Abstract
Breast cancer risk is higher among obese women and women with diabetes. Adiponectin is a protein exclusively secreted by adipose tissue, circulating levels of which have been associated with breast cancer risk. Whether genetic variants within the adiponectin pathway are associated with breast cancer risk is unknown. To explore the association of genetic variants of the adiponectin (ADIPOQ) and adiponectin receptor 1 (ADIPOR1) genes with breast cancer risk, we conducted a case control study of female patients with breast cancer and healthy female controls from New York City recruited between 1999 and 2004. We genotyped 733 hospital-based breast cancer cases and 839 controls for 10 haplotype-tagging single nucleotide polymorphisms (SNP) of ADIPOQ and ADIPOR1. Two ADIPOQ SNPs (rs2241766 and rs1501299), which have been associated with circulating levels of adiponectin, were associated with breast cancer risk [rs1501299*GG: odd ratios (OR), 1.80; 95% confidence interval (95% CI), 1.14–2.85; rs2241766*TG: OR, 0.61; 95% CI, 0.46–0.80]. One ADIPOR1 SNP (rs7539542), which modulates expression of adiponectin receptor 1 mRNA, was also associated with breast cancer risk (OR, 0.51; 95% CI, 0.28–0.92). Based on the known function of rs2241766 and rs1501299, we categorized individuals by adiponectin signaling status and found that, when compared with high signalers, intermediate signalers had a 4.16-fold increase in breast cancer risk (95% CI, 0.49–35.19), and low signalers had a 6.56-fold increase in breast cancer risk (95% CI, 0.78–54.89; Ptrend = 0.001). This is the first report of an association between functionally relevant variants of the adiponectin pathway and breast cancer risk. The results warrant further studies of the adiponectin pathway in breast cancer.
Introduction
Breast cancer is the most common malignancy in women in developed countries. In 2007, an estimated 178,480 new cases of breast cancer were diagnosed in the United States (1). Several studies have shown an association between obesity, weight gain, and breast cancer risk (2, 3). Furthermore, there is evidence that weight loss, as well as decrease in fat consumption, may lead to decreased risk for breast cancer (4, 5). There has also been extensive research on the association of diabetes mellitus (DM) and the metabolic syndrome and breast cancer (6). In a recent meta-analysis of 20 case control studies, we reported a 20% increased risk for breast cancer in women with DM (7). In the four cohort studies included in the same meta-analysis, breast cancer risk increased by 24% in patients with DM. The mechanism underlying the increased risk of breast cancer in obese and/or diabetic subjects has been proposed to include changes in levels of estrogens (8, 9), insulin resistance, insulin-like growth factors (IGF), as well as IGF-binding proteins (10, 11).
Several proteins produced by adipose tissue have been studied in relation to breast cancer risk. There is emerging evidence that one of these adipokines, adiponectin, is associated with breast cancer risk (6). Adiponectin, a protein secreted by the adipose tissue, has been found to be an endogenous insulin sensitizer, the circulating levels of which are decreased in obese and diabetic subjects. Moreover, adiponectin has the potential of regulating the secretion of estrogens, tumor necrosis factor (TNF) (12, 13), and IGF (14).
Recently, circulating levels of adiponectin have been found to correlate with breast cancer risk (15–17). Because its levels are inversely correlated with adiposity (13), it has been suggested that decreased levels of adiponectin may explain the increased risk of breast cancer in obesity (13, 18). In fact, several groups have shown that after adjustment for body mass index (BMI), women with higher adiponectin levels had a 65% reduced risk for breast cancer (15, 16, 19). Furthermore, the breast cancer cell lines MCF-7, MDB-MB-231, and T47D were found to express both adiponectin receptors ADIPOR1/R2 (15, 20), and exposure of T47D cells to adiponectin significantly inhibited their proliferation (15).
Several adiponectin polymorphisms have been shown to affect adiponectin levels, and polymorphisms of both the ligand and its type 1 receptor (ADIPOR1) have been associated with risk for insulin resistance, cardiovascular disease, and DM (21–28). However, to date, the association of these polymorphisms with breast cancer risk has not been studied. In this study of breast cancer cases and controls, we systematically evaluated selected haplotype-tagging single nucleotide polymorphisms (SNP) in genes encoding adiponectin and its type I receptor in relation to breast cancer risk.
Materials and Methods
Study participants
As part of institutional review board-approved protocols, we collected blood samples from 733 consecutive female patients admitted to Memorial Sloan-Kettering Cancer with a diagnosis of breast cancer. Recruitment of cases occurred in two phases, the first one between January 1, 1998 and December 31, 1999, the second one occurred between December 1, 2002 and January 31, 2004. All breast cancer cases were histologically confirmed at Memorial Sloan-Kettering Cancer Center. Information regarding sex, age, age at breast cancer diagnosis, and ethnic status was recorded. In a subset of 152 patients, information on estrogen receptor (ER) and progesterone receptor (PR) status as assessed by immunohisto-chemistry was collected at the time of case collection. A sample of 839 healthy female volunteers from New York City ages 20 to 87 years was recruited between January 1, 2003 and December 31, 2004. The last date of follow-up is September 30, 2006. None of the controls had any personal history of cancer at the time of blood donation. This was ascertained by a questionnaire completed by each healthy volunteer. The controls were matched to cases on gender and geographic region and, thus, are representative of the source population where the cases come from. Although age categories were obtained for all participants, exact age information was not available for some controls. However, in that subgroup of individuals, which represents 7.9% of controls, the age ranged from 20 to 40 years (Table 1). None of the controls had any personal history of cancer as ascertained by a questionnaire administered at the time of enrollment. Ethnic status for cases and controls was self-defined. All cases and controls signed an informed consent. All personal identifiers from both cases and controls were permanently removed. The study was approved by the Institutional Review Board.
Table 1.
Characteristics of breast cancer cases and controls
| Cases, n(%) | Controls, n(%) | |
|---|---|---|
| Age (y) | ||
| 20–40 | 114 (16.2) | 27 (7.9) |
| 41–50 | 186 (26.4) | 80 (23.5) |
| 51–60 | 185 (26.3) | 117 (34.3) |
| 61–70 | 130 (18.5) | 73 (21.4) |
| 71+ | 89 (12.6) | 44 (12.9) |
| Race | ||
| White | 563 (76.8) | 801 (95.5) |
| Black | 68 (9.3) | 19 (2.3) |
| Hispanic | 33 (4.5) | 10 (1.2) |
| Asian | 22 (3.0) | 2 (0.2) |
| Unknown | 47 (6.4) | 7 (0.8) |
| rs266729 | ||
| GG | 49 (7) | 57 (7.1) |
| CG | 293 (40) | 296 (37) |
| CC | 385 (53) | 448 (55.9) |
| rs822395 | ||
| CC | 86 (12) | 113 (13.8) |
| AC | 314 (43.9) | 352 (43) |
| AA | 316 (44.1) | 354 (43.2) |
| rs822396 | ||
| GG | 19 (2.7%) | 33 (4) |
| AG | 187 (26.1%) | 210 (25.6) |
| AA | 510 (71.2) | 577 (70.4) |
| rs2241766 | ||
| TT | 524 (73.6) | 520 (64.9) |
| TG | 167 (23.5) | 252 (31.5) |
| GG | 21 (2.9) | 29 (3.6) |
| rs1501299 | ||
| TT | 50 (7) | 80 (9.8) |
| TG | 289 (40.3) | 316 (38.5) |
| GG | 379 (52.8) | 424 (51.7) |
| rs2232853 | ||
| CC | 425 (59.2) | 533 (65.6) |
| TC | 250 (34.8) | 213 (26.2) |
| TT | 43 (6) | 67 (8.2) |
| rs12733285 | ||
| TT | 100 (14) | 126 (15.6) |
| TC | 294 (41.1) | 315 (39) |
| CC | 321 (44.9) | 366 (45.4) |
| rs1342387 | ||
| AA | 201 (28.4) | 209 (25.9) |
| AG | 362 (51.1) | 419 (51.9) |
| GG | 145 (20.5) | 180 (22.3) |
| rs7539542 | ||
| GG | 117 (16.2) | 111 (13.8) |
| GC | 308 (42.7) | 361 (44.8) |
| CC | 297 (41.1) | 334 (41.4) |
| rs10920531 | ||
| CC | 264 (37) | 279 (34.2) |
| AC | 329 (46.1) | 394 (48.3) |
| AA | 121 (16.9) | 143 (17.5) |
DNA isolation
DNA from whole blood lymphocytes was extracted using the QIAamp DNA Blood Mini kit and was stored at −20°C until use for genotyping. All DNA samples underwent whole genome amplification using the Illustra Genomiphi V2 DNA Amplification kit (GE Healthcare). The samples were stored at −20°C.
Selection of SNPs
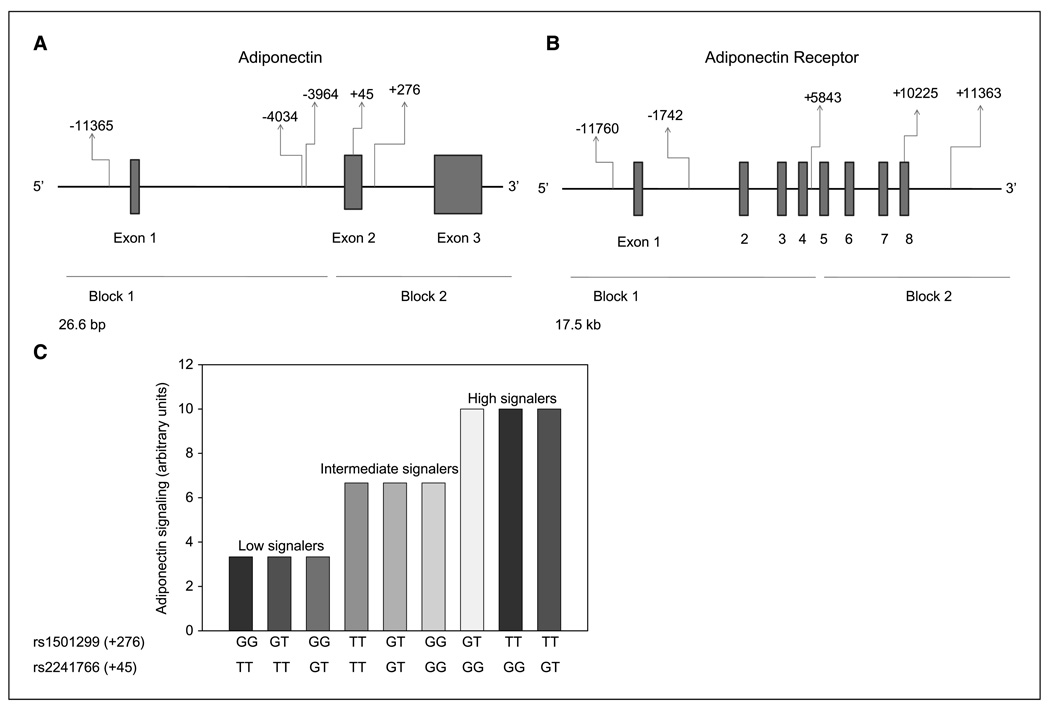
Ten haplotype-tagging SNPs were selected for genotyping (Table 2). The adiponectin gene has >10 SNPs (22, 27) and two linkage disequilibrium blocks with a block boundary between −2,049 and −450 (Fig. 1A; ref. 24). We selected to genotype rs266729 (5′ flanking region), rs822396 (intron 1), and rs822395 (intron 1) to tag block 1 and rs1501299 (exon 2) and rs2241766 (intron 2) to tag block 2 (Fig. 1A) as these are the five most common SNPs and have been studied more extensively by others as to their functionality and relation to diseases such as DM (21–24). ADIPOR1 has >28 SNPs and two linkage disequilibrium blocks (Fig. 1B; ref. 26). One block extends from the 5′ flanking region to intron 4 and the other is located at the 3′ end of the gene. Based on this, we selected five common SNPs for genotyping (Table 2). For block 1, we selected the following tagging SNPs: rs2232853 (5′ flanking region), rs12733285 (intron 1), and rs1342387 (intron 4; Fig. 1B). For block 2, we selected rs7539542 (exon 8) and rs10920531 (3′ flanking region; Fig. 1B). These SNPs were selected because they correspond to both linkage disequilibrium blocks.
Table 2.
Haplotype-tagging SNPs for the ADIPOQ and ADIPOR1 genes
| SNPs | Position | Gene | Position | Block |
|---|---|---|---|---|
| rs266729 | −11365 C→G | ADIPOQ | 5′ flanking | 1 |
| rs822395 | −4034 A→C | ADIPOQ | Intron 1 | 1 |
| rs822396 | −3964 A→G | ADIPOQ | Intron 1 | 1 |
| rs2241766 | +45 T→G | ADIPOQ | Exon 2 | 2 |
| rs1501299 | +276 G→T | ADIPOQ | Intron 2 | 2 |
| rs2232853 | −11760 C→T | ADIPOR1 | 5′ flanking | 1 |
| rs12733285 | −1742 C→T | ADIPOR1 | Intron 1 | 1 |
| rs1342387 | +5843 G→A | ADIPOR1 | Intron 4 | 1 |
| rs7539542 | +10225 C→G | ADIPOR1 | Exon 8 | 2 |
| rs10920531 | +11363 A→C | ADIPOR1 | 3′ flanking | 2 |
Figure 1.
A, adiponectin gene (ADIPOQ) with haplotype blocks and haplotype-tagging SNPs. B, Adiponectin receptor 1 gene (ADIPOR1) with haplotype blocks and haplotype-tagging SNPs. C, genotypic combinations of adiponectin functional variants and their predicted adiponectin signaling levels. X axis, various genotypic combinations of adiponectin genotypes. Y axis, predicted level of adiponectin signaling based on studies in humans expressed in arbitrary units.
Genotyping
Genotyping for all 10 SNPs was performed by Taq man SNP allelic discrimination, by means of an ABI 7900HT (Applied Biosystems). Results were ascertained by using the software SDS 2.3 (Applied Biosystems). All results were automatically called. A total of 5% of samples were genotyped in duplicate and showed 100% concordance.
Statistical analysis
The distributions of genotypes of each SNP between cases and controls were compared by using χ2 tests. Genotypes of each SNP were first compared separately. To examine the effect of allele penetrance, we combined genotypes of each SNP that carry less common allele variant in both homozygotes and heterozygotes to compare the other homozygous alleles in case control analysis. Univariate and multivariate unconditional logistic regression models were used to estimate crude and adjusted odd ratios (OR). In the multivariate logistic models, each SNP was further adjusted for age, race, and other SNPs in the same gene. In the overall study sample, age was adjusted as a categorical variable (≥50 years old and <50 years old). Alex Hsi performed the statistical analyses.
Similar statistical methods were used for analyses of signaling assessment, ER, and PR receptor. In signaling assessments, genotypes of studied SNPs were categorized as high, intermediate, and low signalers based on the known functional status. Rs1501299 (+276) GG is associated with decreased levels of serum adiponectin (29, 24). Rs2241766 (+45) TT has also been associated with decreased levels of serum adiponectin (29, 30). We therefore classified individuals who had 276*GG/45*TT, 276*GT/45*TT, and 276*GG/45*GT as low signalers; individuals with 276*TT/45*TT, 276*GT/45*GT, and 276*GG/45*GG as intermediate signalers; and individuals with 276*GT/45*GG, 276*TT/45*GG, and 276*TT/45*GT genotypes as high signalers (Fig. 1C). For ER and PR studies, genotype distributions of SNPs were analyzed based on comparison of ER and PR status within breast cancer cases. Statistical analysis of the data were performed with SAS 9.1 (SAS Institute). Bonferroni adjustment for multiple comparisons was also performed. We present both unadjusted and Bonferroni adjusted P values in the Tables.
Results
Haplotypes of ADIPOQ and breast cancer risk
Of the five haplotype-tagging SNPs selected for the analysis two of them (rs2241766 and rs1501299), both tagging block 2 of ADIPOQ were found to be significantly associated with breast cancer risk. Rs2241766 and rs1501299 have been found to alter adiponectin levels (21, 23, 25, 27) and are also associated with obesity (31), risk of insulin resistance, hypertension, and cardiovascular disease (21, 22, 24, 25). The high-expressing rs2241766 G allele (GG and GT genotypes) was associated with decreased breast cancer risk [OR, 0.64; 95% confidence interval (95% CI), 0.49–0.83; Table 4]. Breast cancer population attributable risk (PAR) for the low-expressing rs2241766 TT genotype is 70 per 1,000 individuals. The low expressing rs1501299 G allele was associated with increased breast cancer risk (TG: OR, 1.59; 95% CI, 1.03–2.48; GG: OR, 1.80; 95%, CI, 1.14–2.85; Table 3). Breast cancer PARs for the rs1501299 TG and GG genotypes are 76 and 75 per 1,000 individuals, respectively.
Table 4.
Crude and adjusted ORs (95% CI) by ADIPOQ and ADIPOR1 SNP genotypes
| SNPs | Crude OR | Adjusting for age* , other SNPs†, and race |
|
|---|---|---|---|
| rs266729 | CC | 1.00 | 1.00 |
| GG/CG | 1.13 (0.92–1.38) | 1.21 (0.96–1.53) | |
| rs822395 | CC | 1.00 | 1.00 |
| AA/AC | 1.17 (0.87–1.58) | 1.11 (0.77–1.59) | |
| rs822396 | GG | 1.00 | 1.00 |
| AA/AG | 1.54 (0.87–2.73) | 1.31 (0.70–2.46) | |
| rs2241766 | TT | 1.00 | 1.00 |
| GG/TG | 0.66 (0.53–0.83)‡ | 0.64 (0.49–0.83)‡ | |
| rs1501299 | TT | 1.00 | 1.00 |
| GG/TG | 1.44 (1.00–2.09) | 1.49 (0.99–2.25) | |
| rs2232853 | CC | 1.00 | 1.00 |
| TT/TC | 1.31 (1.07–1.62) | 1.46 (1.13–1.90)§ | |
| rs12733285 | TT | 1.00 | 1.00 |
| CC/TC | 1.14 (0.86–1.51) | 1.35 (0.92–1.97) | |
| rs1342387 | GG | 1.00 | 1.00 |
| AA/AG | 1.11 (0.87–1.42) | 1.15 (0.82–1.60) | |
| rs7539542 | GG | 1.00 | 1.00 |
| CC/C G | 0.83 (0.62–1.09) | 0.57 (0.35–0.94)‖ | |
| rs10920531 | CC | 1.00 | 1.00 |
| AA/AC | 0.89 (0.72–1.09) | 0.81 (0.57–1.15) |
Age adjustment was done by using categorical age groups (age >50 y and age <50 y).
Adjustment for SNPs was only done with SNPs from the same gene.
P < 0.001 without Bonferroni adjustment; P < 0.005 with Bonferroni adjustment.
P < 0.01 without Bonferroni adjustment; P < 0.05 with Bonferroni adjustment.
P < 0.05 without Bonferroni adjustment; 0.05 < P < 0.10 with Bonferroni adjustment.
Table 3.
Crude and adjusted ORs (95% CI) of breast cancer by ADIPOQ and ADIPOR1 SNP genotypes
| SNPs | Crude OR | OR adjusting for age* , SNPs†, and race |
|
|---|---|---|---|
| rs266729 | GG | 1.00 | 1.00 |
| GC | 1.15 (0.76–1.74) | 1.05 (0.65–1.68) | |
| CC | 1.00 (0.67–1.50) | 0.88 (0.55–1.42) | |
| rs822395 | CC | 1.00 | 1.00 |
| AC | 1.17 (0.85–1.61) | 1.03 (0.68–1.55) | |
| AA | 1.17 (0.85–1.61) | 0.97 (0.61–1.54) | |
| Rs822396 | GG | 1.00 | 1.00 |
| GA | 1.55 (0.85–2.81) | 1.39 (0.67–2.87) | |
| AA | 1.53 (0.86–2.73) | 1.65 (0.77–3.53) | |
| rs2241766 | TT | 1.00 | 1.00 |
| TG | 0.66 (0.52–0.83)‡ | 0.61 (0.46–0.80)‡ | |
| GG | 0.72 (0.41–1.28) | 0.63 (0.32–1.25) | |
| rs1501299 | TT | 1.00 | 1.00 |
| TG | 1.46 (0.99–2.16) | 1.59 (1.03–2.48)§ | |
| GG | 1.43 (0.98–2.09) | 1.80 (1.14–2.85)§ | |
| rs2232853 | CC | 1.00 | 1.00 |
| CT | 1.47 (1.18–1.84)‡ | 1.67 (1.23–2.26)‖ | |
| TT | 0.81 (0.54–1.21) | 1.02 (0.60–1.76) | |
| rs12733285 | TT | 1.00 | 1.00 |
| TC | 1.18 (0.87–1.60) | 1.16 (0.78–1.73) | |
| CC | 1.11 (0.82–1.50) | 1.19 (0.77–1.85) | |
| rs1342387 | AA | 1.00 | 1.00 |
| AG | 0.90 (0.71–1.14) | 0.74 (0.53–1.03) | |
| GG | 0.84 (0.63–1.12) | 0.72 (0.46–1.12) | |
| rs7539542 | GG | 1.00 | 1.00 |
| GC | 0.81 (0.60–1.09) | 0.59 (0.36–0.98)§ | |
| CC | 0.84 (0.62–1.14) | 0.51 (0.28–0.92)§ | |
| rs10920531 | CC | 1.00 | 1.00 |
| CA | 0.88 (0.71–1.10) | 0.80 (0.56–1.14) | |
| AA | 0.89 (0.67–1.20) | 0.61 (0.35–1.06) |
Age adjustment was done by using categorical age groups (age >50 y and age <50 y).
Adjustment for SNPs was only done with SNPs from the same gene.
P < 0.001 without Bonferroni adjustment; P < 0.005 with Bonferroni adjustment.
P < 0.05 without Bonferroni adjustment; 0.05 < P < 0.10 with Bonferroni adjustment.
P < 0.01 Without Bonferroni ajustment; P < 0.05 with Bonferroni adjustment.
Haplotypes of ADIPOR1 and breast cancer risk
Of the five haplotype SNPs genotyped, we found that rs2232853 CT genotype (OR, 1.67; 95% CI, 1.23–2.26) and the combination of rs7539542 GC (OR, 0.59; 95% CI, 0.36–0.98) and CC genotypes (OR, 0.57; 95% CI, 0.35–0.94) were significantly associated with breast cancer risk (Table 3 and Table 4). Studies on ADIPOR1 show that haplotype-tagging SNPs of the second block and especially rs7539542, are mostly associated with CAD and DM (26, 32). Moreover, rs7539542 GC and CC have been associated with higher mRNA levels compared with GG (26). Breast cancer PAR for individuals that harbor the rs7539542 GG genotype is 8 per 1,000 individuals.
Analysis according to age and ER and PR status
We analyzed our results according to ER and PR status among cases for which data were available. One genotype (rs1342387*CC) was significantly associated with ER positivity (OR, 4.09, 1.39–12.02; data not shown). However, the confidence intervals are wide given the small number of cases available for this analysis. The lack of clear associations between functional SNPs and hormone receptor status may not be a surprise because at present, there is no clearly shown association between adiponectin levels and ER status (4, 16, 19). Another explanation is the limited power of our analysis. There was also no association with age (data not shown).
Analysis according to signaling assessment
When comparing low signalers to intermediate and high signalers, we found that individuals who had been a priori categorized as intermediate signalers had 46% decreased breast cancer risk, and high signalers had 85% decreased risk for breast cancer (Ptrend = 0.001; refs. 21, 24, 29, 30; Table 5). When we used high signalers as our reference group, we found that intermediate signalers had a 4.16-fold increase in breast cancer risk (95% CI, 0.49–35.19) and low signalers had a 6.56-fold increase in breast cancer risk (95% CI, 0.78–54.89; Ptrend = 0.001). PAR for intermediate signalers was 25% and 38% for low signalers.
Table 5.
Adjusted OR for adiponectin signaling status
| Genotype combinations | n (cases/controls) | OR (95% CI) | OR (95% CI) adjusted for age | |
|---|---|---|---|---|
| Low signalers | 276*GG/45*TT | 581/598 | 1.0 | 1.0 |
| 276*GT/45*TT | ||||
| 276*GG/45*GT | ||||
| Intermediate signalers | 276*TT/45*TT | 124/191 | 0.67 (0.52–0.86) | 0.64 (0.49–0.83) |
| 276*GT/45*GT | ||||
| 276*GG/45*GG | ||||
| High signalers | 276*GT/45*GG | 1/8 | 0.13 (0.02–1.03) | 0.15 (0.02–1.28) |
| 276*TT/45*GG | ||||
| 276*TT/45*GT | ||||
| Ptrend | 0.001 | 0.001 |
Discussion
In this study, we found that two functionally relevant adiponectin polymorphisms, +45 T→G (rs2241766) and +276 G→T (rs 1501299), were significantly associated with breast cancer risk. Furthermore, a polymorphism of ADIPOR1, +10225 C→G (rs7539542), which has been shown to alter mRNA levels of the receptor (with the GG and CG polymorphisms increasing mRNA levels; ref. 26), was also significantly associated with breast cancer risk. This study shows that adiponectin signaling, as assessed by a combination of functionally relevant SNPs, may predict breast cancer risk.
High BMI has been associated with postmenopausal breast cancer (33, 34). The prevailing theory behind the association between obesity and breast cancer is based on the increased levels of estrogens and/or insulin resistance observed in obese women (35, 36). The established association between breast cancer and insulin resistance and obesity has drawn interest for adipokines, a group of proteins synthesized in the adipose tissue (4, 13, 37).
Adiponectin is secreted by adipose tissue, and its levels are inversely correlated to BMI. Several studies have shown that low levels of adiponectin are directly associated with increased breast cancer risk (11, 13, 15, 16, 18, 19). Adiponectin can act as a growth inhibitor for breast cancer cell lines. More specifically, adiponectin has been shown to inhibit the growth of the breast cancer cell lines MDA-MB-231 (38) and T47D (15). Furthermore, it was shown that in MCF-7 cells, growth stimulation with estradiol was suppressed by the presence of adiponectin (20). Several theories exist as to the link between adiponectin and breast carcinogenesis. Adiponectin is inversely correlated with insulin levels and is also associated with IGF-binding proteins, which have been associated with breast cancer (39, 40). Adiponectin has also been shown to inhibit the production of TNF-α in macrophages and its actions in endothelial cells (41), and has been found to bind several growth factors, such as fibroblast growth factor and platelet-derived growth factor-beta polypeptide, which can induce cell proliferation. Adiponectin also inhibits the activation of nuclear factor-κB, which is involved in breast cancer development (42). One of the downstream elements in the adiponectin pathway is 5′-AMP-activated protein kinase (AMPK; refs. 43, 44). Once activated, AMPK targets mammalian target of rapamycin (44), protein kinase B (43), and the c-Jun-NH2- kinase, and signal transducers and activators of transcription 3 pathways that are involved in breast carcinogenesis (45).
There are several factors that have been shown to regulate adiponectin levels. A Mediterranean diet (46) or a diet rich in whole grain and fat (47) have been shown to increase adiponectin levels. Also, better glycemic control can increase adiponectin levels (47). Physical activity has also been shown to influence adiponectin levels. In Zucker Diabetic Fatty rats, a rat model of DM, exercise was associated with a 28% increase in adiponectin levels (48).
Polymorphisms in the adiponectin gene have also been shown to correlate with adiponectin serum levels. Rs266729 (−11365 C→G) has been found, although not consistently, to correlate with adiponectin levels with the GG genotype being associated with decreased adiponectin levels (25). Rs1501299 (+276 G→T) has also been shown to correlate with adiponectin levels with the TT genotype having increased levels of adiponectin compared with GG (24). Finally, rs2241766 (+45 T→G) has been associated with adiponectin levels with the GG correlating with increased adiponectin levels compared with TT (23).
In this study, we found that two functionally significant adiponectin polymorphisms, rs1501299 and rs2241766, were significantly associated with breast cancer risk. More specifically, rs2241766 TG, which increases circulating levels of adiponectin, was associated with a 39% decreased risk for breast cancer (OR, 0.61; 95% CI, 0.46–0.80), whereas the homozygous GG genotype also showed a trend to decreased breast cancer risk (OR, 0.63; 95% CI, 0.32–1.25). Furthermore, rs1501299 TG and GG, which have been shown to decrease levels of circulating adiponectin, were associated with a 59% (OR, 1.59; 95% CI, 1.03–2.48) and 80% (OR, 1.80; 95% CI, 1.14–2.85) increased risk for breast cancer, respectively.
Given the previously shown functional significance of two of the SNPs tested (rs1501299 and rs2241766), we elected to divide patients into high, intermediate, and low signalers. Our hypothesis was that high signalers, i.e., individuals with higher predicted levels of adiponectin and/or expression of its receptor, would have a lower risk of breast cancer compared with low signalers. The findings provide support for our hypothesis in that individuals with intermediate and high adiponectin signaling had significantly lower breast cancer risk (OR, 0.64 and 0.15, respectively).
Haplotype-tagging SNPs for ADIPOR1 have also been shown to alter mRNA levels. More specifically, rs7539542 GG is associated with 30% to 40% lower ADIPOR1 mRNA levels than heterozygotes or CC homozygotes (26). This SNP has also been associated with risk for DM and CAD (26, 32). In our analysis, we found that rs7539542 CC and CG, which are associated with higher mRNA levels, i.e., more adiponectin receptor 1, are associated with a 43% lower risk for breast cancer.
A limitation in our study is the lack of exact age at the time of blood draw for a small portion of our controls as only age range was available. To address this, we did analyses controlling for age. We also conducted sensitivity analyses using hypothetical models to show that the effect of lack of detailed age information in a portion of our samples was negligible. It is possible that age differences in cases and controls affected the allele frequencies observed. Nonetheless, this would be expected to create a bias toward the null hypothesis because it would overestimate the deleterious allele frequency in controls given that a fraction of younger women who would have developed breast cancer were not removed from the control group. Thus, the younger mean age of controls could have resulted in a bias toward the null hypothesis, resulting in a weaker association. Furthermore, inaccurate information with respect to any variable classification would result in “random misclassification,” which would also be expected to result in suppression of effect estimates with a trend toward the null hypothesis. Other limitations of this study are the lack of information on confounding factors such as BMI and family history of breast cancer. However, it has been shown that the association of adiponectin with breast cancer is independent of BMI (15, 16, 19), and the presence of a strong family history of the disease would only weaken the power of our study. Our study also has several strengths. It includes a relatively large number of cases and controls from the same geographic location. The selection of our SNPs was based on previous studies, and the SNPs significantly associated with breast cancer are functionally significant. Although exposure was assessed in the context of a case control study, it is impossible that breast cancer would have changed SNP classification, which is already determined at birth. Thus, this study does fulfill the “time sequence” criterion for causality. This in association with existing epidemiologic evidence and biological plausibility support a causal association between these SNPs and risk for breast cancer.
There is currently only limited information on the allelic frequency of some of the SNPs studied in various ethnic groups.6 For example, the allelic frequency of rs2241766 has only been studied in 752 anonymous unrelated Japanese volunteers. There is no additional information on the allelic frequency in other populations besides the previously cited reports. With respect to rs2232853, the C:T allelic frequency differs between Caucasians (0.675:0.325), Asians (0.956:0.044), and sub-Saharan Africans (0.950:0.050). Finally, for rs7539542, the C:G allelic frequency also differs between Caucasians (0.712:0.288) and Asians (0.310:0.690), but there is no information on its frequency in other ethnic groups.
To our knowledge, this is the first study reporting an association of polymorphisms of adiponectin and its type I receptor with breast cancer risk. If confirmed in subsequent studies, our findings suggest that genetic variants of the adiponectin and adiponectin receptor 1 alter breast cancer risk. Our findings confirm the important role of adiponectin in breast cancer. In the future, we may be able to identify a population of breast cancer patients with low adiponectin levels, either due to genetic predisposition and/or environmental factors, who may benefit from adiponectin therapy. If these exciting results can be confirmed in other studies, the adiponectin axis may emerge as an important modifier of breast cancer risk.
Acknowledgments
Grant support: Walter S. Mander Foundation, Chicago, IL, grants CA112520 and CA108741 from the National Cancer Institute; the Jeannik M. Littlefield-AACR Grant in Metastatic Colon Cancer Research; Lynn Sage grant, ASCO career development award; and a discretionary grant from Beth Israel Deaconess Medical Center.
Footnotes
Conflict of Interest: None of the authors have conflicts of interest.
References
- 1.Jemal A, Siegel R, Ward E, et al. Cancer Statistics, 2006. CA Cancer J Clin. 2006;56:106–130. doi: 10.3322/canjclin.56.2.106. [DOI] [PubMed] [Google Scholar]
- 2.Wolk A, Gridley G, Svensson M, et al. A prospective study of obesity and cancer risk (Sweden) Cancer Causes Control. 2001;12:13–21. doi: 10.1023/a:1008995217664. [DOI] [PubMed] [Google Scholar]
- 3.Huang Z, Hankinson SE, Colditz GA, et al. Dual effects of weight and weight gain on breast cancer risk. JAMA. 1997;278:1407–1411. [PubMed] [Google Scholar]
- 4.Harvie M, Howell A, Vierkant RA, et al. Association of gain and loss of weight before and after menopause with risk of postmenopausal breast cancer in the Iowa women’s health study. Cancer Epidemiol Biomarkers Prev. 2005;14:656–661. doi: 10.1158/1055-9965.EPI-04-0001. [DOI] [PubMed] [Google Scholar]
- 5.Prentice RL, Caan B, Chlebowski RT, et al. Low-fat dietary pattern and risk of invasive breast cancer: the Women’s Health Initiative Randomized Controlled Dietary Modification Trial. JAMA. 2006;295:629–642. doi: 10.1001/jama.295.6.629. [DOI] [PubMed] [Google Scholar]
- 6.Vona-Davis L, Howard-McNatt M, Rose DP. Adiposity, type 2 diabetes and the metabolic syndrome in breast cancer. Obes Rev. 2007;8:395–408. doi: 10.1111/j.1467-789X.2007.00396.x. [DOI] [PubMed] [Google Scholar]
- 7.Larsson SC, Mantzoros CS, Wolk A. Diabetes mellitus and risk of breast cancer: a meta-analysis. Int J Cancer. 2007;121:856–862. doi: 10.1002/ijc.22717. [DOI] [PubMed] [Google Scholar]
- 8.McTiernan A, Wu L, Chen C, et al. Relation of BMI and physical activity to sex hormones in postmenopausal women. Obesity Silver Spring. 2006;14:1662–1677. doi: 10.1038/oby.2006.191. [DOI] [PubMed] [Google Scholar]
- 9.McTiernan A, Ulrich C, Slate S, Potter J. Physical activity and cancer etiology: associations and mechanisms. Cancer Causes Control. 1998;9:487–509. doi: 10.1023/a:1008853601471. [DOI] [PubMed] [Google Scholar]
- 10.Pazaitou-Panayiotou K, Kelesidis T, Kelesidis I, Kaprara A, Blakeman J, Vainas I, et al. Growth hormone-binding protein is directly and IGFBP-3 is inversely associated with risk of female breast cancer. Eur J Endocrinol. 2007;156:187–194. doi: 10.1530/EJE-06-0611. [DOI] [PubMed] [Google Scholar]
- 11.Eliassen AH, Tworoger SS, Mantzoros CS, Pollak MN, Hankinson SE. Circulating insulin and c-peptide levels and risk of breast cancer among predominately premenopausal women. Cancer Epidemiol Biomarkers Prev. 2007;16:161–164. doi: 10.1158/1055-9965.EPI-06-0693. [DOI] [PubMed] [Google Scholar]
- 12.Ouchi N, Kihara S, Arita Y, et al. Novel modulator for endothelial adhesion molecules: adipocyte-derived plasma protein adiponectin. Circulation. 1999;100:2473–2476. doi: 10.1161/01.cir.100.25.2473. [DOI] [PubMed] [Google Scholar]
- 13.Rose DP, Komninou D, Stephenson GD. Obesity, adipocytokines, and insulin resistance in breast cancer. Obes Rev. 2004;5:153–165. doi: 10.1111/j.1467-789X.2004.00142.x. [DOI] [PubMed] [Google Scholar]
- 14.Chabrolle C, Tosca L, Dupont J. Regulation of adiponectin and its receptors in rat ovary by human chorionic gonadotrophin treatment and potential involvement of adiponectin in granulosa cell steroidogenesis. Reproduction. 2007;133:719–731. doi: 10.1530/REP-06-0244. [DOI] [PubMed] [Google Scholar]
- 15.Korner A, Pazaitou-Panayiotou K, Kelesidis T, et al. Total and high-molecular-weight adiponectin in breast cancer: in vitro and in vivo studies. J Clin Endocrinol Metab. 2007;92:1041–1048. doi: 10.1210/jc.2006-1858. [DOI] [PubMed] [Google Scholar]
- 16.Miyoshi Y, Funahashi T, Kihara S, et al. Association of serum adiponectin levels with breast cancer risk. Clin Cancer Res. 2003;9:5699–5704. [PubMed] [Google Scholar]
- 17.Snoussi K, Strosberg AD, Bouaouina N, Ben AS, Helal AN, Chouchane L. Leptin and leptin receptor polymorphisms are associated with increased risk and poor prognosis of breast carcinoma. BMC Cancer. 2006;6:38. doi: 10.1186/1471-2407-6-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mantzoros C, Petridou E, Dessypris N, et al. Adiponectin and breast cancer risk. J Clin Endocrinol Metab. 2004;89:1102–1107. doi: 10.1210/jc.2003-031804. [DOI] [PubMed] [Google Scholar]
- 19.Chen DC, Chung YF, Yeh YT, et al. Serum adiponectin and leptin levels in Taiwanese breast cancer patients. Cancer Lett. 2006;237:109–114. doi: 10.1016/j.canlet.2005.05.047. [DOI] [PubMed] [Google Scholar]
- 20.Dieudonne MN, Bussiere M, Dos SE, Leneveu MC, Giudicelli Y, Pecquery R. Adiponectin mediates anti-proliferative and apoptotic responses in human MCF7 breast cancer cells. Biochem Biophys Res Commun. 2006;345:271–279. doi: 10.1016/j.bbrc.2006.04.076. [DOI] [PubMed] [Google Scholar]
- 21.Filippi E, Sentinelli F, Romeo S, et al. The adiponectin gene SNP+276G>T associates with early-onset coronary artery disease and with lower levels of adiponectin in younger coronary artery disease patients (age <or = 50 years) J Mol Med. 2005;83:711–719. doi: 10.1007/s00109-005-0667-z. [DOI] [PubMed] [Google Scholar]
- 22.Hara K, Boutin P, Mori Y, et al. Genetic variation in the gene encoding adiponectin is associated with an increased risk of type 2 diabetes in the Japanese population. Diabetes. 2002;51:536–540. doi: 10.2337/diabetes.51.2.536. [DOI] [PubMed] [Google Scholar]
- 23.Heid IM, Wagner SA, Gohlke H, et al. Genetic architecture of the APM1 gene and its influence on adiponectin plasma levels and parameters of the metabolic syndrome in 1,727 healthy Caucasians. Diabetes. 2006;55:375–384. doi: 10.2337/diabetes.55.02.06.db05-0747. [DOI] [PubMed] [Google Scholar]
- 24.Menzaghi C, Ercolino T, Di PR, et al. A haplotype at the adiponectin locus is associated with obesity and other features of the insulin resistance syndrome. Diabetes. 2002;51:2306–2312. doi: 10.2337/diabetes.51.7.2306. [DOI] [PubMed] [Google Scholar]
- 25.Qi L, Doria A, Manson JE, et al. Adiponectin genetic variability, plasma adiponectin, and cardiovascular risk in patients with type 2 diabetes. Diabetes. 2006;55:1512–1516. doi: 10.2337/db05-1520. [DOI] [PubMed] [Google Scholar]
- 26.Soccio T, Zhang YY, Bacci S, et al. Common haplotypes at the adiponectin receptor 1 (ADIPOR1) locus are associated with increased risk of coronary artery disease in type 2 diabetes. Diabetes. 2006;55:2763–2770. doi: 10.2337/db06-0613. [DOI] [PubMed] [Google Scholar]
- 27.Vasseur F, Helbecque N, Dina C, et al. Singlenucleotide polymorphism haplotypes in the both proximal promoter and exon 3 of the APM1 gene modulate adipocyte-secreted adiponectin hormone levels and contribute to the genetic risk for type 2 diabetes in French Caucasians. Hum Mol Genet. 2002;11:2607–2614. doi: 10.1093/hmg/11.21.2607. [DOI] [PubMed] [Google Scholar]
- 28.Crimmins NA, Martin LJ. Polymorphisms in adiponectin receptor genes ADIPOR1 and ADIPOR2 and insulin resistance. Obes Rev. 2007;8:419–423. doi: 10.1111/j.1467-789X.2007.00348.x. [DOI] [PubMed] [Google Scholar]
- 29.Kang ES, Park SY, Kim HJ, et al. The influence of adiponectin gene polymorphism on the rosiglitazone response in patients with type 2 diabetes. Diabetes Care. 2005;28:1139–1144. doi: 10.2337/diacare.28.5.1139. [DOI] [PubMed] [Google Scholar]
- 30.Yang WS, Tsou PL, Lee WJ, et al. Allele-specific differential expression of a common adiponectin gene polymorphism related to obesity. J Mol Med. 2003;81:428–434. doi: 10.1007/s00109-002-0409-4. [DOI] [PubMed] [Google Scholar]
- 31.Loos RJ, Ruchat S, Rankinen T, Tremblay A, Perusse L, Bouchard C. Adiponectin and adiponectin receptor gene variants in relation to resting metabolic rate, respiratory quotient, and adiposity-related phenotypes in the Quebec Family Study. Am J Clin Nutr. 2007;85:26–34. doi: 10.1093/ajcn/85.1.26. [DOI] [PubMed] [Google Scholar]
- 32.Qi L, Doria A, Giorgi E, Hu FB. Variations in adiponectin receptor genes and susceptibility to type 2 diabetes in women: a tagging-single nucleotide polymorphism haplotype analysis. Diabetes. 2007;56:1586–1591. doi: 10.2337/db06-1447. [DOI] [PubMed] [Google Scholar]
- 33.Trentham-Dietz A, Newcomb PA, Egan KM, et al. Weight change and risk of postmenopausal breast cancer (United States) Cancer Causes Control. 2000;11:533–542. doi: 10.1023/a:1008961931534. [DOI] [PubMed] [Google Scholar]
- 34.Wenten M, Gilliland FD, Baumgartner K, Samet JM. Associations of weight, weight change, and body mass with breast cancer risk in Hispanic and non-Hispanic white women. Ann Epidemiol. 2002;12:435–434. doi: 10.1016/s1047-2797(01)00293-9. [DOI] [PubMed] [Google Scholar]
- 35.Hankinson SE, Willett WC, Manson JE, et al. Alcohol, height, and adiposity in relation to estrogen and prolactin levels in postmenopausal women. J Natl Cancer Inst. 1995;87:1297–1302. doi: 10.1093/jnci/87.17.1297. [DOI] [PubMed] [Google Scholar]
- 36.Holmes MD, Willett WC. Does diet affect breast cancer risk? Breast Cancer Res. 2004;6:170–178. doi: 10.1186/bcr909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stephenson GD, Rose DP. Breast cancer and obesity: an update. Nutr Cancer. 2003;45:1–16. doi: 10.1207/S15327914NC4501_1. [DOI] [PubMed] [Google Scholar]
- 38.Rose DP, Connolly JM, Liu XH. Effects of linoleic acid on the growth and metastasis of two human breast cancer cell lines in nude mice and the invasive capacity of these cell lines in vitro. Cancer Res. 1994;54:6557–6562. [PubMed] [Google Scholar]
- 39.Kaaks R, Toniolo P, Akhmedkhanov A, et al. Serum C-peptide, insulin-like growth factor (IGF)-I, IGF-binding proteins, and colorectal cancer risk in women. J Natl Cancer Inst. 2000;92:1592–1600. doi: 10.1093/jnci/92.19.1592. [DOI] [PubMed] [Google Scholar]
- 40.Moschos S, Chan JL, Mantzoros CS. Leptin and reproduction: a review. Fertil Steril. 2002;77:433–444. doi: 10.1016/s0015-0282(01)03010-2. [DOI] [PubMed] [Google Scholar]
- 41.Ouchi N, Shibata R, Walsh K. AMP-activated protein kinase signaling stimulates VEGF expression and angiogenesis in skeletal muscle. Circ Res. 2005;96:838–846. doi: 10.1161/01.RES.0000163633.10240.3b. [DOI] [PubMed] [Google Scholar]
- 42.Wang Y, Lam KSL, Xu JY, et al. Adiponectin inhibits cell proliferation by interacting with several growth factors in an oligomerization-dependent manner. J Biol Chem. 2005;280:18341–18347. doi: 10.1074/jbc.M501149200. [DOI] [PubMed] [Google Scholar]
- 43.Goldstein BJ, Scalia R. Adiponectin: a novel adipokine linking adipocytes and vascular function. J Clin Endocrinol Metab. 2004;89:2563–2568. doi: 10.1210/jc.2004-0518. [DOI] [PubMed] [Google Scholar]
- 44.Luo Z, Saha AK, Xiang X, Ruderman NB. AMPK, the metabolic syndrome and cancer. Trends Pharmacol Sci. 2005;26:69–76. doi: 10.1016/j.tips.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 45.Miyazaki T, Bub JD, Uzuki M, Iwamoto Y. Adiponectin activates c-Jun NH2-terminal kinase and inhibits signal transducer and activator of transcription 3. Biochem Biophys Res Commun. 2005;333:79–87. doi: 10.1016/j.bbrc.2005.05.076. [DOI] [PubMed] [Google Scholar]
- 46.Mantzoros CS, Williams CJ, Manson JE, Meigs JB, Hu FB. Adherence to the Mediterranean dietary pattern is positively associated with plasma adiponectin concentrations in diabetic women. Am J Clin Nutr. 2006;84:328–335. doi: 10.1093/ajcn/84.1.328. [DOI] [PubMed] [Google Scholar]
- 47.Mantzoros CS, Li T, Manson JE, Meigs JB, Hu FB. Circulating adiponectin levels are associated with better glycemic control, more favorable lipid profile, and reduced inflammation in women with type 2 diabetes. J Clin Endocrinol Metab. 2005;90:4542–4548. doi: 10.1210/jc.2005-0372. [DOI] [PubMed] [Google Scholar]
- 48.de Lemos ET, Reis F, Baptista S, et al. Exercise training is associated with improved levels of C-reactive protein and adiponectin in ZDF (type 2) diabetic rats. Med Sci Monit. 2007;13:BR168–BR174. [PubMed] [Google Scholar]