Abstract
MAML1 is a transcriptional coregulator originally identified as a Notch coactivator. MAML1 is also reported to interact with other coregulator proteins, such as CDK8 and p300, to modulate the activity of Notch. We, and others, previously showed that MAML1 recruits p300 to Notch-regulated genes through direct interactions with the DNA–CSL–Notch complex and p300. MAML1 interacts with the C/H3 domain of p300, and the p300–MAML1 complex specifically acetylates lysines of histone H3 and H4 tails in chromatin in vitro. In this report, we show that MAML1 potentiates p300 autoacetylation and p300 transcriptional activation. MAML1 directly enhances p300 HAT activity, and this coincides with the translocation of MAML1, p300 and acetylated histones to nuclear bodies.
INTRODUCTION
The protein Mastermind like-1 (MAML1) was cloned on the basis of homology to the Drosophila Mastermind (1), a neurogenic gene genetically linked to Notch function (2–4). When the intracellular domain (ICD) of Notch interacts with the conserved DNA-binding protein CSL (CBF1 in vertebrates, Suppressor of Hairless in Drosophila and Lag-1 in Caenorhabditis elegans), the formation of a corepressor CSL complex is disrupted, and coactivators, such as PCAF, GCN5 (5), p300 (6) and MAML1 (7,8) are recruited. MAML1 binds to the ankyrin (ANK) repeat region of Notch ICD, stabilizes the interaction between Notch ICD and CSL and functions as a coactivator for all four Notch receptors (1). The MAML family comprises MAML2 and MAML3, and all of the MAML proteins appear to function specifically in Notch signaling (7,8). The highest sequence homology among MAML proteins exists within the N-terminal domain of MAML1, which contains two alpha helices. This polypeptide binds CSL and Notch in a binding pocket formed by CSL and the ANK domain of Notch, as revealed by crystal structures of the DNA-bound CSL–Notch–MAML1 complex (9,10). MAML1 has also been shown to function as a coactivator for MADS box transcription enhancer factor (MEF) 2C (11), p53 (12) and β-catenin (13).
The exact mechanism of how MAML1 functions as a coactivator of Notch is not fully understood. However, MAML1 has been shown to be important for recruitment of different coregulators, such as the histone acetyltransferase (HAT) p300 and the cyclin-dependent kinase (CDK) 8. Recruitment of CDK8 by MAML1 leads to phosphorylation of Notch1 and subsequent degradation by the Fbw7/Sel10 ubiquitin ligase (14). It has previously been reported that MAML1 potentiates Notch ICD-mediated transcription from chromatin templates in vitro by recruiting p300 to a DNA–CSL–Notch complex (15,16). Moreover, MAML1 and p300 have been shown to co-localize on the Hes1 promoter in cell culture (14). MAML1 directly interacts with both p300 and histones, and the p300-MAML1 complex specifically acetylates histone H3 and H4 tails in chromatin in vitro (17). In addition to acetylating histones, p300 also acetylates MAML1, which requires a proline repeat motif at residues 83–90 (PAPAAPAP) in MAML1, which is conserved between p53 (18) and MAML1 (17).
In addition to a centrally located 380-residue HAT domain (see Figure 1A), the p300 protein contains several well-defined protein interaction domains that regulate transcription by bridging the basal transcription machinery with various transcription factors that bind DNA with sequence specificity (19). This protein–DNA interaction is further facilitated by p300 acetylation of the transcription factors and modulation of the chromatin structure by p300 acetylation of histones (20). The p300 HAT domain, like that of CBP, acetylates itself and several other domains in p300 in vitro (21–24). Although autoacetylation, so far, has not been studied to a similar extent as p300 acetylation of other proteins, recent studies report that autoacetylation regulates p300 acetyltransferase activity on histone tails and other substrates, which modulates protein–protein interactions and transcription (22,25). Interestingly, the APC/C (anaphase-promoting complex/cyclosome) subunits APC5 and APC7 (26), and GAPDH (glyceraldehydes-3-phosphate dehydrogenase) (27) have been reported to stimulate p300 autoacetylation by directly interacting with p300.
Figure 1.
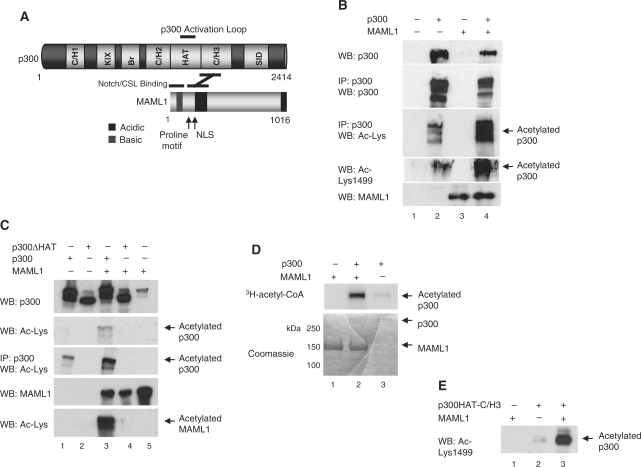
MAML1 enhances p300 autoacetylation. (A) Schematic of p300 and MAML1 domains. (B) and (C) p300 autoacetylation is regulated by MAML1 in vivo. Plasmids expressing p300 or p300ΔHAT, as described in the figures, and MAML1 were cotransfected into 293 cells, whole-cell extracts prepared, and the p300 protein was immunoprecipitated with a p300 antibody. Proteins were separated by SDS–PAGE and detection of the p300 protein and acetylation p300, and MAML1, were monitored by western blotting with antibodies recognizing p300, MAML1 or acetylated lysines in p300. (D) In vitro acetylation of p300. Recombinant full-length proteins of p300 and MAML1 (indicated at the top) were incubated with 3H-acetyl-CoA, and reaction products were separated by SDS–PAGE and visualized by autoradiography (top panel). Coomassie blue-stained SDS-gel showing p300 and MAML1 proteins used in the acetylation assay (lower panel). (E) Recombinant p300 and MAML1 (indicated at the top) were incubated with cold acetyl-CoA, and reaction products were separated by SDS–PAGE and visualized by western blotting with an antibody recognizing acetylated Lys1499 in p300.
Previous studies have shown that p300 contains a proteolytically sensitive loop region (residues 1520–1560, see Figure 1A) in the HAT domain that is conserved between human and other mammalian homologs. This activation loop serves an autoinhibitory function and can be modified by autoacetylation of several of its lysines, which enhances p300 HAT activity (22). The p300Δloop protein (deletion of amino acids 1523–1554) functions as a more potent coactivator for the androgen receptor as compared with wild-type p300, further connecting this activation loop with regulation of activity (22). In this report we have investigated how MAML1 can influence p300 activity. We found that full-length MAML1 enhances p300 autoacetylation in the HAT domain, which requires the C/H3 domain of p300, but not the activation loop of p300. We also found that full-length MAML1 potentiates p300-dependent transcription coactivation, and is even more pronounced for p300 lacking the activation loop. Finally, we found that MAML1 enhances p300 HAT activity and this coincides with the translocation of full-length MAML1, p300 and acetylated histones to nuclear bodies.
MATERIALS AND METHODS
Plasmids
cDNAs encoding MAML1 1–1016, MAML1 1–625 and MAML1 1–300 were amplified with PCR and subcloned into pcDNA-3.1 (Invitrogen), Flag-tagged pcDNA3.1 and pBIND (Promega) expression vectors. pCI-Flag-p300 and pCI-Flag-p300Δloop (Δaa1523–1554) constructs were as described previously (22). Expression plasmids CMVß-p300-CHA and CMVß-p300-CHA-Δaa1737–1836 (C/H3 domain mutant) was generously provided by Dr E. Treuter and Dr J. DeCaprio, respectively. pCI-Flag-p300ΔHAT (Δaa 1472–1522) was a gift from Dr J. Boyes and Flag-p300HAT-C/H3 (aa 1195–1810) was a gift from Dr R.G. Roeder. pVR1012-Gal4-p300 was generously provided by Dr N. Perkins. pcDNA3.1/myc-His-Notch1 ICD (aa 1760–2556) was a gift from Dr T. Kadesch.
Cell lines
The f-MAML1 cell line was made by transfecting 293 cells with a f-MAML1-pcDNA3.1 vector (geneticin selection).
Expression and purification of proteins
FLAG-tagged proteins for in vitro acetylation assays were expressed in Sf9 cells via baculovirus and purified as described previously (17).
Protein acetylation assays
For p300 autoacetylation in vitro, 0.5 μg of MAML1 proteins were incubated with 50 ng of p300 and 3 μM 3H-acetyl-CoA in 20 μl reaction volume in HAT buffer (50 mM Tris–HCl, pH 8.0, 50 mM KCl, 5% glycerol, 0.1 mM EDTA, 1 mM DTT, 1 mM PMSF, 10 mM sodium butyrate) at 30°C for 1 h, and the reaction mixture was subjected to SDS–PAGE and autoradiography. In addition, 2 μg of FLAG-MAML1 and 1 μg of FLAG-p300 (1195–1810) were added to HAT buffer and 20 μM cold acetyl-CoA. The reaction mixture was incubated at 30°C for 30 min and acetylation of Lys1499 analyzed by western blot.
HAT assay
In vitro HAT assays were conducted in a 30 μl reaction volume, at 0.1 M HEPES, 0.2 mM EDTA, 0.1 mg/ml BSA and 2 mM DTT. First, a synthetic peptide of the 15 N-terminal residues of histone H4 was added to reaction tubes with peptide final reaction concentrations of 10 and 60 μM. Purified MAML1 protein, or control buffer, was added with MAML1 final reaction concentrations of 0, 100, 200 and 400 nM. Recombinant full-length p300, or control buffer, was added with p300 final reaction concentrations of 0 and 5 nM. These reaction tubes were allowed to equilibrate for 10 min at 30°C. Then, 14C-acetyl-CoA was added to each tube with a final concentration of 20 mM. The reactions were allowed to proceed for 7 min at 30°C, and quenched with 6 μl of 6× tricine loading buffer. Aliquots of each reaction were analyzed for total radioactivity alongside a 14C-BSA standard using a scintillation counter. Aliquots of each reaction were analyzed for acetylation activity by separating with gel electrophoresis on a 16% Tris–tricine gel, alongside a 14C-BSA standard, followed by quantification with a phosphorimager and subtraction of background. The rate of acetylation of the H4–15 peptide was calculated in nM/min, and the low level of acetylation in the no-p300 control for each peptide and MAML1 concentration condition subtracted. The experiment was repeated exactly, run on a separate gel, and data from the duplicates were averaged and standard deviations calculated. Curves were fit using a standard ligand–receptor interaction. Data at 10 μM peptide were qualitatively similar to those at 60 μM peptide, however, the larger error prevented a precise comparison.
Transient transfections and immunoprecipitation
293 cells were transiently transfected with 700 ng GAL4-p300, 1 μg MAML1 and the reporter plasmid pG5-luc using Lipofectamin 2000 transfection reagent (Invitrogen). After 48 h, cells were lysed in 250 μl Roche lysis buffer and after centrifugation the levels of luciferase were measured. The bars in the Figures 2G, 4B and 5D represent mean values obtained from at least three independent experiments. For in vivo acetylation experiments, 293 cells were transiently transfected with pcDNA-MAML1 and pCMV-p300-HA using Lipofectamine 2000 (Invitrogen) and treated with 10 mM sodium butyrate after 32 h. The cells were harvested 48 h after transfection, and lysed in RIPA lysis buffer (25 mM Tris–HCl pH 7.6, 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS), and immunoprecipitation of p300 was performed with an antibody recognizing the p300 N-terminus (Santa Cruz N-15) coupled to Protein A Sepharose (GE Healthcare) following the manufacturer's protocol. The input and IP (immunoprecipitation) samples were analyzed by western blotting using the following antibodies: p300 (N-15), acetylated-Lysine antibody (Cell Signaling), MAML1 (Bethyl Laboratories), acetylated lysine 1499, acetylated histone H3 (Ac-H3) and acetylated histone H4 (Ac-H4) (Upstate Biotechnology), histone H3 (Abcam) and GAPDH (Ambion). For siRNA transfections we used DharmaFECT 1 siRNA reagents, and predesigned MAML1 SMARTpool set of 4 siRNA, from Dharmacon.
Figure 2.
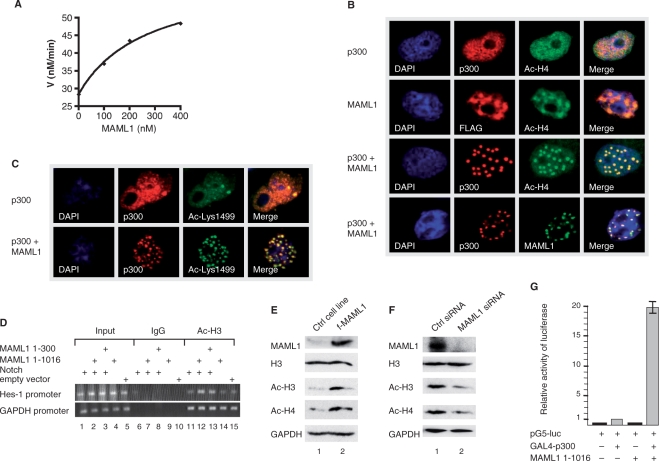
The p300 HAT activity and coactivator function is potentiated by MAML1. (A) 5 nM recombinant full-length p300, MAML1 and 60 µM synthetic histone H4 N-terminal tail 15-mer peptide were allowed to pre-equilibrate for 10 min, followed by a 7 min reaction at 30°C with 20 mM 14C-acetyl-CoA. HAT reactions were separated by Tris–tricene gel electrophoresis alongside 14C-BSA, acetylation levels quantified with a phosphoimager, and background subtracted. This was performed in duplicate and averages are shown. The curve is calculated using the equation of a standard binding isotherm, with apparent Kd of 270 ± 60 nM and apparent maximal rate stimulation of 2.2 ± 0.13-fold. (B) Vectors expressing p300 and FLAG-MAML1, as indicated in the figures, were cotransfected into Cos7 cells and after 24 h the cells were immunostained with antibodies recognizing p300, FLAG-tag and acetylated histone H4 or (C) acetylated lysine 1499 in p300. (D) U2OS cells were transfected with Notch1 ICD and MAML1 1–1016 or 1–300 plasmids. Chromatin precipitation was done with an antibody recognizing acetylated H3 and a control IgG antibody, followed by PCR analysis. Whole-cell extracts were prepared from (E) f-MAML1 and a control (Ctrl) cell line or (F) U2OS cells transfected with 100 nM siRNA MAML1 or Ctrl siRNA. Detection of the MAML1 protein, acetylated histone H3 and histone H4, and GAPDH, were monitored by western blotting with antibodies recognizing MAML1, acetylated H3 (Ac-H3), acetylated H4 (Ac-H4) or GAPDH. (G) MAML1 potentiates p300 activity in vivo. 293 cells were cotransfected with a luciferase reporter containing five Gal4-binding sites and vectors expressing Gal4-p300 and MAML1 full-length protein 1–1016.
Figure 4.
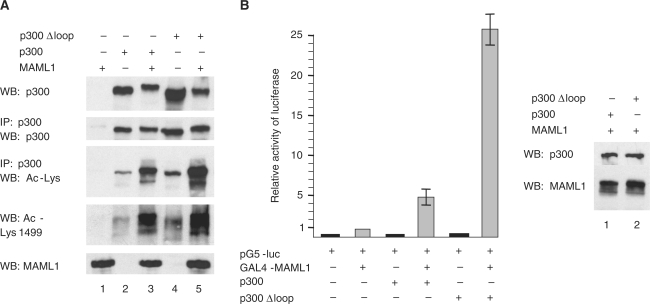
The activation loop in p300 is not involved in MAML1-dependent p300 autoacetylation. (A) 293 cells were transfected with MAML1, p300 and p300Δloop and the lysates from the cells were subjected to SDS–PAGE. MAML1, p300 and the acetylation of p300, were visualized in western blotting with antibodies recognizing MAML1, p300, acetylated lysines or acetylated Lys1499 in p300. (B) The p300Δloop protein function as a more potent coactivator of MAML1. 293 cells were cotransfected with a luciferase reporter containing five Gal4-binding sites and vectors expressing Gal4-MAML1, p300 or p300Δloop.
Figure 5.
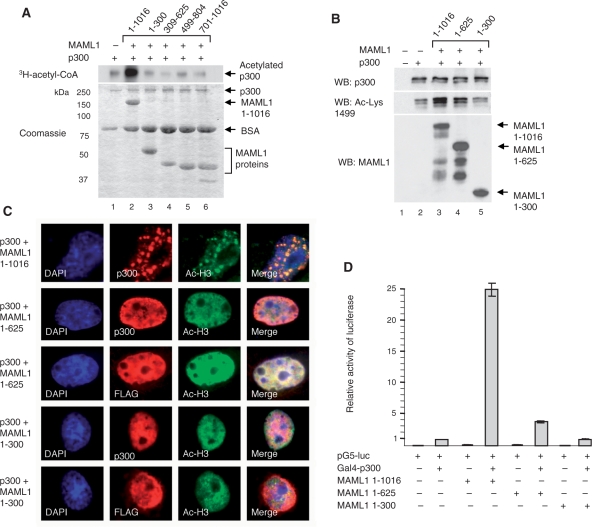
MAML1-dependent p300 activation requires full-length MAML1 protein. (A) In vitro acetylation of p300. Recombinant MAML1 proteins (indicated at the top) were incubated with p300 and 3H-acetyl-CoA, and reaction products were separated by SDS–PAGE and visualized by autoradiography (top panel). Coomassie blue-stained SDS-gel showing p300 and MAML1 proteins used in the acetylation assay (lower panel). (B) Plasmids expressing p300 and MAML1 1–1016, 1–625 or 1–300 were cotransfected into 293 cells and whole-cell extracts prepared. Proteins were separated by SDS–PAGE and monitored by western blotting with antibodies recognizing p300, MAML1 or acetylated Lys1499 in p300. (C) Cos-7 cells were transfected with p300-HA and FLAG-MAML1 expression plasmids as indicated in the figure. Twenty-four hours after transfection cells were immunostained with antibodies recognizing p300, FLAG-tag and acetylated histone H3. (D) Vectors expressing Gal4-p300 and MAML1 1–1016, 1–625 or 1–300, as indicated in the figure, were cotransfected with a luciferase reporter containing five Gal4-binding sites into 293 cells.
ChIP assay
ChIP assays were performed following a modified version of Upstate ChIP protocol. 300 ng of Notch, 1 μg of MAML1 1–1016 or 1–300, and empty vector to a final plasmid amount of 3 μg, were transfected using TransIT-LT1 transfection reagent (Mirus) into U2OS cells. Twenty hours after transfection, cells were cross-linked with 1% formaldehyde for 10 min at 37°C, washed two times with ice cold PBS and lysed in 500 μl NE buffer. The chromatin was shared by diagenode sonication for 12 min at 4°C, centrifuged at 13 000 r.p.m. for 10 min, and diluted 10 times with dilution buffer. Four micrograms of Acetyl-H3 (Upstate Biotechnology) and control IgG antibodies (Santa Cruz Biotechnology) were used for immunoprecipitation. The Hes-1 or GAPDH promoter regions were amplified with PCR (35 cycles) by sequence-specific primers.
Immunostaining
Cos-7 cells were seeded on glass slides (Menzel-Glaser) the day before transfection. Confluent cells of 60–80% were transfected using TransIT-LT1 transfection reagent (Mirus) and incubated for 24 h. Cells were washed (three times for 3 min, otherwise indicated) with ice-cold PBS and fixed with 4% paraformaldehyde for 20 min at 4°C. Cells were washed with PBS and permeabilized with 1% Triton X-100 (Sigma) in PBS at RT. Slides were washed and blocked with 3% BSA (Sigma) in PBS for an hour at RT. After PBS wash the slides were incubated with primary antibodies diluted in 3% BSA with 0.2% Triton X-100 in PBS, overnight at 4°C. Next day, slides were washed four times for 5 min and incubated with FITC-conjugated rabbit or Cy3-conjugated mouse antibodies (Jackson ImmunoResearch Laboratories) diluted in PBS for 2 h at RT. Slides were washed four times for 5 min and incubated with 0.1 μg/ml DAPI (Sigma) in PBS for 20 s. After three washes with PBS, slides were mounted with Immuno-Mount medium (Thermo) and analyzed with fluorescence microscope coupled with AxioCam digital camera (MRm), using Axio Vision software (Zeiss).
RESULTS
MAML1 enhances p300 autoacetylation
When we previously reported that p300 acetylates MAML1 (17), we also noticed that in our in vitro acetylation assay p300 became more acetylated in the presence of MAML1. To see if MAML1 increases p300 autoacetylation in cell culture, plasmids expressing MAML1 and p300 or p300ΔHAT were cotransfected into 293 cells. Whole-cell extracts were prepared, and the p300 protein was immunoprecipitated with a p300 antibody. p300 and MAML1 were detected by western blotting with antibodies recognizing MAML1, p300 or acetylated lysines in p300. As shown in Figure 1B, the acetylation of p300 is strongly enhanced in the presence of MAML1 (compare lanes 2 and 4). Interestingly, acetylation of Lys1499, a lysine shown to regulate p300 acetylation of histones (22), is also increased. The p300 autoacetylation is totally dependent on the p300 HAT domain. When the p300 protein lacks the HAT domain, there is no detectable acetylation (Figure 1C, lanes 2 and 4), which suggests that MAML1 itself does not acetylate p300 or recruit other HATs.
To analyze whether MAML1 is directly responsible for potentiating p300 autoacetylation, we performed in vitro acetylation assays. Recombinant p300 and MAML1 were incubated with 3H-acetyl-CoA and reaction products visualized by autoradiography. Our data show that MAML1 strongly potentiates p300 autoacetylation in vitro (Figure 1D, compare lanes 2 and 3). In addition, recombinant p300 (amino acids 1195–1810) and MAML1 were incubated with cold acetyl-CoA, and acetylation monitored by western blotting with an antibody recognizing acetylation of Lys1499 in p300. As shown in Figure 1E, acetylation of p300 Lys1499 is strongly increased in the presence of MAML1 (compare lanes 2 and 3).
The p300 HAT activity and coactivator function is potentiated by MAML1
Autoacetylation of p300, including at lysine 1499, has been shown to regulate the p300 HAT activity (22). Therefore, we performed an in vitro HAT assay, to see if MAML1 directly affects the HAT activity. Recombinant full-length p300, MAML1 and histone H4 tail peptide were incubated with 14C-acetyl-CoA for 7 min. The acetylation reaction was quenched, separated on a Tris–tricene gel and acetylation levels quantified with phosphoimager. As shown in Figure 2A, MAML1 significantly enhances p300 acetylation of histone H4 tails, the p300 HAT activity is dependent on the concentration of MAML1 and the apparent Kd for MAML1 is 270 ± 60 nM.
In addition, we investigated if MAML1 regulates p300 and histone acetyl-lysine localization in cell culture. Cos7 cells were cotransfected with p300-HA and FLAG-MAML1 and immunostained with antibodies recognizing p300, FLAG-tag and acetylated histones H4 (Figure 2B) and H3 (Supplementary Data, Figure 2B), and in all cases data for H4 and H3 were consistent. In the absence of transfected MAML1, p300 is present throughout the nucleus and histones are acetylated (Figure 2B, row 1). In the absence of transfected p300, there are low levels of acetylated histones in the nucleus that colocalize with MAML1 (row 2). When cells are transfected with both p300 and MAML1, MAML1 strongly directs acetylation of histone H4 in nuclear bodies. p300, MAML1 and acetylated histones all colocalize to these nuclear bodies (rows 3 and 4). To see if the directing of p300 to nuclear bodies by MAML1 is correlated with hyperacetylated (and more activated) p300, we investigated if MAML1 confers increased acetylation of p300 Lys1499 in nuclear bodies. Vectors expressing p300-HA and FLAG-MAML1 were cotransfected into Cos7 cells and immunostained for acetylated p300 Lys1499. As shown in Figure 2C, MAML1 transfection not only induces the translocation of p300 to nuclear bodies, it also leads to increased acetylation of Lys1499 in p300 in cell culture.
ChIP assays were performed to examine whether acetylation of histones in response to MAML1 takes place at the promoter region of the known Notch target gene, Hes-1. U2OS cells were transfected with Notch1 ICD and MAML1 1–1016 or 1–300. Immunoprecipitation was done with an antibody recognizing acetylated H3 and a control IgG antibody. Our data in Figure 2D shows that the control IgG antibody immunoprecipitation of the endogenous Hes-1 promoter (containing two CSL binding sites) is not visible in Notch or MAML1 (lanes 6–10) transfected cells. Anti-Acetyl-H3 precipitation of the Hes-1 promoter is detectable in Notch1 ICD (lane 11) and MAML1 1–1016 (lane 14) transfected cells and significantly enhanced when Notch and MAML1 1–1016 are cotransfected (lane 12). Cotransfection of Notch1 ICD and MAML1 1–300 gives an intermediate effect (lane 13). The histone H3 acetylation events at the Hes-1 promoter are specific since the levels of acetylation are equal at the GAPDH promoter in the absence or presence of Notch and MAML1 transfection (lanes 11–15). These data show that HATs recruited by the Notch–MAML1 complex acetylate H3 at specific promotor regions at a greater level than when Notch or MAML1 are supplied alone.
After establishment of a 293 cell line that stably expresses Flag-tagged MAML1 (f-MAML1) (Figure 2E, lane 2), we investigated if the level of global histone acetylation would be affected in these cells. Lysates were prepared from f-MAML1 cells and a 293 control (Ctrl) cell line, and the levels of acetylated histones were analyzed by western blotting with antibodies recognizing acetylated H3 and H4. We found that acetylation of histone H3 and histone H4 are significantly increased in the MAML1-expressing cell line (Figure 2E, compare lanes 1 and 2), while the histones are equally well expressed in both cell lines (see histone H3, lanes 1 and 2). The expression level of GAPDH, which is not a MAML1 target, is also similar in both cell lines (lanes 1 and 2). The in vivo function of MAML1-dependent histone acetylation was further explored in U2OS cells by using siRNA techniques. As shown by immunoblot in Figure 2F, MAML1 expression was effectively eliminated by MAML1 specific siRNA treatment (lane 2), while the expression of histones (see histone H3, lanes 1 and 2) or GAPDH (lanes 1 and 2) were not affected. MAML1 siRNA resulted in a significant reduction of the global histone H3 (2.7-fold) and histone H4 (2.0-fold) acetylation level compared to that observed with control siRNA-treated cells (compare lanes 1 and 2). Since we found that MAML1 enhances p300 autoacetylation, and HAT activity, we also investigated if MAML1 increases p300 transcriptional activity of a reporter gene in cell culture. 293 cells were cotransfected with a plasmid containing five GAL4 sites upstream of a luciferase gene and plasmids expressing GAL4–p300 and MAML1 1–1016. The level of transactivation by GAL4–p300 was significantly enhanced (20-fold) when cotransfected with MAML1 (Figure 2G).
The p300 C/H3 domain regulates p300 autoacetylation
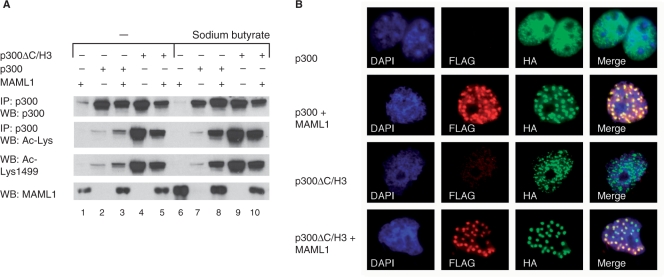
We have previously shown with an in vitro binding assay that MAML1 interacts directly with the p300 C/H3 domain (amino acids 1647–1818, see Figure 1A), suggesting that this domain in p300 could be important for the MAML1-dependent increase in p300 autoacetylation. We therefore transfected 293 cells with MAML1 and p300 or p300ΔC/H3 to perform a cell culture acetylation assay. MAML1 and p300 proteins, and acetylation of p300, were detected by western blotting with antibodies recognizing MAML1, p300, or acetylated lysines in p300. We found that there is much greater autoacetylation of p300ΔC/H3 protein than wild-type p300 (Figure 3A, compare lanes 2 and 4, 7 and 9). There is a robust increase in autoacetylation of wild-type p300 in the MAML1 cotransfected cells (compare lanes 2 and 3), which is further enhanced when the HDAC inhibitor sodium butyrate is included in the assay (compare lanes 7 and 8). However, there is no MAML1-dependent increase in autoacetylation of the p300ΔC/H3 protein (compare lanes 4 and 5, 9 and 10). Furthermore, we found that p300 Lys1499 autoacetylation, which we showed above to be activated in vitro by MAML1, is also constitutively autoacetylated in p300ΔC/H3. However, we could not detect any MAML1-dependent p300 autoacetylation at this residue in our in vivo assay (Figure 3A, row 3).
Figure 3.
The p300 C/H3 domain regulates p300 autoacetylation. (A) Plasmids expressing p300 or p300ΔC/H3 and MAML1 were cotransfected into 293 cells, whole-cell extracts prepared, and the p300 protein was immunoprecipitated with a p300 antibody. Proteins were separated by SDS–PAGE and monitored by western blotting with antibodies recognizing p300, MAML1 or acetylated lysines in p300. (B) p300-HA and FLAG-MAML1 expression plasmids, as indicated in the figures, were cotransfected into Cos7 cells and after 24 h the cells were immunostained with antibodies recognizing the HA-tag or FLAG-tag.
In addition, we investigated if MAML1 could affect p300ΔC/H3 translocation in cells. FLAG-MAML1 was cotransfected with p300-HA or p300ΔC/H3-HA expression plasmids into Cos7 cells and after 24 h the cells were immunostained with antibodies recognizing the HA-tag or FLAG-tag. While wild-type p300 is evenly distributed in the nucleus (Figure 3B, row 1), and translocates to nuclear bodies in the presence of MAML1, where the two proteins colocalize (row 2), p300ΔC/H3 is distributed in nuclear bodies regardless of MAML1 (row 3). However, coexpression of MAML1 further enhances translocation of p300ΔC/H3 proteins to nuclear bodies, indicating that there could still be a functional interaction between MAML1 and p300 without the C/H3 domain, perhaps through another cofactor.
MAML1-dependent p300 autoacetylation is not regulated by the activation loop in p300
p300 has previously been shown to contain an autoinhibitory activation loop which regulates the autoacetylation, and HAT activity, of p300 (22). To see if this loop could be involved in controlling MAML1-dependent p300 autoacetylation, 293 cells were transfected with MAML1 and wild-type p300 or p300 lacking the activation loop (p300Δloop). Lysates were prepared from the cells and the levels of MAML1, p300 and p300 acetylation were analyzed by western blotting with antibodies recognizing MAML1, p300, acetylated lysines or acetylated Lys1499 in p300. As shown in Figure 4A, p300 appears to be significantly more acetylated when lacking the activation loop (compare lanes 2 and 4). However, the MAML1-dependent increase in p300 autoacetylation is similarly proportionately enhanced with wild-type versus loop-deleted p300 proteins (compare lanes 2 and 3, 4 and 5). Since acetylation of p300 wild-type is greater in the presence of MAML1 than acetylation of p300Δloop in the absence of MAML1 (compare lanes 3 and 4), these data argue that MAML1 activation of p300 autoacetylation is independent of this HAT domain regulatory loop. Moreover, in an in vivo binding assay, MAML1 interacts equally well with the p300 and p300Δloop proteins (data not shown).
The p300Δloop protein has previously been shown to function as a more potent coactivator for the androgen receptor than wild-type p300 (22). To see if the p300Δloop protein would function as a more potent coactivator of MAML1, 293 cells were cotransfected with a luciferase reporter containing five Gal4-binding sites and vectors expressing Gal4-MAML1 1–1016 and wild-type p300 or p300Δloop. As shown in Figure 4B, MAML1-induced coactivation is greatly enhanced by p300Δloop relative to wild-type p300. The western blot in Figure 4B shows that the p300 proteins are equally well expressed in 293 cells so we infer that the p300 activation loop deletion and MAML1 synergize to stimulate transcription.
MAML1-dependent p300 autoacetylation and p300 transcriptional coactivator function is optimized by full-length MAML1 protein
Data from our ChIP assay (see Figure 2D) show that full-length MAML1 protein, and to a lesser extent MAML1 1–300, cooperates with Notch1 ICD to stimulate histone H3 acetylation at the Hes-1 promoter. To investigate which domains in MAML1 are required to enhance p300 autoacetylation, we performed an in vitro acetylation assay, where recombinant MAML1 proteins were incubated with p300 and 3H-acetyl-CoA. The full-length MAML1 1–1016 protein strongly enhances p300 autoacetylation (Figure 5A, compare lanes 1 and 2), while MAML1 1–300, 309–625, 499–804 and 701–1016 are not sufficient to stimulate autoacetylation (lanes 3–6).
We next investigated whether full-length MAML1 protein is required to increase p300 autoacetylation in cell culture. 293 cells were transfected with p300 together with MAML1 1–1016, 1–625 or 1–300. Acetylated p300 was detected by western blotting with an antibody recognizing acetylated Lys1499 in p300. As shown in Figure 5B, the autoacetylation of p300 is strongly enhanced in the presence of MAML1 1–1016 (compare lanes 2 and 3), to a lesser extent in the presence of MAML1 1–625 (lane 4), and not significantly by MAML1 1–300 (lane 5). In addition, Cos7 cells were transfected with p300-HA and FLAG-MAML1, to determine which MAML1 domains modulate histone acetylation in nuclear bodies by p300 in cell culture. Immunostaining of the cells with antibodies recognizing p300, FLAG-tag and acetylated histone H3 shows that MAML1 1–1016 induces the relocation of histone acetylation to nuclear bodies (Figure 5C, row 1), whereas MAML1 1–625 (rows 2 and 3) and MAML1 1–300 (rows 4 and 5) fails to do so. To investigate if MAML1 full length is needed to affect p300 transcriptional activity in cell culture, plasmids expressing Gal4-p300 and various MAML1 domains were cotransfected with a luciferase reporter into 293 cells. As shown in Figure 5D, MAML1 1–1016 strongly enhances p300 transcriptional coactivation, while MAML1 1–625 only moderately enhances p300 activity. MAML1 1–300 does not contribute significantly to p300 activation.
DISCUSSION
p300 has previously been reported to enhance transcriptional activation for various activators, and to work in synergy with MAML1 in Notch-mediated transcription (15,16). We show in this report that MAML1 enhances p300 activity, and have investigated the molecular mechanisms and function for this interplay. When we previously reported that p300 acetylates MAML1 (17), we also noticed that p300 became more acetylated in the presence of MAML1. In our in vitro acetylation assay, MAML1 does not seem to have any intrinsic acetyltransferase activity (data not shown). However, the p300 protein has been shown to undergo autoacetylation (22). Our acetylation assay in cell culture shows that the MAML1-dependent increase in p300 autoacetylation is totally dependent on the HAT domain of p300.
MAML1 has previously been shown to stimulate translocation of p300 to nuclear bodies (16). Nuclear bodies are punctuate structures found in the nuclei of certain cells, and they are suggested to be involved in a variety of cellular functions, such as transcriptional regulation and chromatin organization, post-translational modifications of proteins and identification and storage of proteins (28). Consistently, we show that in the presence of MAML1, p300, acetylated histones and MAML1 itself colocalize to nuclear bodies. The levels of p300 autoacetylation have been shown to correlate with p300 HAT activity, in particular autoacetylation of Lys1499 appears to stimulate p300 acetylation of histones (22). We provide evidence that MAML1-enhanced p300 autoacetylation, which includes acetylation of Lys1499, leads to an increase in p300 HAT activity in vitro. MAML1 directs p300-dependent acetylation of histone H3 and H4 in nuclear bodies, and acetylation of Lys1499 in p300 is higher in the nuclear bodies in the presence of MAML1. Thus, our data suggest that the nuclear bodies might be important for p300 modification by acetylation, and perhaps storage of a pool of acetylated, and active, p300. It remains, however, to be investigated if transcription of p300-regulated genes also occurs in the nuclear bodies.
The MAML1 protein contains various domains, and the N-terminal domain interacts with p300. We found that MAML1 1–625 increases p300 autoacetylation, although not as strongly as MAML1 1–1016, suggesting that multiple domains between residues 300–1016 in MAML1 are important for p300 autoacetylation. However, our data show that only MAML1 full length, and none of the truncated MAML1 proteins tested, including amino acids 1–300 and 1–625, is sufficient to fully potentiate p300-dependent transcription coactivation and to direct histone acetylation to nuclear bodies. This data is consistent with a previous report by Fryer et al. describing that full-length MAML1 induces translocation of p300 to nuclear bodies, and it was suggested that the MAML1 protein contains a C-terminal region that is important for in vivo transcriptional activation (16). This latter point is supported by our observation that in cell culture, full-length MAML1 enhances Notch-dependent histone acetylation at a promotor region to a greater extent than does MAML1 1–300.
p300 has previously been shown to contain an autoinhibitory loop that regulates acetylation, and activity, of p300 (22). We confirm here that p300 shows increased acetylation when lacking this activation loop. We also found that, in cell culture, MAML1 potentiates p300-dependent transcription coactivation to a greater extent when p300 lacks the activation loop. However, the MAML1-dependent increase in p300 autoacetylation is not dependent on the loop, which suggests that other domains in p300 can function as targets for regulating p300 activity. We have previously mapped MAML1 binding to p300 C/H3 domain in p300 (17). The C/H3 domain is adjacent to the p300 HAT domain, and a potential domain for regulating HAT activity. It is possible that the C/H3 domain may interact with the negatively charged HAT surface through electrostatic interactions as proposed for the p300 activation loop (29). In this article we describe that autoacetylation of p300ΔC/H3 is enhanced versus wild-type p300, and its ability to localize to nuclear bodies is facilitated relative to p300 wild-type. Furthermore, without the C/H3 domain, p300 is autoacetylated independently of MAML1. The mechanism behind this is not clear, but these observations suggest that a corepressor might interact with the C/H3 domain. For example, HDAC1 has been reported to bind to the p300 C/H3 domain (30). It is possible that MAML1 binding to the C/H3 domain, relieves the corepressor, which leads to enhanced p300 activity. Another theory is that there is no corepressor interacting at the C/H3 domain, but rather MAML1 binding to C/H3 induces a conformational change in the HAT and/or C/H3 domains, which do not affect the autoinhibitory loop, but activates p300. In our in vitro acetylation assay, recombinant Flag-MAML1 enhances autoacetylation (Lys1499) of recombinant p300 (Flag-HAT-C/H3) affinity-purified from Escherichia coli. Moreover, Kraus and collaborators showed with affinity-purified recombinant proteins in an in vitro acetylation assay, that deletion of the C/H3 domain leads to a strong increase in HAT activity (23). Their, and our, data supports the model that it is rather the C/H3 domain itself that represses the p300 HAT activity, and its inhibitory effect can be relieved by interactions with other proteins. It is not clear how the p300ΔC/H3 protein translocates to the nucleus by itself, if autoacetylation per se is sufficient for nuclear body translocation, and how MAML1 can stimulate this translocation. We speculate that a functional interaction might exist between MAML1 and p300 without the C/H3 domain, perhaps through multiple MAML1 binding sites in p300, or through yet another cofactor.
p300 autoacetylation might play a role in recruitment of coregulator proteins to a promoter, acetylation of histones to modulate chromatin structure, pre-initiation (PIC) assembly, transcriptional elongation and promoter clearance. The increased p300 transcriptional coactivation observed in the presence of MAML1, when autoacetylation of the p300 HAT domain is increased, is probably due at least in part to increased acetylation of histones. It has been demonstrated that acetylation of p300 leads to a conformational change that removes the autoinhibitory loop resulting in enhanced HAT activity (22). It could be possible that MAML1 binding might induce a similar conformational change.
It is also possible that the MAML1-dependent increase in p300 autoacetylation leads to p300 dissociation from promoter regions. We have previously shown that p300 binds less strongly to MAML1 in the presence of acetyl-CoA, which could be due to acetylation of MAML1 or increased p300 autoacetylation (17). We also reported previously that MAML1 can bind to histone tails in vitro (17) and speculated that the previously described p300 interaction with histone tails (31) might be important for the p300 coactivator function at Notch-regulated promoters. By using defined in vitro assays, Black and colleagues showed that (i) autoacetylation of p300 causes it to dissociate from GAL4–VP16–Mediator complex (ii) pre-acetylated p300 does not bind to GAL4-VP16-Mediator complexes and (iii) inhibition of the p300 HAT activity prevents p300 from dissociation. They concluded that transcription requires dissociation of p300, and reported that p300 dissociates from a tetracycline-inducible, VP16-responsive promoter in vivo prior to accumulation of transcript, and suggested the possibility that autoacetylation of p300 is controlling the dissociation also in vivo (32).
Black and colleagues also provided evidence that p300 autoacetylation regulates the competitive binding between p300 and TFIID to the Mediator complex, and showed that increased recruitment of p300 in the presence of Mediator resulted in elevated histone acetylation (32). Their data is consistent with previous reports describing a cooperative effect in transcription between Mediator and p300 on chromatin templates in vitro activated by HNF4α (33), PPARγ (34) and ERα (35). MAML1 has been shown to directly interact with the CDK8 kinase, a subunit in the Mediator complex (16), and MAML1 stimulates transcription from DNA templates (15), suggesting that MAML1 binds to the Mediator complex. MAML1 might, through direct interactions with both p300 and Mediator, coordinate the recruitment of these cofactors to promoter regions. Thus, MAML1 may stimulate p300 autoacetylation so that p300 can dissociate from a promoter region and Mediator can then be recruited by MAML1. We have previously hypothesized that it is the p300-interacting proteins that determine the acetylation pattern of histones (17). Based on our new data and from other studies, it seems clear that interacting proteins can regulate p300 activity by controlling p300 autoacetylation. Future directions will focus on investigating the specificity in timing and termination of MAML1 enhanced p300 autoacetylation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Swedish Research Council; the Swedish Cancer Society and the Swedish Childreńs Cancer Foundation (to A.E.W.); US National Institutes of Health (to P.A.C.); Fellowship from the Swedish Children's Cancer Foundation (to M.J.L.). Funding for open access charge: Swedish Research Council.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
Drs J. Boyes, J. DeCaprio, T. Kadesch, N. Perkins, R.G. Roeder and E. Treuter are acknowledged for the gifts of expression plasmids described in the Methods section.
REFERENCES
- 1.Wu L, Aster JC, Blacklow SC, Lake R, Artavanis-Tsakonas S, Griffin JD. MAML1, a human homologue of Drosophila mastermind, is a transcriptional co-activator for NOTCH receptors. Nat. Genet. 2000;26:484–489. doi: 10.1038/82644. [DOI] [PubMed] [Google Scholar]
- 2.Bettler D, Pearson S, Yedvobnick B. The nuclear protein encoded by the Drosophila neurogenic gene mastermind is widely expressed and associates with specific chromosomal regions. Genetics. 1996;143:859–875. doi: 10.1093/genetics/143.2.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smoller D, Friedel C, Schmid A, Bettler D, Lam L, Yedvobnick B. The Drosophila neurogenic locus mastermind encodes a nuclear protein unusually rich in amino acid homopolymers. Genes Dev. 1990;4:1688–1700. doi: 10.1101/gad.4.10.1688. [DOI] [PubMed] [Google Scholar]
- 4.Yedvobnick B, Smoller D, Young P, Mills D. Molecular analysis of the neurogenic locus mastermind of Drosophila melanogaster. Genetics. 1988;118:483–497. doi: 10.1093/genetics/118.3.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kurooka H, Honjo T. Functional interaction between the mouse notch1 intracellular region and histone acetyltransferases PCAF and GCN5. J. Biol. Chem. 2000;275:17211–17220. doi: 10.1074/jbc.M000909200. [DOI] [PubMed] [Google Scholar]
- 6.Oswald F, Tauber B, Dobner T, Bourteele S, Kostezka U, Adler G, Liptay S, Schmid RM. p300 acts as a transcriptional coactivator for mammalian Notch-1. Mol. Cell Biol. 2001;21:7761–7774. doi: 10.1128/MCB.21.22.7761-7774.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu L, Sun T, Kobayashi K, Gao P, Griffin JD. Identification of a family of mastermind-like transcriptional coactivators for mammalian notch receptors. Mol. Cell Biol. 2002;22:7688–7700. doi: 10.1128/MCB.22.21.7688-7700.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin SE, Oyama T, Nagase T, Harigaya K, Kitagawa M. Identification of new human mastermind proteins defines a family that consists of positive regulators for notch signaling. J. Biol. Chem. 2002;277:50612–50620. doi: 10.1074/jbc.M209529200. [DOI] [PubMed] [Google Scholar]
- 9.Wilson JJ, Kovall RA. Crystal structure of the CSL-Notch-Mastermind ternary complex bound to DNA. Cell. 2006;124:985–996. doi: 10.1016/j.cell.2006.01.035. [DOI] [PubMed] [Google Scholar]
- 10.Nam Y, Sliz P, Song L, Aster JC, Blacklow SC. Structural basis for cooperativity in recruitment of MAML coactivators to Notch transcription complexes. Cell. 2006;124:973–983. doi: 10.1016/j.cell.2005.12.037. [DOI] [PubMed] [Google Scholar]
- 11.Shen H, McElhinny AS, Cao Y, Gao P, Liu J, Bronson R, Griffin JD, Wu L. The Notch coactivator, MAML1, functions as a novel coactivator for MEF2C-mediated transcription and is required for normal myogenesis. Genes Dev. 2006;20:675–688. doi: 10.1101/gad.1383706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao Y, Katzman RB, Delmolino LM, Bhat I, Zhang Y, Gurumurthy CB, Germaniuk-Kurowska A, Reddi HV, Solomon A, et al. The notch regulator MAML1 interacts with p53 and functions as a coactivator. J. Biol. Chem. 2007;282:11969–11981. doi: 10.1074/jbc.M608974200. [DOI] [PubMed] [Google Scholar]
- 13.Alves-Guerra MC, Ronchini C, Capobianco AJ. Mastermind-like 1 Is a specific coactivator of beta-catenin transcription activation and is essential for colon carcinoma cell survival. Cancer Res. 2007;67:8690–8698. doi: 10.1158/0008-5472.CAN-07-1720. [DOI] [PubMed] [Google Scholar]
- 14.Fryer CJ, White JB, Jones KA. Mastermind recruits CycC:CDK8 to phosphorylate the Notch ICD and coordinate activation with turnover. Mol. Cell. 2004;16:509–520. doi: 10.1016/j.molcel.2004.10.014. [DOI] [PubMed] [Google Scholar]
- 15.Wallberg AE, Pedersen K, Lendahl U, Roeder RG. p300 and PCAF act cooperatively to mediate transcriptional activation from chromatin templates by notch intracellular domains in vitro. Mol. Cell Biol. 2002;22:7812–7819. doi: 10.1128/MCB.22.22.7812-7819.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fryer CJ, Lamar E, Turbachova I, Kintner C, Jones KA. Mastermind mediates chromatin-specific transcription and turnover of the Notch enhancer complex. Genes Dev. 2002;16:1397–1411. doi: 10.1101/gad.991602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saint Just Ribeiro M, Hansson ML, Wallberg AE. A proline repeat domain in the Notch co-activator MAML1 is important for the p300-mediated acetylation of MAML1. Biochem. J. 2007;404:289–298. doi: 10.1042/BJ20061900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dornan D, Shimizu H, Perkins ND, Hupp TR. DNA-dependent acetylation of p53 by the transcription coactivator p300. J. Biol. Chem. 2003;278:13431–13441. doi: 10.1074/jbc.M211460200. [DOI] [PubMed] [Google Scholar]
- 19.Chan HM, La Thangue NB. p300/CBP proteins: HATs for transcriptional bridges and scaffolds. J. Cell Sci. 2001;114:2363–2373. doi: 10.1242/jcs.114.13.2363. [DOI] [PubMed] [Google Scholar]
- 20.Roth SY, Denu JM, Allis CD. Histone acetyltransferases. Annu. Rev. Biochem. 2001;70:81–120. doi: 10.1146/annurev.biochem.70.1.81. [DOI] [PubMed] [Google Scholar]
- 21.Stiehl DP, Fath DM, Liang D, Jiang Y, Sang N. Histone deacetylase inhibitors synergize p300 autoacetylation that regulates its transactivation activity and complex formation. Cancer Res. 2007;67:2256–2264. doi: 10.1158/0008-5472.CAN-06-3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thompson PR, Wang D, Wang L, Fulco M, Pediconi N, Zhang D, An W, Ge Q, Roeder RG, Wong J, et al. Regulation of the p300 HAT domain via a novel activation loop. Nat. Struct. Mol. Biol. 2004;11:308–315. doi: 10.1038/nsmb740. [DOI] [PubMed] [Google Scholar]
- 23.Kraus WL, Manning ET, Kadonaga JT. Biochemical analysis of distinct activation functions in p300 that enhance transcription initiation with chromatin templates. Mol. Cell Biol. 1999;19:8123–8135. doi: 10.1128/mcb.19.12.8123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ogryzko VV, Schiltz RL, Russanova V, Howard BH, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 25.Karanam B, Wang L, Wang D, Liu X, Marmorstein R, Cotter R, Cole PA. Multiple roles for acetylation in the interaction of p300 HAT with ATF-2. Biochemistry. 2007;46:8207–8216. doi: 10.1021/bi7000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Turnell AS, Stewart GS, Grand RJ, Rookes SM, Martin A, Yamano H, Elledge SJ, Gallimore PH. The APC/C and CBP/p300 cooperate to regulate transcription and cell-cycle progression. Nature. 2005;438:690–695. doi: 10.1038/nature04151. [DOI] [PubMed] [Google Scholar]
- 27.Sen N, Hara MR, Kornberg MD, Cascio MB, Bae BI, Shahani N, Thomas B, Dawson TM, Dawson VL, Snyder SH, et al. Nitric oxide-induced nuclear GAPDH activates p300/CBP and mediates apoptosis. Nat. Cell Biol. 2008;10:866–873. doi: 10.1038/ncb1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bernardi R, Pandolfi PP. Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat. Rev. Mol. Cell Biol. 2007;8:1006–1016. doi: 10.1038/nrm2277. [DOI] [PubMed] [Google Scholar]
- 29.Liu X, Wang L, Zhao K, Thompson PR, Hwang Y, Marmorstein R, Cole PA. The structural basis of protein acetylation by the p300/CBP transcriptional coactivator. Nature. 2008;451:846–850. doi: 10.1038/nature06546. [DOI] [PubMed] [Google Scholar]
- 30.Simone C, Stiegler P, Forcales SV, Bagella L, De Luca A, Sartorelli V, Giordano A, Puri PL. Deacetylase recruitment by the C/H3 domain of the acetyltransferase p300. Oncogene. 2004;23:2177–2187. doi: 10.1038/sj.onc.1207327. [DOI] [PubMed] [Google Scholar]
- 31.An W, Roeder RG. Direct association of p300 with unmodified H3 and H4 N termini modulates p300-dependent acetylation and transcription of nucleosomal templates. J. Biol. Chem. 2003;278:1504–1510. doi: 10.1074/jbc.M209355200. [DOI] [PubMed] [Google Scholar]
- 32.Black JC, Choi JE, Lombardo SR, Carey M. A mechanism for coordinating chromatin modification and preinitiation complex assembly. Mol. Cell. 2006;23:809–818. doi: 10.1016/j.molcel.2006.07.018. [DOI] [PubMed] [Google Scholar]
- 33.Malik S, Wallberg AE, Kang YK, Roeder RG. TRAP/SMCC/mediator-dependent transcriptional activation from DNA and chromatin templates by orphan nuclear receptor hepatocyte nuclear factor 4. Mol. Cell Biol. 2002;22:5626–5637. doi: 10.1128/MCB.22.15.5626-5637.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wallberg AE, Yamamura S, Malik S, Spiegelman BM, Roeder RG. Coordination of p300-mediated chromatin remodeling and TRAP/mediator function through coactivator PGC-1alpha. Mol. Cell. 2003;12:1137–1149. doi: 10.1016/s1097-2765(03)00391-5. [DOI] [PubMed] [Google Scholar]
- 35.Acevedo ML, Kraus WL. Mediator and p300/CBP-steroid receptor coactivator complexes have distinct roles, but function synergistically, during estrogen receptor alpha-dependent transcription with chromatin templates. Mol. Cell Biol. 2003;23:335–348. doi: 10.1128/MCB.23.1.335-348.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.