Abstract
The use of chemically synthesized short interfering RNAs (siRNAs) is currently the method of choice to manipulate gene expression in mammalian cell culture, yet improvements of siRNA design is expectably required for successful application in vivo. Several studies have aimed at improving siRNA performance through the introduction of chemical modifications but a direct comparison of these results is difficult. We have directly compared the effect of 21 types of chemical modifications on siRNA activity and toxicity in a total of 2160 siRNA duplexes. We demonstrate that siRNA activity is primarily enhanced by favouring the incorporation of the intended antisense strand during RNA-induced silencing complex (RISC) loading by modulation of siRNA thermodynamic asymmetry and engineering of siRNA 3′-overhangs. Collectively, our results provide unique insights into the tolerance for chemical modifications and provide a simple guide to successful chemical modification of siRNAs with improved activity, stability and low toxicity.
INTRODUCTION
The use of RNA interference (RNAi)-based strategies has recently become the technique of choice to silence gene expression in mammalian cell culture. In typical strategies, 21-bp dsRNA molecules, termed short interfering RNAs (siRNAs), with perfect complementarity to the target RNA are used as experimental triggers of RNAi (1). Upon delivery into the cell, siRNAs are incorporated into the RNA-induced silencing complex (RISC) containing the signature component of the RNAi machinery, Argonaute 2 (Ago2) (2–4). The siRNA strand containing the thermodynamically less stable 5′-end is preferentially incorporated as the guiding strand of RISC (5,6), while the non-guiding sense strand (SS) of the siRNA duplex is cleaved by Ago2 and liberated (7–9). This generates the activated RISC* containing the guiding antisense strand (AS), which binds the complementary target RNAs and leads to their cleavage by Ago2 (10). Whereas unmodified siRNAs are highly efficient in cell culture, chemical modification is generally considered a prerequisite for fulfilling the potential of siRNAs in vivo and indeed successful experiments in animal models have relied on injection of chemically modified siRNAs (11–13). In particular, the half-life of unmodified siRNAs in vivo is in the range of minutes, but this can be significantly improved by chemical modifications, albeit often at the price of reduced siRNA activity (14–18). In addition, chemical modification of siRNAs may be required for delivery strategies, e.g. by conjugation to lipids (11,19), membrane-penetrating peptides (20) or RNA aptamer sequences (21). Furthermore, modified siRNAs with superior potency will reduce the dose required for gene silencing (14,22,23), and specific chemical modifications can minimize siRNA side-effects, such as the induction of recipient immune responses and inherent off-targeting effects (24–26). Therefore, a primary objective in the optimization of siRNA performance through chemical modification is to identify the type and position of specific alterations that can be introduced without compromising siRNA activity. A direct comparison of the various chemistries used for siRNA optimization is currently hampered by the fact that existing studies evaluate isolated chemistries in a variety of assays and sequence contexts. In addition, multiple chemistries are rarely included in the same siRNA duplex rendering identification of positive or negative synergy impossible.
In this study, we have made a direct comparison of the impact of 21 different types of chemical modifications on siRNA efficiency and cell viability using a total of 2160 siRNAs. The results from this extensive screen provide important insights into where and how siRNA strands can be modified by particular chemistries and how chemistries can be combined; moreover it allows us to formulate design rules to create superior siRNAs in terms of activity, stability and toxicity.
MATERIALS AND METHODS
Cell culture
HeLa cells stably expressing eGFP (HeLa-eGFP) were obtained by transfection with pEGFP-C1 (Clontech Laboratories, USA). A clone of the transfected pool was derived by G418 selection and cultivated in D-MEM (GIBCO-Invitrogen Corporation, USA) containing 10% FBS (foetal bovine serum) and 1% penicillin/streptomycin. The human lung cancer cell line H1299 was grown in RPMI-1640 (GIBCO) containing 10% FBS and 1% penicillin/streptomycin.
Synthesis of oligonucleotides
The phosphoramidites were incorporated into the siRNA sequence through solid-phase DNA/RNA synthesis (27) on an automated DNA/RNA synthesizer (for structures of chemically modified nucleotide monomers, see Figure 1). For a standard RNA synthesis cycle (1–5 µmol scale), O2′-TBDMS protected RNA phosphoramidites and common reagents were used and the stepwise coupling yield of all monomers was >99%. For incorporation of modified nucleotides, a coupling time of 10 min was used. Following standard deprotection, purification and work-up, the composition and purity (>80%) of the resulting oligonucleotides was confirmed by MALDI-MS analysis and ion exchange HPLC. UNA (unlocked nucleic acid), LNA (locked nucleic acid) and ALN (α-L-LNA) modified oligonucleotides were obtained by using commercially available UNA (www.ribotask.com), LNA (www.exiqon.com) and α-L-LNA (www.exiqon.com). The UNA phosphoramidites were synthesized as O2′-TBDMS derivatives by an optimized version of the published procedure for synthesis of the thymine monomer (28). The chemical synthesis of the remaining modified phosphoramidites is previously described; HM (4′-C-hydroxymethyl-DNA) (29), ADA (2′-N-adamantylmethylcarbonyl-2′-amino-LNA) (Babu, B.W. et al., Manuscript in preparation), PYR (2′-N-pyren-1-ylmethyl-2′-amino-LNA) (30), EA (2′-aminoethyl), GE (2′-guanidinoethyl), CE (2′-cyanoethyl) (31,32), AP (2′-aminopropyl) (33), OX (oxetane-LNA) [(34) and references therein], CLNA (2′,4′-carbocyclic-LNA-locked nucleic acid), CENA (2′,4′-carbocyclic-ENA-locked nucleic acid) (35), AENA (2′-deoxy-2′-N,4′-C-ethylene-LNA) (36), OMe (2′-O-methyl), F (2′-fluoro) (22), ANA (altritol nucleic acid) (37), HNA (hexitol nucleic acid) (38), AEM (2′-aminoethoxymethyl) and APM (2′-aminopropoxymethyl) (39).
Figure 1.
Structural overview of chemical modifications investigated.
Evaluation of siRNA effect on eGFP expression and cell viability
A library of 2160 siRNAs was derived by annealing 48 ASs and 45 SSs in Tris–Borate–EDTA buffer (Ambion Applied Biosystem Inc., USA) by heating to 90°C for 2 min and slow cooling to 25°C for 45 min using a Gene Amp-R PCR System 9700 termocycler (Applied Biosystems, USA). HeLa-eGFP were plated in a 384-well format in 50 μl complete medium 24 h prior to transfection with 10 nM siRNAs (final concentration) complexed with 0.17 µl INTERFERin/50 µl medium (Polyplus-transfection SA, France) using an automated robotic platform (EVO-Freedom, TECAN Ltd, Switzerland). Cells were fixed 72 h post-transfection with 3.7% paraformaldehyde (PFA) and stained with a mix of 4,6-diamidino-2-phenylindole (DAPI) and Sytoblue 42 for nuclear and cytosolic counter-stain and imaged using an automated confocal microscope (Opera, PerkinElmer Cellular Technologies Germany GmbH) equipped with a 20× water immersion objective. Images were analysed in a blind manner using automated image analysis software acquiring 10 fields per well corresponding to ∼1000 cells (Acapella, PerkinElmer Cellular Technologies Germany GmbH). Cell number, nuclei size and eGFP intensity data were analysed using the R-Project statistical package (40). The eGFP intensity threshold was calculated by taking the mean of the values detected in the positive controls in each plate plus three times the standard deviation; eGFP values below this threshold point were considered as eGFP down-regulation. The results represent average values from triplicate assays repeated twice. eGFP expression was normalized to cells transfected with siEGFP mismatch control (Dharmacon-, Dharmacon, Inc.) and cell viability was normalized to cells treated with transfection reagent alone.
siRNA stability assay
Annealed siRNAs were incubated at 37°C in 80% FBS in D-MEM (Gibco) for the indicated time. Aliquots of 5 µl (each containing 20 pmol of siRNA) were diluted in 25 µl 1.2× TBE loading buffer (1.2× TBE, 10% glycerol, bromphenol blue) and snap frozen on dry ice. Samples were run on a 6% agarose gel and stained using SYBR Gold® (Invitrogen).
Dual-luciferase assay
The reporter constructs pISOAS-target and pISOSS-target are described previously (41). H1299 cells were plated in 48-well plates in RPMI supplemented with 10% FBS and grown to 40–60% confluency overnight. pISOAS-target and pISOSS-target (25 ng) were co-transfected with 0.2 ng pRluc-N2 (Perkin–Elmer) and siRNA duplexes (5–50 nM final concentration) by simultaneous use of 0.5 µl TransIT-LT1 (Mirus) and 0.5 µl TransIT-TKO (Mirus) according to manufacturer's protocol. Luciferase activity was measured 48 h post-transfection using the ‘Dual-luciferase reporter assay system’ (Promega) according to the manufacturer's protocol on a FLUOstar luminometer (BMG labtech); firefly luciferase signals (sample) were normalized to the renilla luciferase signals (transfection control).
RESULTS
We have evaluated the impact of 21 different types of chemical modifications on the activity and toxicity of a standard 21–22 bp siRNA targeting eGFP using all combinations of 48 ASs and 45 SSs, thereby generating a total of 2160 chemically different siRNA duplexes (see Table 1 for AS and SS sequences). The modifications can be broadly categorized into 2′-substituted RNAs (OMe, F, DNA, AEM, APM, EA, AP, CE, GE), 4′-modified RNAs (HM), Locked RNAs (LNA, ALN, ADA, PYR, OX, AENA, CENA, CLNA) and RNAs with radical modifications of the ribose sugar ring (UNA, ANA and HNA; Figure 1; see ‘Material and Methods’ section for full names). In brief, HeLa cells stably expressing eGFP were transfected with siRNA duplexes (10 nM final concentration), and eGFP levels and cell viability were evaluated 72 h post-transfection.
Table 1.
Overview of chemically modified ASs and SSs
| Name | AS/SS | Modification | eGFP levelsa | Viabilitya | Sequence (5′–3′) |
|---|---|---|---|---|---|
| DHARM1 | AS | OMe | 0.24 ± 0.02 | 0.82 ± 0.11 | ACOMeUUGUGGCCGUUUACGUCGC |
| DO1001 | AS | OMe/EA | 0.20 ± 0.04 | 0.88 ± 0.11 | AOMeCUUGUGGCCGUUUACGUCGCEAU |
| DO1002 | AS | OMe/CE | 0.49 ± 0.06 | 1.01 ± 0.24 | AOMeCUUGUGGCCGUUUACGUCECGCU |
| GS2371 | AS | ANA | 0.41 ± 0.03 | 0.58 ± 0.15 | AANACANAUUGUGGCCGUUUACGUCGC |
| GS2372 | AS | ANA | 0.11 ± 0.01 | 0.59 ± 0.08 | ACUUGUGGCCGUUUACGUCGANACANAU |
| GS2373 | AS | ANA | 1.12 ± 0.08 | 0.73 ± 0.19 | ACUUGANAUGGANACCGANAUUUANAACGANAUCGANACU |
| GS2378 | AS | ANA | 0.25 ± 0.03 | 0.51 ± 0.07 | ACUANAUGUANAGGCCGUUUANAACGUANACGANACANAU |
| GS2537 | AS | HNA/DNA | 0.19 ± 0.02 | 0.57 ± 0.04 | ACUUGUGGCCGUUUACGUHNACHNAGHNATDNA |
| GS2538 | AS | HNA/DNA | 0.21 ± 0.04 | 0.48 ± 0.07 | ACHNAUUGUGGCCGUUUACGUHNACHNAGHNATDNA |
| GS2539 | AS | HNA/DNA | 0.20 ± 0.02 | 0.80 ± 0.12 | ACUHNAUGUGGCCGUUUACGUHNACHNAGHNATDNA |
| GS2540 | AS | HNA/DNA | 0.63 ± 0.08 | 0.71 ± 0.12 | ACHNAUUGUGGCHNACGUUUACGUHNACHNAGHNATDNA |
| GS2544 | AS | AEM/DNA | 0.59 ± 0.06 | 0.94 ± 0.12 | ACUUGUGGCCGUUUACGUAEMCAEMGTDNA |
| GS2549 | AS | APM/DNA | 0.87 ± 0.11 | 0.90 ± 0.19 | ACUUGUGGCCGUUUACGUAPMCAPMGTDNA |
| JC10 | AS | F/OMe | 0.32 ± 0.03 | 0.70 ± 0.28 | AFCOMeUFUOMeGFUOMeGFGOMeCFCOMeGFUOMeUFUOMeAFC OMeGFUOMeCFGOMeCF |
| JC-A1 | AS | AENA | 0.26 ± 0.02 | 0.88 ± 0.18 | ACUAENAUGUGGCCGUUUACGUAENACGC |
| JC-A2 | AS | AENA | 0.63 ± 0.05 | 0.55 ± 0.15 | ACUUAENAGUGGCCGUUUACGUAENACGC |
| JC-A3 | AS | AENA | 0.69 ± 0.11 | 1.05 ± 0.18 | ACUAENAUAENAGUGGCCGUUUACGUAENACGC |
| JC-F1 | AS | CENA | 0.10 ± 0.02 | 0.87 ± 0.17 | ACUCENAUGUGGCCGUUUACGUCENACGC |
| JC-F2 | AS | CENA | 0.18 ± 0.05 | 0.80 ± 0.19 | ACUUCENAGUGGCCGUUUACGUCENACGC |
| JC-F3 | AS | CENA | 0.19 ± 0.04 | 1.08 ± 0.12 | ACUCENAUCENAGUGGCCGUUUACGUCENACGC |
| JC-S1 | AS | CLNA | 0.12 ± 0.01 | 0.80 ± 0.12 | ACUCLNAUGUGGCCGUUUACGUCLNACGC |
| JC-S2 | AS | CLNA | 0.28 ± 0.01 | 0.65 ± 0.16 | ACUUCLNAGUGGCCGUUUACGUCLNACGC |
| JC-S3 | AS | CLNA | 0.31 ± 0.04 | 0.86 ± 0.08 | ACUCLNAUCLNAGUGGCCGUUUACGUCLNACGC |
| JE1001 | AS | EA | 0.13 ± 0.01 | 0.62 ± 0.17 | ACUUGUGGCCGUUUACGUCGEACEAU |
| JH1001 | AS | AP | 0.45 ± 0.09 | 0.78 ± 0.17 | AAPCUUGUGGCCGUUUACGUCGCU |
| JW1186 | AS | HM/LNA | 0.10 ± 0.04 | 0.95 ± 0.22 | ACUUGTHMGGCCGUUUACGUCGLNACLNAU |
| JW1187 | AS | HM/LNA | 0.76 ± 0.13 | 1.01 ± 0.25 | ACTHMUGTHMGGCCGUUTHMACGTHMCGLNACLNAU |
| W006 | AS | LNA | 0.09 ± 0.01 | 0.49 ± 0.11 | ACUUGUGGCCGUUUACGUCGLNACLNAU |
| W010 | AS | LNA | 0.54 ± 0.08 | 0.67 ± 0.07 | ACTLNAUGTLNAGGCCGUUTLNAACGTLNACGLNACLNAU |
| W042 | AS | HM | 0.18 ± 0.03 | 0.44 ± 0.09 | ACUUGUGGCCGUUUACGUCHMGCHMU |
| W047 | AS | ALN | 0.14 ± 0.03 | 0.46 ± 0.04 | ACUUGUGGCCGUUUACGUCGCALNU |
| W053 | AS | 0.14 ± 0.02 | 0.43 ± 0.09 | ACUUGUGGCCGUUUACGUCGC | |
| W054 | AS | HM | 0.33 ± 0.06 | 0.69 ± 0.12 | ACUUGUGGCCGUUUACGUCTHMTHMU |
| W059 | AS | HM | 0.18 ± 0.02 | 0.72 ± 0.14 | ACUUGUGGCCGUUUACGUCGTHMU |
| W068 | AS | ADA/LNA | 0.54 ± 0.11 | 0.64 ± 0.12 | ACUUGTADAGGCCGUUUACGUCGLNACLNAU |
| W075 | AS | LNA | 0.14 ± 0.02 | 0.86 ± 0.15 | ACUUGUGGCCGUUUACGUCLNAGLNAC |
| W095 | AS | ADA/LNA | 0.36 ± 0.04 | 0.78 ± 0.15 | ACUUGUGGCCGUUUACGTADACGLNACLNAU |
| W096 | AS | PYR/LNA | 0.38 ± 0.06 | 0.52 ± 0.15 | ACUUGTPYRGGCCGUUUACGUCGLNACLNAU |
| W097 | AS | PYR/LNA | 0.46 ± 0.05 | 0.73 ± 0.19 | ACUUGUGGCCGUUUACGTPYRCGLNACLNAU |
| W106 | AS | OMe | 1.11 ± 0.12 | 0.73 ± 0.31 | AOMeCOMeUOMeUOMeGOMeUOMeGOMeGOMeCOMeCOMeGOMeUOMeUOMeUOMeAOMeCOMeGOMeUOMeCOMeGOMeCOMeUOMe |
| W123 | AS | UNA/LNA | 0.11 ± 0.02 | 1.08 ± 0.22 | ACUUGUUNAGGCCGUUUACGUCGLNACLNAU |
| W124 | AS | UNA/LNA | 0.16 ± 0.05 | 0.89 ± 0.22 | ACUUNAUGUGGCCGUUUACGUCGLNACLNAU |
| W125 | AS | UNA/LNA | 0.11 ± 0.03 | 0.89 ± 0.13 | ACUUGUGGCCGUUUACGUUNACGLNACLNAU |
| W126 | AS | UNA/LNA | 0.13 ± 0.03 | 0.72 ± 0.15 | ACUUGUGGCCGUUNAUUACGUCGLNACLNAU |
| W127 | AS | UNA | 0.26 ± 0.05 | 0.66 ± 0.12 | ACUUGUGGCCGUUUACGUCGCUNAU |
| W128 | AS | UNA | 0.56 ± 0.11 | 0.80 ± 0.23 | ACUUNAUGUGGCCGUUUACGUCGCUNAU |
| W180 | AS | LNA | 0.27 ± 0.04 | 0.78 ± 0.22 | ACUUGUGGCCGUUUACGUCGLNACLNA |
| W209 | AS | LNA | 1.18 ± 0.23 | 0.88 ± 0.22 | ACTLNAUGTLNAGGCCGUUTLNAACGTLNACGLNACLNA |
| DO003 | SS | EA | 0.10 ± 0.01 | 0.67 ± 0.10 | GACGUAAACGGCCACAAEAGUUC |
| DO004 | SS | GE | 0.12 ± 0.01 | 0.62 ± 0.05 | GACGUAAACGGCCACAAGEGUUC |
| GS2332 | SS | HNA | 0.15 ± 0.01 | 0.55 ± 0.12 | GAHNACGUAAAHNACGGCCAHNACAAHNAGUUC |
| GS2366 | SS | HNA | 0.40 ± 0.06 | 0.65 ± 0.60 | GACGUHNAAAAHNACGGCCAHNACAAHNAGUUCTDNA |
| GS2368 | SS | ANA/DNA | 0.11 ± 0.02 | 0.71 ± 0.11 | GANAAANACGUAAACGGCCACAAGUUCTDNA |
| GS2369 | SS | ANA/DNA | 0.10 ± 0.01 | 0.64 ± 0.18 | GACGUAAACGGCCACAAGANAUANAUCTDNA |
| GS2370 | SS | ANA/DNA | 0.31 ± 0.03 | 0.73 ± 0.22 | GACGUANAAAAANACGGANACCAANACAAANAGUUCTDNA |
| GS2374 | SS | ANA | 0.16 ± 0.03 | 0.60 ± 0.14 | GACGUAAACGGCCACAAGUUANACANAU |
| GS2383 | SS | HNA | 0.11 ± 0.01 | 0.78 ± 0.16 | GACGUAAACGGCCACAAGUHNAUHNAC |
| GS2534 | SS | HNA/DNA | 0.11 ± 0.01 | 0.58 ± 0.15 | GHNAAHNACHNAGUAAACGGCCACAAGHNAUHNAUHNATDNA |
| GS2535 | SS | HNA/DNA | 0.19 ± 0.03 | 0.64 ± 0.09 | GACGUHNAAAAHNACGGHNACCAHNACAAGUUTDNA |
| GS2536 | SS | HNA/DNA | 0.13 ± 0.02 | 0.74 ± 0.12 | GACGUHNAAAAHNACGGHNACCAHNACAAGUHNAUHNATDNA |
| GS2542 | SS | AEM/DNA | 0.11 ± 0.01 | 0.66 ± 0.15 | GACGUAAACGGCCACAAGUAEMUAEMTDNA |
| GS2543 | SS | AEM/DNA | 0.11 ± 0.02 | 0.62 ± 0.20 | GACAEMGUAEMAAACAEMGGCCACAAGUAEMUAEMTDNA |
| GS2547 | SS | APM/DNA | 0.12 ± 0.01 | 0.64 ± 0.18 | GACGUAAACGGCCACAAGUAPMUAPMTDNA |
| GS2548 | SS | APM/DNA | 0.17 ± 0.02 | 0.61 ± 0.08 | GACAPMGUAPMAAACAPMGGCCACAAGUAPMUAPMTDNA |
| JC1 | SS | OX | 0.11 ± 0.01 | 0.63 ± 0.20 | GACGUAAACGGCCACAOXAGOXUUC |
| JC2 | SS | OX | 0.13 ± 0.02 | 0.81 ± 0.19 | GAOXCGOXUAAACGGCCACAAGUUC |
| JC5 | SS | F/OMe | 0.13 ± 0.02 | 0.90 ± 0.19 | GFAOMeCFGOMeUFAOMeAFAOMeCFGOMeGFCOMeCFAOMeCFA OMeAFGOMeUFUOMeCF |
| JE0007 | SS | EA | 0.11 ± 0.02 | 0.79 ± 0.23 | GACGUAAACGGCCACAAGUUEAC |
| JE0008 | SS | EA | 0.11 ± 0.02 | 0.66 ± 0.13 | GACGUAAACGGCCACAAGUEAUC |
| JE010 | SS | EA | 0.13 ± 0.01 | 0.84 ± 0.15 | GACGUEAAAACGGCCACAAGUEAUEAC |
| JW1104b | SS | LNA | GACGUAAACGGCCACAAGUTLNACLNAU | ||
| JW1106 | SS | LNA | 0.12 ± 0.01 | 0.80 ± 0.16 | GACLNAGUAAACLNAGGCCACLNAAAGUTLNACLNAU |
| JW1188 | SS | HM | 0.15 ± 0.02 | 0.62 ± 0.15 | GACGUAAACGGCCACAAGTHMTHMC |
| JW1189 | SS | HM | 0.11 ± 0.02 | 0.74 ± 0.13 | GACGTHMAAACGGCCACAAGTHMTHMC |
| W004+W179 | SS | LNA | 0.13 ± 0.03 | 0.71 ± 0.17 | GACLNAGUAAACLNAG + GCCLNAACLNAAAGUTLNACLNA |
| W007 | SS | DNA | 0.69 ± 0.09 | 1.06 ± 0.19 | GDNAADNACDNAGDNATDNAADNAADNAADNACDNAGDNAGDNACDNA CDNAADNACDNAADNAADNAGDNATDNATLNACLNATDNA |
| W008 | SS | DNA/LNA | 0.41 ± 0.05 | 1.02 ± 0.16 | GDNAADNACLNAGDNATDNAALNAADNAADNACLNAGDNAGDNACLNA CDNAADNACLNAADNAADNAGLNATDNATLNACLNATDNA |
| W009 | SS | DNA/LNA | 0.71 ± 0.06 | 1.04 ± 0.17 | GDNAALNACDNAGLNATDNAALNAADNAADNACDNAGLNAGDNACLNA CDNAALNACDNAALNAADNAGLNATDNATLNACLNATDNA |
| W011 | SS | LNA | 0.12 ± 0.02 | 0.69 ± 0.13 | GACGTLNAAAACGGCCACAAGUTLNACLNAU |
| W013 | SS | LNA | 0.19 ± 0.03 | 0.95 ± 0.13 | GACLNAGTLNAAALNAACLNAGGCCACAAGUTLNACLNAU |
| W037 | SS | LNA | 0.19 ± 0.03 | 0.66 ± 0.11 | GACLNAGUAAACLNAGGCCLNAACLNAAAGUTLNACLNAU |
| W043 | SS | HM | 0.11 ± 0.02 | 0.60 ± 0.14 | GACGUAAACGGCCACAAGUTHMCHMU |
| W044 | SS | HM | 0.12 ± 0.02 | 0.67 ± 0.17 | GACHMGTHMAAACHMGGCCHMACAAGUTHMCHMU |
| W049 | SS | ALN | 0.11 ± 0.02 | 0.64 ± 0.14 | GACGUAAACGGCCACAAGUTALNCALNU |
| W050 | SS | ALN | 0.12 ± 0.02 | 0.68 ± 0.15 | GACALNGUAAACALNGGCCACALNAAGUTALNCALNU |
| W060 | SS | HM | 0.10 ± 0.01 | 0.70 ± 0.11 | GACGUAAACGGCCACAAGUUTHMU |
| W069 | SS | ADA/LNA | 0.15 ± 0.02 | 0.93 ± 0.27 | GACGTADAAAACGGCCACAAGUTADACLNAU |
| W129 | SS | UNA/LNA | 0.20 ± 0.04 | 0.79 ± 0.22 | GACGUUNAAAACGGCCACAAGUTLNACLNAU |
| W130 | SS | UNA/LNA | 0.40 ± 0.05 | 0.78 ± 0.18 | GACGUAAACCUNAGGCCACAAGUTLNACLNAU |
| W131 | SS | UNA | 0.11 ± 0.02 | 0.58 ± 0.16 | GACGUAAACGGCCACAAGUUUUNAU |
| W132 | SS | UNA | 0.13 ± 0.02 | 0.59 ± 0.11 | GACGUUNAAAACGGCCACAAGUUUUNAU |
| W181 | SS | LNA | 0.14 ± 0.02 | 0.65 ± 0.18 | GACLNAGUAAALNACGGCCLNAACLNAAAGUTLNACLNAU |
| W194 | SS | LNA | 0.13 ± 0.03 | 0.45 ± 0.17 | GACGUAAACGGCCACAAGUTLNACLNA |
| W207 | SS | 0.14 ± 0.02 | 0.43 ± 0.09 | GACGUAAACGGCCACAAGUUC |
The name, type of chemical modification, eGFP levels, viability and sequence of the investigated ASs and SSs are given. The following modification abbreviations are used: 2′-O-methyl (OMe), 2′-fluoro (F), 2′-deoxy (DNA), 2′-aminoethoxymethyl (AEM), 2′-aminopropoxymethyl (APM), 2′-aminoethyl (EA), 2′-aminopropyl (AP), 2′-cyanoethyl (CE), 2′-guanidinoethyl (GE), 4′-C-hydroxymethyl-DNA (HM), locked nucleic acid (LNA), alfa-L-LNA (ALN), 2′-N-adamant-1-ylmethylcarbonyl-2′-amino-LNA (ADA), 2′-N-pyren-1-ylmethyl-2′-amino-LNA (PYR), oxetane-LNA (OX), 2′,4′-carbocyclic-ENA-LNA (CENA), 2′,4′-carbocyclic-LNA-locked nucleic acid (CLNA), unlocked nucleic acid (UNA), altritol nucleic acid (ANA) 2′-deoxy-2′-N,4′-C-ethylene-LNA (AENA) and hexitol nucleic acid (HNA). The position of the modification is shown in bold in the oligo sequence.
aeGFP levels and viability is given for the particular strand in combination with an unmodified opposing strand (SS=W207, AS=W053). eGFP levels were normalized to cells transfected with siEGFP mismatch control (Dharmacon, Dharmacon, Inc.) and viability was normalized to cells treated with transfection reagent alone.
bThe JW1104 SS is not included in the large-scale siRNA screen but in subsequent analysis only.
Modifications are well tolerated in the SS
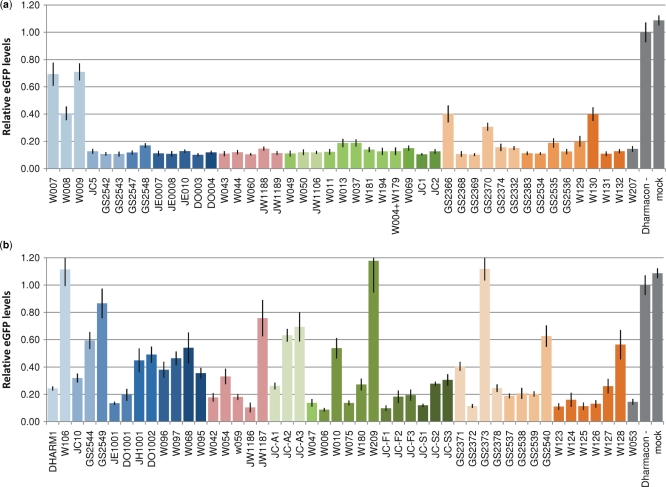
We initially evaluated the impact of chemical modification on siRNA activity for all ASs and SSs in combination with an unmodified RNA strand (SS and AS termed W207 and W053, respectively). Modifications in the SS were well tolerated, and even heavily modified SSs supported knock down (KD) of eGFP levels to below 20% [e.g. HNA (GS2536), AEM (GS2543), APM (GS2548), HM (W044), OMe/F (JC5) and LNA (W037)] (Figure 2a and Table 1). Only in a few cases did introduction of multiple modifications result in reduced silencing potency, as seen for ANA (GS2366), HNA (GS2370), an additional UNA residue (W130) and full substitution using DNA (W007) and DNA/LNA (W008, W009). Bulky modifications were relatively well tolerated in the SS [e.g. ADA (W069), AEM (GS2542, GS2543) and APM (GS2547, GS2548)] as opposed to in the AS (see below). Interestingly, thermodynamic stabilization of the siRNA duplex by LNA could be achieved via SS modification, whereas a similar level of LNA modification in the AS abolished siRNA activity (compare SS W037 and AS W010; Figure 2). This illustrates that the SS can be functionalized by extensive modifications as long as the RNA duplex structure is not grossly distorted.
Figure 2.
Silencing activity of chemically modified SSs and ASs. HeLa-eGFP were transfected with the indicated siRNAs (10 nM concentration) and eGFP levels were evaluated 72 h post-transfection. (a) Silencing activity of modified SSs in combination with the unmodified AS, W053. (b) Silencing activity of modified ASs in combination with the unmodified SS, W207. Colour-code: 2′-OH substituted oligos (blue), 4′-modified oligos (pink), 2′-locked oligos (green) and ribose ring modified oligos (orange).
Position-dependent tolerance for AS modification
Modification of the AS had a strong impact on siRNA silencing activity with the most efficient ASs being only modestly modified (Figure 2b and Table 1). Accordingly, extensive modification reduced activity [e.g. HM (JW1187), LNA (W010), ANA (GS2373) and HNA (GS2540)] with the notable exception of the fully OMe/F-substituted AS (JC10). We generally found bulky modifications to be poorly tolerated in both the AS seed region [e.g. ADA (W068), PYR (W096), Figure 2b; AEM and APM in position 3, data not shown] and the 3′-end [e.g. ADA (W095), PYR (W097), AEM (GS2549), APM (GS2544), OMe/EA (DO1002)], presumably due to helix distortion or altered strand selection.
The AS seed region (positions 2–8) guides the initial target recognition by RISC* (42), and modifications in this region should therefore influence silencing activity. We found that single modifications in the seed region were generally well tolerated for non-bulky groups that do not strongly influence the thermodynamic stability [e.g. HNA (GS2538, GS2539), ANA (GS2378), OMe- (DHARM1), CENA (JC-F1, JC-F2, JC-F3) and HM (JW1186), Figure 2b and Supplementary Figure 1]. In contrast, modifications that stabilize the seed region, such as LNA (Supplementary Figure 1) and AENA (JC-A1, JC-A2 and JC-A3; Figure 2b) had a negative effect on silencing activity, presumably by altering strand selection or seed–target interactions. The strongly destabilizing UNA was better tolerated in the seed, but reduced silencing activity when introduced near the 5′-end where it could destabilize AS–target interactions (Figure 2b, compare W124 and W123, and Supplementary Figure 1). We also found some sensitivity to modification in the central positions of the AS, which must be capable of perfect base-pairing to the target to allow cleavage by Ago2 (7); UNA (W126) and alternating OMe/F (JC10) were well tolerated, while HNA and ANA impaired silencing (GS2373 vs. GS2378 and GS2540, Figure 2b). As expected, most modifications were well tolerated in the AS 3′-region [e.g. UNA (W125), HNA (GS2537), HM (W042), LNA (W075) F/OMe (JC10)] and 3′-overhang [e.g. ANA (GS2372), HNA (GS2537), ALN (W047), EA (JE1001), single HM (W059) and LNA (W006)], although UNA- (W127), double HM- (W054) and LNA modifications (W180) decreased silencing activity slightly (Figure 2b).
In conclusion, modification of the AS must take the functional regions into account and should preferably be restricted to the peripheral regions.
Identification of highly efficient siRNAs
We next investigated what chemical modifications could enhance siRNA activity beyond that of an unmodified siRNA duplex. We found two ASs (W006, JC-F1) and six SSs (DO003, GS2369, JC1, W60, GS2542 and GS2534) that performed significantly better than unmodified RNA when paired to an unmodified opposing strand (Figure 2b). However, the efficiency of a SS in combination with an unmodified ASs (W053) does not necessarily allow the prediction of its behaviour in combination with other ASs. Our data from 2160 siRNAs provide valuable insight into how chemically different ASs and SSs can be combined to generate highly modified and functional siRNAs. We identified a total of 134 highly efficient (HE) siRNAs exhibiting enhanced silencing activity compared with unmodified siRNA (Table 2 and Supplementary Table 1). Interestingly, the most efficient siRNA (JW1186-W043) maintained stronger silencing than unmodified siRNAs (W053-W207) even at a lower concentration (3 nM, Supplementary Figure 2), demonstrating a higher potency that may prove important for in vivo applications. Hereby our data show that several types of chemical modifications can indeed enhance siRNA activity beyond that of an unmodified siRNA even in short-term cell-culture experiments.
Table 2.
Highly efficient siRNA duplexes
| AS | SS | eGFP levels | Viability | Rank |
|---|---|---|---|---|
| JW1186 | W043 | 0.06 ± 0.01 | 0.62 ± 0.12 | 1 |
| W123 | W131 | 0.06 ± 0.02 | 1.05 ± 0.08 | 2 |
| JW1186 | W131 | 0.06 ± 0.01 | 0.66 ± 0.20 | 3 |
| JC-S1 | DO003 | 0.06 ± 0.01 | 0.65 ± 0.13 | 4 |
| W123 | W043 | 0.06 ± 0.01 | 0.89 ± 0.20 | 5 |
| GS2372 | GS2383 | 0.06 ± 0.01 | 0.60 ± 0.15 | 6 |
| JC-F1 | W131 | 0.07 ± 0.01 | 0.64 ± 0.17 | 7 |
| W123 | JW1189 | 0.07 ± 0.01 | 0.92 ± 0.25 | 8 |
| GS2372 | DO003 | 0.07 ± 0.01 | 0.47 ± 0.14 | 9 |
| JC-S1 | JC1 | 0.07 ± 0.01 | 0.62 ± 0.13 | 10 |
| W053 | W207 | 0.09 ± 0.01 | 0.73 ± 0.14 | 380 |
| Dharmacon – | – | 1.00 ± 0.07 | 0.78 ± 0.10 | – |
| Mock | – | 1.09 ± 0.04 | 1.00 ± 0.15 | – |
| W006 | W004+W179 | 0.09 ± 0.01 | 0.57 ± 0.16 | 35 |
| JW1186 | W004+W179 | 0.12 ± 0.02 | 0.90 ± 0.10 | 238 |
| W123 | W037 | 0.13 ± 0.02 | 1.01 ± 0.18 | 276 |
| W006 | W037 | 0.13 ± 0.02 | 0.65 ± 0.20 | 326 |
| JC10 | W004+W179 | 0.32 ± 0.06 | 0.88 ± 0.11 | 983 |
| W010 | W004+W179 | 0.34 ± 0.07 | 0.46 ± 0.27 | 1028 |
Relative silencing activity (eGFP levels), viability (# nuclei) and activity ranking of the most efficient siRNA duplexes. Upper panel: top 10 performing siRNA duplexes. Lower panel: selected siRNAs with high activity and LNA modifications in the SS for enhanced serum stability. The unmodified siRNA (W053-W207) is highlighted in bold.
Rescue of AS activity by optimal SSs
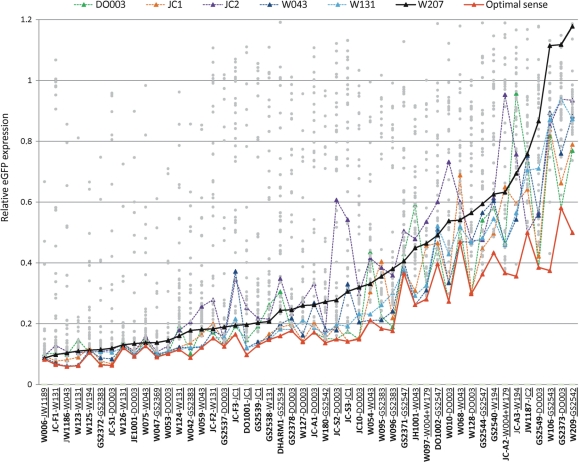
We found only modestly modified ASs among the HE siRNAs (Figure 2b; Tables 2 and 3), yet high levels of siRNA modification would be preferable for many in vivo applications. Interestingly, the activity of most ASs in our screen could be improved by combining the ASs with specific ‘optimal’ SSs [Figure 3, compare ‘optimal SS’ (red line) and unmodified SS (W207, black line)]; the most prominent effect was seen for extensively modified ASs [e.g. OMe (W106), LNA (W209) and ANA (GS2373)]. The activity of certain ASs was improved by several optimal SSs (e.g. W097, GS2549, JC-A2 and GS2544) whereas other ASs were only rescued by one or few SSs (e.g. W209 by GS2542, JC-A3 by W194 and W106 by GS3543, Figure 3 and Supplementary Table 1). Conversely, certain ASs allowed the use of heavily modified SSs that failed to support RNAi when combined with an unmodified AS; the fully LNA/DNA substituted SS W009, produced a potent ∼80% KD in combination with the UNA-modified AS W123 but gave only a 30% KD in combination with unmodified AS W053 (Supplementary Table 1). These observations imply that the reduced activity often observed upon extensive siRNA modification can be compensated by careful matching of specifically modified SSs and ASs. However, our data also identifies SSs that act as more ‘general optimal SSs’ to improve the performance of many ASs and are therefore broadly applicable in siRNA design. In particular, SSs modified in the 3′-overhang by UNA and HM (W131 and W043, respectively) or destabilized in the 3′-end by EA and OX (DO003 and JC1, respectively) produced a strong KD in combination with many ASs (Table 3 and Figure 3).
Table 3.
Highly efficient SSs and ASs
| Strand name | AS/ SS | Sequence (5′–3′) | Percentage in HE siRNAs/ top 3 siRNAs | # impr. ASs/ avr. impr. (%) | Remark |
|---|---|---|---|---|---|
| DO003 | SS | GACGUAAACGGCCACAAEAGUUC | 7/15 | 23/27 | Destabilized 3′-end favours AS selection |
| JC1 | SS | GACGUAAACGGCCACAOXAGOXUUC | 7/10 | 22/25 | Destabilized 3′-end favours AS selection |
| W043 | SS | GACGUAAACGGCCACAAGUTHMCHMU | 6/13 | 25/17 | Disfavoured 3′-overhang favours AS selection |
| W131 | SS | GACGUAAACGGCCACAAGUUUUNAU | 7/11 | 21/13 | Disfavoured 3′-overhang favours AS selection |
| GS2383 | SS | GACGUAAACGGCCACAAGUHNAUHNAC | 6/7 | 17/26 | Disfavoured 3′-overhang favours AS selection |
| JW1189 | SS | GACGTHMAAACGGCCACAAGTHMTHMC | 6/5 | 14/21 | Disfavoured 3′-overhang favours AS selection |
| W004+W179 | SS | GACLNAGUAAACLNAG + GCCLNAACLNAAAGUTLNACLNA | 2/4 | 4/115 | Stabilizing SS for highly modified ASs |
| W006 | AS | ACUUGUGGCCGUUUACGUCGLNACLNAU | 24/– | – | HE ASs, favoured 5′-overhang |
| JE1001 | AS | ACUUGUGGCCGUUUACGUCGEACEAU | 15/– | – | HE ASs, favoured 5′-overhang |
| W123 | AS | ACUUGUUNAGGCCGUUUACGUCGLNACLNAU | 13/– | – | HE ASs, favoured 5′-overhang |
| GS2372 | AS | ACUUGUGGCCGUUUACGUCGANACANAU | 9/– | – | HE ASs, favoured 5′-overhang |
| JC-S1 | AS | ACUCLNAUGUGGCCGUUUACGUCLNACGC | 9/– | – | HE ASs, thermodynamically asymmetric |
| W047 | AS | ACUUGUGGCCGUUUACGUCGCALNU | 9/– | – | HE ASs, favoured 5′-overhang |
| JW1186 | AS | ACUUGTHMGGCCGUUUACGUCGLNACLNAU | 8/– | – | HE ASs, favoured 5′-overhang |
| JC-F1 | AS | ACUCENAUGUGGCCGUUUACGUCENACGC | 7/– | – | HE ASs, thermodynamically asymmetric |
The name, sequence, statistical performance and remarks are given for the most efficient SSs and ASs. ‘Percentage in HE siRNA’ refers to the percentage of the 134 HE siRNAs containing the particular AS/SS. ‘Percentage in top 3 siRNAs’ refers to the summed representation of the particular SS among the three most efficient SSs for each AS (in %). ‘#impr. ASs/avr. improvement’ refers to the number and average improvement (%) of ASs whose activity is significantly enhanced by the particular SS as compared with the unmodified SS (W207). Modifications are highlighted in bold in the sequences.
Figure 3.
Optimal SSs enhance the activity of ASs. Relative eGFP expression of HeLa-eGFP cells transfected with all investigated ASs (X-axis, ASs name given in bold) in combination with all 45 SSs (represented by grey dots) or with selected SSs (coloured triangles/lines). The activity of most ASs in combination with the unmodified SS, W207 (represented by black triangles/line) can be enhanced in combination with a specific, optimal SS for each AS (red triangles/line; name of the particular optimal SS for each AS is underlined). Furthermore the activity of many ASs is enhanced by the thermodynamically asymmetric SSs, DO003 (dark green triangles) and JC1 (orange triangles), as well as the 3′-overhang modified SSs, W043 (dark blue triangles) and W131 (light blue triangles). In contrast, the thermodynamically asymmetric SS, JC2, decreases the activity of many ASs (purple triangles).
Strategies for enhancing siRNA activity
The HE siRNA duplexes were found to harbour modifications either in the 3′-overhang (e.g. W006, JE1001, GS2372, W043, W047, W060, W131), within the siRNA body (e.g. JC-F1, DO003, JC-S1, JC1, GS2369) or in both regions (e.g. GS2383, JW1189, GS2542, GS2534) (Tables 2 and 3), and we therefore speculated that two mechanisms were responsible for enhancing siRNA activity: (i) Modification of the siRNA body to introduce thermodynamic asymmetry to favour AS incorporation into RISC. (ii) Modification of SS and AS 3′-overhangs to enhance serum stability or affect strand selection by RISC.
Destabilization of SS 3′-ends enhances silencing efficiency
Given that strand selection during RISC loading, and thereby AS activity, is dependent on the thermodynamic profile of the siRNA duplex we investigated the impact of modifications on thermodynamic asymmetry. The chemical modifications used in this study have both stabilizing (OMe, F/OMe, HNA, ANA, ALN, LNA) and destabilizing (DNA, AEM, APM, OXE, EA, AP, CE, UNA) properties. We found 15 of the 20 most efficient siRNA duplexes to have chemical modifications favouring AS incorporation by altering the thermodynamic profile. W123 contained a 5′-end destabilizing UNA, whereas JC-F1 and JC-S1 contained CENA and CLNA resulting in stabilization of the AS 3′-end (see Tables 2 and 3 and Supplementary Table 1). Nearly a third of the HE siRNAs (38/134) contained these three ASs suggesting that altering the thermodynamic asymmetry through AS modification is a major determinant of siRNA activity.
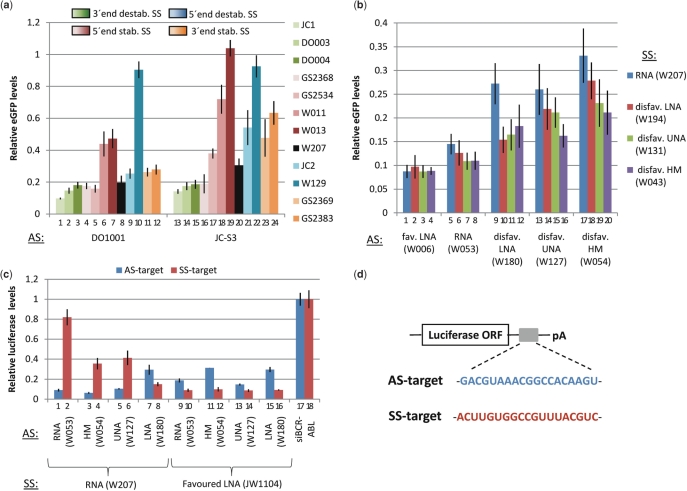
A similar trend was found among the HE SSs; the 3′-end OX-destabilized JC-1 and the 3′-end EA destabilized DO003 improved the performance of most ASs (Figure 3, orange and green, respectively, and Figure 4a, columns 1, 2, 13,14) and resulted in an average 25% improvement in activity for 22 and 23 ASs, respectively (Table 3). Furthermore, JC1 and DO003 were highly represented among the 134 HE siRNAs and among the top three SSs for each AS (Table 3). Conversely, we found both SS 3′-end stabilization (e.g. GS2369 and GS2383; Figure 4a, columns 11, 23 and 12, 24) and SS 5′-end destabilization (e.g. JC2 and W129; Figure 4a, columns 9, 21 and 10, 22) to negatively influence the activity of many ASs. The stabilization of the SS 5′-end should improve siRNA performance, and moderate 5′-end stabilization (e.g. GS2368; Figure 4a, columns 4, 16) did improve the performance of most ASs; however more stabilizing modifications (e.g. multiple HNAs (GS2534; Figure 4a, columns 5, 17), single and extensive LNA modifications [(W011 and W013; Figure 4a, columns 6, 18 and 7, 19)] impaired silencing. This suggests that, although favouring AS selection, extensive thermodynamic stabilization of an siRNA duplex is detrimental to silencing activity. In accordance, the activity of stabilized ASs (e.g. W106, W209 and GS2373) was enhanced both by the 3′-end destabilized SSs (DO003 and JC1) and by the 5′-end destabilized JC-2 although this SS should disfavour AS incorporation into RISC based on thermodynamic asymmetry (Figure 3 and Supplementary Table 1). This highlights that siRNA thermodynamic stability should fall within a range compatible with the components of the RNAi machinery, and that both siRNA thermodynamic asymmetry and stability are critical parameters to consider during siRNA design. Based on our screen, relative destabilization of the SS 3′-end (using DO003 and JC1) represented the more reliable strategy to improve siRNA performance unless high stability of the duplex is specifically required.
Figure 4.
Improvement of siRNA performance by introduction of additional thermodynamic asymmetry and modification of 3′-overhangs. (a) The performance of ASs (exemplified by the representative ASs DO1001 and JC-S3) can be modified by altering the overall thermodynamic profile of the siRNA duplex by introduction of chemical modification in the SS. (b) The siRNA activity is influenced by the nature of the AS and SS 3′-overhangs. ASs modified in the 3′-overhang by LNA, UNA and HM have significantly lower activity than unmodified and LNA-modified AS in combination with the RNA SS (blue). This loss of activity can be partly rescued by using SSs with the disfavoured overhangs LNA, UNA or HM. (c) The LNA-LNA-RNA motif is a favoured 3′-overhang motif. The silencing activity of both the AS (on the AS-target, blue) and SS (on the S-target, red) is shown for the unmodified, HM-, UNA- and LNA-modified ASs in combination with unmodified and LNA-modified SSs. (d) Overview of the luciferase reporters used to evaluate the silencing activity of both the AS (denoted ‘AS-target’) and SS (denoted ‘SS-target’).
Modification of 3′-overhangs influences siRNA activity
The observation that several highly efficient siRNAs (e.g. W006-W060, W006-W043 and W006-W131) are modified in the 3′-overhangs only, highlights the importance of overhangs for silencing activity (Figure 2, Tables 2 and 3). In fact, ASs modified exclusively in the 3′-overhang by LNA (W006), EA (JE1001), ALN (W047) and ANA (GS2372) were found in half (66/134) of the HE siRNAs. The impact of 3′-overhang modification was highly chemistry dependent; the UNA- (W127), LNA- (W180) and HM-modified (W054) ASs showed a significantly reduced silencing activity whereas ASs modified by ANA (GS2372), EA (JE1001) and ALN (W047) were equally efficient to unmodified RNA (W053) (Figure 2b). Interestingly, the LNA-modified overhang in W006 (5′-LNA-LNA-RNA-3′) containing an additional 3′-RNA residue resulted in significantly improved silencing (Figure 2b; see the Discussion section below).
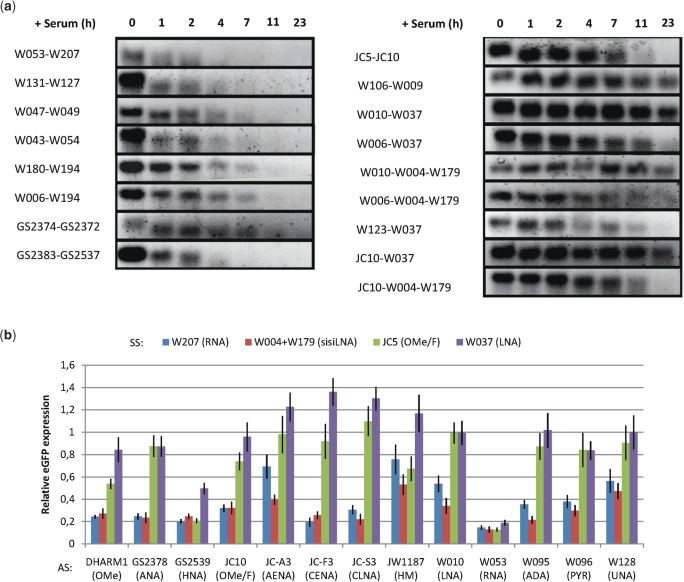
We speculated that the influence of 3′-overhang modifications could result from differences in siRNA serum stability; however, no clear correlation between stability and silencing activity was observed for overhang-modified siRNAs (Figures 2 and 5 and data not shown). Instead modification of the 3′-overhang was found to influence strand selection, as modifications disfavoured in the AS 3′-overhang [e.g. HM (W054), UNA (W127), LNA (W180); Figures 2b and 4b] were found to favour AS selection when incorporated into SS 3′-overhangs [HM (W043), LNA (W194) and UNA (W131); Figure 4b]. In detail, the silencing activity of the LNA-modified AS W180 (Figure 4b, column 9) was enhanced to a level indistinguishable from unmodified siRNA (Figure 4b, column 5) when paired to a SS with disfavoured overhangs, such as LNA (W194), UNA (W131) and HM (W043) (Figure 4b, columns 10–12). Similar SS 3′-overhang effects were seen for the UNA-modified AS W127 (Figure 4b, columns 13–16) and HM-modified AS W054 (Figure 4b, columns 17–20). The HM modification was particularly strongly disfavoured as the HM-modified W054 was a significantly poor AS (Figure 4b, column 17), while the HM-modified SS W043 had the most potent rescue effect on ASs with disfavoured overhangs (Figure 4b, columns 8, 12, 16, 29). This suggests that HM-modified overhangs may be more strongly disfavoured than UNA- (W131) and LNA-modified (W194) SS and can therefore be broadly used in SS design to favour AS-strand selection.
Figure 5.
Enhancing serum stability of siRNAs with minor loss of activity. (a) The biostability of modified siRNAs was evaluated by incubation in 80% FBS. While a low level of chemical modification results in only modest increase in stability (left panel), more extensive and full substitutions result in dramatically improved siRNA stability (right panel). The incubation time is given in hours (h). (b) The eGFP levels of cells transfected with modified ASs paired with either unmodified SS (W207), LNA-modified segmented SS (W004+W179), fully OMe/F substituted SS (JC5) and LNA-modified SS (W037) are given. The segmented LNA-modified SS (W004+W179) generally prevented the loss in activity imposed by the LNA-modified unsegmented SS (W037).
Modified overhangs alter strand selection by RISC
To further investigate the impact of 3′-overhang modifications on altered strand selection, we evaluated the activity of both strands in the siRNA duplex using reporters containing a perfect target site for either the SS or AS downstream of a luciferase open reading frame (Figure 4d). We expected disfavoured modifications in the AS 3′-overhang [LNA (W180), UNA (W127), HM (W054)] to increase incorporation of the unmodified SS (W207) into RISC*, thereby leading to higher SS silencing activity. Indeed, the modest silencing effect of the SS (W207) in an unmodified duplex (5 nM concentration) was significantly increased when combined with an AS with a disfavoured overhang (Figure 4c, compare column 2 with 4, 6, 8). We therefore suggest that disfavoured modifications in 3′-overhangs can be used to favour the incorporation of opposing strands, irrespective of the thermodynamic asymmetry of the siRNA duplex.
Intriguingly, we found the AS W006 to be significantly more efficient than an unmodified AS (W053) when paired to W207 suggesting the 3-nt 5′-LNA-LNA-RNA-3′ overhang motif to be favoured for RISC loading even over the natural RNA–RNA 3′-overhang (Figures 2b and 4b). In agreement, a SS (JW1104) with the 5′-LNA-LNA-RNA-3′ overhang motif exhibited significantly enhanced activity compared with the unmodified SS (W207) (Figure 4c, compare columns 2 and 10) and a similar effect was obtained when combining JW1104 with the disfavoured ASs W180 (LNA), W127 (UNA) and W054 (HM) (Figure 4c, compare columns 4, 6, 8 and 12, 14, 16). We found other highly efficient ASs modified exclusively in the 3′-overhang by EA (JE1001), ANA (GS2372) and ALN (W047) (Table 3) to be favoured during RISC loading as they lowered the activity of the opposing SS (W207) as compared with the unmodified AS W053 (Supplementary Figure 3). Consequently, favoured overhangs such as the 5′-LNA-LNA-RNA-3′ motif and others can be broadly utilized to favour AS incorporation into RISC*, thereby enhancing siRNA potency.
Enhancing siRNA serum stability by chemical modification
The identification of highly nuclease-resistant siRNAs is a key concern for in vivo applications and great efforts have been applied to enhance stability by chemical modification (14,16,22,43). Like other studies, we find siRNA serum stability to be positively correlated with the level of RNA modification for most chemistries. While substitution of 3′-overhangs led to a modest increase in serum stability (e.g. UNA, ALN, HM, LNA, ANA; Figure 5a, left panel), partial or full modification of the siRNA body led to efficient stabilization (OMe/F, OMe, DNA/LNA LNA; Figure 5a, right panel). Although a large number of substitutions within an siRNA does indeed enhance stability, highly modified siRNAs generally displayed poor silencing. Fully OMe/F-substituted siRNAs have previously been reported to be both highly stable and more potent than standard siRNAs (30); however, in our hands a similar siRNA (JC5-JC10) exhibited high stability, but produced a very modest 26% KD (Figure 5a and Supplementary Table 1). This highlights the importance of identifying other strategies for siRNA stabilization that support high silencing activity. Interestingly, we found that LNA-mediated stabilization of selected positions within the siRNA duplex led to similar or even enhanced stability as compared with fully substituted duplexes (Figure 5a, compare W010-W037, W010-W004-W179, JC10-W037 with JC5-JC10 and W106-W009). In fact, the partially LNA-modified W006-W037 and W010-W004-W179 exhibited higher stability than the fully OMe/F-substituted JC5-JC10, while retaining a potent 87% and 66% KD activity, respectively (Table 2 and Figure 5a). Moreover, we found that the sisiRNA design (41) that utilizes an LNA-stabilized segmented SS (W004 + W179) may be broadly applied to introduce LNA-stabilization with minor impact on activity. The sisiRNA constructs W010-W004-W179 and JC10-W004-W179 retained both high silencing activity and serum stability when modified extensively with LNAs, whereas an intact LNA-modified SS (W037) was detrimental to siRNA activity (W010-W037 and JC10-W037, Figure 5). In fact, the sisiRNA design allowed the LNA-stabilization of several differentially modified ASs without major loss of silencing activity (Figure 5b). This implies that siRNA stabilization is preferentially achieved by introducing few LNA modifications, preferentially using the sisiRNA design, rather than generating fully substituted duplexes. Furthermore, selected modifications allow further functionalization of the siRNA duplex through modification of the remaining nucleotide positions.
Reducing siRNA toxicity through chemical modification
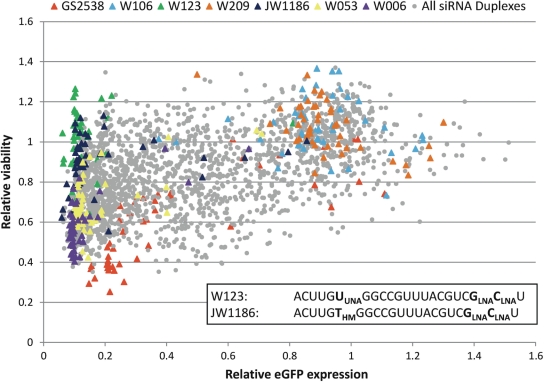
Chemical modifications have previously been shown to affect the toxicity of siRNAs (44) and we therefore evaluated the viability of the siRNA transfected cells after 72 h (as number of nuclei per well relative to mock transfections). The majority of modified SSs resulted in a similar high degree of viability in combination with unmodified RNA AS whereas the viability varied more dramatically among the modified ASs in combination with the RNA SS (Table 1). We found cell viability to correlate with siRNA potency for the majority of ASs (Supplementary Figure 4) with only a few non-functional siRNAs resulting in low cell viability (Figure 6). This suggests that the observed siRNA toxicity arises from efficient ASs interfering with the endogenous RNAi pathway. Indeed, plotting the relative eGFP levels versus relative viability allowed us to identify distinct subgroups of ASs; the unmodified W053, and the LNA-modified W006 displayed high silencing potency and moderate cell viability (Figure 6, yellow and purple triangles, respectively), the OMe- (W106) and heavily LNA- (W209) modified ASs had poor activity and high viability (Figure 6, light blue and brown triangles, respectively), while the HNA-modified GS2538 had high activity and very low viability (Figure 6, red triangles). Interestingly, silencing activity and viability of particularly the UNA- (W123) but also the HM- (JW1186) modified ASs were both high in combination with most SSs (Figure 6, green and dark blue triangles, respectively). Hereby our data shows that although most potent siRNAs cause reduced viability, it is possible to generate highly potent and non-toxic siRNAs by selecting particular chemical modifications.
Figure 6.
Identification of highly efficient siRNAs with low toxicity. Scatter plot showing target cell viability versus eGFP expression (siRNA activity) for all tested siRNAs (grey dots) and for selected ASs in combination with all 45 SS (coloured dots). Silencing activity correlates with toxicity for most siRNAs. The highly active, unmodified (W053, yellow triangles), HNA- (GS2538, red triangles) and LNA- (W006, purple triangles) modified ASs have high activity and poor viability, whereas heavily LNA- (W209, light brown triangles) and OME- (W106, light blue triangles) modified ASs have poor activity, but high viability. In contrast, the UNA- (W123, green triangles) and HM- (JW1186, dark blue triangles) modified ASs have both high activity and viability.
DISCUSSION
Optimization of siRNA performance by chemical modification has been addressed in several studies (14–18,22,23,31,41); however, in most cases cross-study comparisons of chemistries are difficult. In the present study, we have directly compared the impact of 21 types of chemical modifications on siRNA activity and cell viability using 2160 chemically different siRNA duplexes, thus providing unique insight into the ways specific chemistries can be combined to generate siRNAs with superior properties. As a result we have identified 134 siRNA duplexes displaying higher silencing activity than standard siRNAs. The majority of these highly efficient siRNAs are only modestly modified, and especially the AS exhibited a limited and position-dependent tolerance for modification. This is expected as the AS must be bound at the 3′-end by Ago2 during RISC loading (42,45), guide initial target interactions in the seed region (positions 2–8) (42,46,47) and be perfectly base paired in the central region to support Ago2-mediated target cleavage (7,42). Modification of the SS was generally well tolerated, and this strand would therefore be useful as an entry route for extensive modifications in the siRNA duplex.
Our data highlights that improvement of siRNA performance by chemical modifications should focus primarily on ensuring preferential loading of the intended AS into RISC*, as chemical modifications affecting the thermodynamic asymmetry of the siRNA duplex or the recognition of 3′-overhangs were the major contributors to improved silencing activity in our screen. Previous results have demonstrated the strand containing the least stable 5′-end to be preferentially utilized as the guide strand in RISC* (5,6). In agreement, we found chemical modifications leading to relative destabilization of the AS 5′-end to be highly represented among the most potent siRNA duplexes. In contrast, strong destabilization of the AS seed region reduces siRNA silencing activity presumably by weakening its interactions with the target mRNA. AS selection can also be favoured by incorporation of stabilizing modifications in the SS 5′-end or AS 3′-end, and accordingly previous studies have successfully incorporated LNA into SS 5′-ends to favour AS selection (17,23). Although our results support this observation, we found a large increase in thermostability to reduce siRNA activity, and we therefore suggest favouring AS selection by destabilization of the SS 3′-end, unless high stability of the siRNA duplex is specifically required.
Previous studies have given little attention to the choice of siRNA 3′-overhangs, and chemical modification of 3′-overhangs appears to be generally well tolerated [this study and (18,41,48)]. Interestingly, our study shows that chemical modification of siRNA 3′-overhangs can be utilized to favour AS selection during RISC loading irrespective of the thermodynamic asymmetry of the duplex. We have identified 3′-overhang motifs that are either favoured (5′-LNA-LNA-RNA-3′ and others) or disfavoured (5′-RNA-HM-3′ and 5′-RNA-UNA-RNA-3′) by the RNAi machinery, and we therefore propose the incorporation of disfavoured overhangs in the SS and favoured overhang in the AS for optimal siRNA performance.
For many applications, particularly in vivo, extensive modification is required to enhance siRNA stability or support delivery strategies. Improved resistance towards nucleases has been a primary goal of siRNA modification and several highly stabilized siRNAs have already been developed, albeit often at the expense of activity [this study and (14,16,22,49)]. Interestingly, we found that the poor activity of many modified ASs can be partially rescued by combination with an ‘optimal’ SS (Figure 3), suggesting that heavily modified ASs are incompatible with recognition by the RNAi machinery rather than incompetent for target cleavage. Hereby our data provides unique insight into synergistic effects between chemistries that maximize the level of modification and minimize the loss of activity.
Chemical modification of siRNA overhangs could provide protection against exonucleases and was found to elicit a modest increase in serum stability, while resistance towards endonucleases was achieved by substituting multiple positions in the siRNA body (Figure 5). We evaluated the serum stability of the heavily modified, yet functional siRNAs in our screen and found that partially LNA-substituted JC10-W004-W179 and W010-W004-W179 exhibited stabilities similar to highly and fully modified siRNAs. This suggests that enhancement of siRNA stability need not focus on substituting all positions of the siRNA duplex, but can be restricted to few selected positions. We speculate that resistance towards endonucleases could be influenced by the thermodynamic stability of the duplex; in this regard, the segmented sisiRNA design (Table 1: SS W004-W179) provides a convenient way of introducing highly stabilizing LNA modifications in a configuration well tolerated by the RNAi machinery (41). Importantly, this design allows the generation of more functionalized siRNA duplexes in which other positions could be modified to minimize siRNA off-target effects and toxicity or favour delivery strategies. It is noteworthy that extensive stabilization of siRNAs should only be applied when explicitly needed, e.g. upon injections of naked siRNA in vivo. When using siRNAs shielded by transfection reagents in cell culture or even in vivo, chemical modification should primarily aim at maximizing silencing activity as exemplified by the 134 HE siRNAs identified in this study.
It is interesting that the most highly active siRNA duplexes, including the unmodified siRNA (W053-W207), have a negative impact on cell viability compared with mock transfections. Only 26 out of 134 highly efficient siRNAs resulted in viabilities indistinguishable from mock controls, and among these, the UNA- (W123) and HM- (W1186) modified ASs were represented 15 and 6 times, respectively, indicating that very few ASs are both highly active and non-toxic. SiRNA toxicity may have several sources ((50–52) but we suspect that highly active siRNA duplexes interfere with endogenous RNAi pathways as most non-functional siRNAs in our screen are non-toxic. This type of competition between exogenous RNAi substrates and endogenous miRNAs has been observed both in cell culture (53) and in vivo (54). Alternatively, the toxicity may arise from off-target effects due to a miRNA-like behaviour on mRNAs with 6–7 nt matches with the AS/SS seed region (25). Interestingly, the efficient, non-toxic ASs W123 and JW1186 both contain modifications within their seed region, and we speculate that this may lower the potential for off-target effects by destabilizing miRNA-like interactions. Indeed, recent work has demonstrated that siRNA off-targeting can be minimized by a slight destabilization of the siRNA–target interaction by the introduction of DNA in the AS seed (55).
In summary, our results have allowed us to formulate the following guidelines for enhancing siRNA activity through chemical modifications:
siRNA overhangs can be chemically modified to favour AS incorporation into RISC*; the SS should contain disfavoured overhangs such as UNA, HM and LNA-LNA, whereas the AS should contain favoured overhang motifs such as LNA-LNA-RNA.
In order to favour AS incorporation into RISC* the siRNA duplex should preferably be destabilized by modifications in the SS 3′-end rather than stabilized in the SS 5′-end. Additionally, the AS should not contain strongly destabilizing/stabilizing/bulky modifications within the 5′-end of the seed region.
The use of single UNA and HM modifications in the AS seed region increases cell viability upon siRNA transfection.
Enhanced serum stability can be obtained by selective thermodynamic stabilization of a few positions rather than full substitution of the entire siRNA duplex. The sisiRNA design may provide a convenient way to introduce strong LNA stabilization for many types of AS.
As demonstrated here, using the above guidelines to combine various types of chemical modifications will lead to the generation of superior siRNAs with high activity, high stability and low toxicity.
SUPPLEMENTARY DATA
Supplementary Data is available at NAR Online.
FUNDING
EU-FP6 RIGHT project (no. LSHB-CT-2004-005276); the Danish National Research Foundation. Funding for open access charge: EU-FP6 RIGHT project (no. LSHB-CT-2004-005276).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Antje Niederlein for assistance on the image analysis of the data obtained by the screen, Kerstin Korn, Jan Wagner and Sara Christ for the support with robotics and automation that enabled the screen of the whole siRNA combinations. We also thank Hannelore Brill and Stefan Bernhardt for technical assistance during oligonucleotide synthesis. The siRNA screen was conducted at the Technology Development Studio at the Max Planck Institute of Molecular Cell Biology and Genetics.
REFERENCES
- 1.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 2.Hammond SM, Boettcher S, Caudy AA, Kobayashi R, Hannon GJ. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science. 2001;293:1146–1150. doi: 10.1126/science.1064023. [DOI] [PubMed] [Google Scholar]
- 3.Martinez J, Patkaniowska A, Urlaub H, Luhrmann R, Tuschl T. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell. 2002;110:563–574. doi: 10.1016/s0092-8674(02)00908-x. [DOI] [PubMed] [Google Scholar]
- 4.Maniataki E, Mourelatos Z. A human, ATP-independent, RISC assembly machine fueled by pre-miRNA. Genes Dev. 2005;19:2979–2990. doi: 10.1101/gad.1384005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 6.Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 7.Leuschner PJ, Ameres SL, Kueng S, Martinez J. Cleavage of the siRNA passenger strand during RISC assembly in human cells. EMBO Rep. 2006;7:314–320. doi: 10.1038/sj.embor.7400637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rand TA, Petersen S, Du F, Wang X. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell. 2005;123:621–629. doi: 10.1016/j.cell.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 9.Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123:607–620. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- 10.Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 11.Soutschek J, Akinc A, Bramlage B, Charisse K, Constien R, Donoghue M, Elbashir S, Geick A, Hadwiger P, Harborth J, et al. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature. 2004;432:173–178. doi: 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]
- 12.Morrissey DV, Lockridge JA, Shaw L, Blanchard K, Jensen K, Breen W, Hartsough K, Machemer L, Radka S, Jadhav V, et al. Potent and persistent in vivo anti-HBV activity of chemically modified siRNAs. Nat. Biotechnol. 2005;23:1002–1007. doi: 10.1038/nbt1122. [DOI] [PubMed] [Google Scholar]
- 13.Morrissey DV, Blanchard K, Shaw L, Jensen K, Lockridge JA, Dickinson B, McSwiggen JA, Vargeese C, Bowman K, Shaffer CS, et al. Activity of stabilized short interfering RNA in a mouse model of hepatitis B virus replication. Hepatology. 2005;41:1349–1356. doi: 10.1002/hep.20702. [DOI] [PubMed] [Google Scholar]
- 14.Choung S, Kim YJ, Kim S, Park HO, Choi YC. Chemical modification of siRNAs to improve serum stability without loss of efficacy. Biochem. Biophys. Res. Commun. 2006;342:919–927. doi: 10.1016/j.bbrc.2006.02.049. [DOI] [PubMed] [Google Scholar]
- 15.Layzer JM, McCaffrey AP, Tanner AK, Huang Z, Kay MA, Sullenger BA. In vivo activity of nuclease-resistant siRNAs. RNA. 2004;10:766–771. doi: 10.1261/rna.5239604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chiu YL, Rana TM. siRNA function in RNAi: a chemical modification analysis. RNA. 2003;9:1034–1048. doi: 10.1261/rna.5103703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mook OR, Baas F, de Wissel MB, Fluiter K. Evaluation of locked nucleic acid-modified small interfering RNA in vitro and in vivo. Mol. Cancer Ther. 2007;6:833–843. doi: 10.1158/1535-7163.MCT-06-0195. [DOI] [PubMed] [Google Scholar]
- 18.Amarzguioui M, Holen T, Babaie E, Prydz H. Tolerance for mutations and chemical modifications in a siRNA. Nucleic Acids Res. 2003;31:589–595. doi: 10.1093/nar/gkg147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lorenz C, Hadwiger P, John M, Vornlocher HP, Unverzagt C. Steroid and lipid conjugates of siRNAs to enhance cellular uptake and gene silencing in liver cells. Bioorg. Med. Chem. Lett. 2004;14:4975–4977. doi: 10.1016/j.bmcl.2004.07.018. [DOI] [PubMed] [Google Scholar]
- 20.Muratovska A, Eccles MR. Conjugate for efficient delivery of short interfering RNA (siRNA) into mammalian cells. FEBS Lett. 2004;558:63–68. doi: 10.1016/S0014-5793(03)01505-9. [DOI] [PubMed] [Google Scholar]
- 21.McNamara JO, 2nd, Andrechek ER, Wang Y, Viles KD, Rempel RE, Gilboa E, Sullenger BA, Giangrande PH. Cell type-specific delivery of siRNAs with aptamer-siRNA chimeras. Nat. Biotechnol. 2006;24:1005–1015. doi: 10.1038/nbt1223. [DOI] [PubMed] [Google Scholar]
- 22.Allerson CR, Sioufi N, Jarres R, Prakash TP, Naik N, Berdeja A, Wanders L, Griffey RH, Swayze EE, Bhat B. Fully 2′-modified oligonucleotide duplexes with improved in vitro potency and stability compared to unmodified small interfering RNA. J. Med. Chem. 2005;48:901–904. doi: 10.1021/jm049167j. [DOI] [PubMed] [Google Scholar]
- 23.Elmen J, Thonberg H, Ljungberg K, Frieden M, Westergaard M, Xu Y, Wahren B, Liang Z, Orum H, Koch T, et al. Locked nucleic acid (LNA) mediated improvements in siRNA stability and functionality. Nucleic Acids Res. 2005;33:439–447. doi: 10.1093/nar/gki193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vollmer J, Jepsen JS, Uhlmann E, Schetter C, Jurk M, Wader T, Wullner M, Krieg AM. Modulation of CpG oligodeoxynucleotide-mediated immune stimulation by locked nucleic acid (LNA) Oligonucleotides. 2004;14:23–31. doi: 10.1089/154545704322988021. [DOI] [PubMed] [Google Scholar]
- 25.Jackson AL, Burchard J, Leake D, Reynolds A, Schelter J, Guo J, Johnson JM, Lim L, Karpilow J, Nichols K, et al. Position-specific chemical modification of siRNAs reduces “off-target” transcript silencing. RNA. 2006;12:1197–1205. doi: 10.1261/rna.30706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ui-Tei K, Naito Y, Zenno S, Nishi K, Yamato K, Takahashi F, Juni A, Saigo K. Functional dissection of siRNA sequence by systematic DNA substitution: modified siRNA with a DNA seed arm is a powerful tool for mammalian gene silencing with significantly reduced off-target effect. Nucleic Acids Res. 2008;36:2136–2151. doi: 10.1093/nar/gkn042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beaucage SL, Caruthers MH. Current Protocols in Nucleic Acid Chemistry. New Jersey, US: John Wiley & Sons, Inc.; 2003. [DOI] [PubMed] [Google Scholar]
- 28.Nielsen P, Dreioe LH, Wengel J. Synthesis and evaluation of oligodeoxynucleotides containing acyclic nucleosides: introduction of three novel analogues and a summary. Bioorg. Medl Chem. 1995;3:19–28. doi: 10.1016/0968-0896(94)00143-q. [DOI] [PubMed] [Google Scholar]
- 29.Thrane HFJ, Regner M, Wengel J. Novel linear and branched oligodeoxynucleotide analogues containing 4′-C-(hydroxymethyl)thymidine. Tetrahedron. 1995;51:10389–10402. [Google Scholar]
- 30.Sorensen MD, Petersen M, Wengel J. Functionalized LNA (locked nucleic acid): high-affinity hybridization of oligonucleotides containing N-acylated and N-alkylated 2′-amino-LNA monomers. Chem. Commun., 2003:2130–2131.. doi: 10.1039/b307026c. [DOI] [PubMed] [Google Scholar]
- 31.Odadzic D, Bramsen JB, Smicius R, Bus C, Kjems J, Engels JW. Synthesis of 2′-O-modified adenosine building blocks and application for RNA interference. Bioorg. Med. Chem. 2008;16:518–529. doi: 10.1016/j.bmc.2007.09.019. [DOI] [PubMed] [Google Scholar]
- 32.Smicius R, Engels JW. Preparation of zwitterionic ribonucleoside phosphoramidites for solid-phase siRNA synthesis. J. Org. Chem. 2008;73:4994–5002. doi: 10.1021/jo800451m. [DOI] [PubMed] [Google Scholar]
- 33.Haas J, Engels JW. A novel entry to 2′-O-aminopropyl modified nucleosides amenable for further modifications. Tetrahedron Lett. 2007;48:8891–8894. [Google Scholar]
- 34.Pradeepkumar PI, Cheruku P, Plashkevych O, Acharya P, Gohil S, Chattopadhyaya J. Synthesis, physicochemical and biochemical studies of 1′,2′-oxetane constrained adenosine and guanosine modified oligonucleotides, and their comparison with those of the corresponding cytidine and thymidine analogues. J. Am. Chem. Soc. 2004;126:11484–11499. doi: 10.1021/ja048417i. [DOI] [PubMed] [Google Scholar]
- 35.Srivastava P, Barman J, Pathmasiri W, Plashkevych O, Wenska M, Chattopadhyaya J. Five- and six-membered conformationally locked 2′,4′-carbocyclic ribo-thymidines: synthesis, structure, and biochemical studies. J. Am. Chem. Soc. 2007;129:8362–8379. doi: 10.1021/ja071106y. [DOI] [PubMed] [Google Scholar]
- 36.Varghese OP, Barman J, Pathmasiri W, Plashkevych O, Honcharenko D, Chattopadhyaya J. Conformationally constrained 2′-N,4′-C-ethylene-bridged thymidine (aza-ENA-T): synthesis, structure, physical, and biochemical studies of aza-ENA-T-modified oligonucleotides. J. Am. Chem. Soc. 2006;128:15173–15187. doi: 10.1021/ja0634977. [DOI] [PubMed] [Google Scholar]
- 37.Allart B, Kamran K, Rosemeyer H, Schepers G, Hendrix C, Rothenbacher K, Seela F, Van Aerschot A, Herdewijn P. D-Altritol Nucleic Acids (ANA): Hybridisation properties, stability, and initial structural analysis. Chem. Eur. J. 1999;5:2424–2431. [Google Scholar]
- 38.Hendrix C, Helmut R, Verheggen I, Van Aerschot A, Herdewijn P. 1, 5-anhydrohexitol oligonucleotides: synthesis, base pairing and recognition by regular oligodeoxyribonucleotides and oligoribonucleotides. Chem. Eur. J. 1997;3:110–120. [Google Scholar]
- 39.Bobkov GV, Mikhailov SN, Van Aerschot A, Herdewijn P. Phosphoramidite building blocks for efficient incorporation of 2′-O-aminoethoxy(and propoxy)methyl nucleosides into oligonucleotides. Tetrahedron. 2008;64:6238–6251. [Google Scholar]
- 40.R_Development_Core_Team. Vienna, Austria: R Foundation for Statistical Computing; 2008. R: a language and environment for statistical computing. Available at http://www.R-project.org. [Google Scholar]
- 41.Bramsen JB, Laursen MB, Damgaard CK, Lena SW, Babu BR, Wengel J, Kjems J. Improved silencing properties using small internally segmented interfering RNAs. Nucleic Acids Res. 2007;35:5886–5897. doi: 10.1093/nar/gkm548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang Y, Sheng G, Juranek S, Tuschl T, Patel DJ. Structure of the guide-strand-containing argonaute silencing complex. Nature. 2008;456:209–213. doi: 10.1038/nature07315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Braasch DA, Jensen S, Liu Y, Kaur K, Arar K, White MA, Corey DR. RNA interference in mammalian cells by chemically-modified RNA. Biochemistry. 2003;42:7967–7975. doi: 10.1021/bi0343774. [DOI] [PubMed] [Google Scholar]
- 44.Hornung V, Guenthner-Biller M, Bourquin C, Ablasser A, Schlee M, Uematsu S, Noronha A, Manoharan M, Akira S, de Fougerolles A, et al. Sequence-specific potent induction of IFN-alpha by short interfering RNA in plasmacytoid dendritic cells through TLR7. Nat. Med. 2005;11:263–270. doi: 10.1038/nm1191. [DOI] [PubMed] [Google Scholar]
- 45.Song JJ, Liu J, Tolia NH, Schneiderman J, Smith SK, Martienssen RA, Hannon GJ, Joshua-Tor L. The crystal structure of the Argonaute2 PAZ domain reveals an RNA binding motif in RNAi effector complexes. Nat. Struct. Biol. 2003;10:1026–1032. doi: 10.1038/nsb1016. [DOI] [PubMed] [Google Scholar]
- 46.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 47.Ma JB, Yuan YR, Meister G, Pei Y, Tuschl T, Patel DJ. Structural basis for 5′-end-specific recognition of guide RNA by the A. fulgidus Piwi protein. Nature. 2005;434:666–670. doi: 10.1038/nature03514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chiu YL, Rana TM. RNAi in human cells: basic structural and functional features of small interfering RNA. Mol. Cell. 2002;10:549–561. doi: 10.1016/s1097-2765(02)00652-4. [DOI] [PubMed] [Google Scholar]
- 49.Czauderna F, Fechtner M, Dames S, Aygun H, Klippel A, Pronk GJ, Giese K, Kaufmann J. Structural variations and stabilising modifications of synthetic siRNAs in mammalian cells. Nucleic Acids Res. 2003;31:2705–2716. doi: 10.1093/nar/gkg393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sioud M. Innate sensing of self and non-self RNAs by Toll-like receptors. Trends Mol. Med. 2006;12:167–176. doi: 10.1016/j.molmed.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 51.Marques JT, Devosse T, Wang D, Zamanian-Daryoush M, Serbinowski P, Hartmann R, Fujita T, Behlke MA, Williams BR. A structural basis for discriminating between self and nonself double-stranded RNAs in mammalian cells. Nat. Biotechnol. 2006;24:559–565. doi: 10.1038/nbt1205. [DOI] [PubMed] [Google Scholar]
- 52.Sledz CA, Holko M, de Veer MJ, Silverman RH, Williams BR. Activation of the interferon system by short-interfering RNAs. Nat. Cell Biol. 2003;5:834–839. doi: 10.1038/ncb1038. [DOI] [PubMed] [Google Scholar]
- 53.Castanotto D, Sakurai K, Lingeman R, Li H, Shively L, Aagaard L, Soifer H, Gatignol A, Riggs A, Rossi JJ. Combinatorial delivery of small interfering RNAs reduces RNAi efficacy by selective incorporation into RISC. Nucleic Acids Res. 2007;35:5154–5164. doi: 10.1093/nar/gkm543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Grimm D, Streetz KL, Jopling CL, Storm TA, Pandey K, Davis CR, Marion P, Salazar F, Kay MA. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- 55.Ui-Tei K, Naito Y, Takahashi F, Haraguchi T, Ohki-Hamazaki H, Juni A, Ueda R, Saigo K. Guidelines for the selection of highly effective siRNA sequences for mammalian and chick RNA interference. Nucleic Acids Res. 2004;32:936–948. doi: 10.1093/nar/gkh247. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.