Kunkel TA. Saccharomyces cerevisiae DNA polymerase δ: high fidelity for base substitutions but lower fidelity for single- and multi-base deletions. J. Biol. Chem. 2005;280:29980–29987. doi: 10.1074/jbc.M505236200. [DOI] [PubMed] [Google Scholar]
- 56.Carlson KD, Washington MT. Mechanism of efficient and accurate nucleotide incorporation opposite 7,8-dihydro-8-oxoguanine by Saccharomyces cerevisiae DNA polymerase eta. Mol. Cell Biol. 2005;25:2169–2176. doi: 10.1128/MCB.25.6.2169-2176.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee DH, Pfeifer GP. Translesion synthesis of 7,8-dihydro-8-oxo-2′-deoxyguanosine by DNA polymerase eta in vivo. Mutat. Res. 2008;641:19–26. doi: 10.1016/j.mrfmmm.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang Y, Yuan F, Wu X, Rechkoblit O, Taylor JS, Geacintov NE, Wang Z. Error-prone lesion bypass by human DNA polymerase η. Nucleic Acids Res. 2000;28:4717–4724. doi: 10.1093/nar/28.23.4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Maga G, Villani G, Crespan E, Wimmer U, Ferrari E, Bertocci B, Hubscher U. 8-Oxo-guanine bypass by human DNA polymerases in the presence of auxiliary proteins. Nature. 2007;447:606–608. doi: 10.1038/nature05843. [DOI] [PubMed] [Google Scholar]
- 60.Avkin S, Livneh Z. Efficiency, specificity and DNA polymerase-dependence of translesion replication across the oxidative DNA lesion 8-oxoguanine in human cells. Mutat. Res./Fund. Mol. Mech. Mutagen. 2002;510:81–90. doi: 10.1016/s0027-5107(02)00254-3. [DOI] [PubMed] [Google Scholar]
- 61.Fortune JM, Stith CM, Kissling GE, Burgers PM, Kunkel TA. RPA and PCNA suppress formation of large deletion errors by yeast DNA polymerase delta. Nucleic Acids Res. 2006;34:4335–4341. doi: 10.1093/nar/gkl403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Young LC, Hays JB, Tron VA, Andrew SE. DNA mismatch repair proteins: potential guardians against genomic instability and tumorigenesis induced by ultraviolet photoproducts. J. Invest. Dermatol. 2003;121:435–440. doi: 10.1046/j.1523-1747.2003.12450.x. [DOI] [PubMed] [Google Scholar]
- 63.Zhong X, Pedersen LC, Kunkel TA. Characterization of a replicative DNA polymerase mutant with reduced fidelity and increased translesion synthesis capacity. Nucleic Acids Res. 2008;36:3892–3904. doi: 10.1093/nar/gkn312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pavlov YI, Frahm C, McElhinny SAN, Niimi A, Suzuki M, Kunkel TA. Evidence that errors made by DNA polymerase alpha are corrected by DNA polymerase delta. Curr. Biol. 2006;16:202–207. doi: 10.1016/j.cub.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 65.Bebenek K, Matsuda T, Masutani C, Hanaoka F, Kunkel TA. Proofreading of DNA polymerase η-dependent replication errors. J. Biol. Chem. 2001;276:2317–2320. doi: 10.1074/jbc.C000690200. [DOI] [PubMed] [Google Scholar]
- 66.Donlin MJ, Patel SS, Johnson KA. Kinetic partitioning between the exonuclease and polymerase sites in DNA error correction. Biochemistry. 1991;30:538–546. doi: 10.1021/bi00216a031. [DOI] [PubMed] [Google Scholar]
- 67.Shamoo Y, Steitz TA. Building a replisome from interacting pieces: sliding clamp complexed to a peptide from DNA polymerase and a polymerase editing complex. Cell. 1999;99:155–166. doi: 10.1016/s0092-8674(00)81647-5. [DOI] [PubMed] [Google Scholar]
- 68.Chabes A, Georgieva B, Domkin V, Zhao X, Rothstein R, Thelander L. Survival of DNA damage in yeast directly depends on increased dNTP levels allowed by relaxed feedback inhibition of ribonucleotide reductase. Cell. 2003;112:391–401. doi: 10.1016/s0092-8674(03)00075-8. [DOI] [PubMed] [Google Scholar]
- 69.Sabouri N, Viberg J, Goyal DK, Johansson E, Chabes A. Evidence for lesion bypass by yeast replicative DNA polymerases during DNA damage. Nucleic Acids Res. 2008;36:5660–5667. doi: 10.1093/nar/gkn555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kunkel TA. Biological asymmetries and the fidelity of eukaryotic DNA replication. Bioessays. 1992;14:303–308. doi: 10.1002/bies.950140503. [DOI] [PubMed] [Google Scholar]
- 71.Kunz BA, Kohalmi SE, Kunkel TA, Mathews CK, McIntosh EM, Reidy JA. International Commission for Protection Against Environmental Mutagens and Carcinogens. Deoxyribonucleoside triphosphate levels: a critical factor in the maintenance of genetic stability. Mutat. Res. 1994;318:1–64. doi: 10.1016/0165-1110(94)90006-x. [DOI] [PubMed] [Google Scholar]