Abstract
We use cryo-electron microscopy (cryo-EM) to study the 3D shapes of 94-bp-long DNA minicircles and address the question of whether cyclization of such short DNA molecules necessitates the formation of sharp, localized kinks in DNA or whether the necessary bending can be redistributed and accomplished within the limits of the elastic, standard model of DNA flexibility. By comparing the shapes of covalently closed, nicked and gapped DNA minicircles, we conclude that 94-bp-long covalently closed and nicked DNA minicircles do not show sharp kinks while gapped DNA molecules, containing very flexible single-stranded regions, do show sharp kinks. We corroborate the results of cryo-EM studies by using Bal31 nuclease to probe for the existence of kinks in 94-bp-long minicircles.
INTRODUCTION
In 2004 the DNA mechanics community was alerted by an influential paper stating that the spontaneous cyclization of very short DNA molecules containing as few as 94 bp occurs up to 105 times more easily than predicted by the prevailing theories of DNA bending (1). In that paper, Cloutier and Widom (C&W) proposed that thermal motion easily leads to spontaneous localized buckling where double-stranded DNA exits its elastic regime and becomes very flexible. The formation of such sites, better known as kinks, was predicted theoretically in 1975 by Crick and Klug (2) and consisted of sites where the stacking between consecutive base pairs was broken. However, kinks were expected to appear only very rarely in unstressed DNA at 30°C, i.e. under conditions of circularization applied by C&W (3,4). Therefore, the surprisingly high efficiency of DNA cyclization reported by C&W was very puzzling. The paper by C&W initiated many experimental, numerical and theoretical studies addressing the question of DNA mechanics with a particular emphasis on DNA bending (5–17). Some of these studies support C&W's interpretation (5,12) while some others contest it and point out possible problems with the experimental setup (8,16,17).
Aiming to resolve the controversy concerning spontaneous DNA kinking, we used cryo-electron microscopy (cryo-EM) to directly observe the shapes of 94-bp-long DNA minicircles of the same sequence as that used by C&W. If spontaneously occurring hyperflexible kinks help to cyclize 94-bp-long DNA molecules, then, after the cyclization, such kinks should be maintained in highly bent DNA and should be detectable by direct observations using cryo-EM. To have a positive control for the effect of hyperflexible sites on the shape of DNA minicircles, we also observed DNA minicircles with the same sequence but containing two 2-nt-long gaps oppositely located along the circumference of DNA minicircles (Figure 1). Such short gaps were shown to behave like very flexible sites in DNA (18). We also studied molecules containing two nicks (scissions in sugar-phosphate backbones of each of the strands) placed 180° apart along the circumference of the minicircles. Nicks can facilitate kinking of strongly bent DNA. Therefore, nicked DNA molecules would be expected to show the presence of kinks more frequently than covalently closed minicircles but less frequently than gapped minicircles (18).
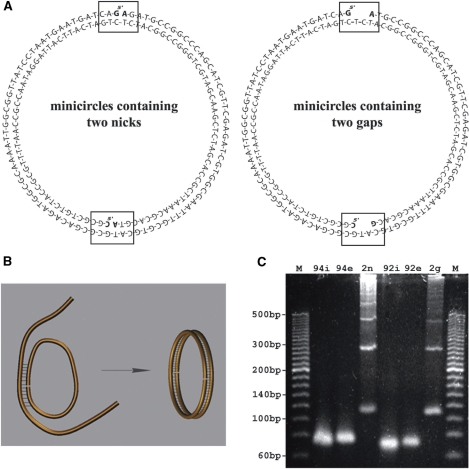
Figure 1.
DNA minicircles, their sequence and their formation by annealing of very long cohesive ends (17). (A) The sequence of doubly nicked and doubly gapped minicircles. The rectangular frames and bold font of the bases flanking the nicks or gaps are given to indicate the exact position of the nicks and gaps. (B) A schematic presentation of the principle of circularization by annealing of very long cohesive ends. (C) The agarose gel analysing the efficiency of formation of DNA minicircles. M—marker lines with 20-bp ladder, lines 2, 3 and 4—substrates and annealing products of minicircles with two nicks, lines 5, 6 and 7—substrates and annealing product of minicircles with two gaps. The 94i, 94e, 92i and 92e indicate 94- or 92 nt-long strands corresponding to the internal (i) and external (e) strands shown in (A). The gel lines 2n and 2g contain products of annealing reactions intended to produce doubly nicked (2n) and doubly gapped (2g) DNA minicircles.
Cryo-EM has proven to be the method of choice to observe the shape of DNA molecules. This method is free of possible artifacts caused by adsorption and/or by staining (19,20). The DNA specimen before cryo-vitrification consists of a DNA solution in a given buffer forming thin layers spanning micrometer large holes in supporting films on EM grids (20,21). Upon immersion of the specimen into liquid ethane that was pre-cooled to the temperature of about –160°C, the cooling speed is so high that the thin layers of aqueous solutions (30–50 nm thick) vitrify without initiating ice crystal formation and DNA molecules are almost instantly immobilized (21). Upon cryo-vitrification the specimens are transferred in a cryo-specimen holder to a pre-cooled electron microscope and observed without devitrification. The imaging contrast is provided by the electron density difference between the DNA and the surrounding vitrified solution. Therefore, possible artifacts due to staining are eliminated. To reconstruct the axial path of the observed DNA minicircles in 3D, we work with pairs of stereoimages that correspond to two different projection views of the vitrified thin layer with suspended DNA minicircles where between the two projections the specimen is tilted by 30° with respect to the incident electron beam (22,23). The resolution and contrast of such images is sufficient to trace the axial path of the analysed DNA molecules and such images have been used previously to study the shapes of 178-bp-long DNA minicircles (23). The development of specialized software dedicated to cryo-EM DNA imaging permits reconstructions of DNA shape with increased accuracy and objectivity (24) when compared to earlier methodology (22). Recently, we have developed a method for sorting reconstructed 158-bp-long DNA minicircles according to their shape similarity (25). Here we use this method to compare the shapes of 94-bp-long DNA minicircles that are covalently closed (i.e. where both strands are uninterrupted) with those that either contain nicks or 2-nt-long gaps.
MATERIALS AND METHODS
DNA constructs
The DNA minicircles studied here are identical in their sequence to the 94-bp-long minicircles that, according to Cloutier and Widom (1), circularized 105 times more easily than would be expected based on a standard DNA flexibility model. This sequence was derived from the sea urchin 5S rDNA nucleosome positioning sequence (26). The 94- and 92-nt-long oligonucleotides were purchased from Microsynth in PAGE purified form. To produce preparative amounts of the minicircles, we used the method of DNA circularization by annealing of-long cohesive ends (17) as is illustrated in Figure 1. For the annealing we either used two complementary 94-nt-long 5′-phosphorylated oligonucleotides that after annealing in 0.5 STE buffer (50 mM NaCl, 5 mM Tris, 0.5 mM, EDTA, pH 8) formed DNA minicircles with two nicks separated from each other by 47 nt (Figure 1), or we used two 92-nt-long oligonucletides that formed DNA minicircles that contained two 2-nt-long gaps oppositely placed along the circumference of the circle (Figure 1). The concentration of DNA was set to about 0.7 μM of each strand. For the annealing, the solution was heated to 94°C and then slowly cooled down to room temperature. For the cryo-EM observation of the doubly nicked and doubly gapped DNA minicircles, the annealed DNA was ethanol precipitated, washed with 70% ethanol and resuspended in water to have the DNA in a low salt solution. HPLC analysis indicated that salt concentration in the resuspended samples corresponded to ∼ 20 mM NaCl. Alternatively, after the DNA resuspension in water, we added magnesium acetate to the final concentration of 2.5 mM. The DNA concentration for cryo-EM preparations was set to about 200 µg/ml. For cryo-EM studies of nicked or gapped DNA minicircles it was not necessary to separate them from their multimeric forms since it was easy to recognize the size of monomeric DNA minicircles on the micrographs. In addition, we produced covalently closed DNA minicircles by the ligation of minicircles with nicks. Since ethanol precipitation of small DNA minicircles results in a significant material loss, we limited the number of precipitation steps to just one, which was performed after the ligation and Bal31 digestion. For this reason, the ligation reactions were performed by adding T4 DNA ligase (New England BioLabs) and 1/10 volume of 10 times concentrated ligation buffer (500 mM Tris–HCl, 100 mM MgCl2, 10mM ATP, 100 mM DTT, pH 7.5) to the finished annealing reactions containing two complementary 94-nt-long 5′-phosphorylated oligonucleotides. Typical ligation reactions were performed at 20°C, for 24 h and contained about 36 μg of annealed DNA and 2000 units of T4 DNA ligase in a volume of 0.9 ml.
To eliminate unligated circles, we used the Bal31 nuclease (New England BioLabs) to digest all non-covalently closed DNA. We verified that the Bal31 digestion worked well when we simply added 1/2 of the final volume of twice concentrated Bal31 reaction buffer (20 mM Tris–HCl, 600 mM NaCl, 12 mM CaCl2, 10 mM MgCl2, 1 mM EDTA, pH 8.0) to the finished ligation reactions. Typical Bal31 digestions contained about 35 μg of DNA and 100 units of Bal31 nuclease in a volume of 2 ml and the digestions lasted for 60 min at 30°C.
We used gel electrophoresis in the presence of ethidium bromide to verify that Bal31 digestion eliminated all nicked minicircles. After Bal31 digestion, the DNA was deproteinized and ethanol precipitated and then resuspended at a concentration of about 200 µg/ml either in water or in 2.5 mM magnesium acetate.
Cryo-EM
DNA minicircles in low salt solutions or in solutions containing 2.5 mM MgCl2 were applied to grids with holey carbon film and, after brief blotting, were plunged into liquid ethane that was pre-cooled to the temperature of about –160°C. After cryo-vitrification, the specimen was maintained at the temperature of liquid nitrogen during transfer and EM imaging in Philips CM12 electron microscope. The images were taken at the magnification of 45 000 using a Gatan CCD camera. For the stereo-pairs of images, the specimen holder was tilted by 30° between the two exposures.
3D reconstruction of DNA minicircles’ shapes
To reconstruct the 3D shape of the DNA minicircles out of aligned stereo-pairs of micrographs, we used the program developed by Jacob et al. (24). The program requires that the user helps the program to localize the corresponding images of the same DNA molecule by clicking several points along the visible axial path of the DNA on both stereo images. The program then constructs a 3D path that maximally correlates with the signal of the two images, taking into account that the images are projections of the same filamentous object where the tilt angle between the two projections is known. The reconstructed axial paths of the minicircles are expressed as lists of points with their x, y and z coordinates in relation to a given reference point. For further analysis, the reconstructed axial paths are resampled (using the spline function of Matlab) to obtain 150 equally spaced points along each curve.
Shape analysis
The shape distance between two reconstructed axial trajectories is determined by a root mean square deviation (RMSD) minimization procedure where one curve is rotated and translated to allow the best superposition with the second curve. To calculate the RMSD, it is necessary to define the corresponding points along the two compared curves. Our data consists of 3D discrete curves composed each of 150 equally spaced points. Offhand, we do not know the correspondence between the points on the two curves. Therefore, we have to take into account all 150 circular permutations of points and also the two possible orientations. The RMSD calculation is then performed by taking into account all possible rotations and translations to minimize the RMSD between the two set of points on the two curves. The permutation and orientation that gives the smallest RMSD is assumed to produce a consistent mapping between the corresponding points on the two curves and the obtained RMSD is taken as the distance between the two curves (27). This distance value is then used for the shape clustering analysis via the SPIN sorting algorithm (28). Of course, if an underlying sequence effect, for instance a gap, affects the shape of the two reconstructed curves, the RMSD is likely to be minimal when both gaps are correctly mapped to each other.
RESULTS AND DISCUSSION
Cyclization of DNA minicircles
Although linear double-stranded DNA molecules with the same 94-bp sequence as those studied here were reported to circularize surprisingly efficiently for their size, the observed yields of circularization were in fact very low (1). To prepare analytic amounts of these DNA minicircles, we used a cyclization approach based on annealing of very long cohesive ends (17). The principle of the method is shown in Figure 1B. Two complementary linear single strands circularize upon annealing since the ends of the first strand are complementary the central region of the second strand, while the ends of the second strand are complementary to the central region of the first strand. Figure 1A shows the actual sequences we used to obtain the DNA minicircles with two nicks (left side) and with two 2-nt gaps (right side). The gel in Figure 1C shows the efficiency of the method. Despite the relatively high concentrations of DNA in the annealing reaction (0.7 µM of each strand), thus facilitating the formation of multimeric circles, we observed that monomeric circles formed efficiently and usually formed with the yield matching or exceeding the formation of dimeric rings, while trimers, tetramers and higher multimers also formed clearly visible bands. Notice that in the shown gel, the DNA is visualized by fluorescence. Therefore, for the same number of monomeric and dimeric circles, the latter should show a twice-stronger signal on the gel. Notice also that the marker lines contain linear DNA and, therefore, one should expect a slight retardation of circular DNA molecules with respect to the linear size markers.
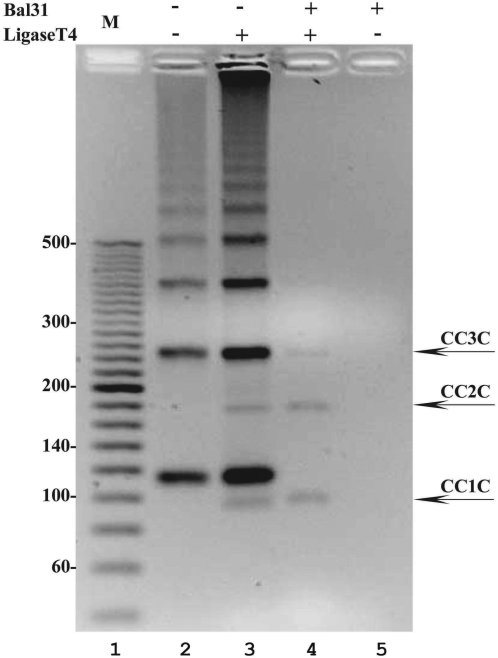
To obtain covalently closed DNA minicircles, we proceeded with ligation of doubly nicked annealed minicircles using T4 DNA ligase. To evaluate the efficiency of the ligation, we ran gels in the presence of ethidium bromide that intercalates to DNA. Since intercalation changes the DNA twist, the covalently closed circles (with both nicks ligated) become torsionally stressed and acquire some writhe, which in turn increases their electrophoretic migration since such molecules are more compact than the torsionally relaxed molecules. In contrast to the covalently closed molecules, the DNA circles with at least one nick dissipate their torsional stress and remain relaxed. Figure 2 shows a gel run in the presence of ethidium bromide. It is visible that the band of monomeric circles splits into two after ligation, where the species with higher electrophoretic migration is only present after ligation. We can therefore conclude that this quicker band corresponds to covalently closed minicircles that acquired writhe as a result of intercalation. To obtain preparations containing only covalently closed circles we eliminated unligated minicircles by the action of Bal31 nuclease. Bal31 nuclease is an exonuclease that can start digestion from a nick site and also shows endonucleolitic activity on ssDNA opposite the site of a pre-existing nick (29). As can be seen in Figure 2, Bal31 nuclease eliminated nicked DNA bands while covalently closed circles were insensitive to Bal31.
Figure 2.
The formation and purification of 94-bp-long covalently closed DNA minicircles. A 2.5% agarose gel run in the presence of ethidium bromide (0.5 µg/ml) reveals that after the ligation reaction of minicircles with nicks, new species appear on the gel (compare lanes 3 with 2). Only these new species resist digestion by Bal31 (lane 4) indicating that they are covalently closed DNA molecules. The electrophoretic migration of these new species is as expected for monomeric, dimeric and trimeric rings that acquired writhe due to ethidium bromide intercalation to covalently closed DNA molecules.
Cryo-EM of DNA minicircles
We studied three categories of DNA minicircles under two different conditions: 94-bp-long DNA minicircles that were covalently closed, doubly nicked or doubly gapped were observed in low salt solution and also in a solution resembling that of a standard ligation reaction. Stacking interactions are relatively weak in low salt solutions (30,31) and therefore kinks could be favoured under these conditions. On the other hand, in low salt solutions charged phosphate groups may not be screened and therefore strong bending may be limited as it would be opposed by intramolecular repulsion (23,30,31). Since C&W postulated that kinks helped the efficient ligation in their experiments (1), we also looked at minicircles under typical ligation conditions that include the addition of millimolar concentrations of magnesium salts. In the presence of millimolar concentrations of magnesium cations the charges of phosphate groups in the DNA backbone are efficiently screened so that the electrostatic repulsion would be attenuated and would not restrict the DNA minicircles from deviating from a circular shape (23). However, as mentioned above, the stacking interactions are stabilized under these conditions and therefore it is difficult to say whether the presence of millimolar concentration of magnesium should favour or disfavour kink formation (31). It needs to be clearly stated that the inadvertent evaporation from thin aqueous layers, that have an extremely high ratio of surface area to volume, makes it difficult to control the salt concentration. The vitrified layers may have up to 2–5 times higher salt concentration than the original solutions (23,32). Our starting low salt solutions contain 20 mM NaCl but the final concentration could reach 100 mM. However even at 50 mM NaCl, it is known that the intersegmental DNA repulsion is screened to a great extent (33).
Figure 3 presents a stereo pair of micrographs showing doubly gapped DNA minicircles that were suspended in a low salt solution. Below the micrographs, a 3D reconstruction of one of those minicircles is presented. It is important to mention that cryo-vitrified molecules do not undergo thermal fluctuations that could change their overall shape. However, chemically identical individual cryo-vitrified DNA minicircles (e.g. doubly gapped 94-bp-long DNA minicircles studied here) differ in their shape due to thermal fluctuations affecting those molecules just before they are immobilized by rapidly increasing viscosity of the aqueous thin layers undergoing cryo-vitrification. We are interested in the preferred minimal energy state of the analysed DNA molecules and we know that the molecules fluctuate around their minimal energy state.
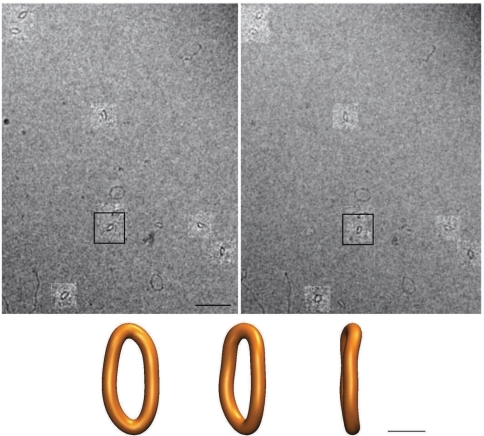
Figure 3.
A pair of stereo micrographs with the highlighted regions showing the corresponding DNA minicircles and the reconstructed 3D shape of one of the imaged minicircles. The imaged preparation contains doubly gapped DNA minicircles that were suspended in a low salt solution. The reconstructed 3D shape (shown in three different orientations) is that of the molecule ‘boxed’ on the micrographs. In addition to monomeric minicircles, the micrographs also reveal the presence of oligomeric circles (e.g. the one above the boxed minicircle) and as well as of longer linear molecules formed during the annealing. The magnification bar on a micrograph corresponds to 40 nm while the bar for the reconstructed shapes corresponds to 5 nm. Notice that the angular difference between the two images is 30° and not 6° that would have been suitable for giving a 3D impression upon direct viewing. However, the 30° difference brings much more information for the computer assisted 3D reconstruction of DNA minicircles’ shapes.
Figure 4 presents a gallery of all reconstructed shapes of the three categories of DNA minicircles observed under the two different conditions. For visual comparison, all reconstructed molecules are shown in the same orientation with respect to three principal axes of rotation (that were calculated assuming that the reconstructed shapes are rigid and have their mass uniformly distributed along the reconstructed centre line of the DNA). The reconstructed molecules are seen along the shortest principal axis of rotation, which would give the highest moment of inertia if the object would be rotated along this axis. The longest axis of rotation, which would give the smallest moment of inertia, is oriented vertically and the middle axis is oriented horizontally. To measure the deviation from circularity of the observed minicircles, we determined the ratio between the longest and middle axis of rotation and called this ratio the ellipticity. In a planar ellipse, these two axes would correspond to the major and minor axis of the ellipse. For a perfectly circular object, the ellipticity is 1 while for ellipses it is bigger than 1 and the more elongated the ellipse is the higher is its ellipticity. Since the minicircles are usually nearly planar, the ratio between the longest and middle axis of rotation is close to the ratio between the major and minor axis of the ellipse that would be obtained upon bringing the minicircles to planarity. We expected that the presence of two kinks oppositely placed along the circumference of a circle would cause the molecules to adopt elliptical shapes with the two kinks located at two apexes of the ellipse. In the absence of kinks, we expected that the energy minimizing shapes should be close to a regular circle.
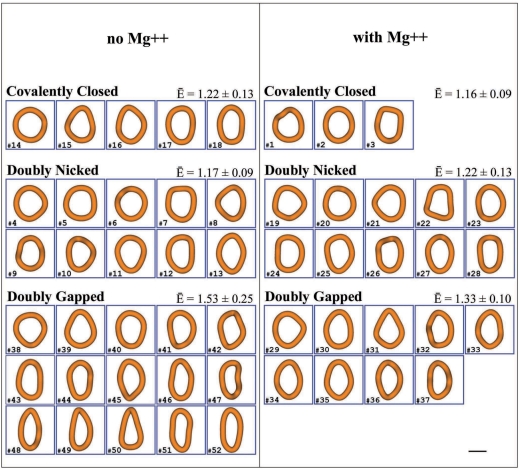
Figure 4.
The gallery of reconstructed shapes of covalently closed, doubly nicked and doubly gapped DNA minicircles that were imaged either in low salt solutions providing moderate screening of electrostatic repulsion between DNA segments (left panel) or in low salt solutions containing in addition millimolar concentrations of Mg2+ and thus providing strong screening of the electrostatic repulsion (right panel). The magnification bar denotes 5 nm. The numbers of individual reconstructed DNA molecules correspond to their entry numbers in the distance matrices presented in Figures 5 and 6. The E-bar symbol indicates the mean ellipticity for a given category of the reconstructed DNA minicircles.
Looking first at samples observed under low salt conditions (left panel of Figure 4) we see that doubly gapped minicircles (left lower part of Figure 4) indeed showed clearly elliptical shapes with a ratio between major and minor axis of 1.53 ± 0.25 (n = 15). Minicircles with two nicks (left middle part of Figure 4) were much more circular and had a mean ellipticity value of 1.17 ± 0.09 (n = 10). The covalently closed minicircles (left upper part of Figure 4), for which we expected the most round shapes, had a mean ellipticity value of 1.22 ± 0.13 (n = 5). However, taking into account their standard deviation (given above), we see that the mean ellipticity values of doubly nicked and covalently closed minicircles are within their respective dispersion range and we can conclude that both categories of minicircles behave similarly. Several studies concluded that nicks do show nearly normal stability of stacking (34) and that stacking at nick sites becomes destabilized only under partially denaturing conditions (31). Since the doubly nicked DNA molecules were on average less elliptical than the covalently closed minicircles, we take it as a strong indication that, under these conditions and for this size of DNA minicircles, the presence of nicks does not provide additional DNA flexibility when compared to covalently closed molecules. Doubly gapped DNA molecules not only showed high mean ellipticity but they also showed large variation of the ellipticity. We believe that this large variation is the consequence of high flexibility of the DNA at both gaps. Very flexible sites allow the molecules to sample a much wider range of configurations than minicircles without very flexible sites.
As previously mentioned, low salt conditions promote electrostatic repulsion between unscreened phosphate groups and this may limit the deviation from circularity in the studied minicircles. Strong electrostatic repulsion could in principle ‘hide’ the presence of kinks in doubly nicked and covalently closed DNA minicircles since, even in the presence of kinks, the energy minimizing shape could be a perfect ring. This a priori correct argument was invalidated, however, by the observation that doubly gapped molecules clearly deviated from circularity when observed in low salt solution. Therefore, the repulsion certainly was not strong enough to maintain the doubly gapped DNA molecules in perfectly circular form. The same should apply to kinks, which are the sites where base stacking is broken. This in turn leads to DNA becoming very flexible (12). For this reason, the presence of sharp bends in gapped molecules demonstrates that if kinks were present in the nicked or covalently closed minicircles, then they should be also visible as sharp bends.
Since Cloutier and Widom postulated that spontaneously appearing kinks facilitated enzymatic ligation of the minicircles, it was important to observe the minicircles under conditions close to those required for the action of DNA ligase, i.e. in the presence of millimolar concentration of magnesium salts. It is possible, for example, that the DNA kinking occurs more easily in the presence of magnesium ions. The right panel in Figure 4 presents the reconstructed shapes of DNA minicircles from preparations that were resuspended in solution containing 2.5 mM MgCl2. We estimate that the final concentration of MgCl2 in the cryo-vitrified specimens was at least 5 mM since even during the automatic plunging that we applied, the thin aqueous layers easily lose more than 50% of their volume due to evaporation (23). Therefore, we estimate that the DNA minicircles presented in the right panel of Figure 4 were imaged when surrounded by a sufficient concentration of magnesium to be compared to the conditions of standard ligation reactions. Similar to the preparations without the added magnesium, we observed that the doubly gapped DNA molecules (lower part of Figure 4) were most elliptical and showed a ratio of the major to minor axis of 1.33 ± 0.10 (n = 9). The minicircles with two nicks (middle part of Figure 4) were more circular than the molecules with two gaps and had a mean ellipticity value of 1.22 ± 0.13 (n = 10). The covalently closed minicircles from the preparations with added magnesium (upper row in the right panel of Figure 4) turned out to be most circular and showed the mean ellipticity value of 1.16 ± 0.09 (n = 3).
Comparing the characteristics of the three types of DNA minicircles observed under conditions of moderate and strong charge screening, it is somewhat surprising that the doubly gapped molecules showed higher ellipticity under moderate charge screening conditions where the repulsion should limit strong bending. However, it is possible that in the presence of 5–10 mM of MgCl2 the bases in the gaps tend to stack and that this limits the bending there (35). Comparing doubly gapped minicircles with doubly nicked and covalently closed minicircles we see that under both conditions (moderate and strong screening of electrostatic repulsion) doubly gapped minicircles are significantly more elliptical than the other two classes and this suggests that these doubly nicked and covalently closed minicircles do not contain stable kinks. To support this observation, we performed statistical tests to see whether the difference in ellipticity between the doubly gapped and remaining DNA molecules is statistically significant. To do this, we grouped together the doubly gapped DNA molecules presented in Figure 4 and compared them with the set of covalently closed and doubly nicked DNA minicircles grouped together. The Wilcoxon statistical test, which does not assume any particular distribution of ellipticity values, rejected the null hypothesis with the P-value of 10–5. Therefore, it is highly unlikely that the observed or higher difference in the mean ellipticity of two groups is the result of random sampling from the same population with unknown dispersion of the ellipticity value. We also performed the Student t test, which assumes that ellipticity values have a Gaussian distribution (which may not be the case), and this gave a P-value of 2 × 10–6 thus showing even stronger rejection of its null hypothesis. More details about the statistical analysis are provided as Supplementary Data.
Comparing the images of the doubly gapped minicircles with the images of the doubly nicked and covalently closed DNA minicircles we propose that the sites of kinks, i.e. the apexes of ellipsoidal minicircles, are the sites where gaps are located. Although we do not have a formal proof that this is the case the following four arguments provide a strong support for this interpretation: (i) Several earlier studies revealed that gaps, including those formed by two missing nucleotides, form very flexible sites that are detectable by gel retardation assay where linear molecules that can be easily bent migrate slower than molecules that are unbent (31). (ii) The high ellipticity was observed in the majority of minicircles with two gaps placed 180° apart on the circular map of the minicircles. However, this was not the case of molecules that were covalently closed or contained two nicks. (iii) When an elastic filament contains a site that is more flexible than the rest of the filament and when this filament is forced to be bent, then the minimal energy state is reached when the bend localizes at the flexible site rather than when the bending is redistributed over the entire filament. (iv) In case of a circular elastic filament with two flexible sites located 180° apart, the minimal energy state is achieved when the two flexible sites are both strongly bent to a similar extent.
Global comparison of shapes, SPIN method
The ellipticity value is a convenient measure of shape provided that the analysed DNA minicircles remain practically planar. However, the flexible sites in the doubly gapped DNA should also permit out of plane bending. Out of plane bending could be also induced by the residual torsional tension in the covalently closed DNA molecules. Therefore, we decided to perform a more global comparison of the shapes of the analysed categories of DNA minicircles. We recently presented a method of global shape analysis for comparing DNA minicircles (25). We applied this method here hoping to better reveal the characteristic differences between the different groups of analysed DNA minicircles. The measure of shape difference or similarity is based on the determination of the root mean square deviation (RMSD) between the corresponding points on the pairs of curves representing the axial paths of the reconstructed DNA minicircles [see ‘Material and Methods’ section for a general description of RMSD values calculation procedure and ref. (27) for more a detailed explanation]. Once we have all of the pair-wise RMSD, we can visualize these in form of the shape-distance matrix where the shape-distance value is expressed in a colour scale.
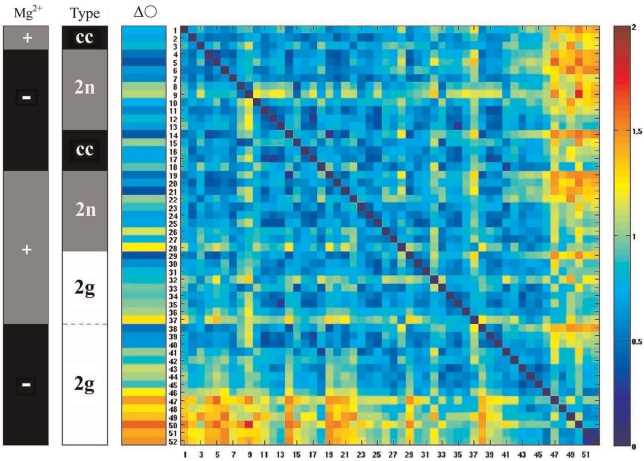
Figure 5 presents the shape-distance matrix between the reconstructed 3D shapes of the 52 individual DNA minicircles belonging to the six different analysed groups. The colour of each square in the distance matrix indicates the RMSD value for a given pair of compared minicircles (the numbers of compared minicircles are listed along X and Y axis and correspond to the numbers indicated in Figure 4). The RMSD colour scale is presented to the right of the distance matrix: blue indicates that the relevant two molecules are similar in their shape while red indicates strong dissimilarities. The black–grey–white vertical bar indicates covalently closed (cc) minicircles as black, doubly nicked (2n) as grey and doubly gapped (2g) as white. In the black-grey vertical bar, grey indicates the molecules that were imaged in solutions containing millimolar concentrations of MgCl2 (+), while black indicates the minicircles imaged in low salt solutions without added MgCl2 (–). Looking at both of the black-grey-(white) vertical bars in Figures 5 and 6, it is possible to trace the origin of every molecule to each of the six groups presented in Figure 4. In addition, the numbers of individual molecules indicated in Figures 5 and 6 are the same as those indicated in the Figure 4, thus allowing us to recognize each individual molecule.
Figure 5.
The colour-coded shape–distance matrix between the reconstructed 3D shapes of 52 individual minicircles belonging to the six different analysed groups. The RMSD colour scale (right) indicates the shape distances between all possible pairwise comparisons of shape. The six different groups were placed in the order of their increasing mean ellipticity and the molecules within each group were also placed in the same order. That order is reflected in Figure 4 by the identification numbers associated to the individual reconstructed DNA molecules. Two black–grey–(white) vertical bars to the left of the distance matrix allow us to trace the origin of individual minicircles to each group (see the main text). The colour vertical bar to the left of the distance matrix (denoted with ΔO) shows the shape distance of every individual analysed minicircle to a perfect circle whose length corresponds to that of the analysed molecules.
Figure 6.
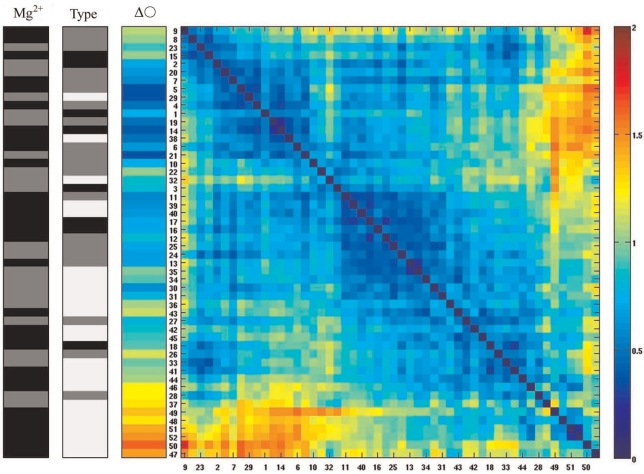
The shape distance matrix after the SPIN ordering procedure (25,28). Notice that in contrast to Figure 5 the order of the molecules was rearranged by the SPIN algorithm. However, gapped molecules are still grouped nearly together indicating that these molecules have, on average, a different shape than doubly nicked and covalently closed DNA molecules (see the main text).
In the distance matrix presented in Figure 5, the analysed groups of molecules were placed in an arbitrary order where individual groups were ordered by increasing mean ellipticity. Within each group the molecules were ordered also with increasing ellipticity. The matrix is symmetric since the order of the molecules along the horizontal (from left to right) and vertical direction (from top to bottom) is the same. The colour vertical bar to the left of the distance matrix shows the shape distances of the individual analysed molecules when compared to a perfect circle. While comparing shape distances of individual DNA minicircles to a perfect circle, it is well visible that differences in ellipticity show a good correlation with the RMSD values, i.e. molecules with small ellipticity show small shape distances to a perfect planar circle, while this distance increases for molecules with high ellipticity. Comparing shape distances between different categories of DNA minicircles we see, for example, that the average shape distance between covalently closed minicircles imaged in solutions containing charge-neutralizing concentrations of divalent cations and doubly gapped minicircles observed in low salt solution is rather large and showed mean RMSD value of 1.05 nm. On the other hand, the RMSD values between molecules belonging to the four different groups of covalently closed and doubly nicked DNA minicircles showed quite small RMSD on average when compared to each other. For example, the average distance between covalently closed DNA minicircles imaged in solution with and without millimolar concentrations of Mg2+ was of 0.76 nm. However, the RMSD comparison also revealed the shape differences between the molecules that were not detected by a simple comparison of the ellipticity values. For instance, the minicircle 9 shows relatively high RMSD value when compared with many molecules with similar ellipticity. A visual inspection of the reconstructed shape of the minicircle 9 revealed very strong bending out of plane. This demonstrates that the ellipticity value is not a sufficient shape descriptor for the molecules that deviate strongly from planarity.
The order of the DNA minicircles entered into the distance matrix of Figure 5 was based on the increasing ellipticity values of the analysed groups of DNA minicircles. We were, however, interested in a more global order based on the sorting and clustering of the molecules according to their overall shape similarity. The SPIN algorithm rearranges the order of the molecules in the shape–distance matrix until values near the diagonal reach a possible minimum (28) and this is achieved when similar molecules (with small shape distance expressed in RMSD) are placed near each other (25). The SPIN algorithm can start from any input order of molecules and will rearrange this order by bringing similar molecules close to each other in the matrix. In particular, we were interested whether doubly gapped DNA molecules will be separated from the rest of the molecules based on their overall shape characteristics. We observed that the SPIN sorting algorithm placed the great majority of doubly gapped DNA molecules (19 out of 24 molecules indicated with white colour in the ‘Type’ bar) close to each other, thus indicating that these molecules show overall similarity of shapes. One of the characteristics of those shapes is high ellipticity. The remaining 5 doubly gapped DNA minicircles were interspersed between covalently closed and doubly nicked molecules. This placement was, however, consistent with the expected behaviour of minicircles with two very flexible sites that due to thermal fluctuations can also adopt more circular shapes as their momentary configurations. We observed that 24 out of 28 covalently closed and doubly nicked molecules (indicated with black and grey colour in the ‘Type’ bar) were placed close together by the SPIN algorithm, indicating that molecules belonging to these categories had essentially the same shape (taking into account thermal fluctuations). One of characteristics of those shapes is low ellipticity. This result indicates that double nicked and covalently closed DNA minicircles have essentially the same quasi-circular shape and that they do not reveal the presence of hyperflexible kinks such as those present in doubly gapped DNA minicircles.
Bal31 nuclease does not reveal the presence of kinks in covalently closed DNA minicircles
As described in ‘Materials and Methods’ section, we have used Bal31 nuclease to purify covalently closed DNA minicircles from nicked circles that were incomplete products of the ligation reactions. Bal31 nuclease has very useful properties for this purpose as it has exonucleolitic activity that can initiate digestion from the nick sites and also has an activity of single-stranded endonuclease that can start digestion of the strand opposing the nick (29). Bal31 endonucleolitic activity can also be used to digest DNA molecules having regions with a destabilized double-helix, such as those appearing in negatively supercoiled DNA (36). More recently Du et al. (17) used Bal31 nuclease to detect kinks in extremely small DNA minicircles. Since kink formation destabilizes the double helix locally, this makes the kink site sensitive to Bal31 nuclease. In their elegant studies, Du et al. (17) analysed Bal31 susceptibility of small DNA minicircles that were formed by the long cohesive ends annealing method followed by ligation. When the DNA minicircles were maintained in torsionally relaxed form then even minicircles having only 85 or 86 bp did not show the presence of kinks. However, torsionally relaxed extremely small DNA minicircles having only 63 base pairs were digested by Bal31 nuclease. This shows that the bending stress in these extremely small DNA minicircles is too large to be accomplished by smooth bending of dsDNA and can only be achieved by the formation of kinks (17). As mentioned above, Du et al. did not detect kinks in 85 and 86-bp minicircles and this puts in question Cloutier and Widom's proposal that invoked kink formation in the cyclization of 94-bp-long minicircles since the bending stress in 85-bp-long DNA minicircles is certainly higher than that in 94-bp-long minicircles. However, Du et al. used a completely different DNA sequence than the sequence selected for its facility of cyclization by C&W. Therefore, one needs to consider the formal possibility that 94-bp-long minicircles with the sequence identical to that used by C&W can form kinks.
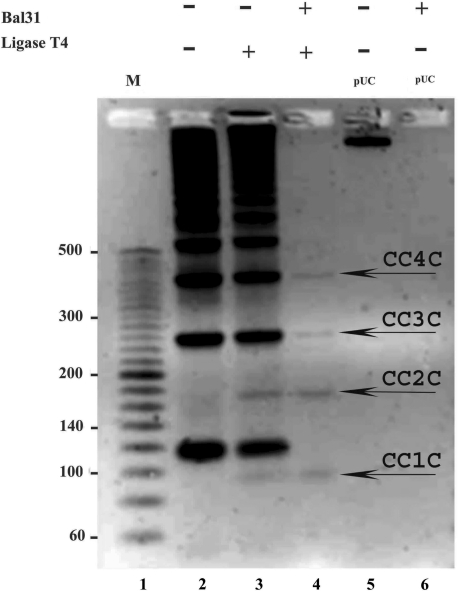
Although we did use Bal31 nuclease to eliminate minicircles with nicks and leave behind the covalently closed ones (see Figure 2) we did not have a positive control where, under our reaction conditions, molecules with locally destabilized DNA helix but without nicks can be eliminated by Bal31 digestion. Negatively supercoiled DNA provides such a control. Figure 7 shows that under the optimal Bal31 reaction conditions recommended by New England Biolabs (10 mM Tris–HCl, 300 mM NaCl, 6 mM CaCl2, 5 mM MgCl2, 0.5 mM EDTA, pH 8.0), covalently closed 94-bp minicircles resist Bal31 action (fourth line of the gel) while the negatively supercoiled plasmid pUC18 gets completely digested (compare lanes 5 and 6). Notice that the agarose gel shown in Figure 7 is run in the presence of ethidium bromide and this permits the electrophoretic separation of covalently closed DNA minicircles in monomeric and higher sizes from the circular forms that are not completely ligated. The separation is due to the fact that ethidium bromide intercalation changes DNA twist and this induces writhing in covalently closed DNA molecules but not in the nicked DNA molecules that remain torsionally relaxed due to their freedom of rotation at the nick sites (37). Notice that Bal31 digestion (the digestion was performed in the absence of ethidium bromide) eliminated nicked forms while the covalently closed DNA circles remained undigested. This applied to 94-bp-long DNA minicircles (CC1C) and also to dimeric (CC2C) and higher forms. The resistance of 94-bp-long DNA minicircles to Bal31 digestion provides the biochemical indication that they do not contain kinks. It is important to stress here that 94-bp-long DNA minicircles were resistant to Bal31 digestion not due to their small size, which could, in principle, make it difficult for the enzyme to be correctly positioned for its action. Nicked 94-bp-long minicircles were completely digested (compare lanes 3 and 4 in Figure 7) and Du et al. have shown that if kinks are present then covalently closed DNA minicircles as small as 63 bp can be digested by Bal31 nuclease (17).
Figure 7.
Bal31 nuclease detects a destabilized duplex DNA structure in covalently closed supercoiled DNA molecules but does not reveal the presence of kinks in 94-bp covalently closed DNA minicircles. A 2.5% agarose gel run in the presence of ethidium bromide (0.5 µg/ml) reveals that while negatively supercoiled DNA (lane 6) and nonligated nicked rings are completely digested by Bal31 nuclease (see lanes 2–4), the covalently closed monomeric, dimerc, trimeric and tetrameric circles (indicated with arrows) remain resistant to action of Bal31 nuclease (compare lanes 4 and 3).
Possible explanation why the cyclization efficiency calculated by Cloutier and Widom was greatly overestimated
By using cryo-EM to observe 94-bp-long DNA minicircles with the identical sequence as those studied by Cloutier and Widom (1), we conclude that these minicircles do not contain hyperflexible sites (kinks) such as those observed in the case of DNA minicircles with 2-nt-long single-stranded regions. To corroborate our cryo-EM studies, we also used Bal31 nuclease to probe those minicircles for the presence of DNA kinks. Du et al. showed earlier that 63-bp DNA minicircles are so strongly bent that they form kinks and become sensitive to Bal31 nuclease, while 85-bp DNA minicircles can accommodate their bending stress without kinks and without becoming sensitive to Bal31 nuclease (17). However, the 94-bp-long minicircles with the sequence selected for their very efficient cyclization by C&W were not previously tested for their sensitivity to Bal31 nuclease. Our tests reveal resistance of those minicircles to Bal31 and therefore indicate the absence of kinks in these minicircles.
If there are no kinks in those 94-bp-long minicircles, why then did C&W conclude that those minicircles cyclize 105 times more easily than predicted according to prevailing theories of DNA bending (1)? Du et al. (8) suggested that in the ligation reactions performed by C&W, the kinetic assumptions underlying the calculation of the J-factor and derived from its cyclization efficiency (38,39) were not met and that led to great overestimation of this efficiency. More specifically, special care needs to be taken to have conditions where the rate of substrate decay (i.e. the dissociation of annealed sticky ends in both monomeric and dimer forms) is much higher than the rate of ligase binding to the joined ends. The Du et al. analysis revealed that for the type of sticky ends and for the concentration of ligase used by C&W, these conditions were not satisfied for the dissociation rate of annealed ends in dimeric molecules. Du et al. performed their own circularization of very small DNA minicircles (105–130-bp-long) taking care that the above mentioned conditions were satisfied and determined that the cyclization efficiency was just as expected according to the standard models of DNA flexibility. However, Du et al. did not analyse the cyclization efficiency of the actual sequence selected for its extremely efficient cyclization by C&W since it is particularly difficult to fulfil the kinetic assumptions needed to determine the J factor for such short DNA duplexes (8). By observing DNA minicircles identical to these studied by C&W, we directly addressed the question of whether circular DNA of such a small size and of this particular sequence contains sites where double-stranded DNA exits its elastic regime and becomes very flexible. Our cryo-EM observations and biochemical testing using Bal31 nuclease did not reveal presence of such DNA deformations.
CONCLUSIONS
The underlying idea of our study was that if the cyclization of 94-bp-long DNA molecules at ambient temperature depends on thermally driven events during which the DNA exits its elastic regime and forms hyperflexible sites (as proposed by C&W), then, after the cyclization, such sites (kinks) should be maintained in the highly bent 94-bp-long DNA and should be detectable by direct observations using cryo-EM and/or by biochemical methods. Since both of those approaches did not detect the presence of kinks, our results suggest that the standard model of DNA flexibility holds and the DNA does not need to exit its elastic regime to circularize 94-bp-long DNA molecules.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Swiss NSF grant 3100A0-116275 (to A.S.) and 205320-112178 (to J.H.M.); American NSF grant DMS#0810415 (to E.J.R.). The Open Access publication charge for this paper has been waived by Oxford University Press — NAR Editorial Board members are entitled to one free paper per year in recognition of their work on behalf of the journal.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Drs Diana Velluto and Conlin O’Neil for HPLC analysis needed to determine the salt concentration in resuspended DNA samples. We thank Prof Jonathan Widom for his constructive comments on the manuscript.
REFERENCES
- 1.Cloutier TE, Widom J. Spontaneous sharp bending of double-stranded DNA. Mol. Cell. 2004;14:355–362. doi: 10.1016/s1097-2765(04)00210-2. [DOI] [PubMed] [Google Scholar]
- 2.Crick FH, Klug A. Kinky helix. Nature. 1975;255:530–533. doi: 10.1038/255530a0. [DOI] [PubMed] [Google Scholar]
- 3.Gueron M, Kochoyan M, Leroy JL. A single mode of DNA base-pair opening drives imino proton exchange. Nature. 1987;328:89–92. doi: 10.1038/328089a0. [DOI] [PubMed] [Google Scholar]
- 4.Russu IM. Probing site-specific energetics in proteins and nucleic acids by hydrogen exchange and nuclear magnetic resonance spectroscopy. Methods Enzymol. 2004;379:152–175. doi: 10.1016/S0076-6879(04)79009-6. [DOI] [PubMed] [Google Scholar]
- 5.Yan J, Marko JF. Localized single-stranded bubble mechanism for cyclization of short double helix DNA. Phys. Rev. Lett. 2004;93:108108. doi: 10.1103/PhysRevLett.93.108108. [DOI] [PubMed] [Google Scholar]
- 6.Wiggins PA, Phillips R, Nelson PC. Exact theory of kinkable elastic polymers. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2005;71:021909. doi: 10.1103/PhysRevE.71.021909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ranjith P, Kumar PB, Menon GI. Distribution functions, loop formation probabilities, and force-extension relations in a model for short double-stranded DNA molecules. Phys. Rev. Lett. 2005;94:138102. doi: 10.1103/PhysRevLett.94.138102. [DOI] [PubMed] [Google Scholar]
- 8.Du Q, Smith C, Shiffeldrim N, Vologodskaia M, Vologodskii A. Cyclization of short DNA fragments and bending fluctuations of the double helix. Proc. Natl Acad. Sci. USA. 2005;102:5397–5402. doi: 10.1073/pnas.0500983102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yan J, Kawamura R, Marko JF. Statistics of loop formation along double helix DNAs. Phys. Rev. E. 2005;71:17. doi: 10.1103/PhysRevE.71.061905. [DOI] [PubMed] [Google Scholar]
- 10.Yuan C, Rhoades E, Lou XW, Archer LA. Spontaneous sharp bending of DNA: role of melting bubbles. Nucleic Acids Res. 2006;34:4554–4560. doi: 10.1093/nar/gkl394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Czapla L, Swigon D, Olson WK. Sequence-dependent effects in the cyclization of short DNA. J. Chem. Theory Comput. 2006;2:685–695. doi: 10.1021/ct060025+. [DOI] [PubMed] [Google Scholar]
- 12.Lankas F, Lavery R, Maddocks JH. Kinking occurs during molecular dynamics simulations of small DNA minicircles. Structure. 2006;14:1527–1534. doi: 10.1016/j.str.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 13.Fogg JM, Kolmakova N, Rees I, Magonov S, Hansma H, Perona JJ, Zechiedrich EL. Exploring writhe in supercoiled minicircle DNA. J. Phys. Condens. Matter. 2006;18:S145–S159. doi: 10.1088/0953-8984/18/14/S01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ruscio JZ, Onufriev A. A computational study of nucleosomal DNA flexibility. Biophys. J. 2006;91:4121–4132. doi: 10.1529/biophysj.106.082099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wiggins PA, van der Heijden T, Moreno-Herrero F, Spakowitz A, Phillips R, Widom J, Dekker C, Nelson PC. High flexibility of DNA on short length scales probed by atomic force microscopy. Nat. Nanotechnol. 2006;1:137–141. doi: 10.1038/nnano.2006.63. [DOI] [PubMed] [Google Scholar]
- 16.Yuan C, Lou XW, Rhoades E, Chen H, Archer LA. T4 DNA ligase is more than an effective trap of cyclized dsDNA. Nucleic Acids Res. 2007;35:5294–5302. doi: 10.1093/nar/gkm582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Du Q, Kotlyar A, Vologodskii A. Kinking the double helix by bending deformation. Nucleic Acids Res. 2008;36:1120–1128. doi: 10.1093/nar/gkm1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Protozanova E, Yakovchuk P, Frank-Kamenetskii MD. Stacked-unstacked equilibrium at the nick site of DNA. J. Mol. Biol. 2004;342:775–785. doi: 10.1016/j.jmb.2004.07.075. [DOI] [PubMed] [Google Scholar]
- 19.Adrian M, ten Heggeler-Bordier B, Wahli W, Stasiak AZ, Stasiak A, Dubochet J. Direct visualization of supercoiled DNA molecules in solution. EMBO J. 1990;9:4551–4554. doi: 10.1002/j.1460-2075.1990.tb07907.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dubochet J, Adrian M, Dustin I, Furrer P, Stasiak A. Cryoelectron microscopy of DNA molecules in solution. Methods Enzymol. 1992;211:507–518. doi: 10.1016/0076-6879(92)11028-h. [DOI] [PubMed] [Google Scholar]
- 21.Dubochet J, Adrian M, Chang JJ, Homo JC, Lepault J, McDowall AW, Schultz P. Cryo-electron microscopy of vitrified specimens. Q. Rev. Biophys. 1988;21:129–228. doi: 10.1017/s0033583500004297. [DOI] [PubMed] [Google Scholar]
- 22.Dustin I, Furrer P, Stasiak A, Dubochet J, Langowski J, Egelman E. Spatial visualization of DNA in solution. J. Struct. Biol. 1991;107:15–21. doi: 10.1016/1047-8477(91)90026-s. [DOI] [PubMed] [Google Scholar]
- 23.Bednar J, Furrer P, Stasiak A, Dubochet J, Egelman EH, Bates AD. The twist, writhe and overall shape of supercoiled DNA change during counterion-induced transition from a loosely to a tightly interwound superhelix. Possible implications for DNA structure in vivo. J. Mol. Biol. 1994;235:825–847. doi: 10.1006/jmbi.1994.1042. [DOI] [PubMed] [Google Scholar]
- 24.Jacob M, Blu T, Vaillant C, Maddocks JH, Unser M. 3-D shape estimation of DNA molecules from stereo cryo-electron micro-graphs using a projection-steerable snake. IEEE Trans. Image Process. 2006;15:214–227. doi: 10.1109/tip.2005.860310. [DOI] [PubMed] [Google Scholar]
- 25.Amzallag A, Vaillant C, Jacob M, Unser M, Bednar J, Kahn JD, Dubochet J, Stasiak A, Maddocks JH. 3D reconstruction and comparison of shapes of DNA minicircles observed by cryo-electron microscopy. Nucleic Acids Res. 2006;34:e125. doi: 10.1093/nar/gkl675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simpson RT, Stafford DW. Structural features of a phased nucleosome core particle. Proc. Natl Acad. Sci. USA. 1983;80:51–55. doi: 10.1073/pnas.80.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Coutsias EA, Seok C, Dill KA. Using quaternions to calculate RMSD. J. Comput. Chem. 2004;25:1849–1857. doi: 10.1002/jcc.20110. [DOI] [PubMed] [Google Scholar]
- 28.Tsafrir D, Tsafrir I, Ein-Dor L, Zuk O, Notterman DA, Domany E. Sorting points into neighborhoods (SPIN): data analysis and visualization by ordering distance matrices. Bioinformatics. 2005;21:2301–2308. doi: 10.1093/bioinformatics/bti329. [DOI] [PubMed] [Google Scholar]
- 29.Legerski RJ, Gray HB, Jr, Robberson DL. A sensitive endonuclease probe for lesions in deoxyribonucleic acid helix structure produced by carcinogenic or mutagenic agents. J. Biol. Chem. 1977;252:8740–8746. [PubMed] [Google Scholar]
- 30.Furrer P, Bednar J, Stasiak AZ, Katritch V, Michoud D, Stasiak A, Dubochet J. Opposite effect of counterions on the persistence length of nicked and non-nicked DNA. J. Mol. Biol. 1997;266:711–721. doi: 10.1006/jmbi.1996.0825. [DOI] [PubMed] [Google Scholar]
- 31.Yakovchuk P, Protozanova E, Frank-Kamenetskii MD. Base-stacking and base-pairing contributions into thermal stability of the DNA double helix. Nucleic Acids Res. 2006;34:564–574. doi: 10.1093/nar/gkj454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cyrklaff M, Adrian M, Dubochet J. Evaporation during preparation of unsupported thin vitrified aqueous layers for cryo-electron microscopy. J. Electron. Microsc. Tech. 1990;16:351–355. doi: 10.1002/jemt.1060160407. [DOI] [PubMed] [Google Scholar]
- 33.Randall GL, Pettitt BM, Buck GR, Zechiedrich EL. Electrostatics of DNA-DNA juxtapositions: consequences for type II topoisomerase function. J. Phys. Condens. Matter. 2006;18:S173–S185. doi: 10.1088/0953-8984/18/14/S03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hagerman KR, Hagerman PJ. Helix rigidity of DNA: the meroduplex as an experimental paradigm. J. Mol. Biol. 1996;260:207–223. doi: 10.1006/jmbi.1996.0393. [DOI] [PubMed] [Google Scholar]
- 35.Mills JB, Vacano E, Hagerman PJ. Flexibility of single-stranded DNA: use of gapped duplex helices to determine the persistence lengths of poly(dT) and poly(dA) J. Mol. Biol. 1999;285:245–257. doi: 10.1006/jmbi.1998.2287. [DOI] [PubMed] [Google Scholar]
- 36.Gray HB, Jr, Ostrander DA, Hodnett JL, Legerski RJ, Robberson DL. Extracellular nucleases of Pseudomonas BAL 31. I. Characterization of single strand-specific deoxyriboendonuclease and double-strand deoxyriboexonuclease activities. Nucleic Acids Res. 1975;2:1459–1492. doi: 10.1093/nar/2.9.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bates AD, Maxwell A. DNA Topology. Oxford: Oxford University Press; 2005. [Google Scholar]
- 38.Shore D, Baldwin RL. Energetics of DNA twisting. I. Relation between twist and cyclization probability. J. Mol. Biol. 1983;170:957–981. doi: 10.1016/s0022-2836(83)80198-3. [DOI] [PubMed] [Google Scholar]
- 39.Shore D, Langowski J, Baldwin RL. DNA flexibility studied by covalent closure of short fragments into circles. Proc. Natl Acad. Sci. USA. 1981;78:4833–4837. doi: 10.1073/pnas.78.8.4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.