Abstract
Neuronal proteins of the BTB/kelch and PDZ domain families interact with different regions of the cytoplasmic C-terminal domain of the GluR6 kainate receptor subunit. The BTB/kelch protein KRIP6 binds within a 58 amino acid segment of GluR6 proximal to the plasma membrane. In contrast, PDZ domain proteins, such as PICK1 and PSD95, interact with the last 4 residues of the GluR6 C-terminus. KRIP6 reduces peak currents mediated by recombinant GluR6 receptors and by native kainate receptors in neurons, whereas PICK1 stabilizes kainate receptors at synapses. Thus, protein–protein interactions at the C-terminal domain of GluR6 are important for regulating kainate receptor physiology. Here, we show by co-clustering and co-immunoprecipitation that KRIP6 interacts with PICK1 in heterologous cells. In addition, we demonstrate a novel modulation of GluR6 receptors by PICK1 resulting in increased peak current and relative desensitization of GluR6-mediated currents, phenotypes opposite to those produced by KRIP6. Importantly, these effects cancel out when KRIP6 and PICK1 are co-expressed together with GluR6. KRIP6 and PICK1 strongly co-cluster and co-immunoprecipitate regardless of the presence of GluR6. Immunofluorescence analysis reveals that GluR6 can either join the KRIP6–PICK1 clusters or remain separate; however, co-expression of KRIP6 reduces the fraction of PICK1 that co-immunoprecipitates with GluR6. Taken together, these results indicate that, in addition to a previously demonstrated direct interaction with the GluR6 C-terminal domain, KRIP6 regulates kainate receptors by inhibiting PICK1 modulation via competition or a mutual blocking effect.
Keywords: PDZ domain, Desensitization, Hippocampus, Immunofluorescence, Patch-clamp
1. Introduction
The BTB/kelch family comprises a large group of highly conserved proteins involved in diverse cellular functions including cytoskeleton interactions, nuclear transcription, and ubiquitination (Stogios et al., 2005; Prag and Adams, 2003). Recent evidence indicates that two neuronally expressed members of the BTB/kelch family, KRIP6 and actinfilin, directly interact with the C-terminal domain of GluR6, a kainate subtype of mammalian glutamate receptors (Laezza et al., 2007a; Salinas et al., 2006). In heterologous expression systems KRIP6 suppresses peak current density and reduces steady-state desensitization of GluR6-mediated currents (Laezza et al., 2007a), whereas actinfilin promotes GluR6 degradation via a Cul-3 mediated ubiquitination process (Salinas et al., 2006). Interestingly, while KRIP6 controls GluR6 functional properties via its N-terminal BTB domain, actinfilin operates via its C-terminal kelch repeats, suggesting that, although KRIP6 and actinfilin belong to the same group of proteins, their molecular mechanism of action might be substantially different. In hippocampal neurons, overexpression of KRIP6 inhibits kainate receptor-mediated currents (Laezza et al., 2007a) and suppression of actinfilin via siRNA increases the number of GluR6-containing synaptic pools (Salinas et al., 2006), indicating that BTB/kelch proteins play important and diverse roles in regulating native kainate receptors.
Kainate receptors comprise homomeric and heteromeric assemblies of the GluR5-7 and KA1–KA2 subunits (Huettner, 2003). Similar to other ionotropic glutamate receptors, of the AMPA and NMDA subfamilies, kainate receptors interact via their cytoplasmic C-termini with PDZ domain proteins (Cognet et al., 2006; Jaskolski et al., 2005; Isaac et al., 2004). Interactions at the extreme C-terminal tail of GluR6 (-ETMA) have been demonstrated for PSD95, PICK1, GRIPa and syntenin 2α (Hirbec et al., 2003, 2002; Mehta et al., 2001; Garcia et al., 1998). In addition, PICK1 has been shown to interact with cytoplasmic domains of AMPA receptor subunit GluR2 that are proximal to the extreme C-terminus (Hanley et al., 2002). Perfusion of soluble peptides competing out PICK1 interaction reduces post-synaptic kainate receptor-mediated currents at the mossy fiber to CA3 terminals, consistent with a role of PDZ domain proteins in regulating stability of kainate receptors at synaptic sites (Hirbec et al., 2003). More recently, it has been demonstrated that PICK1 interaction with kainate receptors is required for kainate receptor-mediated LTD at perirhinal cortex layer II–III synapses, indicating that this interaction has important roles in activity-dependent synaptic remodeling mediated by kainate receptors (Park et al., 2006). Thus, BTB/kelch and PDZ domain proteins are essential in regulating functional aspects of native kainate receptors.
Interestingly, actinfilin has been shown to interact directly with the PDZ domain protein CAP70 in a heterologous expression system and to form a complex with PSD95 in the hippocampus (Chen and Li, 2005; Chen et al., 2002). Importantly, both actinfilin and KRIP6 interact with the C-terminal domain of GluR6 at a site far from the PDZ domain binding region (Laezza et al., 2007a; Salinas et al., 2006). In addition, KRIP6 interaction with GluR6 is unaffected by deletion of the last 4 amino acids of the GluR6 C-tail (-ETMA), which are required for binding to PDZ domains (Laezza et al., 2007a). Thus, BTB/kelch and PDZ proteins might access kainate receptors at spatially segregated regions of the GluR6 C-tail.
Here, we have investigated whether KRIP6 can interact with PDZ domain proteins functionally related to GluR6-containing kainate receptors and whether the presence of a PDZ domain protein affects the previously characterized regulation by KRIP6 of GluR6-mediated currents.
2. Methods
2.1. Cell culture, transfection, and immunocytochemistry
COS-7 cells and HEK 293 cells were maintained in DMEM and 10% FBS. Dr. M. Linder (Washington University, St. Louis, MO) provided HEK 293 cells stably expressing a Myc-tagged GluR6 construct that was kindly donated by Dr. Christophe Mulle (University of Bordeaux, Bordeaux, France). This line was maintained in media containing 500 µg/ml G418 (Invitrogen). All transfections were performed with Lipofectamine 2000 (Invitrogen) following vendor specifications. GluR6a (Q) cDNA was expressed from the pcDNA3.1 vector as previously described (Huettner et al., 1998). N-terminally epitope-tagged forms of KRIP6 were expressed from Myc-pRK5 (obtained from P. Worley) and from pECFP-N1 (Clontech) as previously described (Laezza et al., 2007a). The N-terminally epitope-tagged form of GFP-KRIP6 was obtained by swapping the CFP tag of CFP-KRIP6 with EGFP (Clontech). N-terminally epitope-tagged forms of wild type and of PDZ domain double mutant PICK1 K27A D28A (Myc-PICK1KD/AA) were expressed from Myc-pRK5 (Boudin and Craig, 2001; Boudin et al., 2000) or from EYFP-pRK5 (PICK1 wild type only). To obtain EYFPPICK1, the EYFP coding region (from pEYFP Clontech) was amplified by PCR and subcloned into 5′EcoRI and 3′SalI sites of Myc-PICK1 in pRK5 replacing the c-Myc epitope tag. The N-terminally epitope-tagged mRFP-PICK1 was obtained by PCR amplification of PICK1 (1–416), which was subcloned in frame into 5′BsRGI and 3′NotI sites of mRFP-N1. The C-terminally epitope-tagged form of PSD95-YFP and the N-terminally epitope-tagged form of human Myc-sprouty were as previously described (Sharma et al., 2006; Laezza et al., 2007b). ECFP-N1 was from Clontech. Monomeric RFP, a kind gift of Dr. R. Tsien, was subcloned into the N1 expressing Clontech vector by PCR amplification and replacement of the EYFP coding sequence. The C-terminally epitope-tagged FGF13-GFP was a gift of Dr. D. Ornitz (Washington University, St. Louis, MO).
2.1.1. Immunostaining
Transfected COS-7 cells were fixed with fresh 4% paraformaldehyde and 10% sucrose in phosphate-buffered saline (PBS) for 15 min, rinsed with PBS, permeabilized with PBS containing 0.25% Triton X-100 and incubated in PBS containing 10% BSA for 30 min at 37 °C to block nonspecific staining. The appropriate primary antibodies were applied overnight at room temperature in PBS containing 3% BSA. After washing in PBS, cells were incubated in secondary antibodies (45 min; 37 °C). Coverslips were mounted in elvanol (Tris–HCl, glycerol, and polyvinyl alcohol with 2% 1,4-diazabicyclo [2,2,2] octane).
2.1.2. Antibodies
Primary antibodies were rabbit anti-GFP (1:1000, Molecular Probes); mouse anti-Myc (1:1000, 9E10 clone, Upstate). Secondary antibodies were Alexa 568 anti-mouse IgG1 (1:500), Alexa 647 anti-rabbit or anti-mouse (1:500) (all from Molecular Probes) or Texas Red goat anti-mouse IgG (Jackson Labs).
2.1.3. Image quantification
Images were acquired for each fluorophore using either an Axioplan 2 or an Axio Imager Z1 microscope (both Zeiss, Oberkochen, Germany) with a 63× (1.4 NA) objective or a Nikon Eclipse E600 microscope with a 60× (1.0 NA) objective. Image analysis was performed with MetaMorph software (Universal Imaging) as follows. We first defined the total cell area by manually drawing an outline along the cell perimeter. Then a mask corresponding to KRIP6 or PICK1 clusters was generated by thresholding KRIP6 or PICK1 pixel intensity within the cell boundary. The regions defined by the cell boundary and by the KRIP6 or the PICK1 mask were then applied to the unthresholded KRIP6, PICK1, GluR6 or PSD95 images. The enrichment index was defined as the fluorescence density within the KRIP6 or PICK1 mask divided by the fluorescence density over the entire cell, where fluorescence density = total pixel intensity/region area. Background was numerically subtracted from the pixel intensity values prior to fluorescence density calculation. Monomeric Red fluorescent protein co-transfected with CFP-KRIP6 had an enrichment index of 1.11 ± 0.21 (n = 5), as expected for a uniformly distributed non-polarized protein (Lou et al., 2005; Rivera et al., 2003).
2.2. Immunoprecipitation and Western blot
At 24 h post-transfection, HEK 293 cells were washed twice with phosphate-buffered saline and scraped in 500–800 µl of modified RIPA buffer: 50 mM Tris–HCl (pH 7.4), 1% NP-40, 0.5% Na-deoxycholate, 0.15 M NaCl, 1 mM EDTA, or in the following lysis buffer: 20 mM Tris–HCl, 150 mM NaCl, 1% NP-40 and a protease inhibitor mixture (Calbiochem, protease inhibitor set # 3, 1:50–100). After sonication, the supernatant was centrifuged at maximum speed on a table top centrifuge at 4 °C for 15 min. Supernatants were collected and incubated with rabbit anti-Mycagarose beads (Sigma, St. Louis, MO) for 2 h at 4 °C with agitation. The mixture was washed three times with RIPA buffer, one time with 150 mM NaCl and two times with modified RIPA buffer or simply three times in PBS and 0.1% NP-40 and then 2× sample buffer (BIORAD) containing 5 mM tris(2-carboxyethyl) phosphine (TCEP) was added. Mixtures were heated for 15 min at 65 °C and loaded on a 7.5% or 4–15% polyacrylamide gel (BIORAD). Resolved proteins were transferred to PVDF membranes (Millipore) overnight at 4 °C and blocked in Tris-buffered saline (TBS) with either 5% skim milk and 0.1% Tween-20 or 2% of Amersham blocking reagent. Membranes were then incubated in blocking buffer containing mouse anti-GFP (1:5000, Covance) or rabbit anti-GFP (1:1000, Molecular Probes) to reveal the YFP-PICK1 protein and monoclonal 9E10 anti-Myc (1:1000, Santa Cruz). Washed membranes were incubated with HRP-conjugated secondary antibodies and detected with either SuperSignal Pico chemiluminescent substrate (Pierce) or ECL Advance Western Blotting Detection kit (Amersham). For co-immunoprecipitation experiments described in Fig. 4C, D, either GFP- or CFP-KRIP6 was co-expressed with YFP-PICK1. The densitometry measurements were performed with Quantity One (BIORAD) using the rectangular volume tool.
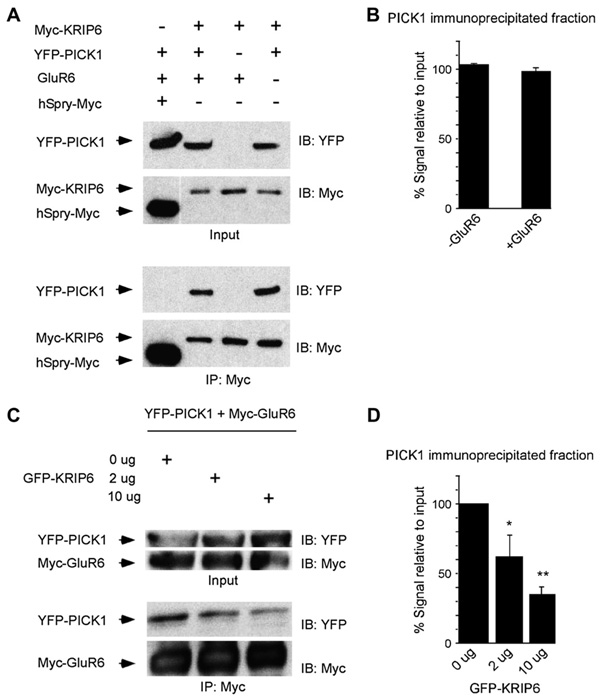
Fig. 4.
KRIP6 co-immunoprecipitates with PICK1 and decreases the interaction between GluR6 and PICK1. (A) Immunoprecipitation with Myc antibody resulted in co-immunoprecipitation of YFP-PICK1 from lysates of HEK 293 cells co-expressing Myc-KRIP6. YFP-PICK1 did not interact with human sprouty-Myc (hSpry-Myc). The input represents 1% of the material used for immunoprecipitation. Following immunoprecipitation with anti-Myc-agarose beads (IP: Myc), immunoblots (IB) were performed with either the anti-YFP or the anti-Myc antibody (right). (B) Densitometry of co-immunoprecipitated YFP-PICK1 expressed as percentage relative to input plotted in the absence or presence of GluR6. Co-expression of GluR6 did not affect the fraction of YFP-PICK1 bound to Myc-KRIP6 (98 ± 2.6% with GluR6 versus 103 ± 0.8% without GluR6, n = 3; p = 0.15, t-test). (C) HEK-Myc-GluR6 cells were transiently transfected with YFP-PICK1 and increasing amounts of GFP-KRIP6 construct. The input represents 5% of the material used for immunoprecipitation. Following immunoprecipitation with anti-Myc-agarose beads (IP: Myc), immunoblots (IB) were performed with either the anti-YFP or the anti-Myc monoclonal antibody (right). Increasing the amount of GFP-KRIP6 reduced the amount of YFP-PICK1 that co-immunoprecipitated with the anti-myc beads (Myc-GluR6). (D) Densitometry of co-immunoprecipitated YFP-PICK1 expressed as percentage relative to input plotted as a function of the GFP-KRIP6 (or CFP-KRIP6, see Section 2) used in the transfections. These analyses revealed that increasing the amount of GFP-KRIP6 significantly reduced the amount of YFP-PICK1 co-precipitating with anti-Myc-agarose beads (*p < 0.05, **p < 0.01; ANOVA post hoc Dunnett).
2.3. Electrophysiology
Cultured cells were perfused with Tyrode’s solution (in mM): 150 NaCl, 4 KCl, 2 CaCl2, 2 MgCl2, 10 glucose, 10 HEPES (pH adjusted to 7.4 with NaOH), at 1–2 ml/min. Whole-cell electrodes filled with (in mM) 140 Cs-glucuronate, 5 CsCl, 5 MgCl2, 10 EGTA, 5 ATP, 1 GTP, 10 HEPES (pH adjusted to 7.4 with CsOH) had an open tip resistance of 1–5 MΩ. The reference electrode was placed in a well filled with internal solution that connected to the bath via an agar bridge equilibrated with 4 MKCl. Currents recorded with an Axopatch 200 A amplifier were filtered at 1 kHz and digitized at 10 kHz. The holding potential for most experiments was −70 to −80 mV.
2.3.1. Drug applications
Control and kainate-containing solutions were applied by local perfusion from a multibarreled pipette to cells under whole-cell clamp. Kainate was dissolved at 300 µM in a control extracellular solution that contained (in mM): 160 NaCl, 10 HEPES, 2 CaCl2 (pH adjusted to 7.4 with NaOH). Drug reservoirs were held under static air pressure (5–10 psi) and solution flow was controlled by computer-gated electronic valves (The Lee Co.) resulting in a solution exchange time constant of 5–15 ms in the whole-cell recording mode. Whole-cell recordings were used for this study to allow meaningful comparison of peak and steady-state currents (Laezza et al., 2007a).
2.4. Statistical analysis
All the results are presented as mean ± SEM. Populations were compared by ANOVA with post hoc comparison or by unpaired t-test or non-parametric Mann–Whitney rank sum test, depending on their variance and distributions (SigmaStat, Systat Software, San Jose, CA). Differences were considered to be significant for p <0.05.
3. Results
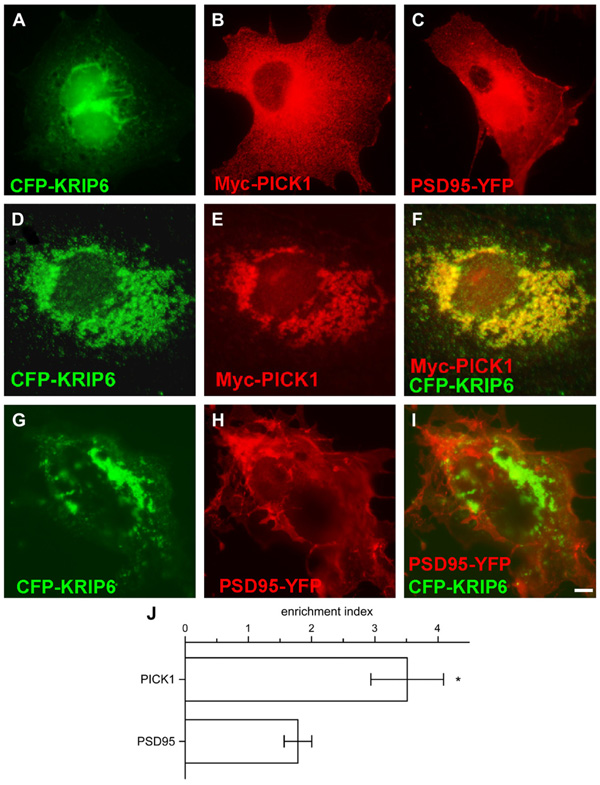
To screen for potential interactions between KRIP6 and PDZ domain proteins functionally related to GluR6-containing kainate receptors, we used fluorescence microscopy of transfected COS-7 cells to analyze the distribution of PICK1 or PSD95 expressed alone or in combination with KRIP6. When expressed alone, Myc-PICK1 or PSD95-YFP appeared dispersed throughout the cellular membranes (Fig. 1B, C), whereas CFP-KRIP6, was either diffusely arrayed or aggregated into perinuclear agglomerates Fig. 1A, as previously described (Laezza et al., 2007a). Notably, PSD95-YFP remained diffusely distributed when co-expressed with CFP-KRIP6 (Fig. 1G–I), whereas Myc-PICK1 co-clustered with CFP-KRIP6 in nearly all cells examined (Fig. 1D–F; n > 80 cells from 3 independent transfections). In addition, co-clustering of PICK1 with KRIP6 was reliably observed by immunofluorescence detection of N-terminal Myc tagged KRIP6 and YFP-PICK1 (data not shown), indicating that the association of the two proteinswas independent of the method used for visualization. Quantitative analysis of fluorescence images revealed that PICK1 (eithermyc or YFP tagged) was more highly concentrated in KRIP6 clusters than PSD95 (PICK1 enrichment index 3.51 ± 0.58, n = 9, versus 1.78 ± 0.22 for PSD95, n = 6; p < 0.02, Mann–Whitney rank sum test) suggesting that a direct interaction with KRIP6 is more likely for PICK1 than for PSD95 (Fig.1J). Interestingly, mutations of PICK1 that eliminate PDZ domain-mediated binding (Xia et al., 1999) did not abolish co-clustering with KRIP6 (Fig. S1, KRIP6 enrichment with PICK1 KDAA, n = 4, 80% of wild type n = 9; p = 0.49; Mann–Whitney rank sum test), suggesting that the interaction of KRIP6 with PICK1 does not require involvement of the PDZ domain.
Fig. 1.
KRIP6 specifically co-clusters with PICK1, but not with PSD95. (A–F) Fluorescence images of COS-7 cells expressing CFP-KRIP6, Myc-PICK1 and PSD95-YFP alone (A–C) or co-expressing CFP-KRIP6 and Myc-PICK1 (D–F) stained with monoclonal mouse anti-Myc antibodies and visualized with Texas Red or Alexa 568 conjugated secondary antibodies (B, E, F). Fluorescence images of COS-7 cells co-expressing CFP-KRIP6 and PSD95-YFP (H, I). CFP images and YFP images in A, D, G and C, H are shown, respectively, in the green and red channels and Myc-PICK1 is shown in the red channel (B, E). Comparing merged images of CFP-KRIP6 and Myc-PICK1 (F) with CFP-KRIP6 and PSD95-YFP (I), it is evident that PICK1, but not PSD95, co-clusters with KRIP6. (J) Bar graph plots the enrichment index (fluorescence density ratio in KRIP6 clusters versus the entire cell) of PICK1 and PSD95 in the presence of KRIP6. PICK1 has an enrichment index of 3.51 ± 0.58 (n = 9) and was significantly (p < 0.02, Mann–Whitney t-test) more enriched in KRIP6 clusters compared to PSD95 (1.78 ± 0.22, n = 6). Data are expressed as mean ± SEM. Scale bar = 5 µm.
To confirm the interaction of KRIP6 and PICK1 biochemically, we tested for co-immunoprecipitation of Myc-KRIP6 and YFP-PICK1 in transiently co-transfected HEK 293 cells. Anti-Myc-agarose beads efficiently immunoprecipitated Myc-KRIP6 from HEK cell extracts and co-immunoprecipitated YFP-PICK1. Control experiments, using an unrelated protein, hSpry-Myc, demonstrated specificity of YFP-PICK1 co-immunoprecipitation by Myc-KRIP6 (Fig. 4A). Thus, PICK1 specifically co-clusters and co-immunoprecipitates with the BTB/kelch protein KRIP6. Interestingly, direct interactions of KRIP6 and of PICK1 with independent regions of the C-terminal domain of GluR6 have been previously demonstrated (Laezza et al., 2007a; Hirbec et al., 2003). Thus, two proteins that interact with separate domains of the C-terminal tail of GluR6, interact with each other, raising the possibility that GluR6 might be functionally regulated in a complex way by a synergistic or competitive interaction between KRIP6 and PICK1.
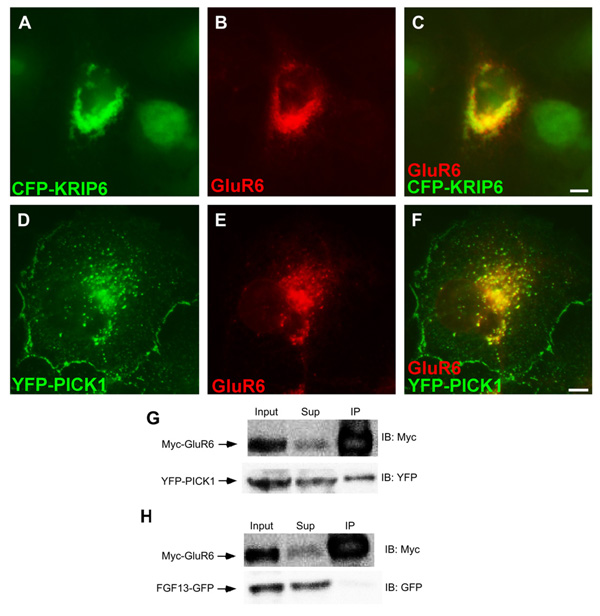
To evaluate the functional consequences of interaction between PICK1 and GluR6 in the absence or presence of KRIP6, we used side-by-side fluorescence analysis and electrophysiological recordings of COS-7 cells transfected with wild type GluR6 alone, GluR6 in pair-wise combination with PICK1 or KRIP6, or GluR6 together with both PICK1 and KRIP6. As shown in Fig. 2D–F, co-expressed YFP-PICK1 and GluR6 formed co-localized puncta and large perinuclear aggregates, a pattern that is significantly reduced if GluR6 is co-expressed with the PICK1 PDZ domain double mutant (PICK1 KDAA 44% of wild type PICK1; p < 0.0001, n = 17 WT, 9 mutant, Mann–Whitney rank sum test) (Fig. S1A). To confirm the interaction with wild type PICK1, HEK cells stably expressing Myc-GluR6 (HEK-Myc-GluR6) were transiently transfected with YFP-PICK1 or with FGF13-GFP, as a negative control, and immunoprecipitations were performed with anti-Myc-agarose beads. As illustrated in Fig. 2G, H, YFP-PICK1, but not FGF13-GFP, co-immunoprecipated with Myc-GluR6. Thus, consistent with previous reports of a direct interaction between PICK1 and the C-terminal domain of GluR6 (Hirbec et al., 2003), PICK1 and GluR6 co-cluster and co-immunoprecipitate in a heterologous expression system. As expected from our earlier study (Laezza et al., 2007a), GluR6 also co-clustered with CFP-KRIP6 in cells co-expressing both proteins (Fig. 2A–C).
Fig. 2.
GluR6 co-clusters with KRIP6 and PICK1 and co-immunoprecipitates with PICK1. (A–F) Fluorescence images of COS-7 cells co-expressing GluR6, stained with a polyclonal anti-GluR6/7 antibody, visualized with an Alexa 568 (B, C) or Alexa 647 (E, F) conjugated secondary antibodies, and either CFP-KRIP6 (A–C) or YFP-PICK1 (D–F). CFP images and YFP images in A, C, D, F are shown in the green channel; GluR6 is shown in the red channel. Merged images (C, F) clearly indicate that GluR6 co-clusters with CFP-KRIP6 and with YFP-PICK1. Quantification of these interactions is illustrated in Fig. 3 panels I, J. Scale bars = 5 µm. (G) Immunoprecipitation with myc antibody resulted in co-immunoprecipitation of YFP-PICK1 from lysates of HEK cells stably expressing Myc-GluR6. Myc-GluR6 did not interact with FGF13-GFP. (H) The input and the supernatant collected after co-immunoprecipitation (Sup) represent 2.5% of the material used for immunoprecipitation. Following immunoprecipitation with anti-Myc-agarose beads (IP: Myc), immunoblots (IB) were performed with either the anti-YFP or the anti-Myc antibody (right).
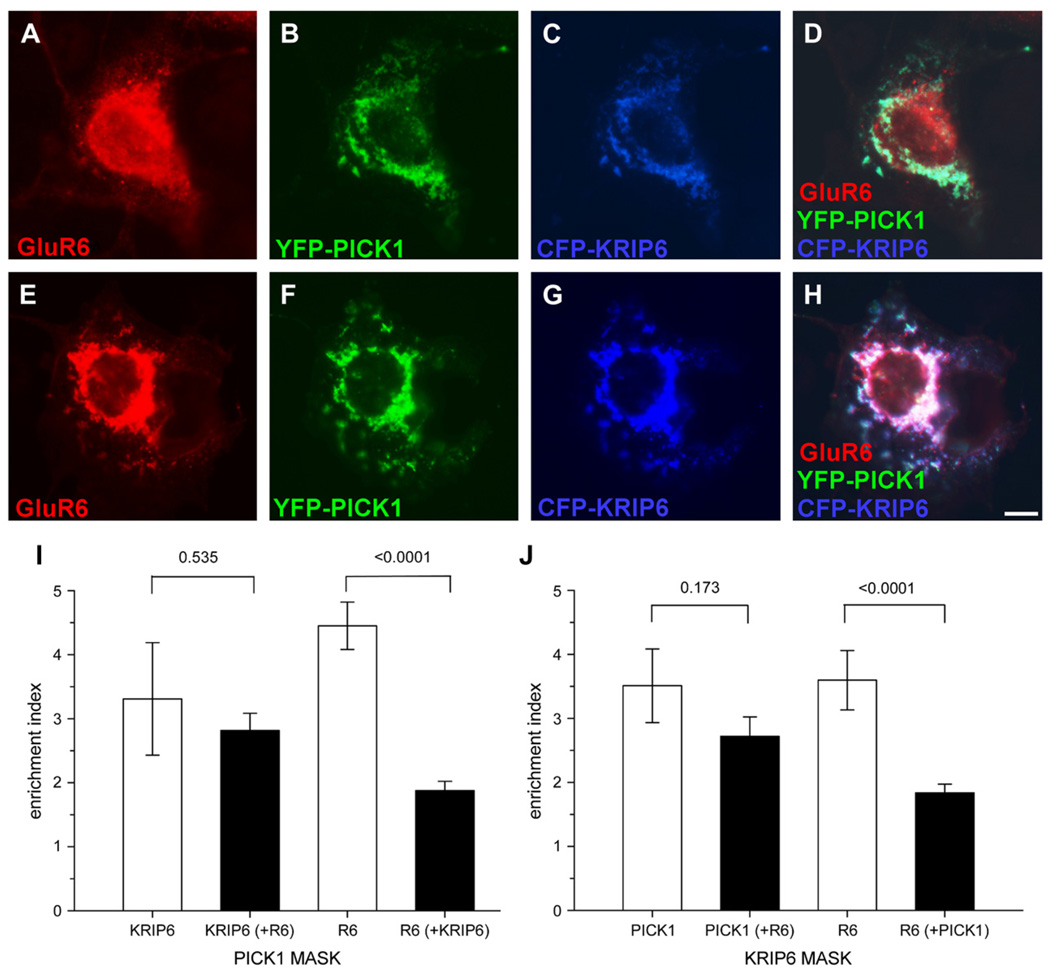
To test whether GluR6, PICK1 and KRIP6 formed triple co-clusters, COS-7 cells were transiently transfected with GluR6, CFP-KRIP6 and YFP-PICK1. In 75% of cells that expressed all three proteins (n = 28) KRIP6 and PICK1 were strongly co-localized, whereas GluR6 remained separate (Fig. 3A–D; GluR6 enrichment index < 2). In the remaining 25% of cells (n = 7) all three proteins were localized together in triple co-clusters (Fig. 3E–H; GluR6 enrichment index > 2). Quantification of pair-wise and triple co-localization is plotted in Fig. 3I and J using either KRIP6 or PICK1 fluorescence as the thresholded mask, respectively (see Section 2). Importantly, KRIP6 and PICK1 co-localization was not significantly reduced by the addition of GluR6. Enrichment of PICK1 with KRIP6 and vice versa were reduced by 23 and 15%, respectively, upon co-expression with GluR6, however, neither change was significant (p = 0.535 and p = 0.173, respectively, n = 9 double, 28 triple co-expression, Mann–Whitney rank sum test). In contrast, GluR6 displayed substantially greater co-localization when expressed pair-wise with either KRIP6 or PICK1 than when all three proteins were co-expressed together (Fig. 3D, I, J). GluR6 enrichment with KRIP6 decreased by 49% when PICK1 was co-expressed (p < 0.0001, n = 12 double, 28 triple co-expression, Mann–Whitney rank sum test), whereas GluR6 enrichment with PICK1 decreased by 58% when KRIP6 was co-expressed (p < 0.0001, n = 17 double, 28 triple co-expression, Mann–Whitney rank sum test). Collectively, these results suggest that the interaction between KRIP6 and PICK1 might be of higher affinity than the respective association of KRIP6 or PICK1 with GluR6.
Fig. 3.
The presence of GluR6 does not affect the interaction between KRIP6 and PICK1. (A–H) Fluorescence images of representative examples of COS-7 cells expressing CFP-KRIP6, YFP-PICK1 and GluR6, stained with polyclonal anti-GluR6/7 antibodies and visualized with Alexa 647 conjugated secondary antibodies. GluR6 images are shown in red (A, E, D, H). YFP (B, F, D, H) and CFP (C, G, D, H) images are shown in green and blue, respectively. From the merged images (D, H) is evident that GluR6 is either distributed independently from PICK1 and KRIP6 (D) or joins PICK1 and KRIP6 clusters (H). (I) Bar graphs plot the enrichment index of KRIP6 co-expressed with PICK1 in the absence (first from left) or in the presence of GluR6 (second from left). The enrichment index of KRIP6 is calculated through a PICK1 MASK and expressed as fluorescence density ratio of KRIP6 in PICK1 clusters versus the whole cell. The enrichment index of KRIP6 was unchanged by co-expression of GluR6 (3.31 ± 0.88, n = 9 versus 2.82 ± 0.27, n = 28, p = 0.535, Mann–Whitney rank sum test). A representative example of KRIP6 and PICK1 co-expression in the absence of GluR6 is shown in Fig. 1A–C. Quantification of GluR6 enrichment in PICK1 clusters, either in the absence (third bar from left) or in the presence of KRIP6 (fourth bar from left) shows that GluR6 association with PICK1 is dramatically reduced in the presence of KRIP6. The enrichment index of GluR6 and PICK1 alone (4.45 ± 0.37, n = 17) was reduced to 1.88 ± 0.14 (n = 28) by co-expression of KRIP6 (p < 0.0001, Mann–Whitney rank sum test). A representative example of GluR6 and PICK1 is shown in Fig. 2D–F. (J) The same data set of panel I is analyzed using a KRIP6 MASK. Consistent with the results shown in panel I, PICK1 co-clustered with KRIP6 equally well in the absence (enrichment index = 3.51 ± 0.58, n = 9) or in the presence (enrichment index = 2.72 ± 0.30, n = 28) of GluR6. In contrast, GluR6 co-clustering with KRIP6 (enrichment index = 3.60 ± 0.46, n = 12) was significantly reduced (p < 0.0001, Mann–Whitney rank sum test) by co-expression of PICK1 (enrichment index = 1.84 ± 0.14, n = 28). A representative example of GluR6 and KRIP6 is shown in Fig. 2A–C. Data are expressed as mean ± SEM. Scale bar = 10 µm.
The PICK1 PDZ domain double mutant (PICK1 KDAA) displayed significantly weaker co-localization with GluR6 as compared to wild type PICK1 (PICK1 KDAA 44% of wild type PICK1; p < 0.0001, n = 9 mutant, 17WT, Mann–Whitney rank sum test), whereas pair-wise co-localization of KRIP6 with PICK1 was not reduced by PDZ domain mutation (PICK1 KDAA 80% of wild type; p = 0.49, n = 4 mutant, 9 WT, Mann–Whitney rank sum test) (Fig. S1). In addition, PICK1 KDAA reduced the co-localization of GluR6 with KRIP6 to a similar extent as wild type PICK1, confirming that the PDZ domain of PICK1 is not necessary to mediate interaction with KRIP6 (41% reduction by PICK1 KDAA; p = 0.015, n = 11 versus 49% reduction by PICK1; p < 0.0001, n = 28, Mann–Whitney rank sum test) (Fig. S1).
To confirm that the association between KRIP6 and PICK1 was unaffected by the presence of GluR6, lysates of HEK 293 cells co-transfected with Myc-KRIP6 and YFP-PICK1, in the presence or absence of GluR6, were immunoprecipitated with an anti-Myc antibody and immunoblotted using anti-Myc and anti-GFP antibodies. As shown in Fig. 4A, B, co-expression of GluR6 did not modify the fraction of YFP-PICK1 that co-immunoprecipitated with Myc-KRIP6. Thus, KRIP6 and PICK1 associate whether or not GluR6 subunits are present; addition of GluR6 does little to enhance or reduce this interaction.
The results illustrated in Fig. 3 demonstrate that the enrichment index of GluR6 and PICK1 decreased in the presence of KRIP6, suggesting that KRIP6 might interfere with the interaction between GluR6 and PICK1. To test for such interference, HEK-Myc-GluR6 cells were transiently transfected with a constant amount of YFP-PICK1 and increasing amounts of GFP- or CFP-KRIP6, and immunoprecipitations were again performed with anti-Myc-agarose beads. Control experiments were also performed on cells transiently transfected with the FGF13-GFP construct, as described above (Fig. 2H). As illustrated in Fig. 4C, substantially less YFP-PICK1 co-immunoprecipitated from cells co-transfected with GFPKRIP6 and YFP-PICK1, compared with cells transfected with the YFP-PICK1 construct alone. Quantification of immunoprecipitated YFP-PICK1 protein in blots from three independent experiments demonstrated that the fraction of YFP-PICK1 co-immunoprecipitating with Myc-GluR6 was progressively reduced as the amount of transfected GFP-KRIP6 increased (Fig. 4D).
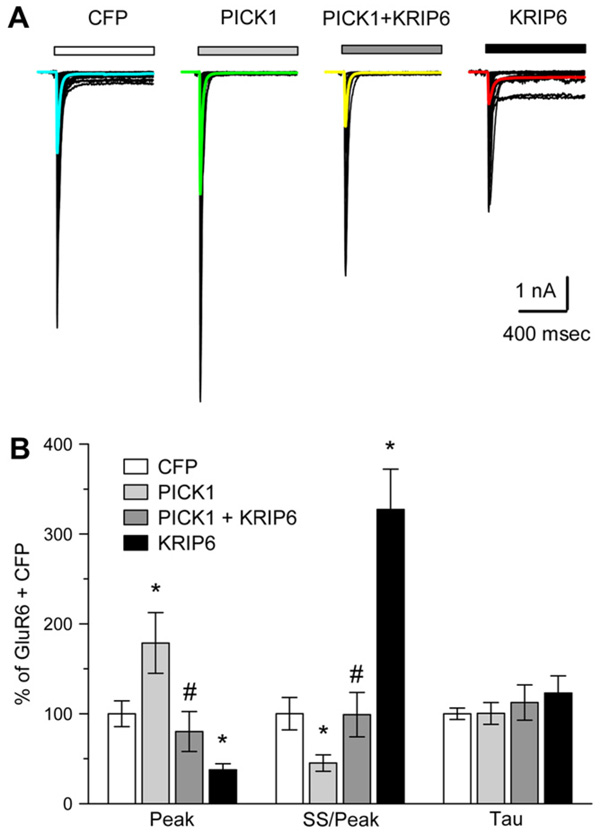
Previous work has shown that interactions with PDZ domain or BTB/kelch proteins can alter the functional properties of channels containing GluR6. For example, co-expression of the PDZ domain protein PSD95 together with GluR6 results in co-clustering of the two proteins and a slight reduction in desensitization of GluR6-mediated currents (Garcia et al., 1998; Bowie et al., 2003). Similarly, KRIP6 interacts with GluR6 and reduces both peak current and steady-state desensitization of homomeric GluR6 channels (Laezza et al., 2007a). In addition, PICK1 is known to bind the C-terminal domain of GluR6 in biochemical assays and to play a role in stabilizing synaptic kainate receptors at mossy fiber inputs to CA3 pyramidal neurons (Hirbec et al., 2003). However, it remains to be determined whether interaction with PICK1 directly regulates the functional properties of currents mediated by GluR6-containing receptors and whether the presence of PICK1 might affect the already identified functional modulation of GluR6 by KRIP6. Using whole-cell patch-clamp recordings, GluR6-mediated currents were evoked by fast application of 300 µM kainate to COS-7 cells co-transfected with mRFP-PICK1 and to control cells co-transfected with CFP (Fig. 5A). As illustrated in Fig. 5B and Table 1, if mRFP-PICK1 was co-expressed with GluR6, kainate-evoked currents exhibited increased peak current density (179 ± 34%, n = 29 cells, p < 0.05, t-test) compared to GluR6 plus CFP alone (n = 95 cells) and increased steady-state desensitization of GluR6-mediated currents, measured as the ratio of steady-state to peak current (45 ± 9% n = 29 cells with mRFP-PICK1 compared to 95 cells with CFP alone, p < 0.05, t-test). As mentioned above, expression of CFP-KRIP6 inhibits GluR6-mediated currents and reduces steady-state desensitization (Laezza et al., 2007a). Thus, interaction with PICK1 modifies functional properties of GluR6-mediated currents, producing phenotypes opposite to those observed upon co-expression of KRIP6 and GluR6.
Fig. 5.
PICK1 and KRIP6 compete for functional regulation of recombinant GluR6. (A) Families of whole-cell currents evoked by 300 µM kainate in COS-7 cells transfected with CFP (47 cells), mRFP-PICK1 (26 cells), mRFP-PICK1 + CFP-KRIP6 (15 cells) or CFPKRIP6 (34). Curves superimposed in color show the average of currents evoked in all cells tested. (B) Peak current, steady-state/peak current, and decay time constant (Tau) are plotted as a percent of the mean values obtained with CFP alone. *Different from CFP alone (p < 0.05, t-test); #different from mRFP-PICK1 alone and from CFP-KRIP6 alone (p < 0.05, t-test), but not different from CFP alone (p > 0.05, t-test).
Table 1.
GluR6 currents in COS cells
| Cells | Peak current (pA) | Density (pA/pF) | Steady-state/peak | Tau (ms) | #Cells |
|---|---|---|---|---|---|
| CFP | −2049 ± 219 | −42.9 ± 5.5 | 3.6 ± 0.6 | 20.3 ± 1.2 | 95 |
| PICK1 | −4097 ± 679 | −76.9 ± 14.5 | 1.6 ± 0.3 | 20.4 ± 2.4 | 29 |
| PICK1 + KRIP6 | −1827 ± 503 | −34.5 ± 9.6 | 3.6 ± 0.9 | 22.8 ± 4 | 16 |
| KRIP6 | −971 ± 141 | −16.2 ± 2.9 | 11.8 ± 1.6 | 25 ± 3.9 | 84 |
All values are means ± SEM.
We next evaluated whether combined expression of both KRIP6 and PICK1 affected the properties of GluR6-mediated currents. Surprisingly, when both KRIP6 and PICK1 were co-expressed, GluR6-mediated currents were indistinguishable from control conditions (Fig. 5A, B, Table 1), displaying peak current density and relative desensitization that were not significantly different from cells transfected with GluR6 and CFP alone (peak current density 89 ± 25% of control, p > 0.1; ss/peak 99 ± 25%, p > 0.1, n = 16 cells, t-test). Thus, co-expression of KRIP6 and PICK1 abrogates the regulation of GluR6 that each protein exerts independently, suggesting a competitive or mutual blocking effect between the two modulatory proteins and their target.
4. Discussion
Our results provide evidence for a specific association between the BTB/kelch protein KRIP6 and the PDZ domain protein PICK1, two proteins that have been previously shown to interact with distinct regions of the GluR6 C-terminal domain. Furthermore, we demonstrate a novel regulation of GluR6-mediated currents by PICK1, consisting of an increase in peak current density and in relative desensitization. Importantly, the changes in GluR6-mediated currents produced by expression of PICK1 are opposite to those caused by KRIP6 (Laezza et al., 2007a). Regulation is abolished when both PICK1 and KRIP6 are expressed together, resulting in currents with properties indistinguishable from GluR6 alone. Fluorescence microscopy revealed co-localization of KRIP6 and PICK1 whether or not GluR6 was also present. When the three proteins were co-expressed KRIP6, PICK1 and GluR6 either co-distributed together or KRIP6 and PICK1 formed clusters lacking GluR6, indicating that the presence of GluR6 does not destabilize the association between KRIP6 and PICK1. Consistent with this observation, the fraction of PICK1 that co-immunoprecipitates with KRIP6 was unaffected by GluR6 and increasing amounts of KRIP6 produce a coordinate decrease in the fraction of PICK1 that co-immunoprecipitates with GluR6. Thus, two proteins with opposing regulatory functions for kainate receptors, in addition to interacting with GluR6, can directly associate with each other and mutually prevent the regulation of kainate receptor physiological properties.
Interactions of BTB/kelch and PDZ domain proteins have been reported for actinfilin with CAP70 and with PSD95. Actinfilin binds to the first and third PDZ domains of CAP70 in vitro and co-immunoprecipitates with PSD95 in the hippocampus (Chen and Li, 2005). The fact that the interaction between CAP70 and actinfilin occurs via PDZ domains suggests that a canonical PDZ interaction is likely to mediate this binding. The results presented here demonstrate that the interaction of KRIP6 is selective for PICK1, but is not detectable for PSD95, another PDZ domain protein (Hung and Sheng, 2002). However, PICK1 mutations that eliminate PDZ domain-mediated binding (Xia et al., 1999) do not affect co-clustering with KRIP6 (Fig. S1), suggesting that the interaction of KRIP6 and PICK1 does not require involvement of the PICK1 PDZ domain.
The phenotype of GluR6-mediated currents that is observed upon expression of PICK1 is a novel finding. Expression of PICK1 causes an increase in peak amplitude and relative desensitization of currents mediated by GluR6 receptors. The effect on current amplitude might result from increased trafficking of receptors to the cell surface or alternatively be explained by a direct change in channel properties. Extensive evidence supports a role of PICK1 in regulating trafficking of AMPA receptors (Hanley, 2006), however, much less is known about the regulation of kainate receptors by PICK1. Hirbec et al. (2003) demonstrated in hippocampal neurons that kainate receptor-mediated EPSCs are reduced in amplitude by intracellular delivery of small peptides competing out the PDZ domain interaction of PICK1 with native kainate receptors, suggesting a role for PICK1 in maintaining the constitutive pool of synaptic receptors. A study by Yan et al. (2004), however, found that deletion of the PDZ binding domain from GluR6 did not affect surface expression in heterologous cells, suggesting that the PDZ domain binding site of GluR6 is not required for delivery or retention of the channel at the cell surface. Our results suggest that binding of PICK1 may directly increase peak current through kainate receptors and that interference with this interaction could reduce the amplitude of synaptic currents without requiring a change in the number of surface receptors. In addition to the change in peak amplitude of evoked current, our study demonstrates that PICK1 increases the relative desensitization of homomeric GluR6 receptors, a phenotype that can only be explained by a direct modulation of channel properties. The underlying molecular changes produced by PICK1, and whether the effects on peak and steady-state currents are independent, remain to be determined. In addition, further work will be needed to establish whether both KRIP6 and PICK1 can interact simultaneously with individual GluR6 subunits (or GluR6-containing kainate receptors), or whether pair-wise binding is mutually exclusive.
Quantitative immunofluorescence analysis and biochemical assays demonstrate that KRIP6 and PICK1 co-localize, with or without GluR6, and co-immunoprecipitate regardless of the presence of GluR6, suggesting that a simultaneous interaction of KRIP6 and PICK1 with GluR6 might occur. The observation of triple KRIP6, PICK1 and GluR6 co-clusters supports this hypothesis. The fact that KRIP6 and PICK1 interact with distinct and spatially segregated residues of the GluR6 C-tail suggests that spatial hindrance between the two proteins might not be a limiting factor for simultaneous accessibility of the GluR6 C-terminal domain. KRIP6 and PICK1 clusters typically are not enriched for GluR6, however, suggesting that the affinity of interaction between KRIP6 and PICK1 might be higher than their reciprocal interaction with GluR6, and also that GluR6 might exist as an unbound free pool in the presence of both PICK1 and KRIP6. Accordingly, our results show that increasing KRIP6 expression together with GluR6 and PICK1 decreases the fraction of PICK1 that co-immunoprecipitates with GluR6.
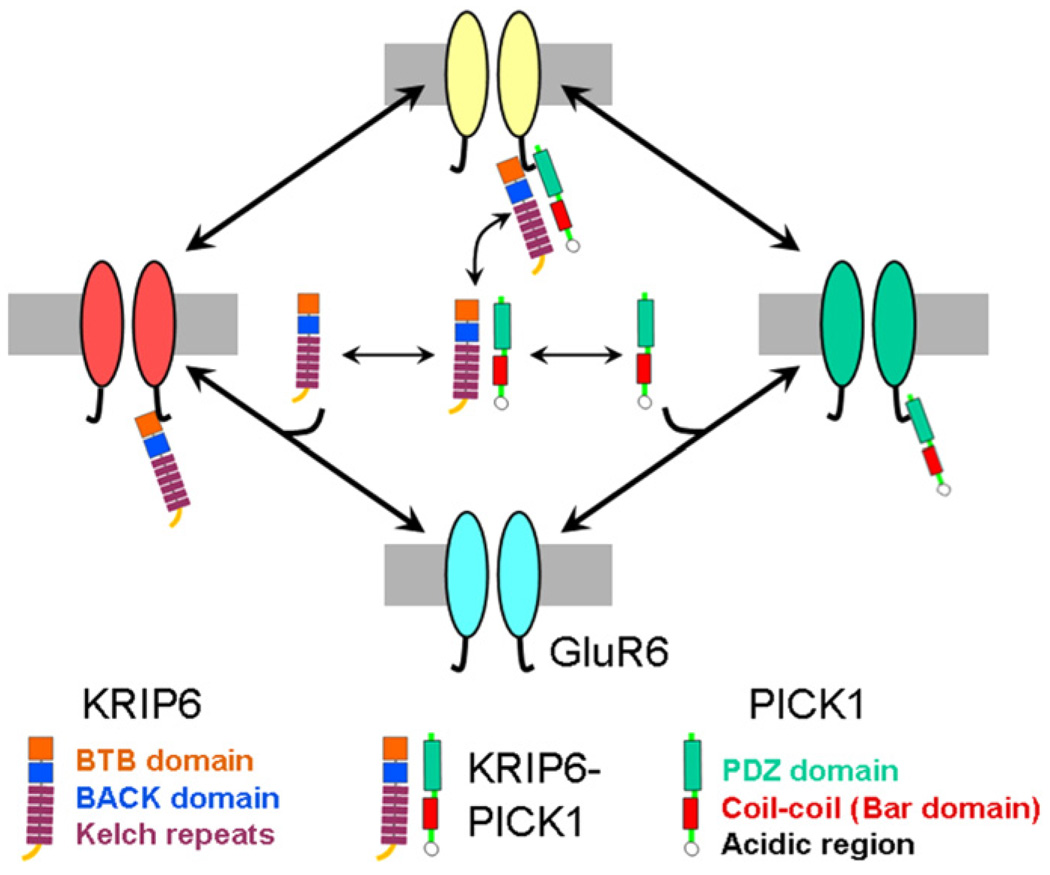
Several different mechanisms might explain the functional phenotype observed when GluR6, PICK1 and KRIP6 are co-expressed. Fig. 6 presents a model depicting all the potential interactions of KRIP6 and PICK1 with each other and with GluR6. The lack of significant change in GluR6-mediated currents relative to control, observed when KRIP6 and PICK1 are co-expressed, might occur if both proteins bound to receptors simultaneously and negated each other’s effect on channel operation, or if the pair-wise association of KRIP6 to PICK1 prevented the productive interaction of either protein with GluR6. Intermolecular interactions of PICK1, via the coil–coil domain (Perez et al., 2001), and of KRIP6, via the N-terminal BTB domain (Laezza et al., 2007a), could contribute to determine the stoichiometry of reciprocal interactions.
Fig. 6.
Model: KRIP6 down regulates GluR6 function by two separate mechanisms. 1. Interaction of KRIP6 with the GluR6 C-terminal region (red pool) directly inhibits peak current and desensitization. 2. Interaction of KRIP6 with PICK1 (yellow pool) prevents the enhancement of peak current and the increase in desensitization produced by PICK1 alone (green pool).
Recent studies have identified a growing number of proteins involved in indirect interactions with kainate receptors (Pinheiro and Mulle, 2006), as well as several which directly bind to subunit C-terminal domains. Such direct interactions have been demonstrated for PDZ domain proteins including PSD95, SAP97 and SAP102, PICK1, GRIP and syntenin 2α (Hirbec et al., 2003, 2002; Garcia et al., 1998). Investigation of the physiological functions that these interactions may serve is at an early stage. In neurons, PICK1 is suggested to play a role in trafficking and stabilization of native kainate receptors within synaptic pools (Hirbec et al., 2003). When co-expressed with GluR6 in heterologous cells PSD95 induces receptor clustering and causes a modest increase in steady-state current (Garcia et al., 1998; Bowie et al., 2003). However, none of the PDZ domain proteins have an exclusive role in regulating kainate receptors, and evidence for robust functional regulation of channel operation by PDZ domain proteins has been lacking. In this respect, KRIP6 differs from these other kainate receptor interacting proteins in that it binds selectively to the C-terminal domain of GluR6 but does not directly interact with other kainate, AMPA, or NMDA receptor subunits. Thus, KRIP6 may function to regulate GluR6-containing kainate receptors preferentially via this direct interaction. Furthermore, KRIP6 is the only protein so far identified that reduces both peak amplitude and desensitization of currents through GluR6 receptors, a phenotype mediated by the BTB domain of KRIP6 (Laezza et al., 2007a).
Previous findings showed that KRIP6 is highly expressed in brain that endogenous KRIP6 co-immunoprecipitates with native GluR6 and that overexpression of KRIP6 significantly reduced peak amplitude of kainate-evoked currents in hippocampal neurons (Laezza et al., 2007a). In the present study, we have provided evidence that extends the regulation of kainate receptors by KRIP6 to an indirect mechanism via interaction with PICK1, suggesting that endogenous KRIP6 could affect native GluR6 kainate receptors by directly binding the GluR6 C-tail or by blocking modulation of GluR6 by PICK1. In addition, increasing amount of KRIP6 decreases the fraction of PICK1 associated with GluR6 (Fig. 4C, D), suggesting that endogenous KRIP6 might act as a dominant negative protein reducing the fraction of PICK1 available to modulate GluR6-containing receptors. Importantly, KRIP6 might also block the functional interaction of PICK1 with other partners, such as GluR5 (Hirbec et al., 2003) and the AMPA subtype, GluR2 (Osten et al., 2000; Dev et al., 1999; Xia et al., 1999), causing more complex effects. Altogether, changes in the levels of endogenous KRIP6 and PICK1 could impact numerous aspects of the kainate receptor physiology by modifying clustering and functional properties of GluR6-containing kainate receptors, protecting or predisposing toward seizures or excitotoxic insults and affecting synaptic remodeling dependent on kainate receptor activity (Pinheiro and Mulle, 2006). Finally, a recent study provides evidence that the BTB/kelch protein KLHL1 plays a role in regulating voltage-gated Ca2+ channels (Aromolaran et al., 2007), raising the interesting possibility that BTB/kelch proteins may serve as general accessory subunits for both ligand- and voltage-gated channels.
Supplementary Material
Acknowledgements
We thank Dr. David Ornitz (Washington University, St. Louis, MO) for sharing equipment and for the gift of FGF13-GFP construct and Dr. Maurine Linder (Washington University, St. Louis, MO) for the HEK 293 cell line stably expressing Myc-GluR6. The Myc-GluR6 construct was kindly provided by Dr. Christophe Mulle (University of Bordeaux, Bordeaux, France). This work was supported by the AES fellowship provided by the Milken Family Foundation (FL), NIH NS30888 (JEH) and NIH NS39286 (AMC).
Appendix. Supplementary data
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.neuropharm.2008.07.021.
References
- Aromolaran KA, Benzow KA, Koob MD, Piedras-Renteria ES. The Kelchlike protein 1 modulates P/Q-type calcium current density. Neuroscience. 2007;145:841–850. doi: 10.1016/j.neuroscience.2006.12.046. [DOI] [PubMed] [Google Scholar]
- Boudin H, Craig AM. Molecular determinants for PICK1 synaptic aggregation and mGluR7a receptor coclustering: role of the PDZ, coiled-coil, and acidic domains. J. Biol. Chem. 2001;276:30270–30276. doi: 10.1074/jbc.M102991200. [DOI] [PubMed] [Google Scholar]
- Boudin H, Doan A, Xia J, Shigemoto R, Huganir RL, Worley P, Craig AM. Presynaptic clustering of mGluR7a requires the PICK1 PDZ domain binding site. Neuron. 2000;28:485–497. doi: 10.1016/s0896-6273(00)00127-6. [DOI] [PubMed] [Google Scholar]
- Bowie D, Garcia EP, Marshall J, Traynelis SF, Lange GD. Allosteric regulation and spatial distribution of kainate receptors bound to ancillary proteins. J. Physiol. 2003;547:373–385. doi: 10.1113/jphysiol.2002.033076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Derin R, Petralia RS, Li M. Actinfilin, a brain-specific actin-binding protein in postsynaptic density. J. Biol. Chem. 2002;277:30495–30501. doi: 10.1074/jbc.M202076200. [DOI] [PubMed] [Google Scholar]
- Chen Y, Li M. Interactions between CAP70 and actinfilin are important for integrity of actin cytoskeleton structures in neurons. Neuropharmacology. 2005;49:1026–1041. doi: 10.1016/j.neuropharm.2005.05.022. [DOI] [PubMed] [Google Scholar]
- Cognet L, Groc L, Lounis B, Choquet D. Multiple routes for glutamate receptor trafficking: surface diffusion and membrane traffic cooperate to bring receptors to synapses. Sci. STKE. 2006;2006:pe13. doi: 10.1126/stke.3272006pe13. [DOI] [PubMed] [Google Scholar]
- Dev KK, Nishimune A, Henley JM, Nakanishi S. The protein kinase C alpha binding protein PICK1 interacts with short but not long form alternative splice variants of AMPA receptor subunits. Neuropharmacology. 1999;38:635–644. doi: 10.1016/s0028-3908(98)00230-5. [DOI] [PubMed] [Google Scholar]
- Garcia EP, Mehta S, Blair LA, Wells DG, Shang J, Fukushima T, Fallon JR, Garner CC, Marshall J. SAP90 binds and clusters kainate receptors causing incomplete desensitization. Neuron. 1998;21:727–739. doi: 10.1016/s0896-6273(00)80590-5. [DOI] [PubMed] [Google Scholar]
- Hanley JG. Molecular mechanisms for regulation of AMPAR trafficking by PICK1. Biochem. Soc. Trans. 2006;34:931–935. doi: 10.1042/BST0340931. [DOI] [PubMed] [Google Scholar]
- Hanley JG, Khatri L, Hanson PI, Ziff EB. NSF ATPase and alph-/beta-SNAPs disassemble the AMPA receptor-PICK1 complex. Neuron. 2002;34:53–67. doi: 10.1016/s0896-6273(02)00638-4. [DOI] [PubMed] [Google Scholar]
- Hirbec H, Francis JC, Lauri SE, Braithwaite SP, Coussen F, Mulle C, Dev KK, Coutinho V, Meyer G, Isaac JT, Collingridge GL, Henley JM. Rapid and differential regulation of AMPA and kainate receptors at hippocampal mossy fibre synapses by PICK1 and GRIP. Neuron. 2003;37:625–638. doi: 10.1016/s0896-6273(02)01191-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirbec H, Perestenko O, Nishimune A, Meyer G, Nakanishi S, Henley JM, Dev KK. The PDZ proteins PICK1, GRIP, and syntenin bind multiple glutamate receptor subtypes. Analysis of PDZ binding motifs. J. Biol. Chem. 2002;277:15221–15224. doi: 10.1074/jbc.C200112200. [DOI] [PubMed] [Google Scholar]
- Huettner JE. Kainate receptors and synaptic transmission. Prog. Neurobiol. 2003;70:387–407. doi: 10.1016/s0301-0082(03)00122-9. [DOI] [PubMed] [Google Scholar]
- Huettner JE, Stack E, Wilding TJ. Antagonism of neuronal kainate receptors by lanthanum and gadolinium. Neuropharmacology. 1998;37:1239–1247. doi: 10.1016/s0028-3908(98)00082-3. [DOI] [PubMed] [Google Scholar]
- Hung AY, Sheng M. PDZ domains: structural modules for protein complex assembly. J. Biol. Chem. 2002;277:5699–5702. doi: 10.1074/jbc.R100065200. [DOI] [PubMed] [Google Scholar]
- Isaac JT, Mellor J, Hurtado D, Roche KW. Kainate receptor trafficking: physiological roles and molecular mechanisms. Pharmacol. Ther. 2004;104:163–172. doi: 10.1016/j.pharmthera.2004.08.006. [DOI] [PubMed] [Google Scholar]
- Jaskolski F, Coussen F, Mulle C. Subcellular localization and trafficking of kainate receptors. Trends Pharmacol. Sci. 2005;26:20–26. doi: 10.1016/j.tips.2004.11.008. [DOI] [PubMed] [Google Scholar]
- Laezza F, Gerber BR, Lou JY, Kozel MA, Hartman H, Craig AM, Ornitz DM, Nerbonne JM. The FGF14(F145S) mutation disrupts the interaction of FGF14 with voltage-gated Na+ channels and impairs neuronal excitability. J. Neurosci. 2007b;27:12033–12044. doi: 10.1523/JNEUROSCI.2282-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laezza F, Wilding TJ, Sequeira S, Coussen F, Zhang XZ, Hill-Robinson R, Mulle C, Huettner JE, Craig AM. KRIP6: a novel BTB/kelch protein regulating function of kainate receptors. Mol. Cell. Neurosci. 2007a;34:539–550. doi: 10.1016/j.mcn.2006.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou JY, Laezza F, Gerber BR, Xiao M, Yamada KA, Hartmann H, Craig AM, Nerbonne JM, Ornitz DM. Fibroblast growth factor 14 is an intracellular modulator of voltage-gated sodium channels. J. Physiol. 2005;569:179–193. doi: 10.1113/jphysiol.2005.097220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta S, Wu H, Garner CC, Marshall J. Molecular mechanisms regulating the differential association of kainate receptor subunits with SAP90/PSD-95 and SAP97. J. Biol. Chem. 2001;276:16092–16099. doi: 10.1074/jbc.M100643200. [DOI] [PubMed] [Google Scholar]
- Osten P, Khatri L, Perez JL, Kohr G, Giese G, Daly C, Schulz TW, Wensky A, Lee LM, Ziff EB. Mutagenesis reveals a role for ABP/GRIP binding to GluR2 in synaptic surface accumulation of he AMPA receptor. Neuron. 2000;27:313–325. doi: 10.1016/s0896-6273(00)00039-8. [DOI] [PubMed] [Google Scholar]
- Park Y, Jo J, Isaac JT, Cho K. Long-term depression of kainate receptor-mediated synaptic transmission. Neuron. 2006;49:95–106. doi: 10.1016/j.neuron.2005.11.035. [DOI] [PubMed] [Google Scholar]
- Perez JL, Khatri L, Chang C, Srivastava S, Osten P, Ziff EB. PICK1 targets activated protein kinase Calpha to AMPA receptor clusters in spines of hippocampal neurons and reduces surface levels of the AMPA-type glutamate receptor subunit 2. J. Neurosci. 2001;21:5417–5428. doi: 10.1523/JNEUROSCI.21-15-05417.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinheiro P, Mulle C. Kainate receptors. Cell Tissue Res. 2006;326:457–482. doi: 10.1007/s00441-006-0265-6. [DOI] [PubMed] [Google Scholar]
- Prag S, Adams JC. Molecular phylogeny of the kelch-repeat superfamily reveals an expansion of BTB/kelch proteins in animals. BMC Bioinform. 2003;4:42. doi: 10.1186/1471-2105-4-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera JF, Ahmad S, Quick MW, Liman ER, Arnold DB. An evolutionarily conserved dileucine motif in Shal K+ channels mediates dendritic targeting. Nat. Neurosci. 2003;6:243–250. doi: 10.1038/nn1020. [DOI] [PubMed] [Google Scholar]
- Salinas GD, Blair LA, Needleman LA, Gonzales JD, Chen Y, Li M, Singer JD, Marshall J. Actinfilin is a Cul3 substrate adaptor, linking GluR6 kainate receptor subunits to the ubiquitin–proteasome pathway. J. Biol. Chem. 2006;281:40164–40173. doi: 10.1074/jbc.M608194200. [DOI] [PubMed] [Google Scholar]
- Sharma K, Fong DK, Craig AM. Postsynaptic protein mobility in dendritic spines: long-term regulation by synaptic NMDA receptor activation. Mol. Cell. Neurosci. 2006;31:702–712. doi: 10.1016/j.mcn.2006.01.010. [DOI] [PubMed] [Google Scholar]
- Stogios PJ, Downs GS, Jauhal JJ, Nandra SK, Prive GG. Sequence and structural analysis of BTB domain proteins. Genome Biol. 2005;6:R82. doi: 10.1186/gb-2005-6-10-r82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J, Zhang X, Staudinger J, Huganir RL. Clustering of AMPA receptors by the synaptic PDZ domain-containing protein PICK1. Neuron. 1999;22:179–187. doi: 10.1016/s0896-6273(00)80689-3. [DOI] [PubMed] [Google Scholar]
- Yan S, Sanders JM, Xu J, Zhu Y, Contractor A, Swanson GT. A C-terminal determinant of GluR6 kainate receptor trafficking. J Neurosci. 2004;24:679–691. doi: 10.1523/JNEUROSCI.4985-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.