Abstract
A number of aminoglycosides have been reported to interact and interfere with the function of various RNA molecules. Among these are 16S rRNA, the group I intron, and the hammerhead ribozymes. In this report we show that cleavage by RNase P RNA in the absence as well as in the presence of the RNase P protein is inhibited by several aminoglycosides. Among the ones we tested, neomycin B was found to be the strongest inhibitor with a Ki value in the micromolar range (35 μM). Studies of lead(II)-induced cleavage of RNase P RNA suggested that binding of neomycin B interfered with the binding of divalent metal ions to the RNA. Taken together, our findings suggest that aminoglycosides compete with Mg2+ ions for functionally important divalent metal ion binding sites. Thus, RNase P, which is an essential enzyme, is indeed a potential drug target that can be used to develop new drugs by using various aminoglycosides as lead compounds.
Keywords: tRNA precursors, tRNA processing
RNase P is an essential endoribonuclease involved in the processing of tRNA precursors—more specifically, the maturation of the 5′ termini of the majority of all known tRNAs in prokaryotes as well as in eukaryotes. In bacteria, RNase P consists of an RNA subunit and a small basic protein, and it has been shown that the catalytic activity of this ribonucleoprotein complex is associated with its RNA subunit (1). The RNA alone is in fact able to cleave a number of substrates correctly in vitro in the absence of the protein. The reaction catalyzed by RNase P requires divalent metal ions, and Mg2+ promotes cleavage most efficiently (see, for example, ref. 2 and references therein). It has been proposed that cleavage proceeds through a trigonal bipyramidal transition state (SN2), where two Mg2+ ions participate in the chemistry of cleavage (ref. 3; see also ref. 4), and recent data are in agreement with this proposal (5, 6). Two metal ions have also been suggested to participate in cleavage by another ribozyme, the group I intron (7–9). Moreover, cleavage by either of these two ribozymes results in a 5′-monophosphate and a 3′-hydroxyl (ref. 2; ref. 10 and references therein).
Several aminoglycosides are known to interact with RNA and interfere with its function. For example neomycin B induces translational misreading, most likely as a result of an interaction with 16S rRNA in the A site of the ribosome (ref. 11 and references therein), and neomycin B inhibits the reactions catalyzed by the hammerhead and the hepatitis delta virus ribozymes (12, 13). Furthermore, this aminoglycoside, as well as several others, inhibits group I intron splicing (14, 15). The inhibitory effect caused by neomycin B has been suggested to be due to displacement of essential divalent metal ions (13, 16, 17). The possibility that neomycin B displaces divalent metal ions, together with the observations that cleavage by RNase P RNA and group I intron show some similarities, prompted us to investigate whether aminoglycosides inhibit the action of RNase P RNA. Our results showed that several members of this class of antibiotics indeed interact with RNase P RNA and interfere with its function. This was shown by using both Escherichia coli RNase P RNA (M1 RNA), alone or in the presence of the E. coli RNase P protein, and Mycoplasma hyopneumoniae RNase P RNA (Hyo P RNA). The data further suggested that the neomycin B competed for specific divalent metal ion binding sites in RNase P RNA.
MATERIALS AND METHODS
Preparation of tRNA Precursors and RNase P RNA.
The tRNASu3Tyr precursor (pSu3) and RNase P RNA gene constructs behind the T7 promoter used in the present study have been described elsewhere (18–20). The pSu3 precursor, the RNase P RNA derived from E. coli (M1 RNA) and from Mycoplasma hyopneumoniae (Hyo P RNA) were generated by run-off transcription of linearized DNA using T7 DNA-dependent RNA polymerase, as described previously (21).
RNase P RNA Assay.
RNase P RNA activity was monitored under our standard assay conditions in buffer I [50 mM Tris⋅HCl, pH 7.2/5% (wt/vol) poly(ethylene glycol) 6000/100 mM NH4Cl/100 mM MgCl2]. To monitor cleavage in the presence of various aminoglycosides the reaction conditions were changed to buffer II [50 mM Tris⋅HCl, pH 7.2/5% (wt/vol) poly(ethylene glycol) 6000/1 mM NH4Cl/10 mM spermidine/10 mM MgCl2]. The final concentrations of aminoglycosides were as indicated. The aminoglycosides used were purchased from Sigma.
Cleavage was performed with an internally labeled substrate as described before (22). The reaction products were separated on 10% (wt/vol) denaturing polyacrylamide gels and visualized with a PhosphorImager (Molecular Dynamics). The concentrations of substrate and ribozyme (M1 RNA or Hyo P RNA) were 5.2 nM and 82 nM, respectively. Cleavage by M1 RNA in the presence of C5 was performed with 5.2 nM pSu3 and 1.6 nM M1 RNA in buffer II as described above. Conditions were adjusted in such a way that measurements were carried out in the linear portion of the curve of kinetics of the cleavage reaction. Cleavage efficiency was plotted as a function of the concentration of the aminoglycoside under study, and Ki was defined as the concentration resulting in 50% inhibition of cleavage activity.
Lead(II)-Induced Cleavage of M1 RNA.
M1 RNA was 3′ end labeled with [32P]pCp according to standard procedures. The RNA so obtained was subjected to lead(II)-induced cleavage in buffer II as described in detail elsewhere (23, 24). Neomycin B (or paromomycin) was added as indicated.
RESULTS
Inhibition of RNase P Cleavage by Neomycin B.
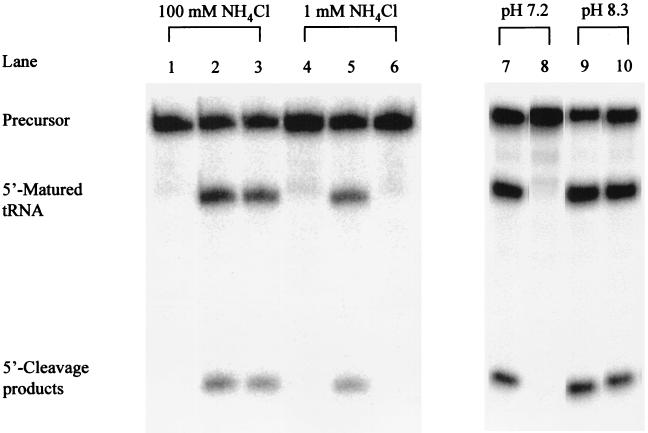
Cleavage by RNase P RNA is usually performed at relatively high concentrations of Mg2+ and/or NH4+ (see, for example, ref. 2 and references therein). Addition of neomycin B to the reaction under our standard assay conditions resulted in no inhibitory effect (Fig. 1). This result is in keeping with a previous report in which the influence of aminoglycosides on RNase P cleavage activity was studied (25). However, it has been suggested that high concentrations of mono and divalent metal ions interfere with the inhibitory action of aminoglycosides (12, 13). We therefore established conditions where both [Mg2+] and [NH4+] were decreased compared with our standard assay conditions. Previous data showed that cleavage can occur at a lower Mg2+ concentration in the presence of spermidine (4). Thus, we added 10 mM spermidine, lowered the Mg2+ concentration to 10 mM, and studied the requirement for NH4+ by titrating its concentration from 1 to 100 mM. We note that these concentrations of spermidine and Mg2+ are more similar to the physiological conditions compared with the more conventional cleavage reaction conditions (ref. 26 and references therein). Our results showed that, irrespective of NH4+ concentration, correct cleavage of pSu3 was observed (Fig. 1; data shown only for cleavage at [NH4+] = 1 mM). The rate of cleavage under these conditions at 1 mM NH4+ was reduced by only a factor of approximately 2 compared with cleavage under our standard assay conditions (data not shown). In the following experiments cleavage was performed at 10 mM Mg2+, 10 mM spermidine, and 1 mM NH4+ (see Materials and Methods).
Figure 1.
Cleavage by M1 RNA alone at 37°C under various conditions as indicated. The final concentrations of reactants and buffer conditions were as outlined in Materials and Methods. The final concentration of neomycin was 1 mM. Lanes 1–3, cleavage in buffer I, where lane 1 = no M1 RNA added, lane 2 = cleavage in the absence of neomycin B, and lane 3 = cleavage in the presence of neomycin B. Time of cleavage for lanes 2 and 3, 0.5 min. Lanes 4–6, cleavage in buffer II, where lane 4 = no M1 RNA added, lane 5 = cleavage in the absence of neomycin B, and lane 6 = cleavage in the presence of neomycin B. Time of cleavage for lanes 5 and 6 = 1 min. Lanes 7–10, cleavage in buffer II at pH values as indicated; lanes 7 and 9 = cleavage in the absence of neomycin and lanes 8 and 10 = cleavage in the presence of neomycin B. Time of cleavage for lanes 7–10 = 3 min.
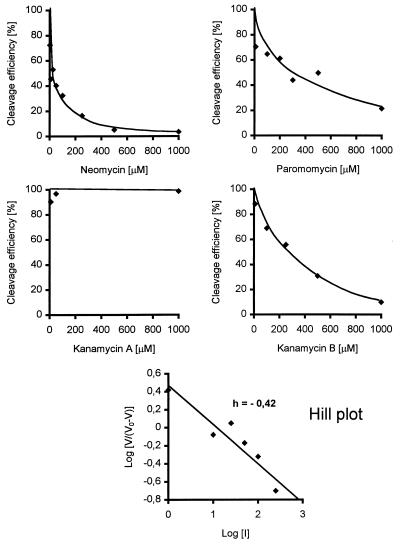
Addition of an increasing concentration of neomycin B under these conditions clearly resulted in an inhibition of M1 RNA activity (Figs. 1 and 2). The inhibition constant, Ki, was determined as described in Materials and Methods to be in the micromolar range (35 μM; Table 1). Similar Ki values were obtained irrespective of whether neomycin B was preincubated with the substrate or with the ribozyme (data not shown). A Hill plot analysis (27) using these data gives a slope (h) < 1 (Fig. 2). This value suggests a negative cooperativity for binding of neomycin B to M1 RNA. Inhibition of cleavage activity by neomycin B of the same order of magnitude was also observed in the presence of the protein subunit (Table 1). This result indicates that the inhibition is not influenced by the presence of the RNase P protein to any significant extent. We also investigated whether neomycin B inhibited cleavage by Hyo P RNA (20). As shown in Table 1, the activity of this RNase P RNA was also inhibited by the addition of this aminoglycoside, with a Ki value approximately 3 times higher than in the case of inhibition of M1 RNA. Taking these results together, we conclude that neomycin B functions as an inhibitor of RNase P cleavage activity. Furthermore, the concentration of neomycin B that caused inhibition of RNase P activity is similar to what has been observed in other systems in which inhibition of various ribozyme activities by neomycin B was studied (Table 1).
Figure 2.
Cleavage efficiency in percent as a function of increasing concentrations of the various aminoglycosides used in this study. Cleavage was performed at 37°C as outlined in the text. (Bottom) Hill plot analysis (27).
Table 1.
Concentrations of aminoglycosides that resulted in 50% inhibition of cleavage by RNase P RNA
| Ribozyme | Aminoglycoside | Ki, μM |
|---|---|---|
| Wild-type M1 RNA | Neomycin B | 35 ± 12 |
| Wild-type M1 RNA + C5 | Neomycin B | 60 ± 16 |
| Wild-type Hyo P RNA | Neomycin B | 100 ± 21 |
| Wild-type M1 RNA | Paromomycin | 190 ± 12 |
| Wild-type M1 RNA | Kanamycin A | ND |
| Wild-type M1 RNA | Kanamycin B | 275 ± 75 |
| M1C254 RNA | Neomycin B | 101 ± 36 |
| M1U255 RNA | Neomycin B | 101 ± 33 |
| Wild-type M1 RNA + C5* | Neomycin B | 60 |
| Hammerhead ribozyme† | Neomycin B | 13.5 |
| Group I intron‡ | Neomycin B | 0.5 |
| HDV self-cleavage RNA§ | Neomycin B | 28 |
The given values are mean ± standard error of several independent experiments. ND, no inhibition detected.
Indicates that we used a precursor that lacked the 3′-terminal CCA-motif (see Fig. 5). This number is an average of two independent experiments, where Ki of experiment 1 was 59 μM and Ki of the second experiment was 60 μM.
This value was taken from Stage et al. (12).
Value from von Ahsen et al. (14).
Value from Rogers et al. (13). HDV, hepatitis delta virus.
Amino Groups Important for Inhibition of RNase P RNA Cleavage.
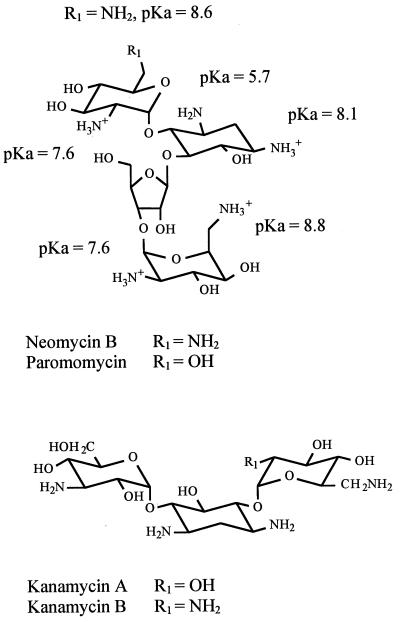
Previous studies have demonstrated that several aminoglycosides inhibit group I intron splicing and the hammerhead cleavage reaction (12, 14, 15). These aminoglycosides differ in their structures at specific positions that have been suggested to play a role in the mechanism of inhibition (see, for example, ref. 28 and references therein). Thus, to investigate whether the ammonium group at the R1 position (Fig. 3) of neomycin B plays a role in the process of inhibition, we studied cleavage by M1 RNA in the presence of paromomycin, which carries a hydroxyl group at this position. Similarly we studied another pair of aminoglycosides, kanamycin A and kanamycin B, where the difference likewise is a substitution of an ammonium group for a hydroxyl as indicated in Fig. 3. The inhibitory effects caused by these aminoglycosides are shown in Fig. 2 and Table 1.
Figure 3.
Structures of the two pairs of aminoglycosides used in this study, neomycin B and paromomycin, and kanamycin A and B. Positions where these aminoglycosides differ are indicated with R1 and the pKa values for the different ammonium groups are given in the figure.
In comparison to inhibition with neomycin B, addition of paromomycin resulted in a weaker inhibition of M1 RNA cleavage activity. The Ki for paromomycin was determined to be 190 μM, suggesting that the ammonium group at the R1 position plays a role in the mechanism of inhibition by neomycin B. In the case of cleavage in the presence of kanamycin A and kanamycin B we observed inhibition only by the latter, whereas addition of the former resulted in no inhibition under the conditions used here, where the highest concentration of aminoglycoside was 1 mM (final). The Ki value for kanamycin B, 275 μM, was significantly higher compared to the Ki values for neomycin B or paromomycin. We conclude that the ammonium group at position R1 (Fig. 3) is important for the inhibitory action by aminoglycosides of the kanamycin type.
An increase in pH suppresses the inhibition caused by neomycin B in other systems. It has been suggested that this suppression is due to a deprotonation of some of the ammonium groups on neomycin B, consequently reducing the magnitude of inhibition (ref. 28 and references therein; Fig. 3). Therefore, to investigate whether this is also the case in the M1 RNA-catalyzed reaction, we studied cleavage in the presence of neomycin B at various pH values. As shown in Fig. 1, we observed a dramatic reduction in the inhibition at higher pH, in keeping with the suggestion that at least some of the ammonium groups have to be protonated to result in an inhibition by neomycin B.
Neomycin B Interferes with Binding of Divalent Metal Ions to RNase P RNA.
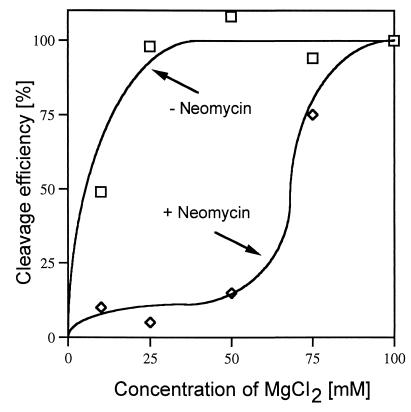
As discussed above, addition of neomycin B did not result in any inhibition of RNase P RNA cleavage at high concentrations of Mg2+ and NH4+. To investigate this phenomenon in more detail we performed cleavage in the presence of neomycin B at various concentrations of Mg2+ in the presence of 10 mM spermidine and 1 mM NH4+. From the results shown in Fig. 4 it is apparent that the inhibition caused by neomycin B is suppressed by an increase in the concentration of Mg2+. A plausible reason is that Mg2+ and neomycin B compete for at least overlapping binding sites in the M1 RNA–substrate complex. We addressed this possibility in the following way.
Figure 4.
Cleavage efficiency in percent as a function of increasing concentration of Mg2+ in the absence and in the presence of neomycin B. Cleavage was performed in buffer II at 37°C as outlined in the text with a final concentration of neomycin B of 1 mM.
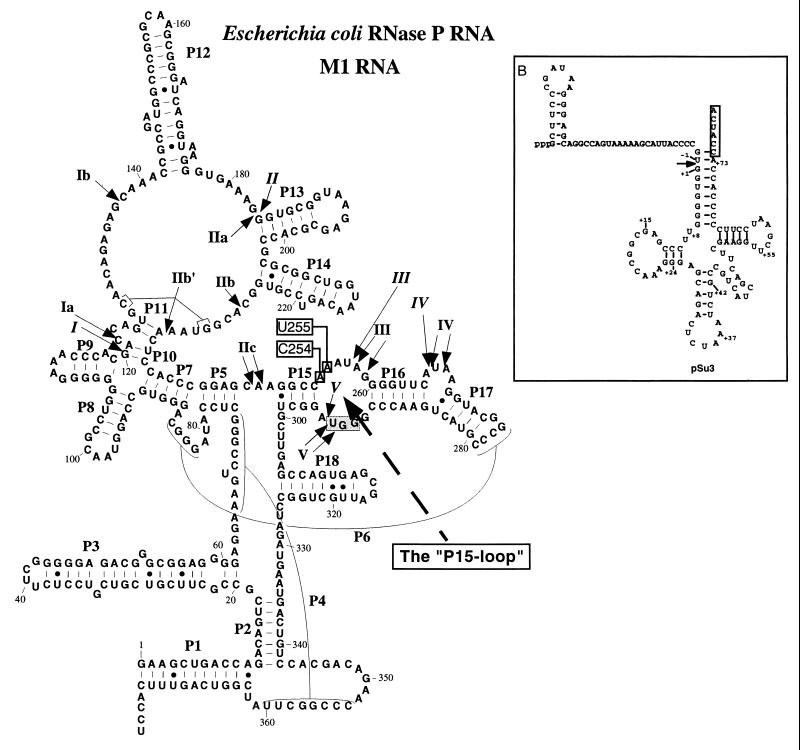
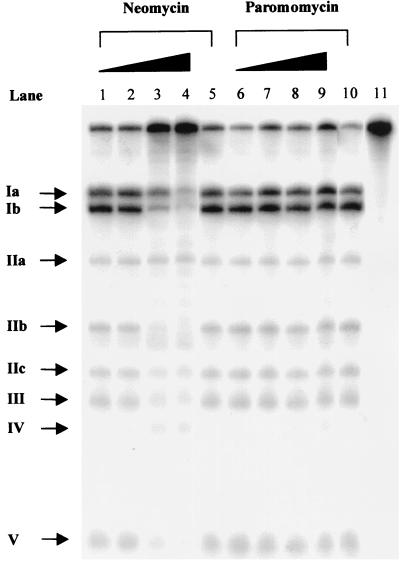
RNase P RNA is cleaved at specific positions in the presence of Pb2+ (or Mg2+), suggesting that divalent metal ions are positioned in the vicinity of these cleavage sites (Fig. 5; refs. 23, 29, and 30). To understand whether the addition of an aminoglycoside interferes with the lead(II)-induced cleavage pattern and thereby divalent metal ion binding we decided to study the Pb2+ cleavage pattern of M1 RNA in the presence of neomycin B or paromomycin. The results are shown in Fig. 6. Clearly, an increase in the concentration of neomycin B inhibits cleavage by Pb2+ at all sites except at sites IIa and IV, whereas addition of paromomycin did not result in any detectable reduction or change in the lead(II)-induced cleavage pattern. We also observed an inhibition of the Pb2+ cleavage pattern of Hyo P RNA in the presence of neomycin B (data not shown). These data demonstrates that the interaction between neomycin B and RNase P RNA influences the site-specific binding of Pb2+. It appears that M1 RNA is cleaved at unspecific positions at higher concentrations of neomycin B (Fig. 6). This finding is not surprising because binding of neomycin B to M1 RNA would most likely result in conformational changes of M1 RNA that as a consequence result in Pb2+-induced cleavage at other positions.
Figure 5.
Illustration of a secondary structure model of RNase P RNA derived from E. coli (M1 RNA; ref. 40). The specific Mg2+-induced cleavage sites are indicated by arrows and italic roman numerals, while the arrows with nonitalic roman numerals indicate the Pb2+-induced cleavage sites. The boxed (shaded box) nucleotides represent residues that are involved in base pairing with the 3′-terminal RCCA sequence of the substrate, the “RCCA-RNase P RNA” interaction (for details see text). The dashed arrow indicates the “P15-loop,” and the M1 RNA derivatives carrying a substitution at position 254 or at position 255 are indicated with open boxes. (Inset) A secondary structure of the tRNA precursor substrate, pSu3, used in this study. The arrow indicate the RNase P cleavage while the boxed residues were deleted in the substrate that lacked the 3′-terminal CCA sequence. The construction and generation of these substrates have been described elsewhere (ref. 41 and references therein).
Figure 6.
Lead(II)-induced cleavage of M1 RNA in the presence and in the absence of increasing concentrations of neomycin B or paromomycin as indicated. The final concentrations of aminoglycoside were 10, 100, 500, and 1000 μM. Lanes 5 and 10 = Pb2+ cleavage of M1 RNA in the absence of aminoglycoside and lane 11 = no Pb2+ or aminoglycoside added. Cleavage was performed as outlined in the text at 37°C and the time of Pb2+ cleavage was 10 min in all cases. The roman numerals refer to the positions of cleavage in M1 RNA as shown in Fig. 5.
Base Substitutions in the P15-Loop Influence Inhibition Caused by Neomycin B.
Two of the divalent metal binding sites, III and V, are located in a functionally important domain of M1 RNA, the P15-loop (Fig. 5). The GGU motif of the P15-loop is involved in base pairing with the 3′-terminal RCCA sequence of the substrate in the ribozyme–substrate (RS) complex (interacting residues underlined; see Fig. 5 and also below; refs. 31 and 32). To investigate the importance of the structural integrity of the P15-loop in relation to inhibition with neomycin B we determined the Ki values for two M1 RNA derivatives, M1C254 RNA and M1U255 RNA (Fig. 5). Changes at these positions were previously shown to influence divalent metal ion binding within the P15-loop as well as cleavage site recognition (24). Cleavage in the presence of neomycin B by any these M1 RNA variants resulted in an almost 3-fold higher Ki value compared with cleavage by wild-type M1 RNA (Table 1). This result indicates that the residues at positions 254 and 255 influence the inhibitory action by neomycin B. Together with the Pb2+ cleavage data, this result suggests that the P15-loop is (or is part of) a neomycin B binding site. The small but significant increase in Ki for these M1 RNA derivatives are in keeping with our previous findings that these substitutions result in modest changes in the Pb2+ cleavage pattern and in the cleavage site recognition process (24). Mutants carrying substitutions in the GGU motif showed very poor cleavage activity under these assay conditions.
To investigate the importance of the interaction between the 3′-terminal RCCA sequence and the P15-loop in the inhibition caused by neomycin B, we determined the Ki value for neomycin B inhibition of the cleavage of a precursor that lacks the 3′-terminal CCA motif by reconstituted RNase P holoenzyme. The data in Table 1 show that cleavage activity on this substrate was inhibited by neomycin B as efficiently as in the case where we used pSu3 as substrate. Thus, it appears that the efficiency of inhibition by neomycin B does not depend on base pairing between the 3′-terminal RCCA sequence of the substrate and the GGU motif of the P15-loop.
DISCUSSION
The aminoglycosides used in the present study are representatives of two families, the neomycin B family and the kanamycin family (Fig. 3). In this report we demonstrate that aminoglycosides of these types inhibit the function of RNase P, and among the ones we tested neomycin B was found to be the strongest inhibitor. The inhibition of RNase P RNA cleavage by neomycin B is sensitive to pH. Furthermore, our data suggest that the ammonium group at the R1 position in both these types of aminoglycosides plays a role in the process of inhibition. These findings are in keeping with previous observations that binding of representatives of these two families of aminoglycosides to other catalytic RNA molecules correlates with the number of positive charges—i.e., protonated ammonium groups (12–14, 28, 33, 34). From our studies it is also apparent that the inhibition caused by neomycin B is sensitive to the Mg2+ concentration (Fig. 4) as well as that it affects binding of divalent metal ions to specific sites in RNase P RNA (Fig. 6). It has been suggested that protonated ammonium groups of neomycin B displace Mg2+ ions that are essential for catalysis (13, 16, 17). Considering these results together with our present data, it is therefore likely that the aminoglycosides used in this study displace Mg2+ ions that are crucial for cleavage by RNase P RNA. An important question is therefore to identify the Mg2+ ions that are displaced by neomycin B, which as a consequence inhibits cleavage. From available data we raise the following intriguing possibilities.
It has been discussed that the chemistry of cleavage catalyzed by RNase P RNA and the group I intron RNA requires the presence of two Mg2+ ions in accordance with the two-metal-ion mechanism model (3, 5–9). Molecular dynamic modeling studies using the hammerhead ribozyme or a group I self-splicing intron have shown that it is possible to dock neomycin B into the center of action of the respective ribozyme. This docking resulted in displacement of specific Mg2+ ions. The intramolecular distances of ammonium groups on the aminoglycoside that have been suggested to be involved in the displacement of functionally important Mg2+ ions varies between 4 and 10 Å (16, 17, 28). Interestingly, in a model of the transition state of the cleavage reaction catalyzed by a group I intron RNA the two Mg2+ ions involved in the chemistry of cleavage are positioned 8.6 Å apart (8). It is therefore plausible that the inhibition of RNase P RNA cleavage by the aminoglycosides used here is the result of displacing the two Mg2+ ions involved in the chemistry of cleavage. Whether any one of these Mg2+ ions is identical to the Mg2+ ions that are positioned in the vicinity of the divalent metal ion cleavage sites in RNase P RNA remains to be established.
RNase P RNA is cleaved at specific positions by divalent metal ions—for example, Mg2+ or Pb2+ (refs. 23, 29, 30; Fig. 5). The lead(II)-induced cleavage is suppressed as a result of an increase in the Mg2+ concentration, suggesting that these divalent metal ions bind if not to the same sites at least to overlapping sites (23, 35). As demonstrated here, neomycin B influenced the lead(II)-induced cleavage pattern in such away that cleavage at specific sites was reduced (Fig. 6 and data not shown). The GGU motif at one of these sites, site V, is engaged in base pairing with the 3′-terminal RCCA sequence of the substrate in the RS complex, the “RCCA–RNase P RNA” interaction (31, 32). The Mg2+ located at site V has been suggested to contribute to function by influencing the establishment and/or stabilization of the RS complex (24, 35). A change in the structure of the P15-loop as a result of base substitutions at position 254 or 255 resulted in a less efficient inhibition by neomycin B (Table 1). We previously showed that changes at these positions influence the Pb2+ cleavage pattern within the P15-loop (24). Thus, displacement of the Mg2+ ions positioned in the vicinity of sites III and V by neomycin B as suggested from our Pb2+ cleavage analysis gives one reason why binding of this aminoglycoside to the P15-loop inhibits cleavage. We also note that the GGU motif is part of an asymmetrical internal loop, and previous studies have shown that such structural elements are aminoglycoside binding sites (36, 37). An interesting possibility is that the P15-loop is part of the active center, and that the Mg2+ ion at site III (and/or V) is involved in the chemistry of cleavage. Binding of an aminoglycoside to this region would interfere with catalysis. This would give a rationale to our finding that the establishment of the RCCA–RNase P RNA interaction does not influence the inhibitory effect caused by neomycin B.
We also have to consider that the region in the vicinity of sites Ia and IIb (Fig. 5) has been suggested to be involved in the interaction with the T-loop/T-stem domain of a tRNA precursor by forming a binding pocket for this region of the substrate (see, for example, ref. 38 and references therein). The presence of correctly coordinated divalent metal ion(s) in this region might play a role in the folding of this binding site and/or stabilize the binding of the substrate. Addition of neomycin B might therefore interfere with the folding of this region, possibly by displacement of the Mg2+ ion(s), which as a consequence would influence RS-complex formation and cleavage. In addition, the crystal structure of yeast tRNAPhe reveals that Mg2+ ions are bound in the D-loop/T-loop domain (39). It is likely that this is also the case for a tRNA precursor. Therefore, these Mg2+ ion binding sites might also be neomycin B binding sites, which as a consequence could affect the RS-complex formation.
Our results clearly suggest that in particular neomycin B binds to M1 RNA and results in a displacement of divalent metal ions. Whether it is enough to displace one of these divalent metal ions by neomycin B to obtain inhibition remains to be seen. Nevertheless, aminoglycosides have been shown to inhibit RNase P cleavage both in the RNA alone reaction and in the presence of C5. This gives us a new tool in our efforts to identify functionally important residues in the RNA subunit of RNase P. For example, as indicated in this report, it is evident that identification of residues that influence aminoglycoside binding can give information about residues that are important for coordination of functionally important Mg2+ ions. Finally, our findings suggest that the ubiquitous RNase P is indeed a potential drug target that can be used to develop new drugs by using various aminoglycosides as lead compounds.
Acknowledgments
We thank our colleagues and Dr. M. Ehrenberg for discussions and S. Rosen for technical assistance. Dr. R. Schroeder is acknowledged for convincing us to initiate this investigation. This work was supported by the Strategic Research Foundation and the Swedish Natural Science Research Council.
ABBREVIATIONS
- Hyo P RNA
Mycoplasma hyopneumoniae RNase P RNA
- pSu3
tRNASu3Tyr precursor
- RS complex
ribozyme-substrate complex
References
- 1.Guerrier-Takada C, Gardiner K, Marsh T, Pace N, Altman S. Cell. 1983;35:849–857. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- 2.Kirsebom L A. Mol Microbiol. 1995;17:411–420. doi: 10.1111/j.1365-2958.1995.mmi_17030411.x. [DOI] [PubMed] [Google Scholar]
- 3.Steitz T A, Steitz J A. Proc Natl Acad Sci USA. 1993;90:6498–6502. doi: 10.1073/pnas.90.14.6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guerrier-Takada C, Haydock K, Allen L, Altman S. Biochemistry. 1986;25:1509–1515. doi: 10.1021/bi00355a006. [DOI] [PubMed] [Google Scholar]
- 5.Warnecke J M, Fürste J P, Hardt W-D, Erdmann V A, Hartmann R K. Proc Natl Acad Sci USA. 1996;93:8924–8928. doi: 10.1073/pnas.93.17.8924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen Y, Li X, Gegenheimer P. Biochemistry. 1997;36:2425–2438. doi: 10.1021/bi9620464. [DOI] [PubMed] [Google Scholar]
- 7.Piccirilli J A, Vyle J S, Caruthers M H, Cech T R. Nature (London) 1993;361:85–88. doi: 10.1038/361085a0. [DOI] [PubMed] [Google Scholar]
- 8.Streicher B, Westhof E, Schroeder R. EMBO J. 1996;15:2556–2564. [PMC free article] [PubMed] [Google Scholar]
- 9.Sjögren A-S, Pettersson E, Sjöberg B-M, Strömberg R. Nucleic Acids Res. 1997;25:648–653. doi: 10.1093/nar/25.3.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cech T R. In: The RNA World. Gesteland R F, Atkins J F, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1993. pp. 239–269. [Google Scholar]
- 11.Wallis M G, Schroeder R. Prog Biophys Mol Biol. 1997;67:141–154. doi: 10.1016/s0079-6107(97)00011-4. [DOI] [PubMed] [Google Scholar]
- 12.Stage T K, Hertel K J, Uhlenbeck O C. RNA. 1995;1:95–101. [PMC free article] [PubMed] [Google Scholar]
- 13.Rogers J, Chang A H, von Ahsen U, Schroeder R, Davies J. J Mol Biol. 1996;259:916–925. doi: 10.1006/jmbi.1996.0369. [DOI] [PubMed] [Google Scholar]
- 14.von Ahsen U, Davies J, Schroeder R. Nature (London) 1991;353:368–370. doi: 10.1038/353368a0. [DOI] [PubMed] [Google Scholar]
- 15.von Ahsen U, Davies J, Schroeder R. J Mol Biol. 1992;226:935–941. doi: 10.1016/0022-2836(92)91043-o. [DOI] [PubMed] [Google Scholar]
- 16.Hermann T, Westhof E. J Mol Biol. 1998;276:903–912. doi: 10.1006/jmbi.1997.1590. [DOI] [PubMed] [Google Scholar]
- 17.Hoch I, Berens C, Westhof E, Schroeder R. J Mol Biol. 1998;282:557–569. doi: 10.1006/jmbi.1998.2035. [DOI] [PubMed] [Google Scholar]
- 18.Vioque A, Arnez J, Altman S. J Mol Biol. 1988;202:835–848. doi: 10.1016/0022-2836(88)90562-1. [DOI] [PubMed] [Google Scholar]
- 19.Kirsebom L A, Svärd S G. Nucleic Acids Res. 1992;20:425–432. doi: 10.1093/nar/20.3.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Svärd S G, Mattsson J G, Johansson K-E, Kirsebom L A. Mol Microbiol. 1994;11:849–859. doi: 10.1111/j.1365-2958.1994.tb00363.x. [DOI] [PubMed] [Google Scholar]
- 21.Milligan J F, Groebe D R, Whiterell G W, Uhlenbeck O C. Nucleic Acids Res. 1987;15:8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mattsson J G, Svärd S G, Kirsebom L A. J Mol Biol. 1994;241:1–6. doi: 10.1006/jmbi.1994.1467. [DOI] [PubMed] [Google Scholar]
- 23.Ciesiolka J, Hardt W-D, Schlegl J, Erdmann V A, Hartmann R K. Eur J Biochem. 1994;219:49–56. doi: 10.1111/j.1432-1033.1994.tb19913.x. [DOI] [PubMed] [Google Scholar]
- 24.Kufel J, Kirsebom L A. J Mol Biol. 1996;263:685–698. doi: 10.1006/jmbi.1996.0608. [DOI] [PubMed] [Google Scholar]
- 25.Vioque A. FEBS Lett. 1989;246:137–139. doi: 10.1016/0014-5793(89)80269-8. [DOI] [PubMed] [Google Scholar]
- 26.Jelenc P C, Kurland C G. Proc Natl Acad Sci USA. 1979;76:3174–3178. doi: 10.1073/pnas.76.7.3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Engel P C. In: Enzymology Labfax. Engel P C, editor. San Diego/BIOS Scientific, Oxford: Academic; 1996. p. 110. [Google Scholar]
- 28.Tor Y, Hermann T, Westhof E. Chem Biol. 1998;5:R277–R283. doi: 10.1016/s1074-5521(98)90286-1. [DOI] [PubMed] [Google Scholar]
- 29.Kazakov S, Altman S. Proc Natl Acad Sci USA. 1991;88:9193–9197. doi: 10.1073/pnas.88.20.9193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zito K, Hüttenhofer A, Pace N R. Nucleic Acids Res. 1993;21:5916–5920. doi: 10.1093/nar/21.25.5916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kirsebom L A, Svärd S G. EMBO J. 1994;13:4870–4876. doi: 10.1002/j.1460-2075.1994.tb06814.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tallsjö A, Kufel J, Kirsebom L A. RNA. 1996;2:299–307. [PMC free article] [PubMed] [Google Scholar]
- 33.Clouet-d’Orval B, Stage T K, Uhlenbeck O C. Biochemistry. 1995;34:11186–11190. doi: 10.1021/bi00035a025. [DOI] [PubMed] [Google Scholar]
- 34.Wang H, Tor Y. J Am Chem Soc. 1997;119:8734–8735. [Google Scholar]
- 35.Kufel J, Kirsebom L A. RNA. 1998;4:777–788. doi: 10.1017/s1355838298970923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zapp M L, Stern S, Green M R. Cell. 1993;74:969–978. doi: 10.1016/0092-8674(93)90720-b. [DOI] [PubMed] [Google Scholar]
- 37.Fourmy D, Recht M I, Blanchard S C, Puglisi J D. Science. 1996;274:1367–1371. doi: 10.1126/science.274.5291.1367. [DOI] [PubMed] [Google Scholar]
- 38.Kirsebom L A. In: The Many Faces of RNA. Eggleston D S, Prescott C D, Pearson N D, editors. London: Academic; 1997. pp. 127–144. [Google Scholar]
- 39.Holbrook S R, Sussman J L, Warrant R W, Church G M, Kim S-H. Nucleic Acids Res. 1977;4:2811–2820. doi: 10.1093/nar/4.8.2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Haas E S, Banta A B, Harris J K, Pace N R, Brown J W. Nucleic Acids Res. 1996;24:4775–4782. doi: 10.1093/nar/24.23.4775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Svärd S G, Kirsebom L A. J Mol Biol. 1992;227:1019–1031. doi: 10.1016/0022-2836(92)90518-o. [DOI] [PubMed] [Google Scholar]