Abstract
Individual RNA aptamers are often used to modulate the function of their target proteins, and multi-valent aptamers have been constructed to enhance their activity. To expand the utility of aptamers in manipulating and controlling biological processes, here we advance a general method for the design and construction of composite aptamers. The resulting molecular constructs resemble proteins in that they can form specific interactions with three or more different partners and be readily integrated into existing protein regulatory networks. As the first embodiment of this method, we created a tetra-valent aptamer that simultaneously binds to two molecules of the Drosophila protein B52 and two copies of streptavidin, thus mimicking the function of an antibody in immunochemical assays. We demonstrated that the performance of this ‘aptabody’ rivals that of a monoclonal antibody against B52 in these assays. While this study was performed in vitro and the composite aptamer we made was intended to mimic an existing protein, the same method can be used to accommodate arbitrary combinations of individual aptamers in composite molecular contexts, and these constructs can be delivered into living cells, where they are able to utilize existing cellular infrastructure for their production and processing.
INTRODUCTION
Proteins are able to play a predominant role in most biological processes largely because an individual protein molecule can bear multiple specific sites recognized by other molecules, including other proteins, which enables them to assemble into networks or complexes. Novel protein-like reagents that can be readily integrated into existing protein networks or complexes of living cells and organisms are highly desirable in order to understand and control biological processes (1). However, the generation and application of novel proteins is difficult, and alien proteins are usually highly antigenic to an organism. Structured, low-antigenic RNA molecules recapitulating the key features of proteins can be created if we possess two experimental capabilities: (i) the ability to generate ligands to individual target molecules, and (ii) the ability to connect and recombine multiple single-site ligands into a composite molecular entity. The first capability has been realized through the applied in vitro evolution process (SELEX) that generates RNA aptamers (2,3). To attain the second capability, here we explore the possibility of stitching RNA aptamers together with other RNA structural or functional units to form molecules with multiple functional sites, which resemble proteins. This allows aptamer-based molecular constructs to function not only as inhibitors by blocking binding sites on proteins, but also as novel connectors.
The recent development of structural nucleic acid nanotechnology provides many examples of composite DNA and RNA molecules, as well as the general principles for their design and construction (4,5). This approach utilizes well-structured components, combined through affinity and structure to achieve structural predictability with a precision (or resolution) of 1 nm or less in the products. However, only a few portable elements and aptamers are structurally well characterized, which makes it difficult to engineer diverse yet specific interactions. On the other hand, although multivalent aptamers, especially dimeric constructs, have been successfully generated by connecting aptamers either covalently or noncovalently (6–8), including three or more aptamers in a single molecular entity still poses significant technical difficulties. In most cases, when more than one functional unit was to be incorporated into one RNA molecule, each unit was encoded by a single segment and these segments were strung together consecutively. A notable and widely used example is the ‘hybrid RNA’ in the yeast three-hybrid system (9). While this and other early studies clearly demonstrated that multivalent RNAs could be designed such that at least two (sometimes three) domains are simultaneously functional, simple concatenation often results in misfolding of individual domains. Alternatively and more reliably, double-stranded stems can be used as points of integration to assemble multiple RNA components. This strategy has been used successfully to generate combined ribozyme-aptamer molecules to implement Boolean logic operations (10,11).
Our method advanced here is a general and convenient scheme of rational modular design using well-characterized structural elements to connect various aptamers with confirmed secondary structures. In contrast to linear concatenation, we utilize two-dimensional graphs to aid our design. While the three-dimensional structure of the resulting construct may not be precisely predictable, it is relatively easy to make sure that each individual aptamer in the composite is correctly folded and functional. To prove this principle, we constructed a composite RNA aptamer molecule that mimics a particular protein in in vitro assays. For an experimentally tractable and objectively comparable definition of function for a generic protein, we took a ‘behavioral’ approach, i.e. determining whether the non-protein molecule is capable of imitating a given protein's ‘behavior’ under conditions defined by the protein. A non-protein can be considered a mimic of the protein if the non-protein is able to interact with all partners of the protein with comparable affinity and specificity, and does not interact with any non-partner of the protein. Based on this definition, we made composite aptamers that mimic antibodies in immunochemical assays. Previously, a DNA ‘aptabody’ had been reported, which is a homodimer of two thrombin-binding aptamers formed through noncovalent linkage (12). Conceptually different from this DNA molecule, our ‘aptabody’ constructs function as connectors between a specific antigen and a generic straptavidin-conjugated secondary reagent. In the present case, the antigen to be detected is the Drosophila splicing factor B52. We used multiple assays to demonstrate that the RNA aptabodies were able to mimic the function of B52 antibodies and their performance was quantitatively comparable to a monoclonal antibody against B52.
MATERIALS AND METHODS
Proteins and antibodies
Full-length B52 protein was expressed in Sf9 cells from a baculovirius expression system and purified by the standard SR protein purification procedure (13,14). To make a His-tagged B52-RNA recognition motifs (RRMs) construct, a BamHI fragment coding for the SR domain was deleted from the B52 cDNA and the remaining cDNA coding for the two RRMs was inserted into the vector pET-16b (Novagen) to form the plasmid pET-RRMs(B52). The protein was produced in BL21(DE3)plysS host cells. Cleared Escherichia coli lysate was prepared under denaturing condition (8 M Urea), and the protein was refolded on the Ni-NTA Superflow resin (Qiagen) with a step-wise urea gradient (4, 2, 1 and 0 M) before elution. Purified streptavidin and its conjugates (ImmunoPure streptavidin, horseradish peroxidase-conjugated streptavidin and Texas Red-conjugated streptavidin) were purchased from Thermo Scientific-Perce Protein Research Products. The monoclonal antibody against B52, Bv32, was described previously (15). The monoclonal penta-His antibody was purchased from Qiagen.
Aptamer and aptabody constructs
Each DNA template for the homodimers and the monomer derivatives of the streptavidin aptamer S1was made through bidirectional extension of an overlapping pair of oligonucleotide primers purchased from Integrated DNA Technologies. The sequences of these primer pairs (forward and reverse) are listed below. Apt(BBS)2: forward-76mer (5′-GTAATACGACTCAC TATAGGGATCGCCGCGGCTGGT CAACCAGGCGACCGCCGC GGCCACAGCGGTGGGCT GGTCA-3′); reverse-78mer (5′-GAATCCCGAAGGAT CCGGGAACGCTGGTGGGCGG TCGCCTGGTTGACCAGCCCACCGCTG TGGCCGCGGCGGTCGCCT-3′). S1: forward-60mer (5′-GTAATACGACTCACTA TAGGGAGTCGACCGACCAGAA TCATGCAAGTGCGTA AGATAGTC-3′); reverse-60mer (5′-ATGAGTCTAGATGT AGACGCACATAATACGC CCCCGGCCCGC GACTATCTTACGCACTTG-3′). S1-45: forward-45mer (5′-GTAATACGACTCA CTATAGGGACCGACCAG AATCATGCAAGTGCG-3′); reverse-45mer (5′-CCCGGCCCGCGACTAT CTTACGCACTTGCATGA TTCTGGTCGGTC-3′). S1-77: forward-58mer (5′-GTAATACGACTCACTATAGG GCCATCGATGCGGCCGCCGAC CAGAATCATGCAAGTGC-3′); reverse-58mer (5′-GGCCATCGATGCGGCCG CCGACCCGCGACTA TCTTACGCACTTGCATGATTCTGGTCG-3′). Apt(S1)2: forward-80mer (5′-GTAATACGACTCACT ATAGGATCCGTGACCGAC CAGAATCATGCAAGTGCG TAAGATAGTCGCGGGT CGGGTCATACTCC-3′); reverse-80mer (5′-GAATCCGCCTCCCGGCC CGCGACTATCTTA CGCACTTGCATGATTCT GGCCGGGAGTATGACCCGACCCG CGACTATCTT-3′). R-06 dimer: forward-62mer (5′-GTAATACGACTCACTAT AGGATCCGTGACGTCAACAC GGTCCCGGACGTGT TGACGTCCATA-3′); reverse-80mer (5′-GAATCCGCCTCCTCAACACGT CCGGGACCGTGTTGAGGAGTATGGAC GTCAACACGTCCGGGAC CGTGTTGACGTCACGG-3′). TAR dimer: forward-65mer (5′-GTAATACGACTCACTATAG GGATCGCCGCCGAGCCCGGGAGCTC GGCGGCCACAGCGGT GGGAGC-3′); reverse-60mer (5′-GAATCCCGAAGGAT CCGGGAACGCTGGTGGGAGCTCC CGGGCTCCCACCGCTGT GGCCGC-3′).
To form the template for the Aptabody Sa-B52, the templates of Apt(BBS)2 and Apt(S1)2 were digested with BamHI and the longer pieces were ligated. The ligation product with correct composition was amplified using the following two primers: T7forward (5′-GTAATACGACTCACTATAGG-3′) and S1reverse (5′-GAATCCGCCTCCCGGCCCGC-3′). The same method was used to generate the templates for Aptabodies R06-B52 and Sa-TAR: the longer pieces in each set of the BamHI digestion products were ligated pair-wisely, Apt(BBS)2 with the R-06 dimer and the TAR dimer with Apt(S1)2.
All RNA constructs were produced by in vitro transcription from double-stranded DNA templates using the MEGAshortscript T7 kit (Ambion). After transcription, they were extracted with phenol and precipitated with ethanol. Before use in binding assays or immunochemical assays, they were reconstituted in binding buffer for the assay, heated to 70°C for 10 min and chilled on ice.
Binding assays
32P-labeled RNA probes were prepared using the MAXIscript T7 in vitro transcription kit (Ambion) and [α-32P]CTP (GE Healthcare). All binding assays were performed in 20-µl volumes in 1× binding buffer. A typical binding-assay mixture with labeled RNA contained about 20 fmol of 32P-labeled RNA probe and different amounts (usually 1–10 pmol) of protein. For B52, the binding buffer contained 50 mM Tris–Cl (pH 7.6), 200 mM potassium acetate, 5 mM MgCl2 and 2.5 mM dithiothreitol (DTT) (14). For streptavidin, the binding buffer contained 50 mM HEPES (pH 7.4), 10 mM MgCl2 and 100 mM NaCl (16). Electrophoretic mobility shift assay (EMSA) was performed at 4°C. The B52 binding mix was run on a 2.2% agarose gel in ¼ TBE buffer (22.25 mM Tris base, 22.25 mM borate, 0.5 mM EDTA); the streptavidin binding mix was run on a 6% or 7% polyacrylamide gel in TGB buffer (25 mM Tris base, 200 mM glycine). For the binding of the aptabody to both B52 and streptavidin, the binding buffer described above for B52 was used (the S1 aptamer binds streptavidin in this buffer as well as in its original buffer).
Dot-blot analysis with aptabody
Serial dilution of B52 was prepared in 1× Binding Buffer [50 mM Tris (pH 7.6), 50 mM NaCl, 50 mM KCL, 15 mM MgCl2] with 1 mM phenylmethanesulfonylfluoride (PMSF), and 2 mM DTT. Two microliters of each protein sample was spotted directly onto a piece of 50 × 6 mm nitrocellulose filter (Schleicher and Schuell). The filter was incubated for 30 min at room temperature in a plastic bag with 1 ml Blocking Buffer [50 mM Tris (pH 7.6), 75 mM NaCl, 75 mM KCl, 15 mM MgCl2, 100 μg/ml yeast total RNA, 2× Denhardt's solution]. Before adding the aptabody the filter was washed twice in Washing Buffer [50 mM Tris (pH 7.6), 75 mM NaCl, 75 mM KCl, 15 mM MgCl2]. The aptabody was prepared in the Binding Buffer, heated to 70°C for 10 min and put on ice for 5 min. Additional components were added so that the final preparation also contains 1 mM PMSF, 2 mM DTT, 100 μg/ml yeast total RNA and 100 u/ml SUPERase•In (Ambion). This aptabody preparation at the concentration of 10 μg/ml was incubated with the filter for 1 h at room temperature, then the filter was washed four times with the Washing Buffer described above. Horseradish peroxidase-conjugated streptavidin was prepared in the Binding Buffer with 100 μg/ml yeast total RNA and 100 u/ml SUPERase•In to reach the final concentration of 1 μg/ml, and incubated with the filter for one hour at room temperature. Afterwards, the filter was washed four times using the Washing Buffer with 0.5% Tween-20. To visualize the dots the filter was incubated with 2 ml of ECL plus substrate (GE Healthcare) for 3–5 min and exposed to film for 1 min.
Western blot analyses
Electrophoresis and transfer were performed using Mini-PROTEAN 3 Electrophoresis Cell and Mini Trans-Blot Module (Bio-Rad). Western blot with antibodies was performed according to a standard protocol (17). When aptabodies were used, this protocol was modified in the following aspects. A re-naturing step was added after transfer: the blot was incubated at room temperature successively in 6 M, 3 M, 1 M and 0.1 M guanidine-HCl, each for 30 min. The Blocking Buffer was the one described above for dot blot with additional 10% glycerol. Other solutions were identical to those used for dot-blot analysis.
Staining Drosophila polytene chromosomes with aptabody
Salivary gland polytene chromosomes were prepared from the Oregon-R strain according to a protocol described previously (15). The heat shock was performed for 30 min at 36.5°C. The staining protocol was based on that of indirect immunofluorescence (15) with the following modifications. PBS was substituted by the Binding Buffer described for dot blot above. The Blocking Buffer was 2× Denhardt's and 200 μg/ml yeast total RNA in Binding Buffer. The primary antibody was substituted by the aptabody at 200 μg/ml, and the secondary antibody by Texas Red-conjugated streptavidin at 10 μg/ml, both prepared in solution as described above for dot blot. The Washing Buffer was also the one used for dot blot with 0.5% Tween-20.
RESULTS
A general scheme for molecular design of composite aptamers
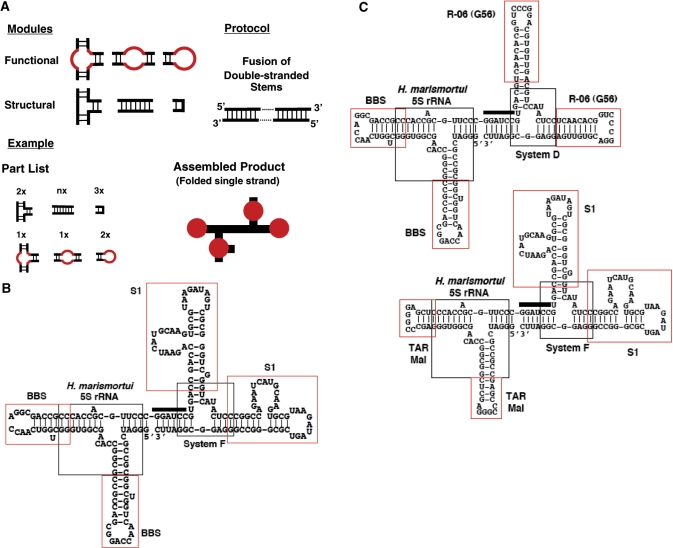
The capability of forming networks requires that protein molecules be able to bear three or more specific binding sites. To recapitulate this feature, we would like to incorporate more than three individual aptamers into a single molecular entity. The major challenge to the design and construction of such multivalent aptamers is to maintain the correctly folded structure of each individual aptamer in the composite molecule. To this end, we have developed a generic method so that multiple aptamers can be joined together combinatorially with the help of additional structural elements. In this method, confirmed secondary structures serve as a starting point for rational modular design. As RNA secondary structure formation causes significantly larger free energy change than that involved in tertiary interactions, the basic properties of the conformational energy landscape of a molecule can be understood at this level (18). As shown in Figure 1A, our method comprises a set of structural and functional modules and a protocol to connect them together. The functional modules, in this case individual RNA aptamers, have known affinities to known targets; the structural modules serve as connectors to link two or more aptamers into a single folded strand.
Figure 1.
Rational modular design of composite RNA aptamers. (A) Depiction of the general scheme. Functional ‘loops’ are combined and organized with the help of structural elements. All elements are connected via double-stranded stems into a putative composite molecule. In the assembled product of the hypothetical example, thick black lines represent double-stranded stems; solid red dots represent individual aptamers. (B) Predicted most stable secondary structure of the Aptabody Sa-B52 by mfold (35). This molecule comprises six parts, including four aptamers and two three-way junctions. Each part is indicated. The thick bar in the middle signifies the sequence corresponding to the BamHI site on the template. (C) Predicted most stable secondary structure of the Aptabodies R06-B52 (upper left) and Sa-TAR (lower right) by mfold, annotated as in (B).
Aptamers can be regarded as functional modules containing functional ‘loops’ in association with different structural elements. Each ‘loop’ is defined by a single function without regard to structure. The three types of functional modules are (Figure 1A, first row from right to left): (a) functional apical ‘loops’ and the associated stem ‘neck’; (b) functional internal ‘loops’ embedded in a stem; and (c) functional internal ‘loops’ constituting the strand-exchange junction of three or more stems. These functional modules can be stitched together with the help of three types of structural modules (Figure 1A, second row form left to right): (a) multibranch junctions with known structures, in particular three-way junctions with known co-axial stacking arrangements of branching stems, are used to organize and present aptamers; (b) stems (usually no longer than one helical turn) consisting of complementary strands that form a double helix, are used like ‘connective tissues’ to adjust local stability and relative orientation between aptamers; and (c) stable small U-turns with known structures, such as hairpins with stable tetra-loops, are used to maintain the continuity of the strand, if necessary. A single protocol of stem connection (Figure 1A, upper right corner) can be applied once or additional times in the process of design, allowing for the combinatorial joining of modules together in a single molecular strand. Once a tentative composite is pieced together using junctions and U-turns, its secondary structure can be predicted using free-energy minimization algorithms (19). If the individual aptamers are predicted to be misfolded, short double-stranded segments can be introduced to fortify local structure until each aptamer is correctly folded in the predicted most stable secondary structure before being tested in binding assays.
We chose to make an ‘aptabody’ as the first embodiment of this scheme because, compared to other proteins, it is relatively easy to define the function of antibodies using standard in vitro immunochemical assays. Although aptamers are often compared to antibodies, the analogy between an aptamer and an (monoclonal) antibody is limited to their specificity and affinity for the target or antigen—an individual aptamer is analogous to a single Fab fragment (20). The aptamer moiety of the DNA ‘aptabody’ for thrombin is only analogous to an F(ab′)2 fragment rather than a full antibody (12). As depicted in Figure 1B, our first RNA aptabody, named Sa-B52, is designed for the detection of the Drosophila B52 protein (15). It is a tetra-valent aptamer in which four aptamers were connected by two three-way junctions: a dimer of a B52 aptamer (14) forms an F(ab′)2 analog, and a dimer of a aptamer for streptavidin (16) forms the ‘Fc-like’ part that would be recognizable by any straptavidin-conjugated secondary reagent. To further demonstrate the ‘plug-and-play’ feature of the method, we also designed two more constructs (Figure 1C). One of them, named ‘R06-B52’, is a di-dimer of the B52 aptamer and an aptamer for the HIV TAR element, R-06 (21,22); the other, named ‘Sa-TAR’, contains two copies of the TAR element and two copies of the streptavidin aptamer. This pair could function like the Aptabody Sa-B52 when they form a complex through the TAR•R-06 interaction.
Construction of an ‘aptabody’
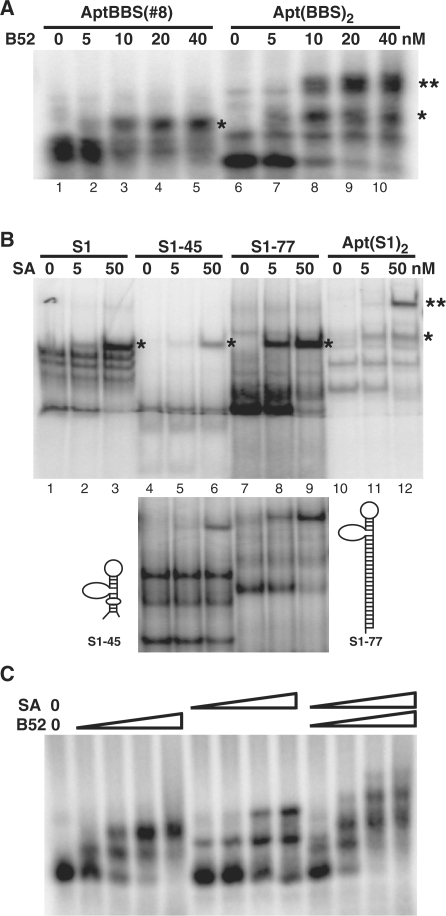
We have previously identified a class of B52 aptamers and through extensive mutational analysis defined a minimized version as a simple stem–loop (14). These aptamers, named BBSs (B52 Binding Sequences), bind to the two RRMs of B52. Our first experiment attempted to make a divalent aptamer and to rigorously demonstrate that each individual aptamer in the dimeric construct is functional and functions independently. We grafted two copies of the minimized B52 aptamer to two stems of the three-way junction of Haloarcula marismortui 5S RNA (23) to form a new construct, Apt(BBS)2. As shown in Figure 2A, we used EMSA to examine the activity of the dimer and found that the aptamer dimer was able to bind two B52-RRMs molecules to produce two shifted bands. When the target was present at low concentration, only one binding site of the dimer was occupied (Figure 2A, lane 7). As the target concentration increases, both binding sites were occupied (Figure 2A, lanes 8–10). In theory, when any macromolecule has two identical binding sites for a ligand, the two sites would appear to have affinities different by a factor of four, with the first site appearing twice as tight as a monomer and the second appearing half as tight, if there is no steric hindrance. Indeed, for our dimeric construct the first binding event showed an apparent affinity higher than the original monomer (Figure 2A, compare lane 3 with lane 8).
Figure 2.
Binding activity of each individual aptamer in a composite molecule. (A) Generating a F(ab′)2-like dimeric aptamer for B52. BBS was the original B52 aptamer [B52 Binding Sequence (14)]. Binding reactions were run on a 2.2% agarose gel. Shifted bands representing single and double occupancy of the aptamer binding sites are indicated by one or two asterisks respectively. (B) Minimization and augmentation of the streptavidin aptamer. S1 was the original full-length streptavidin aptamer (16). In the upper panel all four versions of the aptamer were run on the same polyacrylamide gel (6%, 37.5:1) to obtain better comparison of shifted bands. In the lower panel the same reaction for S1-45 and S1-77 were run on a different gel (7%, 19:1) to obtain better resolution of both shifted and un-shifted bands. The secondary structures of the reduced S1-45 and the augmented S1-77 are sketched on either side of the lower panel. ‘SA’ stands for streptavidin. (C) Confirming the activity of individual aptamers in the di-dimer aptabody. The binding reaction contains increasing concentration of B52, streptavidin, or both, as indicated, ranging from 5 to 50 nM. The gel was 2.2% agarose.
For streptavidin, several aptamers had been published (16). We picked one of these aptamers, designated ‘S1’ in the original publication, for further characterization to define a minimized version that can be incorporated into our construct. Based on the predicted secondary structure of S1, we first deleted 7 nt from the 5′ end and 32 nt from the 3′ end, reducing the 84-mer aptamer to a 45-mer named ‘S1-45’. As shown in Figure 2B, this construct was active, but its affinity to streptavidin seemed to be lower than the original version. The minimized version consists of a stem–loop structure with an apical loop and a side loop. We suspected that it might not be as stably folded as the original. To test this hypothesis, we extended the double-stranded stem at the distal end from the apical loop and corrected an A-C mismatch in the original sequence to form the construct S1-77 (Figure 2B, compare the two sketches of secondary structure). This manipulation not only fully restored the affinity to the original level, but also reduced the formation of alternative structures (as shown by the reduction of protein-indifferent bands generated by S1 and S1-45 in Figure 2B). Next, we designed a dimeric construct Apt(S1)2 according to the same principle used to make Apt(BBS)2. Here the three-way junction, System F, was one previously observed to be extraordinarily stable among 12 similar variants (24). As shown in Figure 2B (lanes 11 and 12), both minimized S1 aptamers were fully functional in this new sequence and structural context; and the apparent affinity of the dimer for first streptavidin is higher than the monomer (Figure 2B, compare lanes 2 and 11) due to increased local concentration of binding sites.
When we designed the sequences for the templates that produced the two dimeric constructs described above, we included one BamHI site near the 3′ end of the Apt(BBS)2 template and another one near the 5′ end of the Apt(S1)2 coding sequence. By digesting these two templates with BamHI and ligating the two longer fragments, we were able to form the template for the Aptabody Sa-B52 depicted in Figure 1. To confirm that each one of the four aptamers in this composite was functional, we performed EMSA for the tetra-valent molecule produced by in vitro transcription to bind B52 and streptavidin either separately or simultaneously. As shown in Figure 2C, the aptabody was able to bind both B52 and streptavidin simultaneously. In addition, this binding assay was also used to measure the relative avidity of these two types of aptamers in the composite. Because in an immunochemical assay the binding of the ‘antigen’ B52 occurs before the binding of streptavidin, the ‘F(ab′)2’ end of the molecule should have higher avidity to B52 than the ‘Fc’ end to streptavidin, thus ensuring the stable formation of B52–aptabody–streptavidin triple complex. We chose the two aptamers, BBS and S1, according to their reported Kd, which were 20 nM and 70 nM, respectively (14,16). As shown in Figure 2C, this difference in their Kd was apparently maintained in the context of the aptabody, as revealed by the relative avidity of the aptabody for B52 and streptavidin (note that at 50 nM B52 binding was saturating but streptavidin binding was not, presumably due to insufficient amount of streptavidin present).
Aptabodies as functional mimics of antibodies in immunochemical assays
There are three broad types of immunochemical assays, in which antigens are presented in different environments. First, in assays like western blot analysis, the antigens are extracted from cells or tissues and immobilized on the surface of solid matrices. Second, to reveal the localization and quantity of antigens, they are fixed in their native but complex cellular context for in situ immunostaining. Third, antigens in solution may be detected by antibody and precipitated out of the aqueous phase. In all three types of assays, the antigen–antibody complex is further recognized by a secondary reagent (usually a secondary antibody) that binds the antibody and bears an additional label or signal for detection or separation. The diversity of antigen display described above often requires different properties of antibodies. A single antibody or its aptabody mimic may not be able to perform equally well in different assays. As aptamers were selected in solution, most commonly in buffers that resemble the physiological condition of the target molecule, it is not guaranteed that they will be functional in other different environments. As our binding assays described in the previous section have already demonstrated the functionality of each individual aptamer (Figure 2A and B) and the simultaneous binding of B52 and streptavidin by the aptabody in solution (Figure 2C), here we focused our tests of the aptabody in several assay formats defined by antibodies, in which B52 is presented in immobilized or fixed forms.
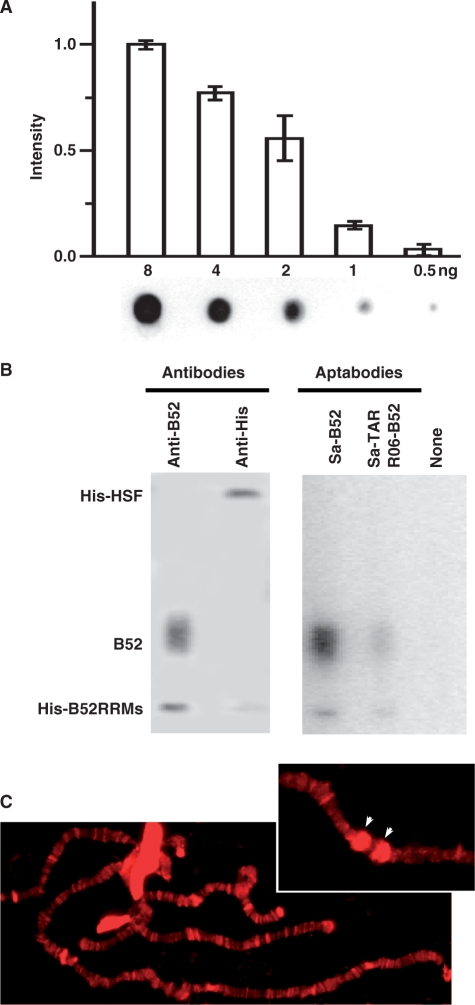
We started our test by asking whether the B52 aptamer in the context of an aptabody were able to recognize B52 on the surface of a membrane, and whether the streptavidin aptamer in this molecule could simultaneously bind horseradish peroxidase (HRP)-conjugated streptavidin. For this purpose, we set up a dot-blot analysis to visualize purified B52 protein spotted directly on nitrocellulose membrane. After testing several different types of blocking buffers and several different concentrations of aptabody, streptavidin and HRP substrate, we established a protocol that is described in detail in the ‘Materials and Methods’ section. As shown in Figure 3A, using this protocol, <5 min of exposure was required to clearly visualize 0.5 ng of B52, and these ‘dots’ would not appear if either B52 or the aptabody was omitted from the protocol (data not shown). In this assay the concentration of aptabody (10 μg/ml, 150 nM) is comparable to the concentration of antibody in a typical western blot assay [1–50 μg/ml (17); the molecular weight of an antibody is roughly twice that of the aptabody]. The HRP-conjugated streptavidin was also used in a concentration similar to that of HRP-conjugated secondary antibody for western blots [1 μg/ml, 10 nM in our protocol compared to 5–0.5 μg/ml (17)]. The sensitivity of this assay was approaching the detection limit of antibodies, which is ∼2 fmol [(17), 0.1 ng for a 50-kDa protein].
Figure 3.
Performance of antibodies in immunochemical assays. (A) Dot-blot analysis. The histogram quantifies three independent assays with the intensity normalized to the highest amount of B52 used (=1.0). Error bars reflect standard deviations. The image of one representative membrane is show under the histogram. (B) Western blot analysis with monoclonal antibodies and aptabodies. The protein sample loaded to each lane was identical. The smear of full-length B52 was possibly caused by heterogeneity of the phosphorylation status in the SR domain. (C) Fluorescence staining of polytene chromosomes. The main panel shows broad distribution of B52 on interbands and puffs. The inset at the upper right corner shows a segment of chromosome 3R after heat treatment, with the cytogenetic loci containing the major heat-shock genes at 87A/C marked by arrowheads.
Compared with the dot blot assay described above, the western blot analysis commonly used in molecular biology is different in two aspects. First, unlike direct spotting, proteins are usually separated on a polyacrylamide gel under denaturing condition before being transferred onto the membrane. Second, while an aptabody is analogous to a monoclonal antibody, in most cases the antibody used for western blot is polyclonal. Therefore, to determine whether an aptabody can faithfully and efficiently mimic the function of an antibody in such an assay, a fair but strict comparison of the performance should be conducted between an aptabody and a monoclonal antibody under the condition defined by the latter. For a western blot with the aptabody, we mixed two different forms of B52 with an unrelated protein, the heat-shock factor (HSF). The B52 protein is composed of two RRMs and an ‘SR domain’ rich in serine and arginine (15). The aptamer binds the two RRMs but not the SR domain (14). The un-tagged full-length B52 used in this assay was purified from Sf9 cells; the RRMs construct was purified from E. coli with a His-tag. The HSF protein also bears a His-tag. Therefore, this trio could be used to compare the aptabody to a monoclonal antibody against B52 (15) as well as a commercially available anti-His antibody. (In a more realistic demonstration of specificity, cell extract or similar samples would have been used, but our un-modified RNA constructs were not designed to withstand RNases that may exist in such samples.)
We compared the performance of antibodies and the Aptabody Sa-B52 in detecting protein samples in the 1–10-ng range. As shown in Figure 3B, both monoclonal antibodies detected their respective antigen and produced signals with comparable intensity. The aptabody detected both forms of B52. Compared to the monoclonal antibody against B52, its signal for the full-length protein was even stronger. While its signal for the RRMs-only construct was weaker, it was stronger than that produced by the anti-His antibody. To further confirm the mechanism of aptabody function, we included in this assay an additional pair of aptabodies—the two constructs depicted in Figure 1C, Aptabodies R06-B52 and Sa-TAR. As shown in the figure, one more lap in the relay from the ‘antigen’ to the signal did not make it fail to detect the ‘antigen’, although the signal is significantly weakened. As in the dot-blot assay, the concentration of aptabodies used here is similar if not identical to the concentration of antibodies.
The B52 protein was originally identified by its unique pattern visualized on the polytene chromosomes of Drosophila salivary glands using its monoclonal antibody in immunofluorescent microscopy (15). After obtaining positive results with the B52 aptabody in western blot analysis, we further tested whether we would be able to detect B52 in situ on these polytene chromosomes. Here we adopted the indirect immunoflurescence protocol by replacing the primary and secondary antibodies with similar amount and concentration of Aptabody Sa-B52 and Texas Red-conjugated streptavidin. The blocking solution was also duly modified in which non-fat milk powder in phosphate buffer was replaced by Denhardt's solution with nonspecific RNA. As shown in Figure 3C, this protocol using aptabody reproduced the banding pattern of B52 distribution formerly generated by B52 monoclonal antibody (15). In particular, we used salivary glands under heat shock to reproduce the characteristic ‘bracketing’ pattern of B52 at the de-condensed heat-shock loci on Chromosome 3R (Figure 3C, upper right corner). When the aptabody was omitted in the protocol, there was no fluorescent signal on the polytene chromosome under either non-heat-shock or heat-shock conditions (data not shown). Taken together, these experiments demonstrated the capability of the aptabody to mimic an antibody in multiple assay formats of immunochemistry.
DISCUSSION
Inside cells, mechanisms bringing two protein molecules together play an important role in cellular regulatory networks (25,26). The same principle can be employed in therapeutic or experimental applications (27,28). For this purpose, we have developed a general method for rational modular design of composite aptamer molecules with multiple valencies and specificities. Individual aptamers have been used like drugs to block protein function (29), and either covalently or noncovalently linked homo-polymeric aptamers have been used to enhance avidity or activate the target (6–8,12,30,31). The utility of aptamers will be dramatically expanded if they can be used as building blocks of molecular interconnectors that create novel connectivity between proteins. As a preliminary study along this line, here we demonstrated in vitro that two unrelated proteins from two different organisms, the splicing factor B52 of Drosophila melanogaster and streptavidin from Streptomyces avidinii, can be bridged by a constructed RNA molecule derived from aptamers for these proteins. This multivalent composite aptamer recapitulated features of antibodies in a single un-modified RNA molecule, and functioned in three standard immunochemical assay formats defined by antibodies. Previously, direct biotin conjugates of aptames were used to detect their target proteins (12,20). But our intention was not to develop a type of reagent that would out-perform the biotinylated aptamers. We chose this simple and even seemingly contrived example in order to allow the underlying idea relevant to RNA structure and function to stand out more clearly.
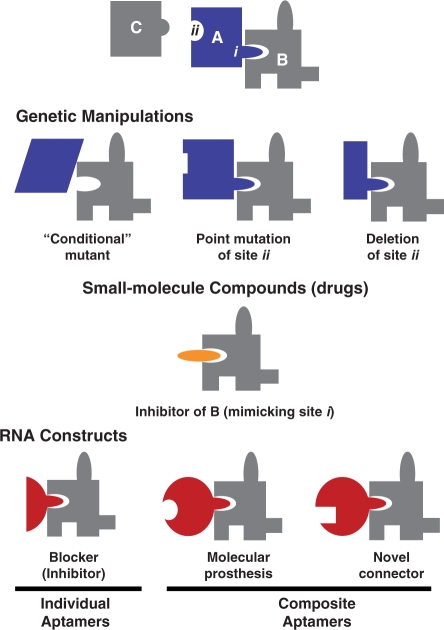
While demonstrating the capability of a composite aptamer to mimic a particular protein under specified conditions is a necessary proof of principle, the utility of this type of constructs is not limited to molecular ‘prostheses’. By splicing together more than one type of RNA aptamer, our method promises to create combinations of binding sites that do not correspond to that of any existing protein. Such ‘protein-like’ molecules can be used to create new connectivity in protein networks. To appreciate the utility of this type of molecular interconnectors, it is helpful to compare them with two commonly used approaches to manipulating and controlling biological processes, as depicted in Figure 4. Here we consider a generic protein ‘A’, whose function as a node in a network is realized through two discrete sites, ‘i’ and ‘ii’. Genetic methods may cause the absence or malfunction of the entire protein (total deletion of the gene or conditional mutants); it may also abolish an individual connection to a protein (deletion or point mutation of site ‘ii’ eliminates its interaction with ‘C’). Small-molecular-weight compounds (drugs), in general, function as inhibitors of protein by blocking active sites. In this case a site ‘i’ mimic inhibits the function of protein ‘B’. Although a few drugs have been developed to induce proximity of two specific proteins (32), there is no general scheme to use genetics or small molecules to create new connectivity between proteins.
Figure 4.
Composite RNA aptamers as functional mimics of protein. A hypothetical protein ‘A’ is used as a generic example, whose function is realized by interaction with proteins ‘B’ and ‘C’ through sites ‘i’ and ‘ii’. RNA aptamers are compared with genetic methods and small molecular compounds for their capability to inhibit, mimic, and modify the function of protein ‘A’ and its partners.
In order to form a network, a ‘protein-like’ molecule should be able to possess at least three specific binding sites: single-site molecules can only form dyadic interactions and double-site molecules can only form linear chains. Moreover, the cellular protein network possesses a scale-free topology and some ‘hub’ proteins interact with many partners through multiple sites (33). Therefore, it is critical for a generic method to provide means to accommodate three or more aptamers in a composite without difficulty. To achieve this goal, we used well-characterized structural elements to organize and present multiple aptamers. A critical feature of our method is the use of three-way junctions. In natural RNAs, three-way junctions occur frequently and often play essential architectural roles. Crystal structures are becoming available for more and more three-way junctions in folded RNAs (23) and general folding rules have been derived for them (34). We incorporate this type of additional structural information into our design at the level of secondary structure to compensate for the lack of information regarding aptamer structure. Our data clearly demonstrated that each of the four aptamers is active in the composite aptabody. The scheme depicted in Figure 1A can be easily executed by a computer program, with further improvements to maximize the base paring, minimize the RNA strand length, and provide alternative arrangements of aptamers in a combinatorial manner. This type of molecule can not only be used in vitro as shown here, but also in vivo. If a ‘protein-like’ composite aptamer is delivered into organisms as a synthetic gene, its production could be regulated temporally and spatially using different promoters (30). In this way a new RNA molecule could be readily integrated into an existing protein network.
FUNDING
A Faculty Research Award Program (FRAP A to H.S.) from the Research Foundation of the State University of New York. Funding for open access charge: College of Arts and Sciences, University at Albany, State University of New York.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
This study was initiated in the laboratory of Dr. J. Lis at Cornell University. His support and encouragement were critical in the early stage of this project. We thank Ms. J. Werner for technical assistance. We are also grateful to Drs. D. Mathews and C. Theimer for their critical comments on the manuscript.
REFERENCES
- 1.Breaker RR. Natural and engineered nucleic acids as tools to explore biology. Nature. 2004;432:838–845. doi: 10.1038/nature03195. [DOI] [PubMed] [Google Scholar]
- 2.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 3.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 4.Seeman NC. At the crossroads of chemistry, biology, and materials: structural DNA nanotechnology. Chem. Biol. 2003;10:1151–1159. doi: 10.1016/j.chembiol.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 5.Jaeger L, Chworos A. The architectonics of programmable RNA and DNA nanostructures. Curr. Opin. Struct. Biol. 2006;16:531–543. doi: 10.1016/j.sbi.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 6.Kim Y, Cao Z, Tan W. Molecular assembly for high-performance bivalent nucleic acid inhibitor. Proc. Natl Acad. Sci. USA. 2008;105:5664–5669. doi: 10.1073/pnas.0711803105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McNamara JO, Kolonias D, Pastor F, Mittler RS, Chen L, Giangrande PH, Sullenger B, Gilboa E. Multivalent 4-1BB binding aptamers costimulate CD8+ T cells and inhibit tumor growth in mice. J. Clin. Invest. 2008;118:376–386. doi: 10.1172/JCI33365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dollins CM, Nair S, Boczkowski D, Lee J, Layzer JM, Gilboa E, Sullenger BA. Assembling OX40 aptamers on a molecular scaffold to create a receptor-activating aptamer. Chem. Biol. 2008;15:675–682. doi: 10.1016/j.chembiol.2008.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.SenGupta DJ, Zhang B, Kraemer B, Pochart P, Fields S, Wickens M. A three-hybrid system to detect RNA-protein interactions in vivo. Proc. Natl Acad. Sci. USA. 1996;93:8496–8501. doi: 10.1073/pnas.93.16.8496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Soukup GA, Breaker RR. Engineering precision RNA molecular switches. Proc. Natl Acad. Sci. USA. 1999;96:3584–3589. doi: 10.1073/pnas.96.7.3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Win MN, Smolke CD. Higher-order cellular information processing with synthetic RNA devices. Science. 2008;322:456–460. doi: 10.1126/science.1160311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hianik T, Porfireva A, Grman I, Evtugyn G. Aptabodies - new type of artificial receptors for detection proteins. Protein Pept. Lett. 2008;15:799–805. doi: 10.2174/092986608785203656. [DOI] [PubMed] [Google Scholar]
- 13.Zahler AM, Lane WS, Stolk JA, Roth MB. SR proteins: a conserved family of pre-mRNA splicing factors. Genes Dev. 1992;6:837–847. doi: 10.1101/gad.6.5.837. [DOI] [PubMed] [Google Scholar]
- 14.Shi H, Hoffman BE, Lis JT. A specific RNA hairpin loop structure binds the RNA recognition motifs of the Drosophila SR protein B52. Mol. Cell Biol. 1997;17:1649–1657. doi: 10.1128/mcb.17.5.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Champlin DT, Frasch M, Saumweber H, Lis JT. Caharacterization of a Drosophila protein associated with boundaries of transcriptionally active chromatin. Genens Dev. 1991;5:1611–1621. doi: 10.1101/gad.5.9.1611. [DOI] [PubMed] [Google Scholar]
- 16.Srisawat C, Engelke DR. Streptavidin aptamers: affinity tags for the study of RNAs and ribonucleoproteins. RNA. 2001;7:632–641. doi: 10.1017/s135583820100245x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harlow E, Lane D. Using Antibodies: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1999. [Google Scholar]
- 18.Tinoco I, Jr, Bustamante C. How RNA folds. J. Mol. Biol. 1999;293:271–281. doi: 10.1006/jmbi.1999.3001. [DOI] [PubMed] [Google Scholar]
- 19.Zuker M. On finding all suboptimal foldings of an RNA molecule. Science. 1989;244:48–52. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]
- 20.Jayasena SD. Aptamers: an emerging class of molecules that rival antibodies in diagnostics. Clin. Chem. 1999;45:1628–1650. [PubMed] [Google Scholar]
- 21.Duconge F, Toulme JJ. In vitro selection identifies key determinants for loop-loop interactions: RNA aptamers selective for the TAR RNA element of HIV-1. RNA. 1999;5:1605–1614. doi: 10.1017/s1355838299991318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van Melckebeke H, Devany M, Di Primo C, Beaurain F, Toulme JJ, Bryce DL, Boisbouvier J. Liquid-crystal NMR structure of HIV TAR RNA bound to its SELEX RNA aptamer reveals the origins of the high stability of the complex. Proc. Natl Acad. Sci. USA. 2008;105:9210–9215. doi: 10.1073/pnas.0712121105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 24.Diamond JM, Turner DH, Mathews DH. Thermodynamics of three-way multibranch loops in RNA. Biochemistry. 2001;40:6971–6981. doi: 10.1021/bi0029548. [DOI] [PubMed] [Google Scholar]
- 25.Austin DJ, Crabtree GR, Schreiber SL. Proximity versus allostery: the role of regulated protein dimerization in biology. Chem. Biol. 1994;1:131–136. doi: 10.1016/1074-5521(94)90002-7. [DOI] [PubMed] [Google Scholar]
- 26.Yik JH, Chen R, Nishimura R, Jennings JL, Link AJ, Zhou Q. Inhibition of P-TEFb (CDK9/Cyclin T) kinase and RNA polymerase II transcription by the coordinated actions of HEXIM1 and 7SK snRNA. Mol. Cell. 2003;12:971–982. doi: 10.1016/s1097-2765(03)00388-5. [DOI] [PubMed] [Google Scholar]
- 27.Crabtree GR, Schreiber SL. Three-part inventions: intracellular signaling and induced proximity. Trends Biochem. Sci. 1996;21:418–422. doi: 10.1016/s0968-0004(96)20027-1. [DOI] [PubMed] [Google Scholar]
- 28.Park SH, Zarrinpar A, Lim WA. Rewiring MAP kinase pathways using alternative scaffold assembly mechanisms. Science. 2003;299:1061–1064. doi: 10.1126/science.1076979. [DOI] [PubMed] [Google Scholar]
- 29.Pendergrast PS, Marsh HN, Grate D, Healy JM, Stanton M. Nucleic acid aptamers for target validation and therapeutic applications. J. Biomol. Tech. 2005;16:224–234. [PMC free article] [PubMed] [Google Scholar]
- 30.Shi H, Hoffman BE, Lis JT. RNA aptamers as effective protein antagonists in a multicellular organism. Proc. Natl Acad. Sci. USA. 1999;96:10033–10038. doi: 10.1073/pnas.96.18.10033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Santulli-Marotto S, Nair SK, Rusconi C, Sullenger B, Gilboa E. Multivalent RNA aptamers that inhibit CTLA-4 and enhance tumor immunity. Cancer Res. 2003;63:7483–7489. [PubMed] [Google Scholar]
- 32.Ho SN, Biggar SR, Spencer DM, Schreiber SL, Crabtree GR. Dimeric ligands define a role for transcriptional activation domains in reinitiation. Nature. 1996;382:822–826. doi: 10.1038/382822a0. [DOI] [PubMed] [Google Scholar]
- 33.Jeong H, Mason SP, Barabasi AL, Oltvai ZN. Lethality and centrality in protein networks. Nature. 2001;411:41–42. doi: 10.1038/35075138. [DOI] [PubMed] [Google Scholar]
- 34.Lescoute A, Westhof E. Topology of three-way junctions in folded RNAs. RNA. 2006;12:83–93. doi: 10.1261/rna.2208106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]