Abstract
Proper recognition and repair of DNA damage is critical for the cell to protect its genomic integrity. Laser microirradiation ranging in wavelength from ultraviolet A (UVA) to near-infrared (NIR) can be used to induce damage in a defined region in the cell nucleus, representing an innovative technology to effectively analyze the in vivo DNA double-strand break (DSB) damage recognition process in mammalian cells. However, the damage-inducing characteristics of the different laser systems have not been fully investigated. Here we compare the nanosecond nitrogen 337 nm UVA laser with and without bromodeoxyuridine (BrdU), the nanosecond and picosecond 532 nm green second-harmonic Nd:YAG, and the femtosecond NIR 800 nm Ti:sapphire laser with regard to the type(s) of damage and corresponding cellular responses. Crosslinking damage (without significant nucleotide excision repair factor recruitment) and single-strand breaks (with corresponding repair factor recruitment) were common among all three wavelengths. Interestingly, UVA without BrdU uniquely produced base damage and aberrant DSB responses. Furthermore, the total energy required for the threshold H2AX phosphorylation induction was found to vary between the individual laser systems. The results indicate the involvement of different damage mechanisms dictated by wavelength and pulse duration. The advantages and disadvantages of each system are discussed.
INTRODUCTION
The cellular response to, and subsequent repair of, DNA damage is a critical cellular function that maintains genome integrity. It is not surprising, therefore, that mutations of many genes involved in DNA damage responses are found to cause human disorders and cancers (1–3). Different types of DNA damage are recognized and processed by specific cellular response pathways to ensure their efficient repair or, if the damage is too severe, apoptotic elimination of the cell occurs (2). For example, crosslinking damage, such as thymine dimers, caused by ultraviolet (UV) light is recognized and processed by the nucleotide-excision repair (NER) pathway; base damage caused by abnormal nucleotide modification, such as oxidation, deamination, or methylation, by the base-excision repair (BER) pathway; and DNA double-strand breaks (DSBs) by non-homologous endjoining (NHEJ), homologous recombination (HR), or single-strand annealing (SSA) pathways. Although many of these DNA damage response/repair factors critical for each pathway have been identified, how these individual factors recognize DNA damage and participate in specific repair pathways in vivo are still not fully understood. In vitro assays to recapitulate certain aspects of different repair pathways have been developed, which were valuable in providing mechanistic insight into the repair protein functions [e.g. DNA strand exchange, D-loop formation, DNA end processing activity, DNA end-joining for DSB repair pathways (4–8) and in vitro base and NER systems (9–11)]. However, the development of methods to analyze molecular changes that occur specifically at the damage sites inside the cell was much needed to understand the DNA damage response pathways in vivo.
A major advance in crosslinking damage studies of the NER pathway came from the use of the pore filter that allows the partial exposure of the cell nucleus to UVC, which allowed cytological detection of factor recruitment, its in vivo kinetics and upstream factor requirements (12). For DSB repair, ionizing radiation-induced focus (IRIF) formation has been and still is widely used as a valuable indicator of factor recruitment and modification at the damage sites. Following ionizing radiation, many factors involved in DSB response/repair form foci, whose colocalization with the DSB marker phosphorylated H2AX (γH2AX) proved the presence of damaged DNA in these foci (13). However, there are other DSB repair factors that do not form IRIF, such as the factors involved in NHEJ repair. Furthermore, it became apparent that visible IRIF formation involves a protein-clustering step secondary to, and distinct from, the initial damage site recruitment of the protein (14). To circumvent these problems, two methods are currently available. One is the chromatin immunoprecipitation (ChIP) method to biochemically detect protein accumulation at the damage sites induced by endonucleases that cut only a limited number of sites in the genome (e.g. HO endonuclease used in yeast, and I-SceI and I-PpoI in mammalian cells) (15–17). The NHEJ factor Ku, which does not form IRIF, was first detected at the damage sites using this method (18). With refinement of the induction of these endonucleases, it is now possible to follow the kinetics of damage recognition to a certain extent although an entire cell population, and not a single cell, must be analyzed and antibodies suitable for ChIP must be available for a given factor. It is also interesting to note that the damage processing appears to differ between the IR- and endonuclease-induced DSBs (19). Thus, the endonuclease-ChIP method is useful but is not without limitations. The second method that complements the shortcomings of IRIF detection and ChIP (but certainly has its own challenges) is the use of a laser microbeam to irradiate a defined submicron region in the cell nucleus allowing single cell analysis of DNA damage recognition/response.
Bonner's group was the first to introduce damage in a small area in the nucleus using an ultraviolet A (UVA) laser in combination with bromodeoxyuridine (BrdU), which made it possible to cytologically analyze both protein accumulation and posttranslational modification(s) at the damage sites in tissue culture cells (13,20). This was based on the previous finding that low-dose UVA, which by itself is not absorbed by DNA, can cause DNA breaks when DNA is pre-sensitized with halogenated nucleotide analogs such as BrdU (21). In fact, γH2AX as DSB marker was defined by this method, which in turn allowed researchers in the field to use γH2AX-positive IRIF as DSB markers (20,22). Since then, a number of laboratories have used the UVA laser of several different wavelengths (337–405 nm) in conjunction with pre-sensitization of DNA with various nucleotide analogs [e.g. BrdU and 5′-iodo-2-deoxyuridine (IdU)] (23–28) or DNA-intercalating dyes (Hoechst) (14,20,29,30) to study the in vivo DSB response. In addition, some laboratories have used green (532 nm) and near-infrared (NIR 800 nm) lasers, which do not require any DNA sensitization (31–37). Although all these systems have been useful in the studies of DSB damage recognition by various factors, it became apparent that contradictory results can be obtained using different laser systems (see below).
We previously utilized a nanosecond (ns) pulsed green laser to induce damage in the cell nucleus, which enabled for the first time the cytological visualization of the damage site targeting of DSB factors that do not form large IRIF, such as NHEJ factors and the sister chromatid cohesion complex cohesin (38). Similar NHEJ factor recruitment was reported using a NIR laser (33). The physiological relevance of these results was supported by the similar results obtained by the endonuclease-ChIP method (16,39–41). However, when an UVA laser was used, these factors were detected only with a higher dose of UVA without DNA sensitization, but not with a low dose UVA with DNA sensitization (25). It was further found that DAPI staining and the DSB response factor 53BP1 recruitment were compromised with the higher dose UVA condition (25), while the green laser-induced damage displayed intact DAPI staining and a robust 53BP1 response (38,42). These results indicated differences of DNA damage induced by different laser systems and prompted us to perform a parallel comparison of laser systems for their proper use and comparison of their results. Here we describe the analysis of 337 nm UVA (with and without BrdU), 532 nm green with two different pulse durations [ns and picosecond (ps)], and femtosecond (fs) 800 nm NIR lasers in terms of the types of DNA damage, corresponding factor recruitment and γH2AX threshold parameters. We also compared SSB/DSB factor recruitment and γH2AX threshold energy for a continuous-wave (cw) 405 nm blue laser with and without BrdU. The similarities and unique characteristics of the laser parameters and damage induced by the different laser systems are compared and the different physical mechanisms of damage production are discussed. These studies should provide an important framework for other investigators who use laser-induced damage to study the DNA repair process.
MATERIALS AND METHODS
Laser systems—the nitrogen (N2) nanosecond (ns) 337 nm (UVA) laser
The N2 laser beam (pulse width: 4 ns, repetition rate: 6 Hz, 337 nm, Spectra physics, Inc., Mountain View, CA), linearly polarized using a polarizer, was relayed to the back aperture of the microscope objective (40×, NA = 1.3) via the epi-fluorescence port of the Zeiss inverted microscope (Axiovert 135) by use of a 1:1 telescopic lens system. An analyzer was mounted on a motorized rotational stage so as to enable control of pulse energy at the focused spot. For fluorescence imaging, light from a mercury lamp was collimated and coupled to the microscope by use of an external dichroic mirror that reflects the UV laser beam but transmits the visible region of the mercury spectrum. The excitation filter was placed in a filter wheel outside the microscope and the dichroic mirror in the filter cube was used to reflect the fluorescence excitation light, the 337 nm laser beam. This dichroic mirror transmitted the emitted fluorescence as well as the transmitted halogen light through the sample. The emission filter blocked the 337nm laser beam. The transmission of the laser beam power at the object plane for the Zeiss UV transmitting Plan-Neofluar 40×/1.3 NA objective was measured to be ∼0.4 using the dual objective method (43). The laser beam was scanned using a XY-scanning mirror introduced in the beam path before the telescopic lens system so as to create a line pattern across each nucleus. The focused laser-scanned area was estimated by multiplying the length of the line by the width (i.e. diffraction-limited spot size = 1.22 λ/NA).
Continuous wave (cw) Blue (405 nm) Diode laser
The cw diode laser (405 nm) pre-coupled to the Olympus confocal microscope (FV 1000), was focused via a 60× (1.2 NA) microscope objective to a diffraction-limited spot size. The power of the laser beam (at the sample plane) was kept at 1.87 mW at the highest (100%) possible throughput. The laser beam was repeatedly scanned at a scanning rate of 40 µs/pixel so as to create a line pattern inside the nucleus. A total power of 1500 nW was used in 1 line scan (20 pixels).
Nd: YAG ns 532 nm (green) laser
The ns Nd: YAG laser (532 nm, repetition rate: 10 Hz, 6 ns; Quantronix-Continuum Lasers, La Mesa, CA) beam was expanded and coupled to the side port of an inverted microscope (Olympus) via mirrors. The beam was focused via a 100× (1.3 NA) microscope objective to a diffraction-limited spot size. The energy of the laser pulse (at the sample plane) was controlled via a change in the current of the pumping source and/or by use of neutral density filters. A green cut-off filter was used to block the 532 nm laser beam from reaching the CCD camera. The sample stage was repeatedly scanned for 2 min with the scanning rate of ∼10 µm/s so as to create a line pattern inside the nucleus.
Nd: YVO4 picosecond (ps) 532 nm (green) laser
The ps Nd: YVO4 laser (532 nm, repetition rate: 76 MHz, 12 ps; Vanguard Laser System, Spectra-Physics, Inc., Mountain View, CA) was expanded and relayed to the back aperture of the microscope objective (the Zeiss Plan-Apochromat, 63×, NA = 1.4) via the epi-fluorescence port of the Zeiss inverted microscope (Axiovert 200). The transmission factor of this objective was measured to be 0.68 using the dual objective method (43). The pulse energy at the focused spot was controlled by the orientation of the Glan-Thompson polarizer (mounted on a motorized rotational stage). The exposure time at a focused spot was controlled by use of a computer-controlled mechanical shutter (Uniblitz). The scanning pattern in the nucleus was generated by the scanning mirror, controlled by in-house developed software on a LabView platform and National Instrument's data acquisition and control board. A green cut-off filter was used to block the 532 nm laser beam from reaching the CCD camera.
Ti: sapphire femtosecond (fs) 800 nm [near infrared (NIR)] laser
The NIR Ti: sapphire laser (∼200 fs, 800 nm, repetition rate: 76 MHz, Coherent Inc., Santa Clara, CA) beam was expanded and coupled to the side port of a Zeiss inverted microscope (Axiovert 200M) via use of mirrors. A XY-scanning mirror was introduced in the beam path to enable scanning of the beam. The fs beam was focused via a 63× (1.4 NA) microscope objective to a diffraction-limited spot. For 800 nm, the transmission factor of the Zeiss Plan-Apochromat 63×/1.4 NA objective was measured to be 0.6, using the dual objective method. The pulse energy at the focused spot was varied by a control on the orientation of the Glan-Thompson polarizer (mounted on a motorized rotational stage). The scanning mirror was controlled by in-house developed software on a LabView platform to create a line pattern across the nucleus. The exposure time at a focused spot (i.e. the number of femtosecond pulses at a focused spot) was controlled by use of a computer-controlled mechanical shutter (Uniblitz).
Laser energy/power measurement
The laser pulse energy (or power) at the object focal plane was determined by measuring the input energy at the back aperture of the objective multiplied by the transmission factor of the objective at that particular wavelength. For the green ps, ns and UV lasers, beam power at the back aperture of the objective was monitored with a power meter/detector (Model S 120 UV, Thorlabs, USA). For the fs NIR laser, a PowerMax power meter (Model PM3, Coherent, USA) was used. In cases where the size of the beam was larger than that of the back aperture of the objective, an aperture (with a diameter the same as that of the microscope objective back aperture) was placed before the detector for power measurement.
Antibodies
Mouse antibodies specific for pyrimidine-pyrimidone (6–4) photoproducts (6-4PP) and Cyclobutane pyrimidine dimer (CPD) (Kamiya Biomedical), 8-oxoguanine (8-oxoG) (Trevigen, Inc.), PARP-1 (Biomol Research Laboratories Inc.), XRCC1 and FEN1 (Gene Tex, Inc.), Ku70 (Novus Biologicals), γH2AX (Upstate, Charlottesville, VA), and phosphor-SMC1 (S957P) (kindly provided by Dr Michael Kastan) were used. Rabbit antibody specific for 53BP1 (Santa Cruz Biotechnology, Inc.) and goat antibody specific for SA2 (Bethyl Laboratories, Inc.) were also used. Donkey anti-mouse IgG and anti-goat IgG conjugated with AlexaFlour 488 and goat anti-rabbit IgG conjugated with AlexaFlour 546 were from Molecular Probes (Invitrogen). Cy3 conjugated goat anti-rabbit IgG was from Jackson ImmunoResearch Laboratories (West Grove, PA).
Cell culture and fixation
HeLa cells were cultured and synchronized as previously described (32). The cells were washed with phosphate-buffered saline (PBS), fixed for 10 min with 4% paraformaldehyde on ice, permeabilized with CSK buffer (10 mM Pipes, pH 7.0, 100 mM NaCl, 300 mM sucrose, and 3 mM MgCl2), 0.5% Triton X-100 for 5 min at 4°C. For PCNA staining, cells were fixed with methanol at −20°C. For CPD, 6-4PP and 8-oxoG staining, following permeabilization, the coverslips were incubated in 2 N HCl for 10 min at 37°C to denature DNA, with three subsequent washes with PBS.
Immunofluorescent staining and image analysis
The staining procedure was described previously (32). Fluorescent images were captured through a 100× Ph3 UPlanFI oil objective (NA 1.3; Olympus) on a microscope (Model IX81; Olympus) with a CCD camera. The experiments were repeated at least three times and each time six to seven cells were examined, which showed consistent results. Furthermore, damaged cells were fixed at different time points [from immediate (2–5 min) to 1 h post irradiation] to ensure that the different results in factor recruitment were not due to different kinetics of recruitment (data not shown). Initially, cell cycle specificity was determined using G1 and S/G2 phase cells (32,38) (data not shown). Since cohesin recruitment is S/G2-specific (32), cohesin staining was performed in the cells synchronized and damaged at S/G2 phase. Recruitment of NER, BER/SSB factors, Ku, and phosphorylation of SMC1 are not cell cycle-restricted, and the experiments were performed in asynchronous cells (38,44–46). S phase synchronization was done by double-thymidine block as previously described (47). G1 phase cells were individually identified by following the mitotic cells as described (38,42).
RESULTS
Different degrees of UV damage are induced by different lasers
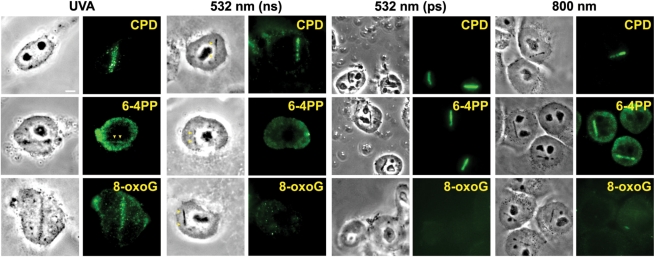
Previously, we used the ns green laser to induce damage in the nucleus and confirmed the recruitment of DSB repair factors, such as the Mre11 complex containing Rad50 and Nbs1, as well as ATM, Ku, BRCA1, PARP1 and γH2AX (32,38). In order to test whether other types of DNA damage are also induced resulting in activation of corresponding repair pathways, damaged cells were stained with antibodies specific for thymine dimers (UV damage), specifically CPD and 6-4PP (Figure 1). The results of the ns green laser system were compared to other lasers, including ns UVA [high-dose relative to the dose required for DSB induction in the presence of BrdU (see Table 1)] without sensitization, fs NIR, and ps green laser systems. The results indicate that ns UVA, ps green, and fs NIR induced both CPD and 6-4PP while ns green generated some CPD, but very little 6-4PP (Figure 1). However, we failed to detect any significant recruitment of NER factors, such as XPC, at green (both ns and ps) and NIR-induced damage sites (data not shown).
Figure 1.
Induction of different types of DNA damage by UVA, ns and ps green, and NIR lasers. At 3–5 min after damage induction by the different lasers indicated at the top, cells were fixed and stained with antibodies specific for CPD, 6-4PP and 8-oxoG. Corresponding brightfield phase contrast images are also shown. Scale bar = 5 µm.
Table 1.
Comparison of threshold energies and specific parameters of five different laser microirradiation conditions necessary for induction of H2AX phosphorylation
| Parameters | UVA/BrDU | UVA | Blue/BrDU | Blue | ns green | ps green | fs NIR |
|---|---|---|---|---|---|---|---|
| Wavelength | 337 nm | 337 nm | 405 nm | 405 nm | 532 nm | 532 nm | 800 nm |
| Pulse width | 4 ns | 4 ns | CW (40 μs/pixel) | CW (40 μs/pixel) | 6 ns | 12 ps | 200 fs |
| Energy (or power)/pulse | 0.004 μJ | 0.04 μJ | 75 nW/pixel | 75 nW/pixel | 0.032 μJ | 0.044 nJ | 0.47 nJ |
| Repetition rate | 6 Hz | 6 Hz | – | – | 10 Hz | 76 MHz, 30 ms | 76 MHz, 10 ms |
| M.O. parameters | 40×/1.3NA | 40×/1.3NA | 60×/1.2NA | 60×/1.2NA | 100×/1.3NA | 63×/1.4NA | 63×/1.4NA |
| Diffraction limited spot size | 316 nm | 316 nm | 412 nm | 412 nm | 499 nm | 464 nm | 697 nm |
| Peak irradiance (W/cm2) | 0.13 × 1010 | 0.13 × 1011 | 0.56 × 102 | 0.56 × 102 | 0.27 × 1010 | 0.22 × 1010 | 0.61 × 1012 |
| Number of pulses/spot | 1 | 1 | 5 | 50 | 85 | 2280000 | 760000 |
| Total energy (or power) delivered = energy (or power)/pulse × no. of pulses per spot × no. of spots in the line (7 μm) | 0.004 μJ × 1 × 20 = 0.08 μJ | 0.04 μJ × 1 × 20 = 0.8μJ | 75 nW × 5 × 20 = 7500 nW | 75 nW × 50 × 20 = 75000 nW | 0.032 μJ × 85 × 14 = 38 μJ | 0.044 nJ × 2280000 × 24 = 2407.7 μJ | 0.47 nJ × 760000 × 12 = 4286 μJ |
SSBs are induced by different lasers, but base damage is induced only by high-dose UVA
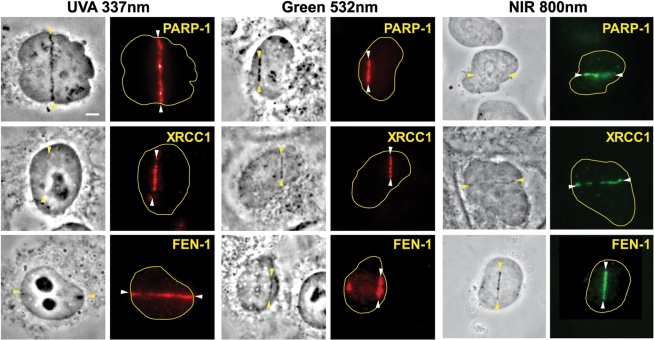
Factors involved in single-strand break (SSB) repair are recruited to the damage sites immediately following damage induction. Those include PARP-1, XRCC1, and FEN-1 (Figure 2). Similar results were reported previously with 365 nm ns UVA (XRCC1, PARP-1, and PCNA), green (PARP-1) and 750 nm fs NIR (PCNA) (32,35,48). Taken together, the results indicate that the process of SSB repair can be studied using ns UVA, ps and ns green, and fs NIR lasers. It should be noted, however, that no significant recruitment of these SSB repair factors were detected at the damage sites induced by the low-dose cw 405 nm laser with BrdU sensitization (Supplementary Figure S2).
Figure 2.
Recruitment of SSB repair and BER factors to the damage sites induced by UVA, green and NIR lasers. Immediately following the damage induction, cells were fixed and stained with antibodies specific for PARP-1, XRCC1 or FEN-1. The location of the induced lesions are indicated by arrowheads. Corresponding brightfield phase contrast images are also shown. Scale bar = 5 µm.
Since the factors described above are also involved in BER, we tested for the presence of base damage in cells damaged by the three different lasers. Interestingly, base damage as represented by 8-oxoG was observed only with high-dose UVA, but not with either the green (both ns and ps) or NIR lasers (Figure 1). Consistent with the presence of base damage, GFP-tagged DNA glycosylases NTH and NEIL2 were recruited to the damage sites, suggesting that base damage generated by the UVA laser is properly recognized (Supplementary Figure 1). The presence of 8-oxoG and DNA glycosylases at the damage sites was also observed previously with a 365 nm UVA laser (48). Therefore, the results indicate that the UVA laser is uniquely suited to study BER pathways in vivo.
The green and NIR lasers are suitable for the detection of DSB factors that do not form IRIF
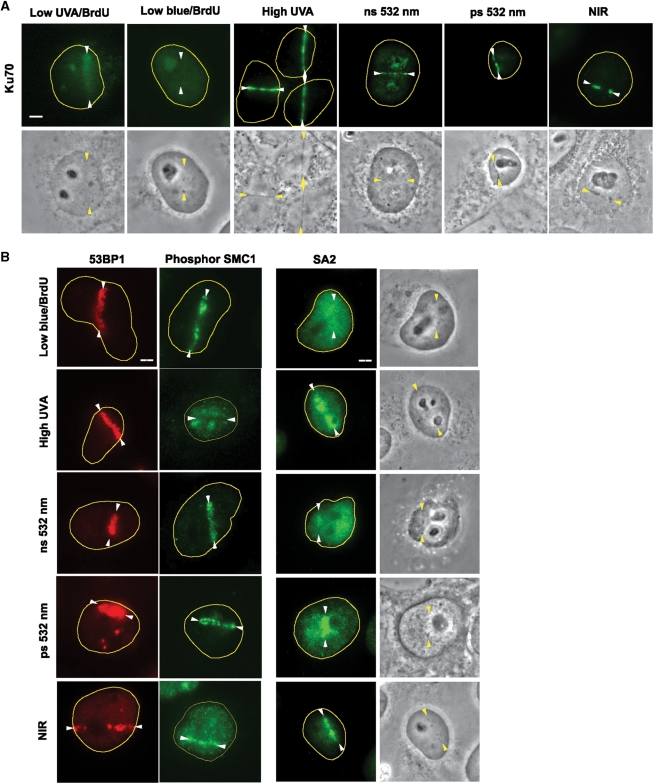
In a previous report, the low-dose UVA laser system, which requires pre-sensitization of DNA with halogenated nucleotide analogs such as BrdU, failed to produce damage that allows visualization of NHEJ factor recruitment, such as Ku (25). Using the low-dose UVA (0.04 μJ/pulse) laser system with BrdU, Ku recruitment was somewhat diffuse but faintly detectable, and no significant Ku signal was detected at the low-dose cw blue laser system with BrdU (Figure 3A). This is in contrast to the highly localized accumulation of Ku at the damage sites using the high-dose (0.27 μJ/pulse) UVA without sensitization, the 532 nm, or the NIR laser systems (Figure 3A) (25,32,33,37,38). NuMA, an abundant nuclear factor, failed to cluster to the damage sites, further supporting the specificity of the observed repair factor recruitment (Supplementary Figure S3). Although high-dose UVA was reported to cause impaired assembly of 53BP1 (25), we found that the high-dose UVA (without BrdU), green and NIR lasers all induced clear recruitment of 53BP1 to the damage sites (Figure 3B). Thus, depending on slight differences in the conditions, the same UVA laser can induce strong or impaired 53BP1 recruitment. This emphasizes the need to clearly define the laser parameters employed.
Figure 3.
DSB responses induced by different laser systems. (A) Ku recruitment to the laser-induced damage sites. Immunostaining of cells damaged by different lasers as indicated using anti-Ku antibody. Lesions are indicated by arrowheads. Corresponding brightfield phase contrast images are also shown. Scale bar = 5 µm. (B) 53BP1 accumulation, hSMC1 phosphorylation and cohesin accumulation at the damage sites. Cells damaged by different lasers as indicated were fixed and stained with antibodies specific for 53BP1, phosphorylated hSMC1 (S966P), or the non-SMC cohesin subunit SA2. Lesions are indicated by arrowheads. Corresponding brightfield phase contrast images for SA2 staining are also shown. Scale bar = 5 µm.
Using the ns green laser, we previously reported the specific recruitment of cohesin to the damage sites, which was later confirmed by ChIP analysis of endonuclease-induced DSB sites (16,32,40,41). Interestingly, although cohesin recruitment was observed with all four laser systems (UVA without BrdU, ns and ps green, and fs NIR), phosphorylation of hSMC1 is impaired at the damage sites induced by high-dose UVA, but not by green and NIR lasers (Figure 3B). The impairment of hSMC1 phosphorylation with the high-dose UVA laser, but not with the green and NIR lasers, suggests that damage induced by high-dose UVA is distinct from damage induced by green and NIR. This is also consistent with the fact that UVA, but not green or NIR, uniquely induces base damage (Figure 1). Taken together with previous results by us and others (25,32,33,38), green and NIR lasers are equally suitable for cytological observation of the recruitment of factors that do not form typical IRIF, while UVA without sensitization appears to cause aberrant DSB responses.
Comparison of γH2AX threshold parameters
To further compare the efficiency of DSB induction, specific parameters and threshold energies necessary for the induction of γH2AX as a function of DSB generation were determined (Table 1). While threshold energy per pulse (0.04 μJ) required for induction of γH2AX is comparable between UVA without BrdU and ns green, the required energy per pulse was much less with NIR and ps green (∼0.4 nJ and 0.044 nJ, respectively). This difference was mitigated by a significantly higher number of pulses per focal spot exposure for the NIR and ps green systems (760 000 pulses per focal spot exposure for NIR and 2.28 million pulses per focal spot exposure for ps green) as opposed to 1 pulse and 85 pulses per focal spot exposure required to induce γH2AX by UVA and ns green, respectively. The requirement for more pulses of the ns green laser compared to UVA may be in part due to the lower peak irradiance of ns green compared to UVA (Table 1).
The total threshold energy per focused laser-scanned area delivered to induce γH2AX was significantly higher with NIR and ps green (∼5000 and 3000 times higher than UVA alone, respectively), whereas ns green was about 50 times higher than UVA alone. In the presence of BrdU, ten-fold less energy was required to induce γH2AX compared to UVA alone, indicating the high efficiency of DSB induction by pre-sensitization of DNA. Similarly, the threshold power to induce γH2AX for cw blue (405 nm) in the presence of BrdU was 375 nW/pixel (five scans of 75 nW/pixel at a power density of 0.56 × 10 W/cm2), similar to the pulsed UVA irradiation, while a 10-fold higher power (3750 nW/pixel) was required to induce γH2AX without BrdU sensitization. However, γH2AX, which can spread to neighboring chromatin regions, may not necessarily be proportional to the number of DSBs induced. The weaker recruitment of Ku and cohesin with low UVA/BrdU and low cw blue/BrdU (Figure 3A) (25) indicates that the number of DSBs induced by UVA with DNA pre-sensitization is significantly lower than that seen with UVA alone, ns and ps green, and NIR.
Since the peak irradiance depends on the amount of energy in the pulse duration, the relative contribution of the various physical mechanisms underlying the induction of (and type of) DNA damage are also expected to depend on the pulse duration in addition to the wavelength. Consistent with this notion, an ∼60-fold higher dose (total energy delivered) was required to elicit a threshold γH2AX signal with ps green compared to ns green, indicating that the pulse duration affects the efficiency (amount of DSB per total energy) of DSB induction (Table 1). This raises the possibility that the mechanism of damage induction may be different between ns and ps green lasers (see ‘Discussion’ section).
Taken together, the results reveal that the efficiency of DSB induction is significantly different between different laser systems, indicative of different mechanisms by which damage is induced.
Comparison of laser systems
We next compared the actual parameters used for our studies and others reported in the literature (Table 2). Our UVA system is comparable to the condition used by Lan et al. for their 365 nm UVA laser that had similar pulse width, repetition rate, focused spot size and peak irradiance (Table 2; [5] and [6]) (48). For UVA with BrdU, an even smaller energy per pulse and overall dose was reported by Mortusewicz et al. (Table 2; [1] and [2]) (49). However, the transmission of the UV transmitting Zeiss Plan-Neofluar 63×/1.25 numerical aperture objective was described as 0.1 in their report, which is in contrast to ∼0.4 measured for the Zeiss UV transmitting Plan-Neofluar 40×/1.3 NA objective using the dual-objective method (43) in our system. This discrepancy may have led to significant under-estimation of the dosimetric values reported in their study (49). For IdU or BrdU sensitized cells, Lukas et al. (23,25) used the similar 337 nm wavelength UVA but required ∼3–4-fold higher total energy and energy per pulse than ours (Table 2; [1] and [3]). This may be because of the larger focused spot size which led to lower peak irradiance in their system. The large spot size is due not only to their use of a low NA objective, but is also due to the non-Gaussian profile of the UVA laser beam in their PALM microdissection system (23). The actual spot size in the equivalent system was measured to be ∼4.2 μm (50), which is six times larger than the diffraction limited spot size. In contrast, nearly diffraction limited spot size (as measured by lesion created on an air-dried red blood cell) was achieved in our system using spatial frequency filtering of the laser beam by use of a pinhole placed in the beam path (Table 2; [1] and [5]). Induction of damage by sensitization with DNA-binding dyes, such as Hoechst, utilized a dye laser that was pumped with a N2 laser to yield a UVA wavelength of 390 nm (Table 2; [4]) (13,29). In this case, the laser pulse energy per dose was significantly higher than those used with our 337 nm UVA system with BrdU (Table 2; [2]).
Table 2.
Comparison of the actual microirradiation parameters used for our studies and others [1] our UVA/BrdU, [2] (49), [3] (23,25), [4] (13,29), [5] our UVA, [6] (48), [7] our ns green (38), [8] (34), [9] our ps green, [10] (33), [11] our NIR
| Parameters | [1] | [2] | [3] | [4] | [5] | [6] | [7] | [8] | [9] | [10] | [11] |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ns N2 UVA/ BrDU | ns N2 UVA/BrDU | ns N2 UVA/ BrdU or IdU | ns N2-dye UVA/Hoechst | ns N2 UVA | ns N2-dye UVA | ns Nd:YAG green | ns Nd:YAG green | ps Nd:YVO4 green | fs Ti: sapphire NIR | fs Ti: sapphire NIR | |
| Wavelength | 337 nm | 337 nm | 337 nm | 390 nm | 337 nm | 365 nm | 532 nm | 532 nm | 532 nm | 800 nm | 800 nm |
| Pulse width | 4 ns | 4 ns | 3 ns | 4 ns | 4 ns | 4 ns | 6 ns | 7 ns | 12 ps | 200 fs | 200 fs |
| Energy/pulse | 0.04 μJ | 0.008 μJa | 0.15 μJb | 0.25 μJ | 0.27 μJ | 0.20 μJ | 0.32 μJ | 0.4 μJ | 0.19 nJc | 0.13 nJ | 0.72 nJ |
| Repetition rate | 6 Hz | 6 Hz | 30 Hz | ∼10 Hz | 6 Hz | 10 Hz | 10 Hz | 10 Hz | 76 MHz, 30 ms | 76 MHz | 76 MHz, 10 ms |
| M.O. parameters | 40×/1.3NA | 63×/1.25NA | 40×/0.6NA | 63×/1.4NA | 40×/1.3NA | 63×/1.4NA | 100×/1.3NA | 100×/1.3NA | 63×/1.4 NA | 63×/1.4 NA | 63×/1.4NA |
| Diffraction limited spot size | 316 nm | 329 nm | 685 nmd | 340 nm | 316 nm | 318 nm | 499 nm | 499 nm | 464 nm | 697 nm | 697 nm |
| Peak irradiance (W/cm2) | 0.13 × 1011 | 0.24 × 1010a | 0.14 × 1011 | 0.69 × 1011 | 0.89 × 1011 | 0.66 × 1011 | 0.27 × 1011 | 0.29 × 1011 | 0.95 × 1010c | 0.17 × 1012 | 0.93 × 1012 |
| Number of pulses/spot | 1 | 1 | 1 | 1 | 1 | 1 | 85 | 10 | 2280000 | ∼502769 (86/13 = 6.6 ms) | 760000 |
| Focused laser-scanned area (μm2) | 2.2 | 3.1 | 4.8 | ∼2.4 | 2.2 | ∼2.2 | 3.5 | ∼3.5 | 3.5 | 4.9 | 4.9 |
| Total energy delivered (μJ) = energy/pulse × no. of pulses per spot × no. of spots in the area | 0.04 μJ × 1 × 20 = 0.8 | 0.008 μJ × 30 = 0.24a | 0.15 μJ × 15 = 2.25b | ∼0.25 μJ × 1 × 14 = 3.5 | 0.27 μJ × 1 × 20 = 5.4 | ∼0.20 μJ × 1 × 20 = 4.0 | 0.32 μJ × 85 × 14 = 380 | 0.4 μJ × 10 × 14 = 56 | 0.19 nJ × 2280000 × 24 = 10397c | 0.13 nJ × 502769 × 13 = 849.68 | 0.72 nJ × 760000 × 12 = 6566 |
Our cw 405 nm blue laser system and irradiation conditions (Table 1) are comparable to a previous study by Lan et al. (51). While the study by Lan et al. required 100 scans (1600 nW/scan) to produce double-strand breaks (detected by an antibody against γH2AX), our threshold was 50 scans (1500 nW/scan) with a power density of 0.56 × 102 W/cm2 which is much smaller than that of the peak power densities used for UVA, green and NIR (Table 2). However, it may be noted that the average power used for ns UVA laser is 0.24 μW (0.04 μJ × 6Hz), which is much smaller than that of the 405 nm laser (1.87 mW). The threshold for γH2AX detection was found to be lower (30–40 scans) if cells were fixed and examined at a later time point (1 h post-irradiation). BrdU pre-treatment led to a significantly reduced threshold (five scans of 75 nW/pixel) by the 405 nm laser, which is similar to that used by Lan et al. (10 scans). The difference between our threshold and that of Lan et al. may be attributed to differences in the fluorescence detectors and parameters (such as gain, sensitivity, exposure time, excitation intensity, etc.) though a similar laser microscope system was used for irradiation.
Mikhailov et al. also used the ns green laser with the pulse width, repetition rate, pulse energy and focusing optics similar to ours (34). Their study had almost the same irradiance (Table 2; [7] and [8]) but less number of pulses (10) per spot to create DNA damage. Therefore, the range of laser energy doses for creating DNA damage by the ns green laser can vary from 50–400 μJ depending on the irradiation mode (e.g. rate of scanning of the stage).
The peak irradiance for our NIR laser (Table 2; [11]) is the highest of all the lasers used. The number of pulses per spot (760 000) and the total energy dose utilized in our system are higher than those used by Mari et al. (33) (Table 2; [10]). In our protocol, the laser beam was on for a 10 ms macropulse (which contained 760 000 200 fs micropulses) and then moved to the next position by a scanning mirror with a delay of ∼100 ms and then switched on again for 10 ms. In contrast, Mari et al. continuously scanned the region of interest with a laser that delivered a 76 MHz beam containing 200 fs micropulses. In the case of our system a significant opportunity for photothermal relaxation exists in between each macropulse that was not present in the study conducted by Mari et al. This may account for the dosimetry differences observed between our study and theirs.
DISCUSSION
Our studies reveal that generation of different degrees of DSB, SSB, crosslinking, and base damage are dictated by different wavelengths, laser energies, pulse frequencies and durations and irradiances, indicating the involvement of different damage mechanisms.
UV crosslinking damage
All three wavelengths induce CPD to a certain extent, but 6-4PP was only seen with the UVA, NIR, and ps green lasers. Surprisingly, the ns green laser, having similar peak irradiance, did not induce detectable 6-4PP. This suggests that cumulative effects (due to high repetition rate) are required for 6-4PP production in the case of the green laser beam. Recently, a similar study assaying both CPD and 6-4PP was performed comparing low-dose UVA (continuous and not pulsed 405 nm diode laser) with Hoechst, fs NIR and UVC (37). Although recombinant NER factors fused to GFP (i.e. GFP-XPC and GFP-XPA) were shown to be detectable at the damage sites induced by the first two lasers, the recruitment was less efficient compared to the UVC laser-induced damage (37). Very little recruitment of endogenous XPC was detected under our conditions (Table 2; [5]), while Lan et al. detected XPC, but not XPA, at the damage sites using UVA (Table 2; [6]) (48). Taken together, the results indicate that unlike the conventional UVC-induced damage, no efficient activation of the NER pathway is associated with damage induced by ns UVA, ns and ps green, and fs NIR despite the presence of detectable crosslinking damage. This may be because the amount of, or possibly the quality of, UV damage induced by these lasers is insufficient to trigger a significant NER repair response compared to the highly effective induction of SSB and DSB responses.
Base damage induction by the UVA laser
Free radicals and reactive oxygen species (ROS) can be formed by various photochemical processes via linear or two-photon absorption. It was shown previously that oxidative DNA base modifications are induced in the wavelength range between 290–470 nm (52). While direct DNA absorption occurs in the UVB range (290–315 nm), an indirect mechanism involving endogenous photosensitizers was suggested for the higher wavelengths (52). Importantly, the induction of base damage appears to drastically decrease above 470 nm (52). This is consistent with our observation that significant base damage is induced by UVA, but not by green or NIR lasers. Efficient recruitment of DNA glycosylases further support the notion that base damage induced by UVA is properly recognized by its processing enzymes for subsequent BER/SSB repair. Importantly, the results indicate that induction of a significant amount of base damage and abnormal DSB responses by UVA (Figures 1 and 3) (25) is wavelength-specific and cannot be attributed simply to the laser dose (i.e. energy delivered).
Detection of non-IRIF forming DSB factors using green and NIR lasers
Ku recruitment to the damage sites is more clearly detectable with the high-dose UVA, green and NIR lasers than with low-dose UVA with BrdU. This can be explained by the differences in the density of DSBs induced by low-dose UVA with BrdU versus others. In the former case, the occurrence of DSBs depends on the efficiency of the incorporation of the sensitizing agents into both strands of DNA in close proximity (otherwise resulting in SSBs). In contrast, Ku recruitment was clearly observed using continuous wave (cw) UVA (405 nm) irradiation in Hoechst-stained cells (37). Hoechst non-specifically binds to the minor groove of DNA and may induce higher number of DSBs in the irradiated area. Despite the large number of DSBs that efficiently recruit Ku, however, we failed to detect any measurable DSBs by the in situ comet assay (53) following ns green laser damage induction, suggesting that damage is clustered to such a small area, without scattering, that it is below the detection sensitivity of the assay (data not shown). The cell viability and normal checkpoint responses in these cells also indicate that highly clustered DSBs, despite the large number, appear to have a less deleterious effect than widely scattered damage. Thus, the biological effect of radiation dose and the number of DSBs are not necessarily comparable between laser microirradiation and conventional nucleus-wide ionizing radiation.
Studies by us and others using the ns green and NIR lasers demonstrated that the factors involved in HR repair, such as Rad51, RPA and Rad54, are recruited to the damage sites even in the G1 phase of the cell cycle (37,38). This is in contrast to clear S/G2-specific accumulation of Rad51 and RPA reported with low-dose UVA with BrdU, which is more similar to the conventional IRIF formation following ionizing radiation (24,25). This can also be explained by the different densities of DSBs at the irradiated regions. It is possible that a high concentration of DSBs induced by green and NIR lasers results in the strong initial recruitment that may negate the necessity to form IRIF, which is secondary to the initial recruitment and requires distinct steps and factors (14). Therefore, low-dose UVA with sensitization is suitable for the analysis of cell cycle-specific IRIF formation but not for detection of non-IRIF forming factors. In contrast, the green and NIR lasers are better suited for detection of the initial recruitment of DSB factors regardless of IRIF formation, but may not be useful for studying the canonical IRIF response.
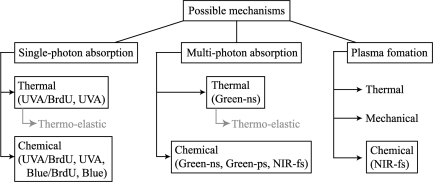
Physical alteration mechanisms
In general, four potential mechanisms for the production of damage to the target structure can be considered: (i) temperature rise produced by linear or two-photon absorption; (ii) generation of large thermo-elastic stresses; (iii) various photochemical processes by linear or two-photon absorption including crosslinking damage and production of free radicals and ROS; and (iv) optical breakdown (plasma formation) produced by a combination of multiphoton and cascade ionization processes, leading to thermal, mechanical and chemical damage (Figure 4).
Figure 4.
Mechanisms of DNA damage by different laser microbeam systems. Three possible damage mechanisms (single-photon absorption, multi-photon absorption, and plasma formation) and their associated thermal, chemical and mechanical effects are listed. Based on the γH2AX-threshold and working laser parameters (e.g. wavelength, pulse duration and frequency, peak irradiance and total input energy), the most likely mechanisms of damage induction by UVA, UVA with BrdU, cw blue, cw blue with BrdU, ns and ps green, and fs NIR are indicated. Although the thermal effect is most likely an important mechanism of damage induction, it is highly restricted temporally and spatially, and is dispersed instantaneously, and therefore any heat-inflicted damage outside of the focused area or any temperature rise in the cell is not expected to occur under the conditions used. Further study is necessary to delineate which mechanisms and/or combination of mechanisms impact each of the laser and biological systems discussed.
Temperature rise produced by linear or two-photon absorption
For a particular wavelength and incident radiant exposure (I0), the temperature rise produced by linear absorption can be calculated by a knowledge of absorption coefficient (μa) at the laser wavelength (λ), density (ρ) and specific heat (c) of the chromosome using the expression ΔT = μaI0/ρc. For example, using the measured absorbance values (54) at 337 nm, μa of chromosome is estimated to be ∼184/cm, ρ is ∼0.38 g/cm3 (55) and c is estimated as ∼1.5 J/g K (56). Therefore, the threshold radiant exposure (51 J/cm2 for 0.04 μJ energy/pulse) will lead to a transient temperature rise of ∼16 463 K by a single 4 ns pulse of UV laser beam (337 nm). This can lead to breakage of bonds and thermal denaturation of DNA, which is found to occur at temperatures higher than 340 K (57). Since the absorption coefficient of BrdU stained chromosomes will be much higher at 337 nm, lower energy per pulse is sufficient to generate a similar transient temperature rise. However, it is important to note that the temperature rise is proportional to the incident radiant exposure and if the spot size at the sample plane is not exactly diffraction limited (as is the case for the nitrogen UVA lasers with non-Gaussian profile), the incident radiant exposure and therefore actual temperature rise can be an order of magnitude smaller. Furthermore, this high temperature will be dissipated over time and space. Thermal diffusion time gives a measure of the confinement of the temperature. For our UVA laser microbeam parameters (λ = 337 nm, NA = 1.3), the thermal diffusion time (Td) was calculated to be ∼50 ns, using the formula: Td = 0.124 λ2/κ (NA)2. Where, thermal diffusivity of water (κ) is taken to be 1.4 × 10−7 m2/s (58). Since the pulse width of the laser beam (4 ns) is much less than the theoretically calculated thermal diffusion time, photo-thermal confinement of the nanosecond UVA laser occurs. The same is true for all of the other pulsed laser beams used in this study, namely ns green, ps green and fs NIR. However, this temperature rise decays inside the chromosome at the center of the focused spot without affecting neighboring regions.
The time-dependent temperature change, after absorption of a pulse by a chromosome (assumed as a thin slab of thickness d) can be approximated by ΔT(t) = ΔT0[1 – exp(–d2/4κt)]. For a 1 µm thick chromosome, only 0.001% of initial temperature-rise (16 463 K) produced by a 4 ns UVA pulse, i.e. 0.16 K will be retained before the delivery of the second pulse (at 6 Hz). This is insignificant for any cumulative temperature rise by the ns UVA laser, especially due to the fact that only one pulse/spot was delivered. This will also hold true for the ns green laser beam operating at 10 Hz. However, the cumulative effects can be important for the high repetition rate (76 MHz) laser beams (ps green and fs NIR). In these cases, cumulative temperature rise may be produced by subsequent pulses, i.e. it is possible that the initial laser pulse alters the absorption properties of the target so that subsequent pulses result in an increase in single photon as well as two-photon absorption leading to subsequent damage to the DNA material. However, evolution of the temperature distribution below optical breakdown threshold (58) shows that only moderate heat accumulation occurs during cell surgery using a high repetition rate (76 MHz) NIR fs laser beam focused by a high NA microscope objective. Calculations based on the given parameters indicate that temperature rise due to single-photon absorption at 337 nm with thermal confinement is dominant for the ns UVA laser with or without sensitization. Similarly, temperature rise due to two-photon absorption at 532 nm with thermal confinement contributes to the ns and ps green laser induced DNA damage (Figure 4).
Generation of large thermo-elastic stresses
Generation of large thermo-elastic stresses by the pulsed laser microbeams can lead to damage to the DNA. Even though the temperature rises due to the ps green and fs NIR lasers are small, the ultra short nature of the pulses may lead to thermo-elastic stresses. The stress relaxation factor (τm), is defined as the ratio of the laser pulse duration (tp) to the time necessary for the stress wave to travel through the heated structure (thickness/speed of sound, i.e. d/ca), provides a measure of the significance of thermoelastic stress (τm = tpca/d). Thus, τm is calculated to be 10.4 (ns UVA), 15.6 (ns green), 0.0312 (ps green) and 0.0005 (fs NIR), using ca of 2600 m/s (59). A value of τm < 1 implies that the laser pulse duration is shorter than the time required for dissipation of thermal stress. Therefore, while thermo-elastic stress would not be very high for the ns UVA and ns green laser, the stress is confined to the focused spot and therefore its magnitudes can be large for ps green and fs NIR laser. However, thermo-elastic tensile stress of only ∼0.014 MPa is estimated to be produced by an individual fs pulse (58), which is too small to cause the DNA damage observed in our study. Furthermore, the threshold irradiance required for thermo-elastic stress confinement for the NIR fs beam has been estimated to be ∼5.0 × 1012 W/cm2 (58), which is five times higher than our threshold (Table 1) and working parameters (Table 2). Thus, for the ns UVA and ns green laser, no stress accumulation leading to DNA damage will occur. For the ps green and fs NIR laser, in spite of satisfaction of the stress confinement criterion, thermo-elastic stresses generated may not be sufficient for induction of DNA damage.
Photochemical processes by linear or two-photon absorption leading to oxidative and crosslinking damage
Free radicals and ROS can be formed by various photochemical processes via linear or two-photon absorption. While UVC and UVB induce damage by direct excitation of DNA, visible light is known to induce one-photon damage indirectly via photochemical reactions in endogenous photosensitizers (60). Furthermore, the 532 nm green ns and ps lasers can produce UV (266 nm) effects by two-photon absorption (61). Similarly, DNA damage using the fs NIR is likely facilitated via nonlinear absorption by DNA through a lower (two or three)-order multi-photon process (35). In fact, the threshold irradiance for our NIR fs beam was higher than the required threshold for photochemical damage previously estimated (i.e. ∼0.26 × 1012 W/cm2) (58). However, since the multi-photon cross-section of cellular chromophores is significantly lower than the cross-section for single-photon excitation (62), the probability of direct photochemical changes by the UVA laser leading to DNA damage is much higher as compared to the green and NIR wavelengths. This is consistent with the lack of base damage induction by green and NIR lasers.
Optical breakdown (plasma formation) leading to thermal, mechanical and chemical damage
Though a three-photon interaction by the NIR fs beam can provide the energy required for excitation (and subsequent DNA damage), the irradiance required for damage will also generate a low-density plasma (free-electrons) which may cause DNA damage. Furthermore, thermal, and mechanical processes can evolve from plasma formation (Figure 4). These may be a combination of multiphoton and cascade ionization processes (58). The threshold irradiance required for low-density plasma formation (also referred to as ‘optical breakdown’) for the NIR fs beam has been estimated to be ∼6.0 × 1012 W/cm2 (58), which is much higher than both the threshold (Table 1) and working (Table 2) parameters used in our studies. The free-electrons generated in the focal volume by plasma formation can induce chemical changes in the micro-irradiated region. Since ∼106 free electrons/pulse are generated (58) in the focal volume at the irradiance that causes 11.8°C/pulse (i.e. peak temperature of 100°C after a series of pulses), free-electron induced chemical changes will dominate any thermal denaturation process of biomolecules such as DNA and/or proteins. Since the required threshold for thermo-elastic stress confinement and/or optical breakdown has been determined to be an order of magnitude higher (58), the primary mechanism of damage by the fs NIR laser in our studies is postulated to be photochemical in nature, where free electrons (low-level plasma) participate in chemical decomposition (bond breaking) along with multi-photon induced chemistry. Indeed, Boudaïffa et al. showed that in contrast to high energy photons, very low energy electron irradiation can induce substantial damage in DNA (SSBs and DSBs) by resonant formation of DNA strand breaking even at electron energies well below the ionization limit of DNA (7.5–10 eV) (63). Thus, for the ps green and fs NIR lasers, there is growing evidence that low-density plasmas can be reliably produced at pulse energies below plasma threshold by multiphoton ionization and therefore photo-damage often involves higher-order photochemical effects or even ionization. Therefore, the irradiance dependence of the observed DNA-damage by the ps green and fs NIR lasers is expected to be of greater slope than that of multiphoton excitation as observed for other photodamage events (64).
For the shorter laser wavelengths (337 nm UVA and both ns and ps 532 nm green) the free-electron density changes slowly with irradiance and therefore, identification of irradiance range for chemical, thermal, and mechanical effects is easier than for NIR wavelengths. Furthermore, these shorter wavelengths provide better spatial resolution (spot size) than the NIR wavelengths (Table 2). The threshold for optical breakdown for the ns green laser is ∼0.8 × 1011 W/cm2 (65), which is higher than our threshold (Table 1) and working (Table 2) values. Similarly, for the ps green laser, the threshold for optical breakdown is ∼0.6 × 1012 W/cm2 (58), which is higher than our threshold and working values. Although the plasma threshold irradiance values are an order of magnitude higher than those used to damage the DNA in this study, the observed DSBs may still be caused by the plasmas for the following reasons. First, the threshold irradiance decreases significantly with increase in NA. Second, the presence of linear absorption (at least for the UVA and green wavelengths) can reduce the threshold irradiance (measured for transparent media) for plasma formation by an order of magnitude (66).
Taken together, based on the laser parameters used, we predict that the prominent mechanism(s) affecting the DNA are (i) for the ns UVA laser: single photon absorption leading to temperature rise and chemical changes, (ii) for the cw 405 nm laser: single photon absorption leading to chemical changes, (iii) for the ns green laser: two-photon absorption leading to thermal and chemical changes, (iv) for the ps green laser: cumulative two-photon absorption effects leading to chemical changes and (v) for the fs NIR laser, photochemical processes emanating from multi-photon processes and low-level plasma formation (Figure 4). It would also be interesting to closely compare the kinetics of each factor's recruitment to see if they are differentially affected by the various damage mechanisms.
In conclusion, our studies revealed varying degrees of DSB, SSB, UV, and base damage generated by UVA with and without BrdU, ns and ps green and NIR lasers. The most efficient DNA damage effects are observed for UVA irradiation in terms of the total energy required for the threshold γH2AX, which are even better with BrdU sensitization since BrdU has an absorption maximum near the laser wavelength used (337 nm). However, careful measurement and comparison of laser parameters indicate the involvement of distinct mechanisms of damage which are dictated by different wavelengths, laser energies, pulse frequency and durations, and irradiances. In order to compare multiple studies, it is imperative that investigators fully describe the laser parameters and the methods used. These are often missing from published papers. Although some DSB factor recruitment and modifications can be observed at the high-dose UVA-induced damage sites, the presence of significant base damage and inconsistent DSB responses distinguish high-dose UVA without sensitization from other laser systems. While a low density of DNA damage induced by UVA with DNA sensitization makes it uniquely suitable for studying IRIF formation, both green and NIR lasers enable efficient detection of the initial recruitment of IRIF-forming factors as well as factors that do not form IRIF and thus are difficult to detect cytologically following conventional DNA damaging methods. Taking advantage of these different laser systems will no doubt further facilitate the molecular understanding of in vivo DNA damage recognition and responses in mammalian cells.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health Laser Microbeam and Medical Program (RR01192); the Air Force Office of Scientific Research (FA9550-04-1-0101); Beckman Laser Institute Inc. Foundation (to M.W.B.); National Institutes of Health (CA100710); Department of Defense Breast Cancer Research Program (DAMD17-03-1-0436 to K.Y.); National Institutes of Health predoctoral fellowship (MBRS-GM-55246 to V.G.-G.). Funding for open access charge: Air Force Office of Scientific Research (FA9550-04-1-0101).
Conflict of interest statement. None declared
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Akira Yasui for comments and providing GFP-Nei12 and GFP-NTH expression plasmids. We also thank Dr Alexander Ball for critical reading of the manuscript.
REFERENCES
- 1.Lehmann AR. DNA repair-deficient diseases, xeroderma pigmentosum, Cockayne syndrome and trichothiodystrophy. Biochemie. 2003;85:1101–1111. doi: 10.1016/j.biochi.2003.09.010. [DOI] [PubMed] [Google Scholar]
- 2.Shiloh Y. ATM and related protein kinases: safeguarding genome integrity. Nat. Rev. Cancer. 2003;3:155–168. doi: 10.1038/nrc1011. [DOI] [PubMed] [Google Scholar]
- 3.Taylor AM. Chromosome instability syndromes. Best Pract. Res. Clin. Haematol. 2001;2001:631–644. doi: 10.1053/beha.2001.0158. [DOI] [PubMed] [Google Scholar]
- 4.Mazin AV, Bornarth CJ, Solinger JA, Heyer WD, Kowalczykowski SC. Rad54 protein is targeted to pairing loci by the Rad51 nucleoprotein filament. Mol. Cell. 2000;6:583–592. doi: 10.1016/s1097-2765(00)00057-5. [DOI] [PubMed] [Google Scholar]
- 5.Paull TT, Gellert M. The 3′ to 5′ exonuclease activity of Mre11 facilitates repair of DNA double-strand breaks. Mol. Cell. 1998;1:969–979. doi: 10.1016/s1097-2765(00)80097-0. [DOI] [PubMed] [Google Scholar]
- 6.Baumann P, Benson FE, West SC. Human Rad51 protein promotes ATP-dependent homologous pairing and strand transfer reactions in vitro. Cell. 1996;87:757–766. doi: 10.1016/s0092-8674(00)81394-x. [DOI] [PubMed] [Google Scholar]
- 7.Baumann P, West SC. DNA end-joining catalyzed by human cell-free extracts. Proc. Natl Acad. Sci. USA. 1998;95:14066–14070. doi: 10.1073/pnas.95.24.14066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Petukhova G, Sung P, Klein H. Promotion of Rad51-dependent D-loop formation by yeast recombination factor Rdh54/Tid1. Genes Dev. 2000;14:2206–2215. doi: 10.1101/gad.826100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kubota Y, Nash RA, Klungland A, Schär P, Barnes DE, Lindahl T. Reconstitution of DNA base excision-repair with purified human proteins: interaction between DNA polymerase beta and the XRCC1 protein. EMBO J. 1996;15:6662–6670. [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Z, Wu X, Friedberg EC. Nucleotide-excision repair of DNA in cell-free extracts of the yeast Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 1993;90:4907–4911. doi: 10.1073/pnas.90.11.4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wood RD, Robins P, Lindahl T. Complementation of the xeroderma pigmentosum DNA repair defect in cell-free extracts. Cell. 1988;53:97–106. doi: 10.1016/0092-8674(88)90491-6. [DOI] [PubMed] [Google Scholar]
- 12.Moné MJ, Volker M, Nikaido O, Mullenders LH, van Zeeland AA, Verschure PJ, Manders EM, van Driel R. Local UV-induced DNA damage in cell nuclei results in local transcription inhibition. EMBO J. 2001;2:1013–1017. doi: 10.1093/embo-reports/kve224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paull TT, Rogakou EP, Yamazaki V, Kirchgessner CU, Gellert M, Bonner WM. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr. Biol. 2000;10:886–895. doi: 10.1016/s0960-9822(00)00610-2. [DOI] [PubMed] [Google Scholar]
- 14.Celeste A, Fernandez-Capetillo O, Kruhlak MJ, Pilch DR, Staudt DW, Lee A, Bonner RF, Bonner WM, Nussenzweig A. Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nat. Cell Biol. 2003;5:675–679. doi: 10.1038/ncb1004. [DOI] [PubMed] [Google Scholar]
- 15.Berkovich E, Monnat RJJ, Kastan MB. Roles of ATM and NBS1 in chromatin structure modulation and DNA double-strand break repair. Nat. Cell Biol. 2007;9:683–690. doi: 10.1038/ncb1599. [DOI] [PubMed] [Google Scholar]
- 16.Potts PR, Porteus MH, Yu H. Human SMC5/6 complex promotes sister chromatid homologous recombination by recruiting the SMC1/3 cohesin complex to double-strand breaks. EMBO J. 2006;25:3377–3388. doi: 10.1038/sj.emboj.7601218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sugawara N, Haber JE. Repair of DNA double strand breaks: in vivo biochemistry. Methods Enzymol. 2006;408:416–429. doi: 10.1016/S0076-6879(06)08026-8. [DOI] [PubMed] [Google Scholar]
- 18.Martin SG, Laroche T, Suka N, Grunstein M, Gasser SM. Relocalization of telomeric Ku and SIR proteins in response to DNA strand breaks in yeast. Cell. 1999;97:621–633. doi: 10.1016/s0092-8674(00)80773-4. [DOI] [PubMed] [Google Scholar]
- 19.Barlow JH, Lisby M, Rothstein R. Differential regulation of the cellular response to DNA double-strand breaks in G1. Mol. Cell. 2008;30:73–85. doi: 10.1016/j.molcel.2008.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rogakou EP, Boon C, Redon C, Bonner WM. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J. Cell Biol. 1999;146:905–915. doi: 10.1083/jcb.146.5.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Limoli CL, Ward JF. A new method for introducing double-strand breaks into cellular DNA. Radiat. Res. 1993;134:160–169. [PubMed] [Google Scholar]
- 22.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 23.Lukas C, Falck J, Bartkova J, Bartek J, Lukas J. Distinct spatiotemporal dynamics of mammalian checkpoint regulators induced by DNA damage. Nat. Cell Biol. 2003;5:255–260. doi: 10.1038/ncb945. [DOI] [PubMed] [Google Scholar]
- 24.Tashiro S, Walter J, Shinohara A, Kamada N, Cremer T. Rad51 accumulation at sites of DNA damage and in postreplicative chromatin. J. Cell Biol. 2000;150:283–291. doi: 10.1083/jcb.150.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bekker-Jensen S, Lukas C, Kitagawa R, Melander F, Kastan MB, Bartek J, Lukas J. Spatial organization of the mammalian genome surveillance machinery in response to DNA strand breaks. J. Cell Biol. 2006;173:195–206. doi: 10.1083/jcb.200510130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C, Lukas J. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell. 2007;131:887–900. doi: 10.1016/j.cell.2007.09.040. [DOI] [PubMed] [Google Scholar]
- 27.Sartori AA, Lukas C, Coates J, Mistrik M, Fu S, Bartek J, Baer R, Lukas J, Jackson SP. Human CtIP promotes DNA end resection. Nature. 2007;450:509–514. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ziv Y, Bielopolski D, Galanty Y, Lukas C, Taya Y, Schultz DC, Lukas J, Bekker-Jensen S, Bartek J, Shiloh Y. Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM- and KAP-1 dependent pathway. Nat. Cell Biol. 2006;8:870–876. doi: 10.1038/ncb1446. [DOI] [PubMed] [Google Scholar]
- 29.Bradshaw PS, Stavropoulos DJ, Meyn MS. Human telomeric protein TRF2 associates with genomic double-strand breaks as an early response to DNA damage. Nat. Genet. 2005;37:193–197. doi: 10.1038/ng1506. [DOI] [PubMed] [Google Scholar]
- 30.Walter J, Cremer T, Miyagawa K, Tashiro S. A new system for laser-UVA-microirradiation of living cells. J. Microsc. 2003;209:71–75. doi: 10.1046/j.1365-2818.2003.01117.x. [DOI] [PubMed] [Google Scholar]
- 31.Gomez-Godinez V, Wakida NM, Dvornikov AS, Yokomori K, Berns MW. Recruitment of DNA damage recognition and repair pathway proteins following near-IR femtosecond laser irradiation of cells. J. Biomed. Optics. 2007;12:020505-. doi: 10.1117/1.2717684. 020501-020503. [DOI] [PubMed] [Google Scholar]
- 32.Kim J-S, Krasieva TB, LaMorte VJ, Taylor AMR, Yokomori K. Specific recruitment of human cohesin to laser-induced DNA damage. J. Biol. Chem. 2002;277:45149–45153. doi: 10.1074/jbc.M209123200. [DOI] [PubMed] [Google Scholar]
- 33.Mari PO, Florea BI, Persengiev SP, Verkaik NS, Brüggenwirth HT, Modesti M, Giglia-Mari G, Bezstarosti K, Demmers JA, Luider TM, et al. Dynamic assembly of end-joining complexes requires interaction between Ku70/80 and XRCC4. Proc. Natl Acad. Sci. USA. 2006;103:18597–18602. doi: 10.1073/pnas.0609061103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mikhailov A, Cole RW, Rieder CL. DNA damage during mitosis in human cells delays the metaphase/anaphase transition via the spindle-assembly checkpoint. Curr. Biol. 2002;12:1797–1806. doi: 10.1016/s0960-9822(02)01226-5. [DOI] [PubMed] [Google Scholar]
- 35.Meldrum RA, Botchway SW, Wharton CW, Hirst GJ. Nanoscale spatial induction of ultraviolet photoproducts in cellular DNA by three-photon near-infrared absorption. EMBO Rep. 2003;12:1144–1149. doi: 10.1038/sj.embor.7400028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen BP, Chan DW, Kobayashi J, Burma S, Asaithamby A, Morotomi-Yano K, Botvinick E, Qin J, Chen DJ. Cell cycle dependence of DNA-dependent protein kinase phosphorylation in response to DNA double strand breaks. J. Biol. Chem. 2005;280:14709–14715. doi: 10.1074/jbc.M408827200. [DOI] [PubMed] [Google Scholar]
- 37.Dinant C, de Jager M, Essers J, van Cappellen WA, Kanaar R, Houtsmuller AB, Vermeulen W. Activation of multiple DNA repair pathways by sub-nuclear damage induction methods. J. Cell Sci. USA. 2007;120:2731–2740. doi: 10.1242/jcs.004523. [DOI] [PubMed] [Google Scholar]
- 38.Kim J-S, Krasieva TB, Kurumizaka H, Chen DJ, Taylor AM, Yokomori K. Independent and sequential recruitment of NHEJ and HR factors to DNA damage sites in mammalian cells. J. Cell Biol. 2005;170:341–347. doi: 10.1083/jcb.200411083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Y, Hefferin ML, Chen L, Shim EY, Tseng HM, Kwon Y, Sung P, Lee SE, Tomkinson AE. Role of Dnl4-Lif1 in nonhomologous end-joining repair complex assembly and suppression of homologous recombination. Nat. Struct. Mol. Biol. 2007;14:639–646. doi: 10.1038/nsmb1261. [DOI] [PubMed] [Google Scholar]
- 40.Ström L, Lindroos HB, Shirahige K, Sjögren C. Postreplicative recruitment of cohesin to double-strand breaks is required for DNA repair. Mol. Cell. 2004;16:1003–1015. doi: 10.1016/j.molcel.2004.11.026. [DOI] [PubMed] [Google Scholar]
- 41.Ünal E, Arbel-Eden A, Sattler U, Shroff R, Lichten M, Haber JE, Koshland D. DNA damage response pathway uses histone modification to assemble a double-strand break-specific cohesin domain. Mol. Cell. 2004;16:991–1002. doi: 10.1016/j.molcel.2004.11.027. [DOI] [PubMed] [Google Scholar]
- 42.Kim J-S, Heale JT, Zeng W, Kong X, Ball AR, Jr, Yokomori K. In situ analysis of DNA damage response and repair using laser microirradiation. Meth. Cell Biol. 2007;82:377–407. doi: 10.1016/S0091-679X(06)82013-3. [DOI] [PubMed] [Google Scholar]
- 43.Misawa H, Koshioka M, Sasaki K, Kitamura N, Masuhara H. Three-dimensional optical trapping and laser ablation of a single polymer latex particle in water. J. Appl. Phys. 1991;70:3829–3835. [Google Scholar]
- 44.Kitagawa R, Bakkenist CJ, McKinnon PJ, Kastan MB. Phosphorylation of SMC1 is a critical downstream event in the ATM–NBS1–BRCA1 pathway. Genes Dev. 2004;18:1423–1438. doi: 10.1101/gad.1200304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Okano S, Lan L, Caldecott KW, Mori T, Yasui A. Spatial and temporal cellular responses to single-strand breaks in human cells. Mol. Cell Biol. 2003;23:3974–3981. doi: 10.1128/MCB.23.11.3974-3981.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Volker M, Moné MJ, Karmakar P, van Hoffen A, Schul W, Vermeulen W, Hoeijmakers JH, van Driel R, van Zeeland AA, Mullenders LH. Sequential assembly of the nucleotide excision repair factors in vivo. Mol. Cell. 2001;8:213–224. doi: 10.1016/s1097-2765(01)00281-7. [DOI] [PubMed] [Google Scholar]
- 47.Gregson HC, Schmiesing JA, Kim J-S, Kobayashi T, Zhou S, Yokomori K. A potential role for human cohesin in mitotic spindle aster assembly. J. Biol. Chem. 2001;276:47575–47582. doi: 10.1074/jbc.M103364200. [DOI] [PubMed] [Google Scholar]
- 48.Lan L, Nakajima S, Oohata Y, Takao M, Okano S, Masutani M, Wilson SH, Yasui A. In situ analysis of repair processes for oxidative DNA damage in mammalian cells. Proc. Natl Acad. Sci. USA. 2004;101:13738–13743. doi: 10.1073/pnas.0406048101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mortusewicz O, Schermelleh L, Walter J, Cardoso MC, Leonhardt H. Recruitment of DNA methyltransferase I to DNA repair sites. Proc. Natl Acad. Sci. USA. 2005;102:8905–8909. doi: 10.1073/pnas.0501034102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vogel A, Lorenz K, Horneffer V, Hüttmann G, von Smolinski D, Gebert A. Mechanisms of laser-induced dissection and transport of histologic specimens. Biophys. J. 2007;93:4481–4500. doi: 10.1529/biophysj.106.102277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lan L, Nakajima S, Komatsu K, Nussenzweig A, Shimamoto A, Oshima J, Yasui A. Accumulation of Werner protein at DNA double-strand breaks in human cells. J. Cell Sci. 2005;118:4153–4162. doi: 10.1242/jcs.02544. [DOI] [PubMed] [Google Scholar]
- 52.Kielbassa C, Roza L, Epe B. Wavelength dependence of oxidative DNA damage induced by UV and visible light. Carcinogenesis. 1997;18:811–816. doi: 10.1093/carcin/18.4.811. [DOI] [PubMed] [Google Scholar]
- 53.Mohanty SK, Rapp A, Monajembashi S, Gupta PK, Greulich KO. Comet assay measurements of DNA damage in cells by laser microbeams and trapping beams with wavelengths spanning a range of 308 nm to 1064 nm. Radiat. Res. 2002;157:378–385. doi: 10.1667/0033-7587(2002)157[0378:camodd]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 54.Cantor KP, Hearst JE. Isolation and partial characterization of metaphase chromosomes of a mouse ascites tumor. Proc. Natl Acad. Sci. USA. 1966;55:642–649. doi: 10.1073/pnas.55.3.642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bahr GF, Golomb HM. Karyotyping of single human chromosomes from dry mass determined by electron microscopy. Proc. Natl Acad. Sci. USA. 1971;68:726–730. doi: 10.1073/pnas.68.4.726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lepock JR, Frey HE, Heynen ML, Senisterra GA, Warters RL. The nuclear matrix is a thermolabile cellular structure. Cell Stress Chaperones. 2001;6:136–147. doi: 10.1379/1466-1268(2001)006<0136:tnmiat>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Darzynkiewicz Z, Traganos F, Sharpless T, Melamed MR. Thermal denaturation of DNA in situ as studied by acridine orange staining and automated cytofluorometry. Exp. Cell Res. 1975;90:411–428. doi: 10.1016/0014-4827(75)90331-6. [DOI] [PubMed] [Google Scholar]
- 58.Vogel A, Noack J, Hüttman G, Paltauf G. Mechanisms of femtosecond laser nanosurgery of cells and tissues. Appl. Phys. B. 2005;81:1015–1047. [Google Scholar]
- 59.Botvinick EL, Venugopalan V, Shah JV, Liaw LH, Berns MW. Controlled ablation of microtubules using a picosecond laser. Biophys. J. 2004;87:4203–4212. doi: 10.1529/biophysj.104.049528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kielbassal C, Roza L, Epe B. Wavelength dependence of oxidative DNA damage induced by UV and visible light. Carcinogenesis. 1997;18:811–816. doi: 10.1093/carcin/18.4.811. [DOI] [PubMed] [Google Scholar]
- 61.Calmettes PP, Berns MW. Laser-induced multiphoton processes in living cells. Proc. Natl Acad. Sci. USA. 1983;80:7197–7199. doi: 10.1073/pnas.80.23.7197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xu C, Zipfel W, Shear JB, Williams RM, Webb WW. Multiphoton fluorescence excitation: new spectral windows for biological nonlinear microscopy. Proc. Natl Acad. Sci. USA. 1996;93:10763–10768. doi: 10.1073/pnas.93.20.10763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Boudaïffa B, Cloutier P, Hunting D, Huels MA, Sanche L. Resonant formation of DNA strand breaks by low-energy (3 to 20 eV) electrons. Science. 2000;287:1658–1660. doi: 10.1126/science.287.5458.1658. [DOI] [PubMed] [Google Scholar]
- 64.Hopt A, Neher E. Highly nonlinear photodamage in two-photon fluorescence microscopy. Biophys. J. 2001;80:2029–2036. doi: 10.1016/S0006-3495(01)76173-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Venugopalan V, Guerra A, III, Nahen K, Vogel A. Role of laser-induced plasma formation in pulsed cellular microsurgery and micromanipulation. Phys. Rev. Lett. 2002;88:078103. doi: 10.1103/PhysRevLett.88.078103. [DOI] [PubMed] [Google Scholar]
- 66.Oraevsky AA, Jacques SL, Esenaliev RO, Tittel FK. Pulsed laser ablation of soft tissues, gels, and aqueous solutions at temperatures below 100 degrees C. Lasers Surg. Med. 1996;18:231–240. doi: 10.1002/(SICI)1096-9101(1996)18:3<231::AID-LSM3>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.