Summary of recent advances
Copper plays a key role in the physiology of methanotrophs. One way that these bacteria meet their high copper requirement is by the biosynthesis and release of high affinity copper-binding compounds called methanobactins. Recent advances in methanobactin characterization include the first crystal structure, detailed spectroscopic analyses, and studies of metal ion specificity. Methanobactin may function in copper uptake, regulation of methane monooxygenase expression, protection against copper toxicity, and particulate methane monooxygenase activity. Methanobactin can extract copper from insoluble minerals and could be important for mineral weathering. Many methanobactin properties are reminiscent of iron siderophores, suggesting a similar mechanism of handling. Methanobactin-like compounds have also been identified in yeast mitochondria, suggesting that these molecules are a more universal phenomenon.
Introduction
Copper is central to the physiology of methanotrophs, gram-negative bacteria that use methane as their sole carbon source [1]. In the first step of carbon assimilation by methanotrophs, methane is oxidized to methanol by methane monooxygenase enzymes (MMOs). All methanotrophs except for one species produce particulate MMO (pMMO), a three-subunit, integral membrane protein complex that is widely believed to contain a copper active site [2]. Under conditions of copper starvation, a few methanotrophs can also express a soluble MMO (sMMO) that contains a diiron active site. In strains that express both pMMO and sMMO, copper in the growth medium represses transcription of the sMMO genes while leading to the expression of pMMO and formation of extensive intracytoplasmic membranes that house pMMO. A number of additional proteins are also differentially expressed as a function of copper concentrations in the growth medium [3]. Given the important role of copper in both metabolic switching and pMMO chemistry, an effective copper acquisition system is critical.
Evidence for a specific copper uptake system was obtained initially in the early 1990s from studies of Methylosinus trichosporium OB3b mutants that constitutively express sMMO and do not express active pMMO [4,5]. The extracellular media from these strains also contains unusually high concentrations of copper. On the basis of these data, Georgiou and coworkers postulated the existence of small copper binding complexes that are excreted by the cells under copper starvation conditions [5]. Over the next decade, several laboratories isolated and partially characterized several such compounds from both M. trichosporium OB3b [6,7] and Methylococcus capsulatus (Bath) [8]. Designated copper binding compounds (CBCs) or copper binding ligands (CBLs), these molecules had molecular masses ranging from 382 to 1218 Da and bound copper ions with high affinity. Issues with homogeneity and degradation precluded determining the chemical composition of these compounds until recently, however. After the development of a successful purification protocol by Graham, DiSpirito and coworkers, the crystal structure of a copper binding compound from M. trichosporium OB3b was reported in 2004, and the molecule was renamed methanobactin (mb) [9–11]. The crystal structure and subsequent biochemical, spectroscopic, and functional studies over the past three years have provided new insight into the properties and function of mb. This review summarizes these recent advances and considers the possibility that mb-like molecules may be a more general phenomenon.
Structure of methanobactin
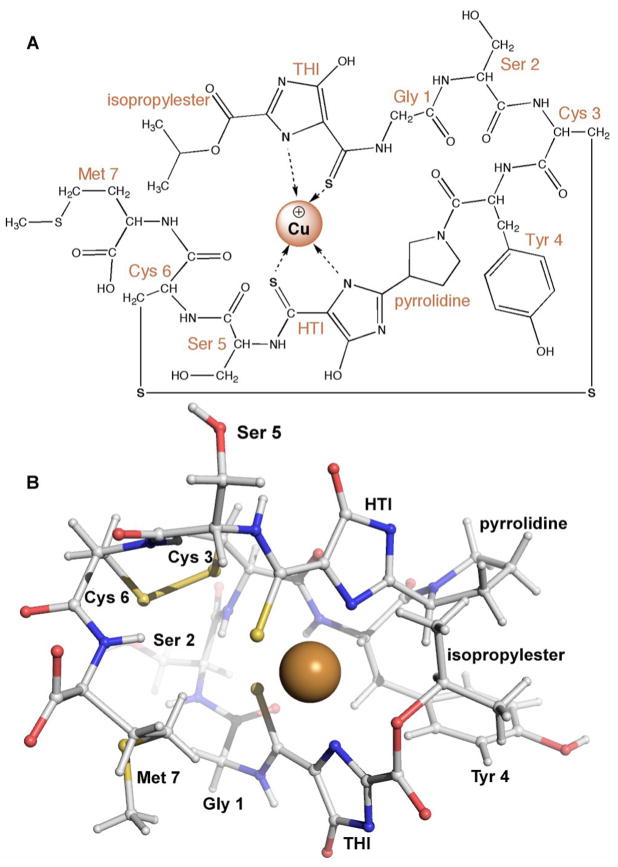
The crystal structure of copper-loaded mb (Cu-mb) from M. trichosporium OB3b revealed a 1217 Da molecule with a chemical composition of C45N12O14H62S5Cu [11]. Both amino acids and non-amino acid functional groups are present with the sequence (N-2-isopropyl ester-(4-thionyl-5-hydroxyimidazolate)-Gly1-Ser2-Cys3-Tyr4-pyrrolidine-(4-hydroxy-5-thionylimidazolate)-Ser5-Cys6-Met7) (Figure 1A). The two cysteine residues form a disulfide bond. The peptidic nature is reminiscent of many iron siderophores [12,13], and led to replacing the term CBC or CBL with methanobactin and referring to it as a chalkophore (chalko is Greek for copper; sidero is Greek for iron). Cu-mb adopts a pyramid-like shape with the copper ion bound at the base of the pyramid (Figure 1B). The copper ion is exposed, but afforded some protection by the isopropylester group. The 4-thionyl-5-hydroxy-imidazolate (THI) and 4-hydroxy-5-thionyl-imidazolate (HTI) moieties each provide one nitrogen and one sulfur ligand, resulting in distorted tetrahedral coordination geometry with Cu-N distances of 2.00 and 2.05 Å and Cu-S distances of 2.38 and 2.39 Å.
Figure 1.
Structure of Cu-mb from M. trichosporium OB3b. (A) Schematic diagram. (B) Ball-and-stick representation of crystal structure (Cambridge Crystallographic Data Center deposition number CCDC 241254). The copper ion is shown as a brown sphere.
By analogy to siderophores, a nonribosomal peptide synthetase (NRPS) [12] may be involved in mb biosynthesis. A NRPS is a multimodular enzyme assembly line that generates peptide products without using an RNA template. The M. capsulatus (Bath) genome encodes two putative NRPS modules [14]. One (MCA2107, 1456 amino acids) includes an adenylation domain, a thiolation domain, and an acetylation domain. The other (MCA1883, 1314 amino acids) contains an N-terminal condensation domain, a central AMP binding domain, and a C-terminal thioesterase domain. There is no evidence linking these gene products with mb biosynthesis, however. Moreover, little is known about iron uptake by methanotrophs, leaving open the possibility that the predicted NRPS produces siderophores.
Spectroscopic properties of methanobactin
The oxidation state of copper in M. trichosporium OB3b Cu-mb has been investigated by several spectroscopic techniques. First, X-ray photoelectron spectroscopy (XPS) suggests the copper is present primarily as Cu(I) [11]. Second, the X-ray absorption near edge spectrum (XANES) shows a 1s→4p transition at 8985 eV diagnostic of Cu(I) and no features attributable to Cu(II) [15]. A weak electron paramagnetic resonance (EPR) signal is observed for M. trichosporium OB3b Cu-mb, but its parameters are more consistent with Cu(II) coordinated by oxygen/nitrogen ligands rather than sulfur ligands. This signal likely arises from adventitiously bound Cu(II) and is similar to spectra reported earlier for heterogeneous M. trichosporium OB3b [6] and M. capsulatus (Bath) [8] preparations. EPR signals attributable to Cu(II) with nitrogen/sulfur coordination are observed upon titration with small amounts of Cu(II), but these signals disappear in < 10 min [9]. Thus, mb always contains Cu(I) regardless of whether it is initially loaded with Cu(II) or Cu(I). How mb reduces Cu(II) to Cu(I) is not known, but possible scenarios include coupling to disulfide bond formation [15] or involvement of the thionyl ligands [10].
The apo form of mb from M. trichosporium OB3b exhibits a distinct optical spectrum with several features in the 250–300 nm range [10,16], including a small peak at 302 nm attributed to the phenoxide form of tyrosine [16] and a feature at 254 nm that may be associated with the two cysteine residues [10]. This feature initially increases in intensity upon copper addition and then decreases when the Cu:mb ratio exceeds 0.25. These changes could indicate disulfide formation between the two cysteines, but have also been proposed to involve charge transfer between the copper and the imidazolate or thionyl groups [16]. Two peaks at 340 or 342 nm and 388 or 394 nm have been assigned to the HTI and THI groups, respectively. The exact energy of these bands, which are affected by light, copper addition, and pH, depends on whether apo mb is isolated directly (as-isolated) [16] or prepared from Cu-mb by dialysis versus EDTA [10].
The two methods of preparing apo mb for spectroscopic analysis also result in slightly different fluorescence spectra. Excitation at 282, 340 or 342, and 388 or 394 nm yields distinct emission peaks [10,16]. The most intense band at 310 nm is observed upon excitation at 282 nm and is due to tyrosine [16]. Excitation at the other wavelengths leads to broad emission bands in the 400–500 nm range. Additional maxima at 610 and 675 nm are observed for the as-isolated, but not the EDTA-treated mb. Similar to what is observed upon iron binding to siderophores, the fluorescence spectrum of mb is quenched upon copper addition, with differences in the degree of quenching observed for the two preparations. Copper also alters the circular dichroism (CD) spectrum of mb [16].
Kinetics and thermodynamics of copper binding by methanobactin
By monitoring absorption changes at 340 nm (THI) upon addition of subsaturating Cu(II) concentrations to M. trichosporium OB3b mb, kobs values of 121 ± 9 s−1 for Cu(II) and 8.27 ± 0.16 s−1 for Cu(I) were obtained. If the absorption was monitored at 394 nm (HTI), the rate was estimated to be > 640 s−1, suggesting that copper is initially coordinated by HTI [16]. Using chelators, the mb copper binding affinity has been estimated to be ~1016 M−1 [7] or more recently, > 8 × 1016 M−1 [16]. Isothermal calorimetry (ITC) studies performed at different Cu(II) to mb ratios gave several binding constants of different magnitudes. On the basis of the ITC data and the effects of different Cu(II):mb ratios on the XPS, optical, fluorescence and CD spectra, DiSpirito and coworkers have proposed a multistep mechanism for the formation of Cu-mb. In their model, Cu(II) initially binds to a mb homodimer and is coordinated by the imidazolate nitrogens and thionyl sulfurs from the two THI groups. Following reduction to Cu(I), the coordination environment is proposed to switch to the four thionyl sulfurs from the two THI and the two HTI groups. Binding of a second Cu(II) ion then results in the final structure observed by crystallography. However, this mechanism is only consistent with data obtained for Cu(II) binding; only a mb monomer appears to be involved for Cu(I).
The ability of mb to bind metal ions besides copper has also been investigated. Addition of excess nickel to mixtures of copper and partially purified M. trichosporium OB3b mb only produced Cu-mb, suggesting a high specificity for copper [7]. More recently, the effects of Ag(I), Au(III), Cd(II), Co(II), Fe(III), Hg(II), Mn(II), Ni(II), Pb(II), U(IV) and Zn(II) on the optical, fluorescence, CD, and XPS spectra of mb were studied [17]. On the basis of this analysis, the metal ions were divided into two classes. The group A metals include Ag(I), Au(III), Hg(II), Pb(II), and U(IV), and are likely coordinated by a mb monomer in a similar fashion to copper. The group B metals include Cd(II), Co(II), Fe(III), Mn(II), Ni(II) and Zn(II), and are proposed to bind two or four mb molecules via the THI groups. According to ITC measurements, the affinity of mb for these alternative metal ions is much lower than its affinity for copper.
Function of methanobactin
Early observations correlating limiting copper concentrations during growth with the presence of mb in spent media [6–8] combined with the similarity to siderophores suggest that mb plays an important role in copper uptake. In recent work, a marked effect was observed upon addition of copper and mb to M. trichosporium OB3b growth medium concomitant with a switch from copper-starved conditions to 10 μM copper. A growth lag typically observed upon a sudden elevation of copper levels was shortened significantly and the cells grew faster [10]. These data are consistent with a role in copper uptake. In addition, mb may reduce copper toxicity. In recent experiments, 16S-rRNA transcript levels were monitored as a function of CuCl2 and mb addition. Only the addition of 1:1 ratios of copper and mb prevented an immediate decrease in 16S-rRNA transcript levels, consistent with a role in suppressing copper toxicity [18]. This protective effect might derive from superoxide dismutase activity, which has been observed for Cu-mb [19].
The ability of mb to facilitate copper uptake from different sources has also been investigated by monitoring the transcript levels of pmoA (pMMO) and mmoX (sMMO) as a function of added copper and mb [18]. When copper was presented as CuCl2, pmoA increased and mmoX decreased regardless of whether exogenous mb was provided. However, when copper was presented as synthetic Cu-doped Fe oxide or Cu-doped borosilicate glass, mb was required to observe the increase in pmoA transcript and the decrease in mmoX transcript. Thus, mb can make copper more available when it is only present as an insoluble mineral. In related work, mb was demonstrated to increase the rate of glass dissolution of Cu-substituted glasses [20]. Thus, methanotrophs may impact mineral weathering via mb production in certain environments. Similarly, siderophore-producing bacteria and siderophores can promote dissolution of iron-containing minerals [21]. These studies represent an important first step toward understanding mb function in nature, not just as it pertains to methanotroph physiology but as a player in the global carbon cycle.
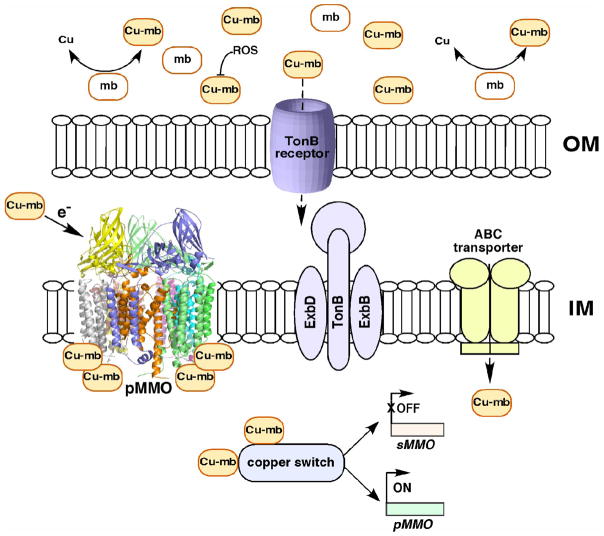
It is not known how methanotrophs internalize Cu-mb. In gram-negative bacteria, TonB-dependant receptors mediate the recognition and uptake of siderophores. Transport across the outer membrane is driven by the cytoplasmic membrane complex, TonB-ExbB-ExbD [22]. This machinery is also utilized for the uptake of cobalamin and a nickel-specific, TonB-dependent system has been identified in Helicobacter pylori [23]. Transport into the cytoplasm is then accomplished by periplasmic binding proteins and ABC transporters. It is conceivable that a similar mechanism exists for the recognition and uptake of Cu-mb (Figure 2). There are six ORFs in the M. capsulatus (Bath) genome that correspond to putative TonB receptors (MCA0440, MCA1957, MCA2074, MCA2180, MCA2321, MCA2751), and one or more of these could be specific for Cu-mb. In addition, the genome encodes a large number of ABC transporters.
Figure 2.
Possible functions of Cu-mb and proposed mechanism of uptake: mb is involved in extracellular copper uptake and is postulated to protect the cells from ROS. Cu-mb in the periplasmic space may be involved in pMMO catalytic activity. In the cytoplasm, Cu-mb may participate in the copper-dependent regulation of MMOs. Cu-mb may be recognized and internalized by integral outer membrane (OM) TonB receptors, inner membrane (IM) embedded TonB-ExbB-ExbD proteins, and ABC transporters.
Finally, there is evidence that mb interacts directly with pMMO. In some preparations, Cu-mb is associated with purified pMMO, and its dissociation decreases enzyme activity [8,19]. Addition of purified M. trichosporium OB3b Cu-mb to M. capsulatus (Bath) cells or membranes containing pMMO increased activity by ~35% as compared to 20% by adding Cu(II) alone [9]. The effects of adding M. trichosporium OB3b Cu-mb to M. trichosporium OB3b pMMO or M. capsulatus (Bath) Cu-mb to M. capsulatus (Bath) pMMO have not been reported. Such data would be useful, especially since it is not known whether the mb molecules from the two methanotrophs have the same chemical composition. The EPR spectrum of pMMO is also affected by Cu-mb, suggesting a possible role in electron transfer to the pMMO copper center(s) [9]. In sum, Cu-mb may function in copper uptake, protection against copper toxicity and resultant reactive oxygen species (ROS), copper-dependent regulation of the two MMOs, and pMMO catalytic activity (Figure 2).
Copper binding compounds from other organisms
In 2004, Winge and coworkers reported that the bulk of copper in the Saccharomyces cerevisiae mitochondrion is present as a soluble, non-proteinaceous, low molecular weight complex, designated CuL. This copper pool is localized to the matrix and is distinct from the two mitochondrial copper-containing enzymes, cytochrome c oxidase and superoxide dismutase [24]. The amount of CuL increases when cells are grown in a high copper medium, and CuL is also found in the cytoplasm. CuL is resistant to protease treatment, cannot be detected by SDS PAGE or mass spectrometry, and is fluorescent with excitation and emission maxima of 220 and 360 nm, respectively. Removal of copper enhances the fluorescence intensity, and quenching can be recovered by addition of Cu(I) preferentially over Cu(II), but not by Zn(II) or Fe(II). Based on competition assays with BCS, CuL is predicted to have an affinity for Cu(I) of only slightly less than 1019 [25]. Thus, many properties of CuL are reminiscent of Cu-mb. Notably, CuL is detected not only in yeast, but also in mouse liver [25]. The plant phytochelatins bear some similarity to Cu-mb and CuL as well. These low molecular weight peptides, also produced by algae and some fungi, have the sequence (γ-GluCys)n-Gly (n = 2–5), and are biosynthesized nonribosomally. Phytochelatin production is stimulated by a number of metal ions, including copper, and complexes with copper, silver, and cadmium have been detected in vivo [26].
Conclusions
The discovery and recent characterization of Cu-mb and the related yeast CuL indicate that the siderophore-mediated strategy of iron uptake and control may be a more general phenomenon. Characterization of Cu-mb from additional species of methanotrophs will provide insight into the diversity of mb structure and what role it may play in interspecies competition for metal ions. Further functional studies are also warranted. Finally, the details of Cu-mb biosynthesis and uptake remain to be unraveled. Methanobactin’s time in the bioinorganic arena has come.
Acknowledgments
Related work in the Rosenzweig laboratory on pMMO is supported by NIH GM070473.
Abbreviations
- CBC
copper binding compound
- CBL
copper binding ligand
- CD
circular dichroism
- Cu-mb
copper-loaded methanobactin
- EPR
electron paramagnetic resonance spectroscopy
- HTI
4-hydroxy-5-thionyl-imidazolate
- ITC
isothermal titration calorimetry
- mb
methanobactin
- MMO
methane monooxygenase
- NRPS
nonribosomal peptide synthetase
- pMMO
particulate methane monooxygenase
- ROS
reactive oxygen species
- sMMO
soluble methane monooxygenase
- THI
4-thionyl-5-hydroxy-imidazolate
- XANES
X-ray absorption near edge spectrum
- XPS
X-ray photoelectron spectroscopy
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
- 1.Hanson RS, Hanson TE. Methanotrophic bacteria. Microbiol Rev. 1996;60:439–471. doi: 10.1128/mr.60.2.439-471.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balasubramanian R, Rosenzweig AC. Structural and mechanistic insights into methane oxidation by particulate methane monooxygenase. Acc Chem Res. 2007;40:573–580. doi: 10.1021/ar700004s. [DOI] [PubMed] [Google Scholar]
- 3.Hakemian AS, Rosenzweig AC. The biochemistry of methane oxidation. Ann Rev Biochem. 2007:76. doi: 10.1146/annurev.biochem.76.061505.175355. [DOI] [PubMed] [Google Scholar]
- 4.Phelps PA, Agarwal SK, Speitel GE, Jr, Georgiou G. Methylosinus trichosporium OB3b mutants having constitutive expression of soluble methane monooxygenase in the presence of high levels of copper. Appl Environ Microbiol. 1992;58:3701–3708. doi: 10.1128/aem.58.11.3701-3708.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fitch MW, Graham DW, Arnold RG, Agarwal SK, Phelps P, Speitel GE, Jr, Georgiou G. Phenotypic characterization of copper-resistant mutants of Methylosinus trichosporium OB3b. Appl Environ Microbiol. 1993;59:2771–2776. doi: 10.1128/aem.59.9.2771-2776.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DiSpirito AA, Zahn JA, Graham DW, Kim HJ, Larive CK, Derrick TS, Cox CD, Taylor AB. Copper-binding compounds from Methylosinus trichosporium OB3b. J Bacteriol. 1998;180:3606–3613. doi: 10.1128/jb.180.14.3606-3613.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Téllez CM, Gaus KP, Graham DW, Arnold RG, Guzman RZ. Isolation of copper biochelates from Methylosinus trichosporium OB3b and soluble methane monooxygenase mutants. Appl Environ Microbiol. 1998;64:1115–1122. doi: 10.1128/aem.64.3.1115-1122.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zahn JA, DiSpirito AA. Membrane-associated methane monooxygenase from Methylococcus capsulatus (Bath) J Bacteriol. 1996;178:1018–1029. doi: 10.1128/jb.178.4.1018-1029.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9•.Choi DW, Antholine WE, Do YS, Semrau JD, Kisting CJ, Kunz RC, Campbell D, Rao V, Hartsel SC, DiSpirito AA. Effect of methanobactin on the activity and electron paramagnetic resonance spectra of the membrane-associated methane monooxygenase in Methylococcus capsulatus. Bath Microbiol. 2005;151:3417–3426. doi: 10.1099/mic.0.28169-0. The possible linkage between Cu-mb and pMMO activity is investigated through activity measurements and EPR spectroscopy. [DOI] [PubMed] [Google Scholar]
- 10•.Kim HJ, Galeva N, Larive CK, Alterman M, Graham DW. Purification and physical-chemical properties of methanobactin: a chalkophore from Methylosinus trichosporiumOB3b . Biochemistry. 2005;44:5140–5148. doi: 10.1021/bi047367r. The purification protocol that resulted in homogeneous, intact, and crystallizable samples of Cu-mb is detailed. [DOI] [PubMed] [Google Scholar]
- 11••.Kim HJ, Graham DW, DiSpirito AA, Alterman MA, Galeva N, Larive CK, Asunskis D, Sherwood PMA. Methanobactin, a copper-aquisition compound from methane oxidizing bacteria. Science. 2004;305:1612–1615. doi: 10.1126/science.1098322. The first crystal structure of Cu-mb reveals its chemical composition and copper coordination geometry. [DOI] [PubMed] [Google Scholar]
- 12.Crosa JH, Walsh CT. Genetics and assembly line enzymology of siderophore biosynthesis in bacteria. Microbiol Molec Biol Rev. 2002;66:223–249. doi: 10.1128/MMBR.66.2.223-249.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drechsel H, Jung G. Peptide siderophores. J Pept Sci. 1998;4:147–181. doi: 10.1002/(SICI)1099-1387(199805)4:3%3C147::AID-PSC136%3E3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 14.Ward N, Larsen O, Sakwa J, Bruseth L, Khouri H, Durkin AS, Dimitrov G, Jiang L, Scanlan D, Kang KH, et al. Genomic insights into methanotrophy: the complete genome sequence of Methylococcus capsulatus (Bath) PLoS Biol. 2004;2:e303. doi: 10.1371/journal.pbio.0020303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hakemian AS, Tinberg CE, Kondapalli KC, Telser J, Hoffman BM, Stemmler TL, Rosenzweig AC. The copper chelator methanobactin from Methylosinus trichosporium OB3b binds Cu(I) J Am Chem Soc. 2005;127:17142–17143. doi: 10.1021/ja0558140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16••.Choi DW, Zea CJ, Do YS, Semrau JD, Antholine WE, Hargrove MS, Pohl NL, Boyd ES, Geesey GG, Hartsel SC, et al. Spectral, kinetic, and thermodynamic properties of Cu(I) and Cu(II) binding by methanobactin from Methylosinus trichosporium OB3b. Biochemistry. 2006;45:1442–1453. doi: 10.1021/bi051815t. A comprehensive investigation of copper binding by mb, including spectroscopic, kinetic, and thermodynamic parameters, is reported. [DOI] [PubMed] [Google Scholar]
- 17.Choi DW, Do YS, Zea CJ, McEllistrem MT, Lee SW, Semrau JD, Pohl NL, Kisting CJ, Scardino LL, Hartsel SC, et al. Spectral and thermodynamic properties of Ag(I), Au(III), Cd(II), Co(II), Fe(III), Hg(II), Mn(II), Ni(II), Pb(II), U(IV), and Zn(II) binding by methanobactin from Methylosinus trichosporium OB3b. J Inorg Biochem. 2006;100:2150–2161. doi: 10.1016/j.jinorgbio.2006.08.017. [DOI] [PubMed] [Google Scholar]
- 18••.Knapp CW, Fowle DA, Kulczycki E, Roberts JA, Graham DW. Methane monooxygenase gene expression mediated by methanobactin in the presence of mineral copper sources. Proc Natl Acad Sci USA. 2007;104:12040–12045. doi: 10.1073/pnas.0702879104. The ability of mb to extract copper from mineral sources is explored, placing mb in the context of the natural environment. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choi DW, Kunz RC, Boyd ES, Semrau JD, Antholine WE, Han JI, Zahn JA, Boyd JM, de la Mora AM, DiSpirito AA. The membrane-associated methane monooxygenase pMMO and pMMO-NADH:quinone oxidoreductase complex from Methylococcus capsulatus Bath. J Bacteriol. 2003;185:5755–5764. doi: 10.1128/JB.185.19.5755-5764.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kulczycki E, Fowle DA, Knapp C, Graham DW, Roberts JA. Methanobactin-promoted dissolution of Cu-substituted borosilicate glass. Geobiology. 2007;5:251–263. [Google Scholar]
- 21.Kalinowski BE, Liermann LJ, Brantley SL, Barnes A, Pantano CG. X-ray photoelectron evidence for bacteria-enhanced dissolution of hornblende. Geochim Cosmochim Acta. 2000;64:1331–1343. [Google Scholar]
- 22.Ferguson AD, Deisenhofer J. TonB-dependent receptors-structural perspectives. Biochim Biophys Acta. 2002;1565:318–332. doi: 10.1016/s0005-2736(02)00578-3. [DOI] [PubMed] [Google Scholar]
- 23.Schauer K, Gouget B, Carriere M, Labigne A, de Reuse H. Novel nickel transport mechanism across the bacterial outer membrane energized by the TonB/ExbB/ExbD machinery. Mol Microbiol. 2007;63:1054–1068. doi: 10.1111/j.1365-2958.2006.05578.x. [DOI] [PubMed] [Google Scholar]
- 24.Cobine PA, Ojeda LD, Rigby KM, Winge DR. Yeast contain a non-proteinaceous pool of copper in the mitochondrial matrix. J Biol Chem. 2004;279:14447–14455. doi: 10.1074/jbc.M312693200. [DOI] [PubMed] [Google Scholar]
- 25•.Cobine PA, Pierrel F, Bestwick ML, Winge DR. Mitochondrial matrix copper complex used in metallation of cytochrome oxidase and superoxide dismutase. J Biol Chem. 2006;281:36552–36559. doi: 10.1074/jbc.M606839200. The copper binding properties of the yeast CuL are reported and CuL is shown to be present in mouse as well. [DOI] [PubMed] [Google Scholar]
- 26.Cobbett C. Phytochelatins and metallothioneins: roles in heavy metal detoxification and homeostasis. Ann Rev Plant Biol. 2002;53:159–182. doi: 10.1146/annurev.arplant.53.100301.135154. [DOI] [PubMed] [Google Scholar]