Abstract
AEBP2 is a zinc finger protein that has been shown to interact with the mammalian Polycomb Repression Complex 2 (PRC2). In the current study, we characterized this unknown protein and tested its potential targeting roles for the PRC2. AEBP2 is an evolutionarily well-conserved gene that is found in the animals ranging from flying insects to mammals. The transcription of mammalian AEBP2 is driven by two alternative promoters and produces at least two isoforms of the protein. These isoforms show developmental stage-specific expression patterns: the adult-specific larger form (51 kDa) and the embryo-specific smaller form (32 kDa). The AEBP2 protein binds to a DNA-binding motif with an unusual bipartite structure, CTT(N)15-23cagGCC with lower-case being less critical. A large fraction of AEBP2's target loci also map closely to the known target loci of the PRC2. In fact, many of these loci are co-occupied by the two proteins, AEBP2 and SUZ12. This suggests that AEBP2 is most likely a targeting protein for the mammalian PRC2 complex.
INTRODUCTION
AEBP2 is a Gli-type zinc finger protein, which was originally identified due to its in vitro binding capability to the promoter region of adipose P2 (aP2) gene encoding a fatty acid-binding protein (1). This initial study revealed that this protein contains three zinc finger units and a novel basic domain, and also that this protein may function as a repressor based on co-transfection reporter assays. Soon afterwards, the homologous protein, called JING (meaning ‘still’), was also isolated from Drosophila (2). According to the results from several studies, JING is involved in border cell migration (2) and development of the central nervous system (3). Genetic studies further suggested that jing may interact with the fly Polycomb Group (PcG) protein complexes (4,5). The potential role of AEBP2 as a component of the PcG complexes has been further strengthened by another series of studies using the mammalian cell line system (6). Human AEBP2 has been co-purified with the mammalian PcG Repression Complex 2 (PRC2), and the subsequent study revealed that the AEBP2 protein can interact with the three core components of PRC2, including EED, SUZ12 and RbAp48, and that the interaction of AEBP2 with these proteins enhances the catalytic activity of the histone methylation activity of the PRC2 complex (7).
Although the core proteins for PRC2 have been identified, the mechanism by which PRC2 is targeted to numerous genomic loci is currently unknown (8). This lack of knowledge is mainly due to the facts that the identified core proteins do not have DNA-binding capability, and that DNA-binding proteins have never been consistently co-purified with the PRC2 (9–12). In Drosophila, however, another Gli-type zinc finger gene, called pho (Pleiohomeotic), has been shown to be a targeting protein for its PcG complexes (13). Recent studies further confirmed the presence of two Pho-containing complexes, INO80 and PhoRC, and PhoRC is now regarded as a new member of PcG complexes based on its repression role through another PcG protein, SFMBT [Sex comb on middle leg-related gene with four mbt domains; (14)]. Along with the other data from several studies, this evidence has long suggested that YY1 (Yin Yang 1), the mammalian homologue of pho, might be a targeting protein for the mammalian PRC2. Nevertheless, this possibility has never been formally demonstrated so far. In that regard, it is intriguing to point out that AEBP2 has both DNA-binding capability and PcG connection. Thus, it has been hypothesized that AEBP2 might be a targeting protein for the mammalian PRC2. However, very little is known about the general aspects of AEBP2, in particular its DNA-binding motif and downstream genes.
As part of the effort of characterizing this largely unknown gene and to test the above possibility, we have conducted a series of comparative genomics and DNA-binding motif studies in the current study. According to the results derived from this study, AEBP2 is an evolutionarily well-conserved protein that is found in all the animals ranging from flying insects to placental mammals. The exon structure of mouse Aebp2 indicates the presence of alternative splicing involving both 5′- and 3′-end exons, and subsequently two major forms of AEBP2 with different protein sizes, 52 and 31 kDa. A series of ChIP cloning experiments using anti-AEBP2 and -SUZ12 antibodies also identified many in vivo target loci that are bound by these two proteins. Subsequent gel shift assays using the sequences obtained from these target loci revealed one potential DNA-binding motif for AEBP2, CTT(N)15-23cagGCC. Also, individual ChIP experiments further demonstrated that a subset of these identified loci are indeed occupied by both the AEBP2 and SUZ12 proteins. These results are consistent with the initial prediction that AEBP2 may be a targeting protein for the mammalian PRC2 complex.
MATERIALS AND METHODS
Global protein sequence alignment
AEBP2- and JING-related sequences were collected from NCBI, UniProt, EMBL and UCSC. ClustalW was used to create protein alignments, and the final outcome was produced and edited using CLC free workbench version 4.0.3 (CLC bio A/S, Denmark). The protein alignment was set using the following parameters: gap opening penalty = 10, gap extension penalty = 0.1.
AEBP2 isoform confirmation through RT–PCR and western blot
Total RNAs were isolated from several tissues of a 3-month-old male mouse using the Trizol RNA isolation kit (Invitrogen). These RNAs were reverse-transcribed using the RT-PCR kit (Invitrogen SuperScript system, Invitrogen). For the 5′-side splicing, the following primer sets were used: mAebp2-a1, 5′-CGGCCAGCGCTACACCCCAAGAACT-3′; mAebp2-a2, 5′-GGGGAGCCGCTGAGCCGCATGGACT-3′; and mAebp2-b, 5′-GAAGCATGCCTGGCACTGGTC-3′. For the 3′-splicing, we used the following primer sets: mAb7-F, 5′-GATACTGCCTTGCTGTTGGACC-3′; mAbU2-R, 5′-TCCATGCCATGTGGACTGCAG-3′, and mAbU3-R, 5′-CTCCACTTCCACCTACAAGGA-3′. The PCR with the mAb-a1 and mAb-b primer set were performed at an annealing temperature of 61°C for 35 cycles. The remaining primer sets were amplified at an annealing temperature of 60°C for 30 cycles. For the detection of the AEBP protein, we prepared crude tissue extracts from the brain and testis of 1-month-old mouse, and also a 14-day-old embryo using the T-PER Tissue Protein Extraction Reagent kit (Cat. 78510, Thermo Scientific). Each extract (10 µg) was separated on 10% SDS–PAGE, transferred on to a PVDF membrane (Hybond-P, Amersham), and incubated with the anti-AEBP2 polyclonal antibody (Cat. 11232-2-AP, Proteintech Group).
AEBP2 GST fusion protein production
Three different GST fusion proteins were produced through cloning different part of the mouse AEBP2 protein (GenBank accession no. NM_178803). To construct these fusion constructs, we have amplified the coding region of AEBP2 using the following primer set: Acidic-mAb-F (5′-ATGGCCGCCGCGCTCGCCGACATG-3′) as a forward primer and mAebp2-b (5′-ATTGCAAATGTCGTTCACTGTTTGCT-3′) as a reverse primer for the fusion construct I, mAebp2-a (5′-ATGGACATAGACAGCACAATTTCCAG-3′) as a forward primer and mAebp2-b (5′-ATTGCAAATGTCGTTCACTGTTTGCT-3′) as a reverse primer for the fusion construct II, and mAebp2-a (5′-ATGGACATAGACAGCACAATTTCCAG-3′) as a forward primer and xbAebp2 (5′-CTGAAGTGTGTGGGTACATGGC-3′) as a reverse primer for the fusion construct III. These products were first subcloned into the pCR4 TOPO vector (Invitrogen) for sequencing verification, and later subcloned into the BamHI and HindIII restriction enzyme sites of the pGEX-4T-1 vector (Amersham Biosciences). The clones were transformed into the BL21 (DE3) competent cells (Strategen). The transformed cells were grown in LB media in 37°C to an absorbance value of 0.56 at 600 nm. The cells were further induced with 0.4 mM IPTG for 4 hours. The cell pellets were first sonicated, and stored in –80°C for later use in gel shift assays.
Electro mobility shift assay (EMSA)
EMSAs were performed as suggested by Promega with alterations in the DNA-binding buffer condition. For most of our EMSAs, we mainly used the NTEN buffer (100 mM NaCl, 1 mM EDTA, 20 mM Tris–Cl pH 6.0, 0.5% NP40). Each reaction contained 10 µg of GST fusion protein along with a given duplex probe (0.007 pmol per reaction), which were labeled with [γ-32 P] ATP. The information regarding the sequences of all the duplex probes used for the current study is available upon request.
ChIP cloning and individual ChIP assays with anti-AEBP2 and anti-SUZ12 antibodies
We performed ChIP cloning experiments using two polyclonal antibodies: AEBP2 (Cat. 11232-2-AP, Proteintech Group) and SUZ12 (Cat. ab12201, Abcam). The brain tissues of a 1-month-old mouse were used as a starting material, and the detailed protocol for our ChIP cloning is available from our previous study (15). DNA products eluted by anti-AEBP2 and SUZ12 ChIP were individually subcloned into the pZErO-2 vector (Invitrogen, Carlsbad, CA). About 200–300 clones were selected and subsequently sequenced using the ABI3130XL automated DNA sequencer (Applied Biosystem).
For individual ChIP experiment, the AEBP2 antibody (20 µl) was added into each fraction (500 µl) of cross-linked and sonicated mouse brain tissue. One mouse brain (1 g) was usually divided into 10 fractions. We followed the protocol of ChIP assay provided by the Upstate company (Upstate Biotech.). The immunoprecipitated DNA was dissolved in 40 µl of TE, and 1 µl of this eluted DNA was used as template DNA for one PCR-based ChIP assay. PCR conditions are as follows: 95°C for 5 min, 40 repetitions of the following cycle of 90°C for 30 s, 60°C for 30 s, 72°C for 30 s, and final extension at 70°C for 15 min. Each reaction included a pair of 25 ng of oligonucleotide primers. The information regarding the sequence of each primer set is available as Supplementary Data 4.
Motif analysis of ChIP-cloned sequences
Both 5′- and 3′-end regions, 4 bp in length, corresponding to the recognition sites for restriction enzymes were removed from each ChIP sequence for motif analyses. Any regions containing repeat elements were also removed from each ChIP sequence using RepeatMasker (http://www.repeatmasker.org/cgi-bin/WEBRepeatMasker). Our motif analyses used a total of 126 and 71 sequences that were obtained from AEBP2 and SUZ12 ChIP-cloned sequences, respectively. A standalone MEME (4.0.0) (http://meme.sdsc.edu/meme/cgi-bin/meme.cgi) was used to derive overrepresented motifs among each set of the ChIP-cloned sequences. We set parameters to ensure that at least a half of the sequences contain potential motifs in each set of ChIP sequences using a ‘-minsites’ parameter. We used the following parameters: -minsites (a half number of the sequences), -minw 5, -nmotifs 3, -revcomp, -dna. MAST (4.0.0) (http://meme.sdsc.edu/meme/cgi-bin/mast.cgi) was also used for the motif occurrence test with default setting.
RESULTS
Identification of AEBP2-related sequences from vertebrates and flying insects
The protein sequence of mouse AEBP2 (GenBank accession no. NP_001005605, 496 amino acids long) was used to identify its related sequences from all available genome sequences. This search identified six AEBP2-related sequences from flying insects, including flies, mosquitoes, honeybees, beetles, and wasp. These insect sequences were previously identified as JING. The same search also identified 22 related sequences from vertebrates, ranging from urochordates to placental mammals: one sea urchin, five fish, one lizard, one chicken, 14 mammal sequences. The average sizes of the predicted ORFs (Open Reading Frames) for these AEBP2-related sequences are as follow: 1744 amino acids for insects’ JING, 450 amino acids for fish's AEBP2, and 500 amino acids for mammalian AEBP2. The sizes of the AEBP2 sequences identified from lizard, frogs, sea urchins, cannot be determined due to the incompleteness of their genome sequences. All these AEBP2-related sequences are available as a Supplementary Data 1.
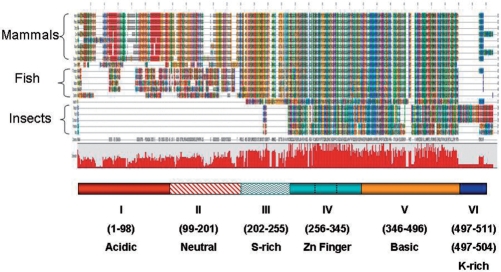
The amino-acid sequences of 20 full-length AEBP2-related sequences were used for global sequence alignment (Figure 1). As shown in Figure 1, the mammalian AEBP2 sequences can be divided into six protein domains: acidic, neutral, serine-rich, zinc finger, basic and lysine-rich domains. Among these six domains, two domains (zinc finger and basic) show the highest levels of amino-acid sequence conservation among all different lineages. The two domains of the mammalian AEBP2 (a.a. 256–496 in mouse AEBP2 Figure 2A) show an average of 38% amino acid sequence identity with those of the insect JING (Figure 2A). The serine-rich domain (a.a. 202–255) also shows high levels of sequence conservation: 83% amino acid sequence identity between the mammal and fish lineage. In contrast, the two domains located in the N-terminal portion of AEBP2 are lineage-specific. Although these two domains show an average 90% amino acid sequence identity between the mammalian species, these domains do not show any obvious similarity to the respective regions of the AEBP2 sequences derived from insects and fish. The acidic domain of mammalian AEBP2 is mainly characterized by arrays of glutamic and aspartic acid residues, whereas the neutral domain is characterized by arrays of glycines and serines. The AEBP2 sequences of the fish lineage also have similar acidic and neutral domains, displaying 38% amino acid sequence identity within the fish lineage. These two domains are localized within one large exon in both mammals and fish (Figure 2B), and the sequences of this exon in both lineages exhibit tandem repeat structure with high CpG densities. As such, many insertions/deletions are detected between the two closely related species of both mammals and fish (data not shown). On the other hand, the lysine-rich domain located in the C-terminus of mouse AEBP2 is found only in mammals, but is well conserved within mammalian species (a.a. 497–511). Overall, mammalian AEBP2 is comprised of six protein domains: four domains are lineage-specific whereas two domains (zinc finger and basic) appear to be well conserved throughout all the lineages.
Figure 1.
Global protein sequence alignment of AEBP2 and JING. AEBP2 and JING protein sequences of different organisms were aligned using the ClustalW program. Different amino acids are represented in different colors and shades. The conservation level of each position is indicated in the graph below the alignment. Six conserved domains are indicated with different colors and patterns. The mouse AEBP2 protein was used as a reference to indicate the position of each conserved domain. The zinc finger and basic domains are the most conserved and show sequence conservation from flying insects to mammals. The zoom-in version of this alignment is available as Supplementary Data 5 or the following website (http://jookimlab.lsu.edu/?q=node/81).
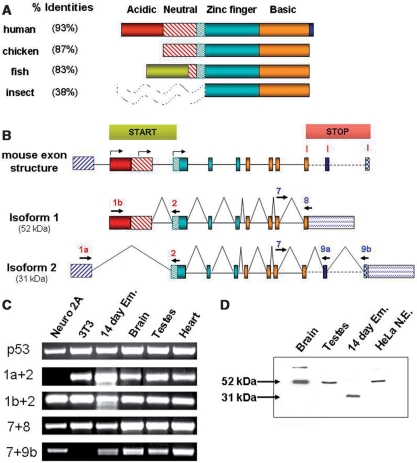
Figure 2.
Exon structure and alternative splicing of the mouse Aebp2 gene. (A) The same colors and patterns as Figure 1 were used to represent different protein domains. The percent identities were calculated through comparing the amino-acid sequences of AEBP2 from individual organisms vs. the mouse. The N terminus of the insects’ JING (dotted-lines) was omitted in this comparison due to the lack of any detectible sequence similarity. The AEBP2 of the fish lineage also has similar acidic and neutral domains, but these do not show any similarity to those of mammalian AEBP2 (indicated with a green box). (B) Isoforms and stage-specific expression of the mouse Aebp2. A total of 11 exons have been found in the mouse Aebp2 so far. Three START codons are indicated: two within Exon 1b and the third one in Exon 2. Three STOP codons are also indicated within exons 8, 9a and 9b. Alternative splicing of these exons could result in at least six different isofoms, including the two major forms detected in this study, Isoform 1 (52 kDa) and Isoform 2 (31 kDa). (C) Alternative splicing confirmed through RT–PCR. This analysis used total RNA from individual tissues, the different amounts of which were normalized to an internal control gene, p53. The combination of exons 1a and 2 is highly expressed in a 14-day-old embryo, which belongs to Isoform 2. (D) Western blot of the mouse AEBP2. Isoform 1 (52 kDa) is dominant in the most tissues examined, while Isoform 2 (31 kDa) was detected only in a 14-day-embryo.
Exon structure and isoforms of mammalian AEBP2
Inspection of all the available cDNA and EST (Expressed Sequence Tag) sequences derived from the mouse Aebp2 gene revealed that mouse Aebp2 is comprised of 11 individual exons that spread over a 60-kb genomic interval in mouse chromosome 6 (Figure 2B). A single exon (exon 1b) encodes both the acidic and neutral domains of mouse AEBP2 (a.a. 1–201). The following three exons (exons 2–4) encode the serine-rich domain and the three zinc finger units (a.a. 201–345), and the next four exons (exons 5–8) encode the basic domain (a.a. 346–496). This exon structure also involves two sets of alternative splicing: one is between the two 5′-end exons (1a and 1b) and the other is between three individual 3′-end exons (8, 9a and 9b). The first alternative splicing yields the two different forms of AEBP2 cDNAs: Isoform 1 with exon 1b and Isoform 2 with exon 1a (Figure 2B). The Isoform 1 cDNA has three potential START codons that are in-frame with the rest of the AEBP2 exons. The first two are located within exon 1b, and the third one is within exon 2. In contrast, exon 1a does not contain any in-frame ATG codon, and thus the Isoform 2 cDNA starting from exon 1a likely uses the third START codon located within exon 2. Since the two potential START codons for Isoform 1 and 2 are separated by 222 amino-acid residues, a large protein size difference is predicted between these two forms. Also, the acidic and neutral domains should be included only in the larger form (Isoform 1), but not in the smaller form (Isoform 2).
The second alternative splicing occurs between several 3′-end exons: Exon 8, 9a and 9b. The first form of the 3′-end alternative splicing simply ends at Exon 8 with its STOP codon and 3′-UTR (Figure 2B). The second form splices out the 3′-UTR of Exon 8, and join only the coding region of Exon 8 (named Exon 8s) to another downstream exon (exon 9a). Exon 9a has an additional 14-amino-acid-long coding region with its STOP codon. In fact, this small peptide region from exon 9a corresponds to the lysine-rich domain that is conserved within every mammalian species (Figures 1 and 2A). The third form of the 3′-end alternative splicing connects exon 8s to another further downstream exon (exon 9b), and this form also has an additional 7-amino-acid-long coding region. The evolutionary conservation of this exon is, however, currently unknown. According to our own survey on ESTs and cDNAs, most cDNAs derived from different tissues start with exon 1b and end with exon 8 (496 a.a. long), but a subset of cDNAs from early embryonic stages start with exon 1a, and end with either exons 8 or 9a (274 or 288 a.a. long). The other combinations of cDNAs are also likely, but the above two forms are believed to be the major forms for mouse AEBP2 cDNAs.
The two sets of alternative splicing predicted from cDNA sequences were tested through RT–PCR-based experiments using total RNAs isolated from several mouse tissues and cell lines (Figure 2C). As shown in Figure 2C (second and third row), the exon combination of 1b and 2 (1b + 2) was detected throughout all the tissues tested. The exon combination of 1a and 2 (1a + 2) was similarly detected in all the samples except the Neuro2A cell line. The overall expression levels of the 1b + 2 combination were higher than those of the 1a + 2 combination. However, this trend was reversed in the embryonic tissues: the expression of the 1a + 2 combination was much higher than that of the 1b + 2 combination. This is also consistent with the fact that all of the available EST clones containing the 1a+2 combination were derived from early embryonic tissues. The detection of these two alternative first exons further suggests the presence of two alternative promoters for mouse Aebp2: one may be responsible for the ubiquitous expression whereas the other one for the embryo-specific expression. We also performed another set of RT–PCR analyses to confirm the presence of the 3′-end alternative splicing (Figure 2C, fourth and fifth row). Since exon 9a is still part of the 3′-UTR of exon 8, we avoided testing the second exon combination (7 + 8s + 9a). We mainly analyzed two different combinations of the 3′-end alternative splicing (7 + 8 and 7 + 8s + 9b). Both exon combinations were detected in most of the tissues examined, but the expression levels of the 7 + 8 combination appear to be higher than those of the 7 + 8s + 9b combination. Overall, RT–PCR analyses indeed confirmed the presence of two alternative splicing, and detected somewhat stage and tissue specificity of these splicing.
We further tested the presence of multiple isoforms of the AEBP2 protein with western blot experiments using a polyclonal antibody raised against the human AEBP2 protein (Figure 2D). This analysis detected two main forms of AEBP2 (52 and 31 kDa, respectively). The larger form (52 kDa) was detected in the HeLa nuclear extracts as well as in the several tissues of the mouse, including brain and testis. Detection of another band in the brain sample was likely caused by non-specific binding to other unknown proteins. The 52-kDa protein appears to correspond to the largest ORF predicted from Isoform 1 cDNAs based on its similar size and ubiquitous expression in most tissues. In the brain of a 14-day-old embryo, however, the same analysis detected only the smaller form (31 kDa). The 31-kDa protein likely corresponds to the ORF derived from Isoform 2 cDNAs based on its smaller size and limited expression in embryonic stages. Due to the limited separation capability of SDS-PAGE gel electrophoresis, however, it is currently unknown whether these two isoforms also have different C-terminal endings, as predicted from the 3′-end alternative splicing. Nevertheless, the above analysis confirmed the existence of two major forms of AEBP2 in vivo: the adult-specific larger form (52 kDa) and the embryo-specific smaller form (31 kDa).
DNA-binding motifs of AEBP2
To characterize DNA-binding motifs for AEBP2, we made three GST-fusion constructs containing different isoforms of mouse AEBP2: Construct I (a.a. 1–496) corresponding to the 52-kDa larger form, Construct II (a.a. 223–496), corresponding to the 31-kDa smaller form, and Construct III (a.a. 223–348), corresponding to a truncated version lacking the basic domain. All of these GST-fusion proteins were successfully expressed in bacteria. However, only the two GST-fusion proteins from Constructs II and III exhibited some levels of DNA-binding activity. The reason for the inactivity of the GST-fusion protein I is currently unknown. We also tested the binding capability of the GST-fusion protein II to the sequences of several individual DNA fragments, which have been derived from ChIP cloning experiments designed to identify the in vivo target loci of the AEBP2. This will be described in detail in the following section. Among these short sequences, one sequence named T1 showed consistently high levels of DNA-binding affinity to the GST-fusion protein II. Thus, we have selected and used this particular sequence as a main probe for our DNA-binding motif assays (Figure 3).
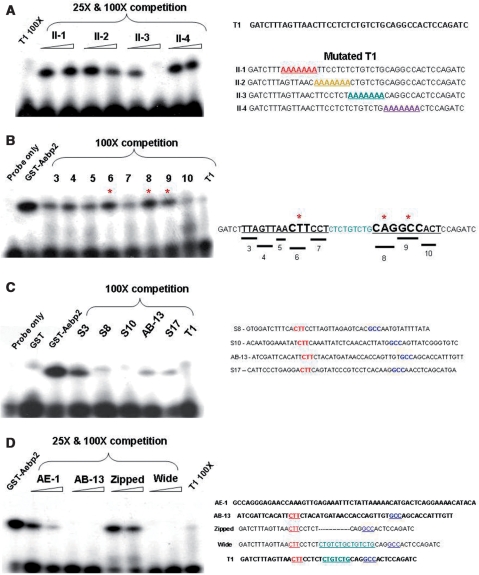
Figure 3.
DNA-binding motifs of AEBP2. Gel shift assays of the GST-AEBP II fusion proteins. (A) Competition assay with the first series of mutants (II-1, II-2, II-3, II-4). Each contains an internal 7-bp-long region substituted with a stretch of As. (B) Competition assay with the second series of mutants. Each contains a 3-bp-region substituted with a stretch of As. Mutants 6, 8 and 9 did not compete well (as indicated with asterisk), and thus these three small regions are thought to be the most critical for the AEBP2 binding. (C) The T1 probe was also competed with other short sequences derived from the ChIP cloning experiment using the AEBP2 antibody. The actual sequences for these ChIP fragments are shown on the right column. (D) Competition assay with different probes: the AE-1 probe from the original sequence that was used to identify the AEBP2 (1), the AB-13 probe from an independent ChIP-derived sequence with high affinity to AEBP2, the Zipped probe with the II-3 region removed and the Wide probe with the II-3 region duplicated.
Several mutant series of the T1 duplex probes were designed and used for our DNA binding motif studies of AEBP2. First, an internal 28-bp-long region of the T1 sequence (8th to 35th position) was divided into four individual 7-bp-long sections, and the sequence of each section was changed into a 7-bp-long stretch of A's (Probe II-1 through 4). Each of these mutant probes was used as a competitor to the P32-labeled T1 probe for gel shift assays (Figure 3A). A shift band was completely abolished in a self-competition experiment, using a 100 to 1 molar ratio of the P32-unlabeled to labeled probes (Figure 3A, Lane 1). Similarly, the third mutant (lanes 6 and 7) competed and abolished the band, indicating that this region is dispensable for the binding activity. In contrast, the three remaining mutants did not compete at all (lanes 2, 3, 4, 5, 8, 9), indicating that the three regions covered by these mutants are important for the binding to AEBP2. This initial series of competition experiments demonstrated that the two regions (8th to 21st and 29th to 35th position) of the T1 sequence are critical for the binding to AEBP2. These two regions were further analyzed using a second series of mutants, each of which has a 3-bp-long stretch of A's (Figure 3B). This series of experiments identified 3 smaller regions showing relatively weak competition (Figure 3B, lanes 6, 8, 9, marked with asterisk), indicating that these three small regions (CTT, CAG, GCC) are likely critical for the binding to AEBP2.
We also performed another independent series of gel shift assays using the DNA sequences derived from the 20 shortest DNA fragments derived from the ChIP cloning experiments using the AEBP2 and SUZ12 antibodies (Figure 4). This survey identified four individual sequences showing high levels of binding affinity to AEBP2 (Figure 3C, S8, S10, AB-13 and S17). Further inspection of the four sequences, averaging 50 bp in length, revealed that all of these sequences share two small motifs with the T1 sequences, CTT and GCC. However, the distance between these two motifs is somewhat variable among these potential binding sites of AEBP2, ranging from 15 to 23 bp in length. Thus, the effect of spacing between motifs was tested using two mutants of the T1 sequences (Figure 3D, lanes 6–9). The spacing region of the T1 sequence was either deleted (Zipped) or duplicated (Wide). The Zipped probe did not compete but the Wide probe competed very well, demonstrating that the spacing between the two motifs requires some minimum distances, but that the size can be variable without any major effect on the binding affinity to AEBP2. We also tested the binding affinity of the original sequence that was used to identify the AEBP2 protein ((1); AE-1, lanes 2 and 3). This sequence competed at some levels, but the binding affinity was much lower than the other sequences used for this study. We repeated the above experiments using GST-fusion protein III, which lacks the basic domain. The results did not show any difference from those of GST-fusion protein II (data not shown). This suggests that the three Gli-type zinc finger motifs shared by both fusion proteins are mainly responsible for the DNA-binding activity of AEBP2. In sum, the above series of experiments identified a DNA-binding motif for AEBP2, which displays an unusual bipartite motif structure, CTT(N)15-23cagGCC with the lowercase bases being less critical for binding.
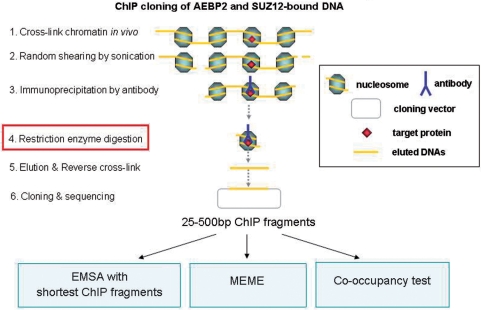
Figure 4.
ChIP cloning scheme and experimental strategies. Our modified ChIP cloning method has one additional step compared to other existing protocols: the restriction enzyme digestion step (step 4) right before the elution step. This allows immediate cloning of shorter ChIP DNA fragments without PCR amplification. Also, this shortening of ChIP fragments further trims other unnecessary long regions from either side of each ChIP fragment while preserving the actual binding site for a given DNA-binding protein. This further facilitates accurate prediction of DNA-binding motifs. The isolated ChIP fragments were subsequently used for the following three experiments. First, we used the sequences derived from the shortest ChIP fragments as probes for our gel shift assays of AEBP2. Second, we used the sequences from the ChIP cloning to identify enriched DNA motifs. Third, we also identified in vivo target loci for both AEBP2 and SUZ12. These confirmed loci were later tested for the co-occupancy by AEBP2 and SUZ12.
ChIP cloning of in vivo target loci bound by AEBP2 and SUZ12 proteins
A series of ChIP cloning experiments were performed to identify in vivo target loci bound by AEBP2 (Figure 4). We previously developed a modified version of ChIP cloning method, which can be used to directly clone very short DNA fragments without PCR amplification (15). This method performs restriction enzyme digestion with 4-bp cutters, such as Sau3AI or Tsp509, directly on the DNA while it is still cross-linked to a target protein as a chromatin complex. This enzyme digestion usually generates much shorter DNA fragments that are compatible with subcloning. Using two polyclonal antibodies against AEBP2 and SUZ12, we have generated two individual libraries containing ChIP-derived DNA fragments. We have sequenced a subset of these two libraries, 250 and 165 clones for AEBP2 and SUZ12, respectively. The average length of the inserts from each library was about 140 bp in length. Individual sequences are available as Supplementary Data 2 and also the associated information can be viewed using a custom track view of the UCSC genome web browser (http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm9&hgt.customText=http://jookimlab.lsu.edu/sites/default/files/Aebp2_bed.txt).
Initial inspection of the sequences from these two libraries derived the following conclusions. First, both libraries contain fractions of repeat sequences, 124/250 for the AEBP2 set and 94/165 for the SUZ12 set. Second, the remaining non-repeat sequences of both sets mapped closely to gene regions of the mouse genome. The list of the genes associated with each set was compared with the list of the mouse genes that are known to be bound by the PRC2 (16). This comparison confirmed that 53 out of the 126 sequences of the AEBP2 set (42%) were derived from the known PcG target loci while 18 of the 71 sequences of the SUZ12 set (25%) came from the PcG target loci. It has been shown that only a small fraction of mammalian genes are controlled by the PRC2 (less than 5% of the entire gene set of mammals) (16). Thus, the observed high levels of enrichments of the PcG downstream genes among the ChIP cloning sets of AEBP2 (42%) and SUZ12 (25%) strongly suggest that both proteins, AEBP2 and SUZ12, are likely involved in the targeting of the PRC2 complex. Some of the notable PcG loci that were identified through our ChIP cloning trials include: Grm8, Abcc3, and Phkb from the AEBP2 set and Pax1, Acvrinp1 and A20Rik from the SUZ12 set. The summary of this comparison is available as Supplementary Data 3.
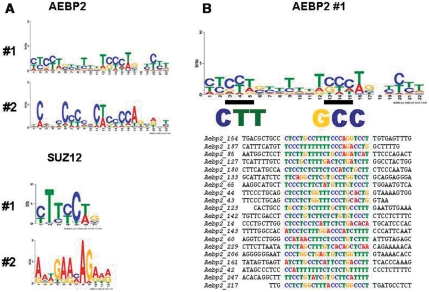
Third, analysis of the non-repeat sequences by the MEME program (http://meme.sdsc.edu/meme/intro.html) revealed the presence of several DNA motifs that were overrepresented within each set of ChIP sequences. The two most significant motifs from each set are shown in sequence logo format (Figure 5). The first motif of the AEBP2 set is 22 bp long, and shared by 77 out of the 126 individual sequences. Interestingly, this motif contains several small regions showing sequence similarity to the two 3-bp-long critical regions, which were shown to be critical for the binding to AEBP2 by our previous gel shift assays, CTT and GCC (Figure 3). We performed another gel shift assays using several AEBP2-ChIP fragments, which are shown in Figure 5B, and the results confirmed again that these ChIP fragments indeed contain the DNA-binding sites for AEBP2 (Supplementary Data 6). The second motif from the AEBP2 set is also 22 bp long, and displays a somewhat similar C-rich consensus sequence as the first motif. This motif is shared by the 56 individual sequences of the AEBP2 set. Similar analyses also identified two motifs from the SUZ12 set, which are shared by 45 and 35 sequences of the total 71 individual sequences, respectively. It is interesting to note that both motifs contain small regions similar to the GAGA motif, which is a frequent DNA motif in the Polycomb Response Element (PRE) of Drosophila (10,17). However, the two motifs from the SUZ12 set are shorter and also shared by fewer of the individual sequences than the two motifs from the AEBP2 set. This suggests that the sequences of the SUZ12 set are more heterogeneous than those of the AEBP2 set. In sum, many in vivo target loci of AEBP2 appear to be derived from the known PcG target regions, and these loci display C-rich sequences with several small motifs, which are reminiscent of the two critical DNA-binding sites of AEBP2.
Figure 5.
Enriched motifs within the AEBP2 and SUZ12 ChIP sequences. Motifs were predicted with each set of AEBP2 and SUZ12-ChIP sequences using the MEME program (http://meme.sdsc.edu/meme4/cgi-bin/meme.cgi). (A) The two most significant motifs identified from each set are shown in the sequence logo format. (B) The most significant motif from the AEBP2 set was shown with the two small motifs, which have been also independently identified through gel shift assays. This most significant motif was also aligned with 20 individual sequences.
Co-occupancy test with AEBP2 and SUZ12 ChIP assays
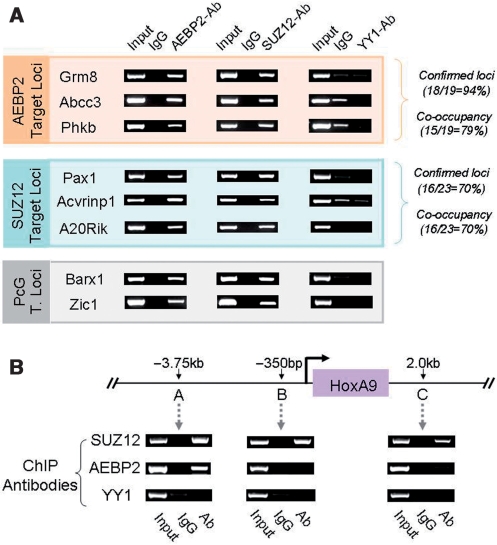
The identified genomic loci by AEBP2 and SUZ12-ChIP cloning were further analyzed using individual ChIP experiments (Figure 6). These individual ChIP experiments were performed to measure what fraction of each library contains genuine in vivo target loci for each protein. According to the results from four different trials, 18 out of 19 tested loci from the AEBP2 set showed consistent enrichment with the AEBP2 antibody, indicating that about 94% of the AEBP2 set likely contains genuine in vivo target loci. A similar test indicated that about 70% of the SUZ12 set (16/23) likely contains in vivo target loci. The representative results from these series of ChIP experiments are shown in Figure 6A and the remaining portion of the results along with other relevant information are also available as Supplementary Data 4.
Figure 6.
Co-occupancy test of the gene loci identified through AEBP2 and SUZ12 ChIPs. (A) Co-occupancy test of the genes identified from AEBP2 and SUZ12-ChIP sequencing. The left panel indicates the genes derived from each round of ChIP sequencing. Grm8, Abcc3 and Phkb were derived from the AEBP2-ChIP sequencing, while Pax1, Acvrinp1 and A20Rik were from the SUZ12-ChIP sequencing. The three loci (Grm8, Abcc3 and Phkb) were first tested through individual ChIP assays using the AEBP2 antibody, and later using another antibody (SUZ12) for the co-occupancy test. This was also repeated for the SUZ12 set (Middle). A subset of the known Polycomb target loci (Barx1 and Zic1) were also included for the co-occupancy test (bottom). (B) Co-occupancy of AEBP2 and SUZ12 on the HoxA9 locus. The co-occupancy of AEBP2 and SUZ12 was only detected at the 3.75-kb upstream region of the transcription start site of HoxA9. We also performed another independent ChIP using the YY1 antibody to test if YY1 is also involved in the targeting of the PRC2 to these loci.
Since AEBP2 is a potential targeting protein for PRC2, we further tested this possibility through performing co-occupancy tests. If the two proteins, AEBP2 and SUZ12, bind to target loci together as a protein complex, many confirmed target loci from one protein (AEBP2) should be also positive with another ChIP experiment using the antibody against the second protein (SUZ12), and vice versa. The results from this co-occupancy test are as follows. Out of the 19 AEBP2 loci tested, 15 (79%) were positive with the SUZ12-ChIP experiments. None of the negative loci from the AEBP2-ChIP were positive with the SUZ12-ChIP. On the other hand, 16 out of the 23 confirmed loci of the SUZ12 set (70%) were also positive with the AEBP2-ChIP. We also extended this co-occupancy test to the known target loci of the PcG complex. Out of the six loci tested, three loci turned out to be positive with both AEBP2 and SUZ12-ChIP experiments (Figure 6A and B; Supplementary Data 4). We also included YY1 ChIP experiments to test if these loci are bound by YY1. None of the tested loci were positive with the YY1-ChIP, suggesting that YY1 may not be involved in the targeting of the mammalian PRC2 to these loci. The HoxA9 locus was analyzed in further detail by including two additional primer sets. The precise PcG target region within this locus is located 3.75-kb upstream of the gene as demonstrated in the high levels of the DNA enrichment by the SUZ12-ChIP experiments (Figure 6B). The two other regions also show some levels of the enrichment with the SUZ12-ChIP, which is consistent with the previous study (7). However, the AEBP2-ChIP showed high levels of the enrichment only at the 3.75-kb upstream region, demonstrating high levels of target selectivity by the AEBP2-ChIP experiment. In sum, the co-occupancy tests revealed that an unusually large fraction of in vivo target loci are co-occupied by both AEBP2 and SUZ12 proteins, further supporting the initial idea that AEBP2 is likely a targeting protein for PRC2.
DISCUSSION
In the current study, we have characterized AEBP2 in terms of evolutionary conservation, genomic structure, DNA binding motifs and potential targeting roles for the Polycomb group repression complex 2 (PRC2). AEBP2 contains two evolutionarily conserved protein domains, the zinc finger and basic domains, and these two domains are also shared by the flying insect protein JING. jing has recently been recognized as a member of the PcG in Drosophila. Mammalian AEBP2 is driven by two alternative promoters and produces at least two major forms of the protein, and these isoforms show developmental stage-specific expression patterns: the adult-specific larger form (52 kDa) and the embryo-specific smaller form (31 kDa). The AEBP2 protein binds to a DNA-binding motif with an unusual bipartite structure, CTT(N)15-23cagGCC. A large fraction of AEBP2's target loci also map closely to the known target loci of the mammalian PRC2. We further confirmed that many of these loci are indeed co-occupied by the two proteins AEBP2 and SUZ12. This supports the prediction that AEBP2 is a targeting protein for the mammalian PRC2 complex.
Global alignment of 20 AEBP2 sequences identified two evolutionarily conserved domains, the zinc finger and basic domains, which are located at the C-terminus of the protein (Figure 1). These two domains maintain very high levels of sequence conservation throughout all the vertebrates, greater than 80% sequence identity in the 280 amino-acid long region. Similar domains are also found even in the flying insect protein JING, sharing an overall 38% sequence identity with AEBP2 (Figure 2A). Although the observed sequence similarity is relatively low, the insects’ jing is thought to be a homolog to vertebrates’ AEBP2 based on the following reasons. First, although the zinc finger domains of both genes are comprised of three typical Gli-type finger units that are quite prevalent in eukaryotic genome, the 2nd finger shows relatively high levels of sequence similarity (63%) between the two groups. Interestingly, the second finger is also three amino acids longer than typical Gli-type finger units (1). Yet, this unique variation is also detected in all the sequences of both AEBP2 and JING proteins (Supplementary Data 1). Second, the basic domain is characterized by a stretch of basic (Lys and Arg) amino-acid residues at the beginning and another stretch of hydrophobic (Leu, Val and Ile) and aromatic (Phe, Tyr and Trp) amino acid residues at the end. According to database search, this unusual domain is only found in the two proteins, AEBP2 and JING. Furthermore, the unique combination of this novel basic domain along with three Gli-type zinc finger units is found again only within these two proteins. Therefore, it is highly likely that vertebrates’ AEBP2 and the flying insects’ JING have been derived from a common ancestor. According to recent genetic studies in flies, the jing locus genetically interacts with several members of PcG members (4). This further suggests that AEBP2 and JING still play a similar role, perhaps in the PcG-mediated repression. If this is the case, the two conserved domains likely play the most central roles for this repression mechanism, DNA binding by the zinc finger units and protein–protein interaction by the basic domain.
One of the unexpected features associated with the Aebp2 gene is the presence of several combinations of alternative splicing, which involve the 5′-end two exons and 3′-end three exons (Figure 2B–D). In principle, six different types of AEBP2 protein isoforms are possible although we have detected only two major forms through western blot analyses. The expression of these two major forms is very developmental stage-specific: the larger form (52 kDa) is mainly detected in adult tissues, whereas the smaller form (31 kDa) is found only in embryonic tissues. This stage-specific expression is thought to be driven by the two different promoters located upstream of the two alternative first exons. According to our own surveys using the EST database, a similar 5′-end alternative splicing of AEBP2 is also detected in other mammals, and the expression of each of the two splicing variants can be easily categorized into either embryonic or adult-specific. In the other vertebrates and invertebrates, however, there appears to be only one first exon for AEBP2 and JING, and also the expression pattern of this cDNA form appears to be spatially and temporally ubiquitous. This suggests that the alternative splicing of AEBP2 and stage-specific expression are unique features found only in mammals. Then, what is the major impetus for the sudden implementation of this alternative splicing for the mammalian AEBP2? This could be explained by the actual products of the alternative splicing: a smaller embryonic form with two conserved domains versus a larger adult form with additional lineage-specific domains (Figure 2). Given the similarities in domain structure between the smaller form and other vertebrate AEBP2 proteins, the smaller form is likely involved in more fundamental biological processes than the larger form, such as determining the pattern and axis of animal body during early development. On the other hand, the larger form with lineage-specific protein domains is likely involved in cellular processes that are more species-specific, such as determining the lineage and location of different cell types within the adult tissues. It is interesting to note that the smaller form with conserved domains participates in earlier developmental processes than the larger form with additional lineage-specific domains. This could be another case of ontogeny recapitulating phylogeny in animal evolution (18). Overall, the alternative splicing and subsequent formation of mammalian AEBP2 isoforms represents a case where alternative splicing has driven functional division and adaptation of genes.
According to DNA-binding motif studies (Figure 3), AEBP2 binds to a consensus sequence with bipartite structure, CTT(N)15-23cagGCC, and this binding is mainly driven by the three zinc finger units. This consensus DNA-binding motif has been further substantiated by the independent observation that the in vivo target loci of AEBP2 also show similar motifs (Figure 5). However, the AEBP2 binding to a bipartite structure motif was unexpected given the fact that the three zinc finger units are juxtaposed right next to each other. This suggests that the recognition of the two smaller motifs within the bipartite motif, which is separated by a spacing, (N)15-23, may be driven by individual zinc finger units of either one or two proteins. The model of single protein binding posits bending of the DNA because of the predicted close proximity between individual fingers, whereas the model involving binding by two proteins hypothesizes potential dimer formation of the AEBP2 protein. We favor the first model based on the following reasons. First, the truncated version of AEBP2, GST-fusion protein III, lacks any domains that could function in dimerization, still showed unchanged binding preference to the bipartite motif (data not shown). Second, the bipartite motif tends to show higher affinity to AEBP2 when the spacing region, (N)15-23, of the bipartite motif is either homopolymeric or polypyrimidine stretch sequences, such as polyA or poly(CT). These types of sequences are known to be common in bending regions of the genome (19). This is further supported by our independent observation that many confirmed target loci of AEBP2 also exhibit polypyrimidine sequence structures (Figure 5). Although we cannot rule out the other possibilities of DNA binding driven by the dimer structure of AEBP2, the above results suggest potential binding of AEBP2 to bent DNA.
The co-occupancy test with the ChIP experiment clearly demonstrated that AEBP2 and SUZ12 bind to similar genomic regions (Figure 6). Since none of the PRC2 core proteins are known to be DNA-binding proteins, this further implicates that AEBP2 may act as a targeting protein for this complex. According to our recent data from a mouse model disrupting the Aebp2 locus (H. Kim et al., unpublished results), some of the known PcG downstream genes are indeed de-repressed in these mutant mice (Supplementary Data 7), further confirming this possibility. Also, the previously reported activity of AEBP2 as a transcriptional repressor supports this possibility (1). However, we do not predict all of the identified target loci of AEBP2 to be PcG target loci based on the following reasons. First, we expect that AEBP2 should be also involved in many other cellular processes besides the predicted PcG-targeting role based on its evolutionary age and also various protein isoforms detected in mammals. YY1 is an example of a similar case: its role has diversified tremendously from its original evolutionarily conserved role in the Polycomb-mediated repression since the split of insects and vertebrates (20,21). Second, as demonstrated in flies, PcG targeting is likely mediated through a combination of several DNA-binding proteins along with critical DNA structures which are yet unrevealed (17). A similar conclusion has been drawn from the current study: although many SUZ12 confirmed loci are also bound by AEBP2, the DNA-binding motifs of AEBP2 were not significantly overrepresented in this pool of the genomic sequences (Figure 5). This suggests that AEBP2 may be one of several DNA-binding proteins involved in the targeting of the mammalian PRC2. In that regard, characterizing the functional contexts of each of the AEBP2 binding to the identified in vivo target loci will be of great interest in the near future.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (R01 GM66225 to J.K.). Funding for open access charge: National Institutes of Health (R01 GM066225).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Drs Jeong Do Kim and Sungryul Yu for their technical help on GST fusion protein expression and western blotting; Jason at the Proteintech Group Inc for his kind advice on the AEBP2 antibody; Jennifer Huang, Jungnam Lee and Kyudong Han for their critical reading of the manuscript.
REFERENCES
- 1.He GP, Kim S, Ro HS. Cloning and characterizing of a novel zinc finger transcriptional repressor. J. Biol. Chem. 1999;274:14678–14684. doi: 10.1074/jbc.274.21.14678. [DOI] [PubMed] [Google Scholar]
- 2.Liu Y, Montell DJ. Jing: a downstream target of slbo required for developmental control of border cell migration. Development. 2001;128:321–330. doi: 10.1242/dev.128.3.321. [DOI] [PubMed] [Google Scholar]
- 3.Sedaghat Y, Miranda WF, Sonnenfeld MJ. The jing Zn-finger transcription factor is a mediator of cellular differentiation in the Drosophila CNS midline and trachea. Development. 2002;129:2591–2606. doi: 10.1242/dev.129.11.2591. [DOI] [PubMed] [Google Scholar]
- 4.Culi J, Aroca P, Modolell J, Mann RS. jing is required for wing development and to establish the proximo-distal axis of the leg in Drosophila melanogaster. Genetics. 2006;173:255–266. doi: 10.1534/genetics.106.056341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McClure KD, Schubiger G. A screen for genes that function in leg disc regeneration in Drosophila melanogaster. Mech. Dev. 2008;125:67–80. doi: 10.1016/j.mod.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-Group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 7.Cao R, Zhang Y. Suz12 is required for both the histone methyltransferase activity and the silencing function of the EED-EZH2 complex. Mol. Cell. 2004;15:57–67. doi: 10.1016/j.molcel.2004.06.020. [DOI] [PubMed] [Google Scholar]
- 8.Kohler C, Villar CBR. Programming of gene expression by Polycomb group proteins. Trends Cell Biol. 2008;18:236–243. doi: 10.1016/j.tcb.2008.02.005. [DOI] [PubMed] [Google Scholar]
- 9.Cao R, Zhang Y. The functions of E(Z)/EZH2-mediated methylation of lysine 27 in histone H3. Curr. Opin. Genet. Dev. 2004;14:155–164. doi: 10.1016/j.gde.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 10.Ringrose L, Paro R. Epigenetic regulation of cellular memory by the Polycomb and trithorax group proteins. Annu. Rev. Genet. 2004;38:413–443. doi: 10.1146/annurev.genet.38.072902.091907. [DOI] [PubMed] [Google Scholar]
- 11.Muller J, Kassis J. Polycomb response elements and targeting of Polycomb group proteins in Drosophila. Curr. Opin. Genet. Dev. 2006;16:476–484. doi: 10.1016/j.gde.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 12.Schwartz Y, Pirrotta V. Polycomb complexes and epigenetic states. Curr. Opin. Cell Biol. 2008;20:266–273. doi: 10.1016/j.ceb.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 13.Brown LJ, Mucci D, Whiteley M, m Dirken ML, Kassis JA. The Drosophila Polycomb group gene pleiohomeotic encodes a DNA binding protein with homology to the transcription factor YY1. Mol. Cell. 1998;1:1057–1064. doi: 10.1016/s1097-2765(00)80106-9. [DOI] [PubMed] [Google Scholar]
- 14.Klymenko T, Papp B, Fischle W, Köcher T, Schelder M, Fritsch C, Wild B, Wilm M, Müller J. A Polycomb group protein complex with sequence-specific DNA-binding and selective methyl-lysine-binding activities. Genes & Dev. 2006;20:1110–1122. doi: 10.1101/gad.377406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang JM, Kim JD, Kim H, Kim J. An improved cloning strategy for chromatin-immunoprecipitation-derived DNA fragments. Anal. Biochem. 2006;356:145–147. doi: 10.1016/j.ab.2006.03.036. [DOI] [PubMed] [Google Scholar]
- 16.Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 17.Ringose L, Rehmsmeier M, Dura JM, Paro R. Genome-wide prediction of polycomb/trithorax response elements in Drosopila melanogaster. Dev. Cell. 2003;5:759–771. doi: 10.1016/s1534-5807(03)00337-x. [DOI] [PubMed] [Google Scholar]
- 18.Gould SJ. Ontogeny and Phylogeny. Cambridge, MA: Harvard University Press; 1977. [Google Scholar]
- 19.Déjardin J, Rappailles A, Cuvier O, Grimaud C, Decoville M, Locker D, Cavalli G. Recruitment of Drosophila Polycomb group proteins to chromatin by DSP1. Nature. 2004;434:533–538. doi: 10.1038/nature03386. [DOI] [PubMed] [Google Scholar]
- 20.Gordon S, Akoryan G, Garban H, Bonavida B. Transcription factor YY1: structure, function, and therapeutic implications in cancer biology. Oncogene. 2006;25:1125–1142. doi: 10.1038/sj.onc.1209080. [DOI] [PubMed] [Google Scholar]
- 21.Kim JD, Faulk C, Kim J. Retrotransposition and evolution of the DNA-binding motifs of YY1, YY2, and REX1. Nucleic Acids Res. 2007;35:3442–3452. doi: 10.1093/nar/gkm235. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.