Abstract
The SNF1/AMPK family of protein kinases is highly conserved in eukaryotes and is required for energy homeostasis in mammals, plants, and fungi. SNF1 protein kinase was initially identified by genetic analysis in the budding yeast Saccharomyces cerevisiae. SNF1 is required primarily for the adaptation of yeast cells to glucose limitation and for growth on carbon sources that are less preferred than glucose, but is also involved in responses to other environmental stresses. SNF1 regulates transcription of a large set of genes, modifies the activity of metabolic enzymes, and controls various nutrient-responsive cellular developmental processes. Like AMPK, SNF1 protein kinase is heterotrimeric. It is phosphorylated and activated by the upstream kinases Sak1, Tos3, and Elm1 and is inactivated by the Reg1-Glc7 protein phosphatase 1. Further regulation of SNF1 is achieved through autoinhibition and through control of its subcellular localization. Here we review the current understanding of SNF1 protein kinase pathways in Saccharomyces cerevisiae and other yeasts.
Keywords: SNF1 protein kinase, Metabolic control, Stress responses, Yeast, AMPK, Glucose
2. INTRODUCTION
SNF1 protein kinase, a founding member of the SNF1/AMPK family, was identified genetically in Saccharomyces cerevisiae in 1981 when the snf1 mutation was recovered in a search for mutants unable to utilize sucrose (snf, sucrose-nonfermenting) (1). The mutant had defects in expression of the SUC2 (invertase) gene (2), and snf1 was later recognized as allelic to ccr1 and cat1 (3, 4). The SNF1 gene was cloned and found to encode the catalytic subunit of a protein-serine/threonine kinase (5, 6). Subsequent studies identified the noncatalytic beta subunits (7–9) and gamma subunit (10, 11). We will refer to the heterotrimeric kinase as SNF1 and to the catalytic subunit as Snf1. In 1994, a cDNA encoding an AMPK catalytic subunit was cloned and was identified as the mammalian ortholog of Snf1 (12–14). This finding resulted in the convergence of yeast and mammalian studies.
SNF1 is required for the yeast cell to adapt to glucose limitation and to utilize alternate carbon sources that are less preferred than glucose, such as sucrose, galactose, and ethanol (15, 16). SNF1 also has roles in various nutrient-responsive, cellular developmental processes, including meiosis and sporulation (1, 17), aging (18), haploid invasive growth (19), and diploid pseudohyphal growth (20). In addition to its primary role in responses to nutrient stress, SNF1 is involved in the cellular responses to other environmental stresses, including sodium ion stress, heat shock, alkaline pH, oxidative stress, and genotoxic stress (21–26). SNF1 affects cellular regulatory processes through a variety of mechanisms, including a major role in control of the genomic transcriptional program and direct effects on the activity of metabolic enzymes.
SNF1 is activated by glucose limitation (23, 27–29) and other stresses (26). SNF1 catalytic activity is regulated by three upstream kinases, Sak1, Elm1, and Tos3 (30–32), by the Reg1-Glc7 protein phosphatase 1 (33, 34), and by autoinhibition (10, 28). In addition, the subcellular localization of SNF1 is highly regulated (35–37).
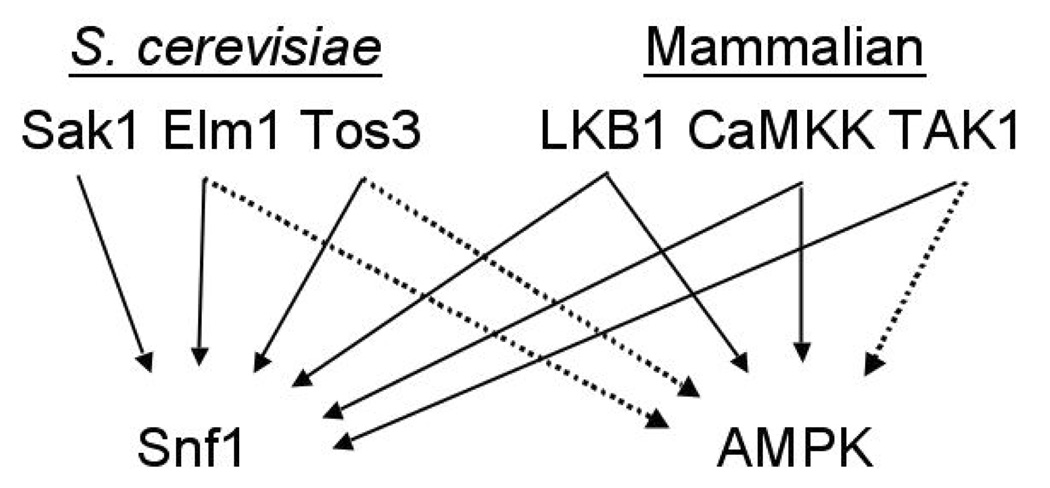
The SNF1/AMPK pathway is remarkably conserved among eukaryotes. Upstream kinases were first identified in yeast (30–32), and Tos3 was shown to also activate AMPK in vitro (30). The catalytic domains of the three yeast kinases are similar to those of Ca2+/calmodulin-dependent protein kinase kinases (CaMKKs) and the tumor suppressor kinase LKB1. This similarity led to the identification of LKB1 as an AMPK-activating kinase (30) and its verification as an authentic upstream kinase in mammalian cells (38–40). Both LKB1 and CaMKK alpha and beta, which were identified subsequently as AMPK kinases (41–43), also function in yeast cells as Snf1-activating kinases (41, 44) (Fig. 1). This conserved cross-species functionality has been further exploited to identify mammalian transforming growth factor-beta activated protein kinase (TAK1) as a Snf1-activating kinase and putative AMPK kinase (45).
Figure 1.
Conservation of SNF1/AMPK kinase cascades. Solid arrows indicate activation in vivo and in vitro. Dotted arrows indicate activation in vitro. Evidence suggests that TAK1 also activates AMPK in vivo (45, 127).
SNF1/AMPK pathways have also been characterized in other yeasts. In the aerobic yeast Kluyveromyces lactis, the homologous KlSnf1 (Fog2) pathway has roles in glucose signaling and carbon source utilization (46, 47). In the pathogenic yeasts Candida albicans and Candida tropicalis, CaSnf1 is essential for viability (48, 49). The pathway has been less well characterized with respect to function in the fission yeast Schizosaccharomyces pombe, but a crystal structure has been determined for a partial heterotrimeric AMPK lacking the catalytic domain (50).
3. SUBUNIT STRUCTURE
Like AMPK, the yeast orthologs are heterotrimeric. The S. cerevisiae genome encodes the catalytic alpha subunit Snf1, three alternate beta subunits, Sip1, Sip2 and Gal83, and the gamma subunit Snf4. There are therefore three alternate SNF1 complexes.
3.1. Snf1 catalytic subunit
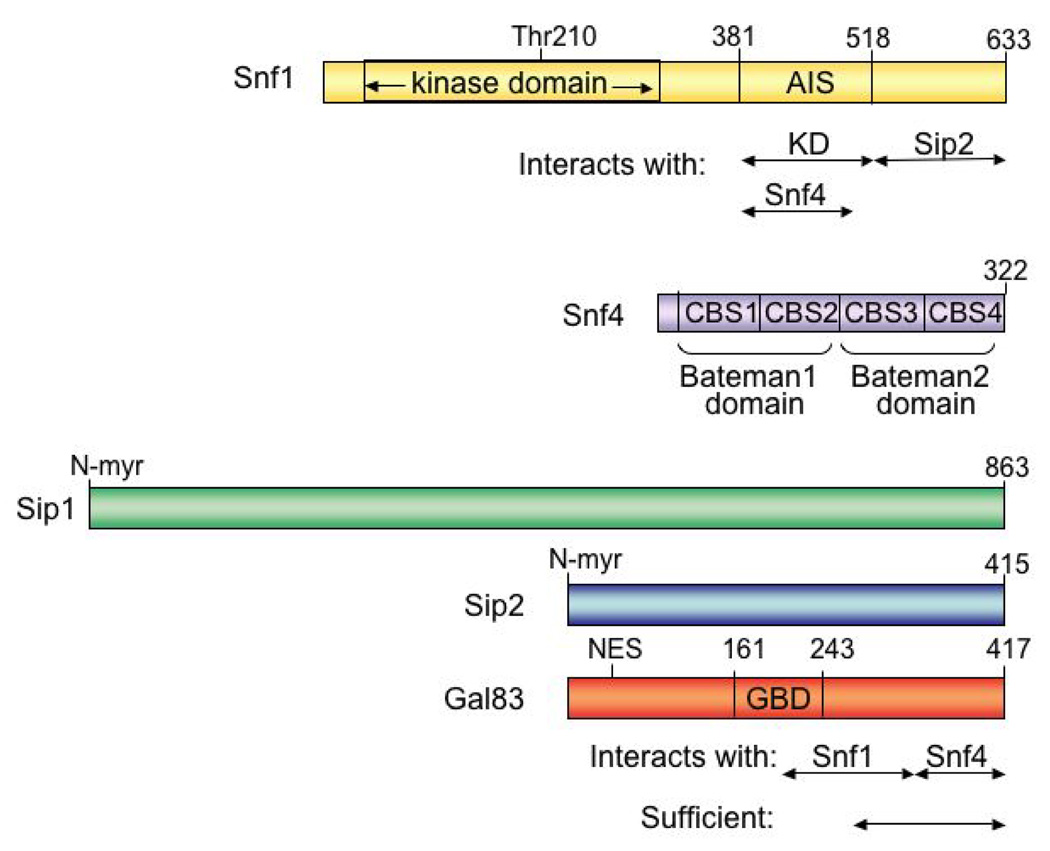
The Snf1 subunit is a constitutively expressed 633 amino acid protein comprising a kinase domain near the N terminus and a C-terminal regulatory region (5, 6, 51) (Fig. 2). Deletion of the C-terminal region bypasses the requirement for Snf4, suggesting that Snf4 antagonizes autoinhibition of the kinase domain by the C terminus (10, 28). The C-terminal region interacts directly with both Snf4 and the kinase domain, and deletion analysis identified residues 392 to 495 of Snf1 as sufficient for interaction with Snf4, and residues 392 to 518 for interaction with the kinase domain (28). Within this overlapping region, alteration of Leu470 to Ser abolishes interaction with Snf4 but maintains interaction with the kinase domain (28). The autoinhibitory sequence (AIS) was further defined by evidence that deletion of residues 381 to 488 bypasses the requirement for Snf4, whereas deletion of residues 381 to 414, which includes an autoinhibitory motif identified in AMPK (52), has a partial effect (53). Alterations in the kinase domain of Gly53 to Arg (10, 54) and Leu183 to Ile (53) partially alleviate the requirement for Snf4. The C-terminal region also interacts with the beta subunits, and deletion analysis identified the C-terminal residues 515 to 633 of Snf1 as sufficient for interaction with Sip2 (55).
Figure 2.
Structure of SNF1 subunits. Numbers, amino acid residues. Arrows, regions mapped by deletion analysis as sufficient for interaction with kinase domain (KD), Snf1, Snf4, or Sip2, as indicated. AIS, autoinhibitory sequence; CBS, cystathionine-beta-synthase repeat; N-myr, N-myristoylation consensus sequence; NES, nuclear export signal; GBD, glycogen binding domain.
Activation of the Snf1 catalytic subunit requires phosphorylation of Thr210 in the activation-loop segment (23, 54), and alteration of Thr210 to Ala, Glu, or Asp results in an inactive kinase (54, 56). Upstream kinases (30, 32) and Reg1-Glc7 protein phosphatase 1 (23, 33, 34) control Thr210 phosphorylation (see section 4). Neither the Snf1 C-terminal region, Snf4, nor the beta subunit is required for phosphorylation of Thr210 or its regulation by glucose signals (23, 30, 57, 58), consistent with genetic evidence regarding function of the truncated Snf1 kinase domain (10, 56).
The crystal structure of the kinase domain of Snf1 has been determined and revealed a dimer in which the activation loop Thr210 is buried (59, 60). Full-length Snf1 was also shown to self-associate in coimmunoprecipitation experiments (60). The crystal structure of a partial trimeric S. pombe AMPK, lacking the kinase domain, also revealed a dimer (50). The biological significance of dimerization remains uncertain.
3.2. Beta subunits
The S. cerevisiae genome encodes three beta subunits, Sip1, Sip2, and Gal83 (Fig. 2). Sip1 and Sip2 were found in a two-hybrid screen as Snf1-interacting proteins (7, 8). Gal83 was originally named for its involvement in regulation of GAL genes (61) and was found to have sequence similarity with Sip2 (9). The beta subunits exhibit considerable functional overlap as each one alone is sufficient for growth under various conditions where SNF1 activity is required (62). The beta subunits contain conserved C-terminal sequences that mediate their interaction with the SNF1 complex (8). Deletion analysis identified residues 198 to 350 of Gal83 and residues 154 to 335 of Sip2 as sufficient for interaction with Snf1 (55). It is now apparent that this region, formerly designated the kinase-interacting sequence (KIS), includes part of the glycogen-binding domain (GBD, see below), as well as Snf1-interacting sequences (Fig. 2). Residues 771 to 863 of Sip1 and residues 332–415 of Sip2 (designated the ASC domain, for association with SNF1 kinase complex) suffice for interaction with Snf4 (8, 55). Consistent with these results, the C-terminal 139 residues of Gal83 (distal to residue 278) are sufficient for beta subunit function (62).
The sequence of the GBD in the AMPK beta1 subunit is conserved in Gal83 (residues 161–243) and Sip2 (residues 163–245), and Gal83 binds glycogen strongly in vitro, whereas Sip2 binds very weakly (63). Alteration of Gal83 residues Trp184 to Ala together with Arg214 to Gln, which abolish glycogen-binding of AMPK beta1, similarly abolished glycogen binding of Gal83, as did the alteration Gly235 to Arg (63), which confers partial glucose-insensitivity to GAL gene expression (9, 61) and function of the Sip4 transcriptional activator (64). In cells, both mutant Gal83 proteins caused upregulation of various SNF1-dependent processes, including glycogen accumulation, expression of RNAs encoding glycogen synthase, haploid invasive growth, the transcriptional activator function of Sip4, and activation of the carbon source-responsive promoter element (63). Moreover, the GBD mutations conferred phenotypes even in the absence of glycogen, in a mutant strain lacking glycogen synthase, indicating that they positively affect SNF1 function by a mechanism that is independent of glycogen binding.
The beta subunits have divergent N termini that confer unique subcellular localization patterns (see section 5). All three are cytoplasmic in conditions of abundant glucose. Upon glucose depletion, Sip1 relocalizes to the vacuolar membrane, Gal83 relocalizes to the nucleus, and Sip2 remains cytoplasmic (35). The beta subunits direct the localization of the Snf1 subunit (35–37).
The abundance of the beta subunits is differently regulated by glucose availability. Gal83 is the major isoform during growth on glucose, and levels of Sip2 increase during shifts to nonfermentable carbon sources. Sip1 is less abundant than either Gal83 or Sip2, and its level remains constant (35, 65). All three isoforms of SNF1 are equally active (66), but consistent with its greater abundance, the Gal83 isoform contributes the most to SNF1 activity in response to glucose limitation (37). Sip2 is N-myristoylated (67), and Sip1 contains an N-myristoylation consensus sequence that is required for its localization to the vacuolar membrane (36).
Although the beta subunits have overlapping functions, they also exhibit various differences. All the beta subunits have distinct roles in haploid invasive growth (68). Sip2 has been implicated in aging (18, 69). Gal83 mediates the interaction of Snf1 with the transcriptional activator Sip4 (62, 64), and possibly with the transcriptional apparatus (70). The different beta subunits also display stress-dependent preferences for activation by Sak1, Tos3, and Elm1 (71).
There is conservation of beta subunits among different yeast species. K. lactis contains only one beta subunit, KlGal83 (Fog1), which has the strongest sequence similarity to Gal83 (46). C. albicans has two beta subunits, one of which is N-myristoylated (72). S. pombe has a beta subunit that is most similar to Sip2. In S. pombe, the crystal structure of the partial trimer shows extensive interactions between the alpha and beta subunits, and the beta subunit contains a highly mobile polar loop at residues 244–255 that covers the pore in the gamma subunit where AMP and ATP bind; the sequence corresponding to this loop is not conserved in S. cerevisiae (50).
3.3. Snf4 subunit
The gene encoding the S. cerevisiae gamma subunit, SNF4, was identified by isolation of a sucrose-nonfermenting mutant (73) and is allelic to CAT3 (74). Snf4 is a constitutively expressed protein of 322 residues that binds the Snf1 and beta subunits, independent of glucose availability (10, 11, 55). Snf4 contains two pairs of cystathionine-beta-synthase (CBS) repeats (75), called Bateman domains (Fig. 2), which in various proteins have been shown to bind adenosine derivatives (76, 77). Unlike AMPK, SNF1 is not allosterically activated by AMP in vitro (14, 27, 29, 76), and Snf4 has a substitution at a residue that is important for the AMP-dependence of AMPK (76). The crystal structure of a dimer of the C-terminal Snf4 Bateman domain has been determined and a possible binding pocket was identified (59). This structure is similar to that of the S. pombe gamma subunit, which binds a single molecule of AMP or ATP in a hydrophobic cleft between CBS repeats 3 and 4 (50). The surface charge of the gamma subunit changes in response to nucleotide binding, suggesting that binding could affect interactions between subunits and thereby regulate catalytic activity (50). It remains to be determined whether S. pombe AMPK is regulated by nucleotides and whether Snf4 binds a ligand.
Snf4 is required for catalytic activity of the heterotrimeric kinase in vivo and in vitro (10, 11, 27). Snf4 binds to the C-terminal regulatory region of Snf1, and genetic studies showed that when this region is truncated, Snf4 is not required for the glucose-regulated function of the remaining Snf1 catalytic domain; these findings indicate that Snf4 counteracts autoinhibition by the AIS (10, 28, 53). The simple model is that Snf4 binds to the AIS when cells are limited for glucose and prevents binding of the AIS to the kinase domain. Consistent with this view, the interaction of overexpressed Snf1 and Snf4 hybrid proteins in the two-hybrid system was strongly inhibited by glucose (28); this inhibition could also reflect, at least in part, glucose inhibition of the nuclear localization of Snf1 (35) as two-hybrid interaction is assayed by lacZ reporter expression. Analysis of mutants lacking Snf4 indicated that it is not required for glucose-regulated phosphorylation of full-length Snf1 on its activation loop Thr210 (23, 58). At least one glucose regulatory mechanism is therefore independent of Snf4. However, recent studies showed that alteration of various residues of Snf4 relieved glucose inhibition of Thr210 phosphorylation and SNF1 activity (S.H. Iram and M.C., unpublished results). Thus, although in the absence of Snf4, phosphorylation of Snf1 Thr210 is glucose regulated, in the context of the SNF1 heterotrimer, Snf4 is required for regulated phosphorylation. These findings indicate that Snf4 has roles in regulating both autoinhibition and the functional interactions of SNF1 with upstream kinases and/or protein phosphatase in response to glucose availability. Finally, it has recently been reported that Snf4 contains a pseudosubstrate sequence that is recognized by Snf1, suggesting that Snf4 also directly inhibits Snf1 (78).
Evidence suggests that a subpopulation of Snf4 is not associated with Snf1. Snf4 is constitutively localized to both the nucleus and the cytoplasm, whereas Snf1 and the beta subunits are excluded from the nucleus during growth in high glucose (35) (see section 5). Ghaemmaghami et al reported a 20-fold excess of Snf4 molecules in the cell relative to other subunits (65), but others have found that Snf1 and Snf4 are present at similar levels (35) and that most, although not all, Snf4 is in complex with Snf1 (58). These observations raise the possibility that Snf4 may have additional functions in the cell independent of its functions in SNF1 protein kinase.
In this regard, it is interesting that Snf4 contains a consensus elongin C binding site and binds to the yeast elongin C, the Elc1 protein (79). Elc1 is a homolog of Skp1, of the Skp1-Cullin-F-box ubiquitin ligase. Mutation of Cys136 in the consensus elongin C-binding sequence relieved glucose inhibition of SNF1 protein kinase; however, deletion of the ELC1 gene did not affect SNF1 catalytic activity (S.H. Iram and M.C., unpublished results). The elc1 mutant displayed no SNF1-related growth defect (79). These results do not support a regulatory role for Elc1 in the SNF1 pathway, but it remains possible that Elc1 affects an unidentified function of Snf4.
4. REGULATION OF SNF1 CATALYTIC ACTIVITY
SNF1 is phosphorylated and activated in response to glucose limitation (23, 27–29), in accord with its central role in adaptation to glucose depletion and utilization of alternate carbon sources, but the signal(s) regulating SNF1 has not yet been identified. As mentioned above, SNF1 is not allosterically activated by AMP (14, 27, 29, 76), but activation correlates with increased cellular AMP:ATP ratios (29). It is important to note that responses are often transient. For example, SNF1 activity is rapidly elevated when cells are transferred from medium containing glucose to sucrose, but after adaptation, minimal SNF1 activity is required for continued growth (80) and, correspondingly, the snf1 mutant exhibits a severe growth defect on sucrose only under nonrespiratory conditions.
Snf1 is also phosphorylated and activated in response to other environmental stresses including sodium ion stress, alkaline pH, oxidative stress, but not treatment with sorbitol or heat shock (26). Some of these responses are also transient; for example, Snf1 is rapidly and strongly activated in response to alkaline stress, but activity returns to basal levels within 1 hr (26). Additionally, Snf1 Thr210 phosphorylation occurs in response to nitrogen limitation, and also to the addition of rapamycin, which implicates the target of rapamycin (TOR) kinase as a negative regulator (81).
The roles of the Snf1-activating kinases and protein phosphatase in regulating phosphorylation of Thr210 are discussed below. It is not yet clear whether the activity of these upstream kinases or the phosphatase, or both, is regulated. Alternatively, signals may be received directly by the kinase complex and affect its accessibility to upstream kinases or phosphatase. Autoinhibition is also an important control point, involving alternate binding of the AIS to the Snf1 kinase domain or to Snf4, as described in section 3.
4.1. Upstream kinases Sak1, Elm1, and Tos3
Three protein kinases with related kinase domains, Sak1, Elm1, and Tos3, activate Snf1 by phosphorylation of Thr210 (30–32). Sak1 (Snf1-activating kinase) was formerly known as Pak1 but was renamed to avoid confusion with the p21-activated kinase family. These upstream kinases are highly similar and exhibit overlapping function as all three must be deleted to abolish SNF1 activity in vivo (30, 32). Phosphorylation of Snf1 Thr210 does not require the Snf1 C-terminal domain, Snf4, or the beta subunits (30, 57, 58). There is no evidence to date that activity of any of the upstream kinases is regulated by glucose signals. Evidence suggests that the three kinases make different contributions to cellular regulation under conditions of different carbon source availability; however, in all cases tested, Sak1 was the major Snf1-activating kinase (30, 37, 71, 82). While Tos3 alone is sufficient for growth on nonpreferred carbon sources, the only condition tested in which the absence of Tos3 causes a significant defect was during continuous growth on glycerol plus ethanol (82). The SNF1 beta subunit also confers some specificity for a particular upstream kinase under different growth conditions (71). Analysis of mutants indicated that Sak1, Tos3, and Elm1 are also required for activation of SNF1 in response to other environmental stress conditions (see section 6.4); each kinase suffices for activation, but again Sak1 has the major role (26).
Sak1 associates with SNF1 to form a stable complex, whereas Tos3 and Elm1 apparently associate with SNF1 more transiently (57). This stable interaction between Snf1 and Sak1 is not regulated by glucose (57), contrary to an earlier report (31). Most of the Sak1 protein in the cell appears to be associated with Snf1, but only a fraction of the total Snf1 is in a complex with Sak1 (57). The nonconserved C-terminal domains of Sak1 and Tos3 and the N terminus of Sak1 are important for SNF1 signaling (83).
Sak1 is found in the cytoplasm in glucose-grown cells, but exhibits some relocalization to the vacuolar membrane in response to glucose depletion, similar to Sip1 (37). Tos3 is cytosolic under both conditions (82). Elm1 localizes to the bud neck (84), consistent with its SNF1-independent roles in cell morphology, septin organization, and cell cycle progression (84–87).
4.2. Reg1-Glc7 protein phosphatase 1
Snf1 catalytic activity is negatively regulated by the Reg1-Glc7 protein phosphatase 1 (PP1); the Glc7 catalytic subunit is directed to SNF1 through the targeting protein Reg1 (33, 34, 56, 88). Different sequences of Reg1 interact with Snf1 and with Glc7 (34, 89). PP1 dephosphorylates the activation loop Thr210 residue during conditions of ample glucose availability (23), and in a reg1 mutant cell, Snf1 catalytic activity is resistant to glucose inhibition (44). Reg1 is cytoplasmic and nuclear excluded in both glucose-grown and glucose-depleted cells(89). It is phosphorylated in response to glucose limitation in a Snf1-dependent manner (34).
Although there is as yet no direct biochemical evidence that the activity of Reg1-Glc7 is regulated, genetic evidence supports a regulatory role for the phosphatase. Expression of the heterologous mammalian AMPK-activating kinases LKB1, CaMKK alpha, or TAK1 in sak1 tos3 elm1 mutant yeast cells restores glucose-regulated SNF1 activity (44, 45). Moreover, the sak1 tos3 elm1 reg1 quadruple mutant expressing mammalian activating kinases showed glucose-insensitive SNF1 activity, and the absence of Reg1 had no effect on activity in glucose-limited cells (44). Although it is possible that the three yeast upstream kinases as well as LKB1 and CaMKK are all similarly glucose regulated, the simple model is that they are constitutively active and that the function of Reg1-Glc7 towards SNF1 is positively regulated by glucose signals that affect either the phosphatase or the SNF1 complex (44).
In similar experiments, in sak1 tos3 elm1 cells expressing mammalian CaMKK alpha, SNF1 was activated by both sodium ion and alkaline stress. These results suggest that these stress signals regulate SNF1 activity by a mechanism that is independent of the upstream kinase.
5. REGULATION OF SUBCELLULAR LOCALIZATION
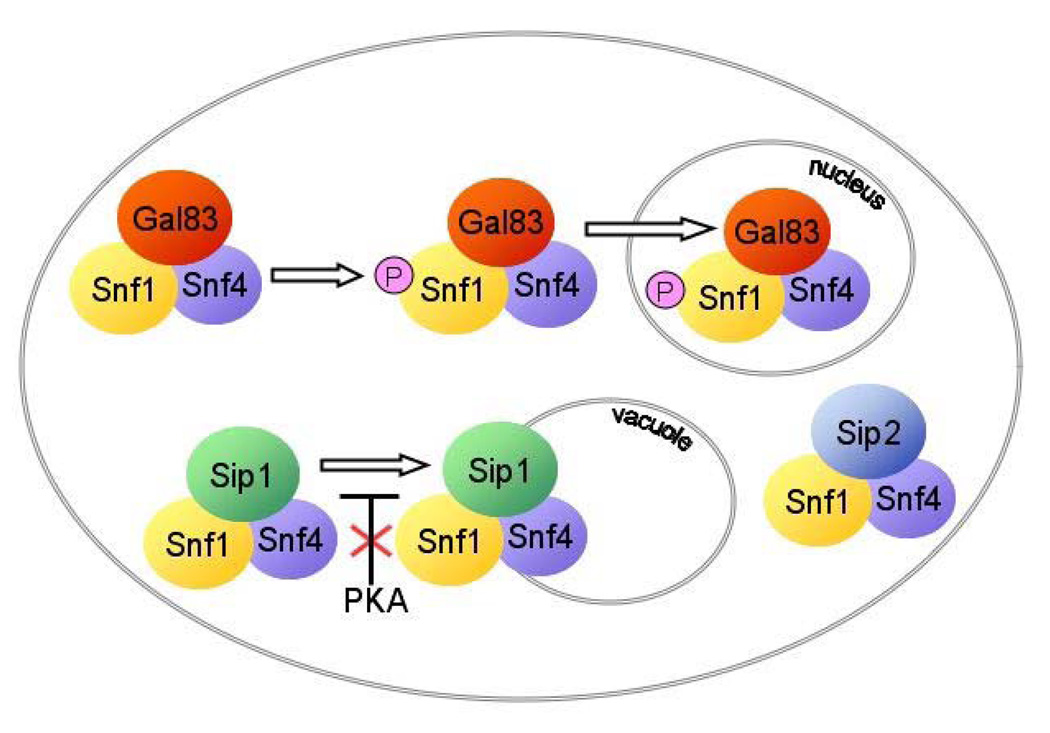
SNF1 is also regulated at the level of subcellular localization, which presumably affects substrate access (Fig. 3). In high glucose, Snf1 and all three beta subunits are cytoplasmic (35). When glucose becomes limiting, the beta subunits take on unique subcellular localization patterns, dependent on their divergent N-terminal sequences (35, 37, 80). Sip2 remains cytoplasmic, while Sip1 relocalizes to the vacuolar membrane, and Gal83 relocalizes to the nucleus. The beta subunits also direct the localization of Snf1. In the sip1 mutant, no vacuolar localization of Snf1 is evident, and in the gal83 mutant, dependent on yeast strain background, Snf1 either fails to accumulate, or accumulates to a lesser extent, in the nucleus. The Snf4 subunit, which is present in excess, is both cytoplasmic and nuclear regardless of nutritional status (35). The effects of environmental stress on localization of the Snf1 and Gal83 subunits has also been examined; both become enriched in the nucleus in response to alkaline pH but not salt stress (26). Sip2 has also been reported to associate with the plasma membrane in cells that have been subjected to a magnetic bead sorting procedure (67).
Figure 3.
Relocalization of alternate forms of SNF1 in response to carbon stress. In conditions of high glucose, all three alternate SNF1 complexes are cytoplasmic. In response to carbon stress, PKA no longer inhibits the localization of Sip1 to the vacuolar membrane. Sip2 remains cytoplasmic. Relocalization of Snf1 protein kinase containing Gal83 to the nucleus in response to carbon stress requires phosphorylation (P) and activation of Snf1 and also a Snf1-independent stress signal that promotes nuclear localization of Gal83.
The localization of Sip1 to the vacuolar membrane is inhibited by protein kinase A (PKA) activity (36). In a mutant lacking the three PKA catalytic subunits, Sip1 is constitutively at the vacuole, independent of glucose availability, and conversely, in a mutant with high PKA activity due to absence of the Bcy1 regulatory subunit, Sip1 is always cytoplasmic. Localization to the vacuolar membrane also requires the N-myristoylation consensus sequence in Sip1 (36).
Gal83 contains a leucine-rich nuclear export signal (NES) in its N terminus, which is required for its cytoplasmic distribution, and export depends on the Crm1 export receptor (80). Nuclear localization of Snf1 and Gal83 depends not only on Gal83 but also on activation of the Snf1 catalytic subunit; evidence indicates that catalytically inactive Snf1 promotes the cytoplasmic retention of Gal83 in glucose-grown cells through its interaction with the C terminus of Gal83 (80). The requirement for activation of Snf1 serves to retain inactive Snf1 in the cytoplasm, where it can be phosphorylated by the activating kinases, none of which is nuclear (37, 82, 84). Nuclear accumulation also requires Sak1, which is the major Snf1-activating kinase, but activation of Snf1 by a heterologous kinase did not rescue the localization defect (44). In contrast, in the snf1 mutant, Gal83 exhibits glucose-regulated nuclear accumulation, and Sak1 is dispensable (37). While phosphorylation of glucose is required for cytosolic localization, evidence suggesting glucose-6-phosphate as a candidate signal regulating Gal83 localization (35) can be accounted for by its inhibitory effects on Snf1 activity (80). Studies of an N-terminal fragment of Gal83, which does not interact with Snf1, showed that addition of 2-deoxyglucose to glucose-limited cells or addition of glucose to cells lacking phosphoglucose isomerase did not cause relocalization from nucleus to cytoplasm (80). The signaling mechanism remains unclear.
6. FUNCTION OF SNF1 PROTEIN KINASE
6.1. Control of transcription in response to carbon stress
SNF1 regulates the transcription of a large set of genes, including those involved in the metabolism of alternative carbon sources, gluconeogenesis, respiration, transport, and meiosis. Genomic expression studies showed that expression of over 400 genes was SNF1-dependent under conditions of limiting glucose, including 29 of the 40 most highly glucose regulated genes (90). The largest class of SNF1-dependent genes functions in transcription and signal transduction, reflecting the central regulatory role of this kinase (90). SNF1 exerts direct effects on transcription through repressors (91, 92), activators (93–95), chromatin (67, 96), and most likely the transcriptional apparatus (70, 97) (Fig. 4). Many genes are coregulated by SNF1 and the activators Adr1 and Cat8 (90). The K. lactis Snf1 and Gal83 homologs are similarly required for transcriptional activation of glucose-repressed genes (46).
Figure 4.
Mechanisms by which SNF1 regulates transcription of glucose-repressed genes. Yeast cells are depicted during growth in high glucose and after exposure to carbon stress. In high glucose, the repressor Mig1 is nuclear and represses transcription of many glucose-repressed genes (GENE) in conjunction with the corepressor Ssn6-Tup1. In response to carbon stress, SNF1 is phosphorylated (P) and moves into the nucleus. Sip4, a representative transcriptional activator, is phosphorylated and activates transcription of a subset of glucose-repressed genes. SNF1 phosphorylates Mig1, which alters its interaction with Ssn6-Tup1 to alleviate repression and promotes its nuclear export. SNF1 is also thought to modify chromatin (depicted by a single orange nucleosome) and affect the RNA polymerase II (Pol II) transcriptional apparatus.
SNF1 affects many glucose-repressed genes through control of the transcriptional repressor Mig1, which functions in conjunction with the corepressor Ssn6 (Cyc8)-Tup1 (98, 99). SNF1 phosphorylates Mig1 (91, 92, 100), which promotes its nuclear export (101). However, evidence suggests that phosphorylation of Mig1 alters its interaction with Ssn6-Tup1 to alleviate repression, but does not release DNA-bound Mig1 from the promoter (102).
SNF1 also regulates various transcriptional activators. SNF1 is required for activation by Cat8 and Sip4, which bind to the carbon source-response element found in gluconeogenic genes (93, 94, 103, 104). Sip4 is phosphorylated in response to glucose limitation, dependent on Snf1 and Gal83 (62, 64, 94). In K. lactis, phosphorylation of KlCat8 is dependent on KlSnf1, and mutation of a consensus SNF1 phosphorylation site in KlCat8 altered glucose regulation of its activation function, as did mutation of the conserved site in S. cerevisiae Cat8 (105). SNF1 promotes the binding of Adr1 to chromatin in the absence of glucose (95). SNF1 affects activation by heat shock transcription factor (HSF) in response to glucose starvation, but not heat stress (106). HSF is a direct target of SNF1 in vitro, and phosphorylation of HSF and binding of HSF to certain low-affinity promoters in response to glucose starvation is Snf1-dependent (107). The stress-response transcription factor Msn2 is another target of SNF1; upon glucose depletion, SNF1 phosphorylates Msn2, thereby inhibiting its nuclear accumulation (108, 109). Finally, SNF1 is required for phosphorylation and nuclear accumulation of Gln3, a GATA transcription factor, in response to glucose starvation, and SNF1 phosphorylates Gln3 in vitro (110).
Genetic evidence suggests that SNF1 has direct effects on the transcriptional apparatus. Recruitment of Snf1 to a promoter results in glucose-regulated transcription, and SNF1 stimulates transcription by RNA polymerase II holoenzyme that has been recruited to an artificial promoter (70). Mutations in SNF4 were recovered as dominant suppressors that restored INO1 transcription caused by a mutant TATA-binding protein (TBP) (111), and Snf1 was found to affect the recruitment of TBP to the INO1promoter (97). Snf1 also affects preinitiation complex formation in vitro (95).
Various lines of evidence implicate SNF1 in regulating modification of chromatin. At the INO1 promoter, it has been reported that SNF1 phosphorylates histone H3 on Ser10, which leads to acetylation of Lys14 by the acetyltransferase Gcn5 and recruitment of TBP, thereby inducing transcription (96), and SNF1-mediated phosphorylation of Ser10 is also required for GAL1 transcription (112). However, another study of the effect of substitution of Ala for Ser10 did not reveal a requirement for histone H3 Ser10 phosphorylation for INO1 transcription (97). Genetic evidence connects SNF1 to Gcn5 in control of HIS3 transcription, and Snf1 phosphorylates Gcn5 in vitro; however, histone H3 Ser10 phosphorylation does not contribute to Gcn5- and SNF1-mediated HIS3 expression(113). In vitro, SNF1 phosphorylates a peptide containing the histone H3 Ser10 site, and this activity increases in aging cells (67). Finally, the role of SNF1 in chromatin binding by Adr1 may reflect direct effects of SNF1 on chromatin (95).
6.2. Control of metabolic enzymes and transporters in response to carbon stress
As part of its function in controlling cellular energy usage, SNF1 regulates the activity of metabolic enzymes involved in fatty acid metabolism and carbohydrate storage (14, 114, 115). SNF1 phosphorylates and inactivates acetyl-CoA carboxylase (Acc1) (27), and thereby inhibits fatty acid biosynthesis during glucose-limiting conditions. Phosphorylation of Acc1 also affects transcription of INO1, a key gene in phospholipid biosynthesis (116). SNF1 is required for accumulation of glycogen, an important storage carbohydrate in yeast (117), and controls the phosphorylation of glycogen synthase (21, 114, 118, 119). SNF1 also appears to affect maintenance of glycogen stores by promoting autophagy (115). SNF1 also controls expression of hexose transporters (120, 121), and in K. lactis, KlSnf1 is involved in the intracellular sorting of the galactose/lactose transporter Lac12 to the plasma membrane (122).
6.3. Roles of SNF1 in response to other environmental stresses
SNF1 is important for responses to many other environmental stresses besides carbon stress. The snf1 mutant shows defects during starvation for other nutrients, including phosphate, sulfate, and nitrogen (21). Notably, diploid mutant cells fail to undergo pseudohyphal differentiation in response to nitrogen limitation (20). Phosphorylation of Snf1 Thr210 is stimulated by nitrogen limitation and negatively regulated by TOR kinase, which has known roles in nitrogen signaling (81).
The snf1 mutant is sensitive to sodium and lithium ion stress (22, 25, 123), other toxic cations such as hygromycin B (25), alkaline pH and heat shock (124), and genotoxic stress caused by hydroxyurea, methylmethane sulfonate, and cadmium (24). The snf1 mutant also shows reduced thermotolerance in stationary phase (21). SNF1 catalytic activity and phosphorylation of Thr210 increase as a result of exposure of cells to sodium ion stress, alkaline pH, oxidative stress, or antimycin A, which inhibits respiratory metabolism, but not sorbitol or heat shock (26). Interestingly, very low levels of SNF1 activity suffice for resistance to some stresses, and in some cases phosphorylation of Thr210 is not essential (24–26). Different stresses have specific effects on SNF1 signaling. For example, SNF1 is regulated differently during adaptation of cells to alkaline pH and NaCl with respect to the time course of activation, and SNF1 becomes enriched in the nucleus in response to alkaline pH but not salt stress (26). Most of the known roles of SNF1 in stress responses involve transcriptional control; for example, the snf1 mutant is defective in induction of ENA1, encoding Na+-ATPase, by alkaline or sodium ion stress (123, 124).
SNF1 also appears to have a role in regulating protective mechanisms against stress in response to glucose starvation. Thus, glucose-depleted cells exhibit SNF1-dependent induction of ENA1 (22), activation of HSF (106, 107), and phosphorylation of the stress-response transcription factor Msn2 (108, 109).
6.4. Roles in regulation of other cellular processes
SNF1 is also central to various nutrient-responsive cellular developmental processes. SNF1 is required for both early and late stages of meiosis (17); the role of SNF1 in entry into meiosis appears to be allowing high-level expression of IME, which encodes a master regulator of meiosis (125). SNF1 is a positive regulator of autophagy, a process for recycling organelles and macromolecules, possibly by regulating Apg1 and Apg13 (115). In addition to its role in diploid pseudohyphal growth, SNF1 is also required for haploid filamentous invasive growth in response to glucose limitation (19) or growth on various nonpreferred carbon sources (126); SNF1 regulates transcription of FLO11, which encodes a cell surface glycoprotein (flocculin) required for adherence, by antagonizing the repressors Nrg1 and Nrg2 (20), and it also affects the filamentation process (68). Finally, SNF1 appears to be involved in the connections between energy metabolism and aging; overexpression of Snf1 and the absence, or lack of N-myristoylation, of Sip2, promote aging, while the absence of Snf4 extends life span (18, 67, 69).
7. PERSPECTIVE
The SNF1 pathway is central to many aspects of cellular regulation in yeast, and genetic analysis of the pathway in this model organism has provided insights relevant to SNF1/AMPK pathways in other eukaryotes. Structural analysis promises deeper understanding of mechanisms affecting function of this kinase, but genetic approaches will continue to be useful in addressing many of the remaining questions regarding its regulation. The identity of the glucose signal(s) is not yet clear, and it remains possible that AMP or ATP regulates SNF1 in vivo. Further studies are needed to determine whether the activity of the Snf1-activating kinases or the Reg1-Glc7 protein phosphatase 1, or both, is regulated, or whether signals to the kinase complex affect its accessibility to the kinases or phosphatase. Autoinhibition is also an important control point, involving alternate binding of the AIS to the Snf1 kinase domain or to Snf4, and the mechanism by which this is regulated is not understood. Moreover, the role of Snf4 in glucose inhibition of phosphorylation has only recently been recognized. The regulation of this kinase is clearly complex, and many more exciting discoveries await us.
ACKNOWLEDGMENTS
This work was supported by a grant from the NIH (GM34095) to M.C.
Abbreviations
- AMPK
AMP-activated protein kinase
- SNF1
Sucrose Nonfermenting 1
- CaMKK
Ca2+/calmodulin-dependent protein kinase kinase
- TAK1
transforming growth factor-beta activated protein kinase
- CBS
cystathionine-beta-synthase
- TOR
target of rapamycin
- AIS
autoinhibitory sequence
- NES
nuclear export signal
- GBD
glycogen binding domain
- HSF
heat shock factor
- TBP
TATA-binding protein
- PKA
protein kinase A
Footnotes
Publisher's Disclaimer: “This is an, un-copyedited, author manuscript that has been accepted for publication in the Frontiers in Bioscience”. Cite this article as appearing in the Journal of Frontiers in Bioscience. Full citation can be found by searching the Frontiers in Bioscience (http://bioscience.org/search/authors/htm/search.htm) following publication and at PubMed (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?CMD=search&DB=pubmed) following indexing. This article may not be duplicated or reproduced, other than for personal use or within the rule of “Fair Use of Copyrighted Materials” (section 107, Title 17, U.S. Code) without permission of the copyright holder, the Frontiers in Bioscience. From the time of acceptance following peer review, the full final copy edited article of this manuscript will be made available at http://www.bioscience.org/. The Frontiers in Bioscience disclaims any responsibility or liability for errors or omissions in this version of the uncopyedited manuscript or in any version derived from it by the National Institutes of Health or other parties.”
REFERENCES
- 1.Carlson M, Osmond BC, Botstein D. Mutants of yeast defective in sucrose utilization. Genetics. 1981;98:25–40. doi: 10.1093/genetics/98.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carlson M, Botstein D. Two differentially regulated mRNAs with different 5' ends encode secreted and intracellular forms of yeast invertase. Cell. 1982;28:145–154. doi: 10.1016/0092-8674(82)90384-1. [DOI] [PubMed] [Google Scholar]
- 3.Ciriacy M. Isolation and characterization of yeast mutants defective in intermediary carbon metabolism and in carbon catabolite derepression. Mol. Gen. Genet. 1977;154:213–220. doi: 10.1007/BF00330840. [DOI] [PubMed] [Google Scholar]
- 4.Zimmermann FK, Scheel I. Mutants of Saccharomyces cerevisiae resistant to carbon catabolite repression. Mol. Gen. Genet. 1977;154:75–82. doi: 10.1007/BF00265579. [DOI] [PubMed] [Google Scholar]
- 5.Celenza JL, Carlson M. Cloning and genetic mapping of SNF1, a gene required for expression of glucose-repressible genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 1984;4:49–53. doi: 10.1128/mcb.4.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Celenza JL, Carlson M. A yeast gene that is essential for release from glucose repression encodes a protein kinase. Science. 1986;233:1175–1180. doi: 10.1126/science.3526554. [DOI] [PubMed] [Google Scholar]
- 7.Yang X, Hubbard EJA, Carlson M. A protein kinase substrate identified by the two-hybrid system. Science. 1992;257:680–682. doi: 10.1126/science.1496382. [DOI] [PubMed] [Google Scholar]
- 8.Yang X, Jiang R, Carlson M. A family of proteins containing a conserved domain that mediates interaction with the yeast SNF1 protein kinase complex. EMBO J. 1994;13:5878–5886. doi: 10.1002/j.1460-2075.1994.tb06933.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Erickson JR, Johnston M. Genetic and molecular characterization of GAL83: its interaction and similarities with other genes involved in glucose repression in Saccharomyces cerevisiae. Genetics. 1993;135:655–664. doi: 10.1093/genetics/135.3.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Celenza JL, Carlson M. Mutational analysis of the Saccharomyces cerevisiae SNF1 protein kinase and evidence for functional interaction with the SNF4 protein. Mol. Cell. Biol. 1989;9:5034–5044. doi: 10.1128/mcb.9.11.5034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Celenza JL, Eng FJ, Carlson M. Molecular analysis of the SNF4 gene of Saccharomyces cerevisiae evidence for physical association of the SNF4 protein with the SNF1 protein kinase. Mol. Cell. Biol. 1989;9:5045–5054. doi: 10.1128/mcb.9.11.5045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carling D, Aguan K, Woods A, Verhoeven AJ, Beri RK, Brennan CH, Sidebottom C, Davison MD, Scott J. Mammalian AMP-activated protein kinase is homologous to yeast and plant protein kinases involved in the regulation of carbon metabolism. J Biol Chem. 1994;269:11442–11448. [PubMed] [Google Scholar]
- 13.Woods A, Munday MR, Scott J, Yang X, Carlson M, Carling D. Yeast SNF1 is functionally related to mammalian AMP-activated protein kinase and regulates acetyl-CoA carboxylase in vivo. J Biol Chem. 1994;269:19509–19515. [PubMed] [Google Scholar]
- 14.Mitchelhill KI, Stapleton D, Gao G, House C, Michell B, Katsis F, Witters LA, Kemp BE. Mammalian AMP-activated protein kinase shares structural and functional homology with the catalytic domain of yeast Snf1 protein kinase. J. Biol. Chem. 1994;269:2361–2364. [PubMed] [Google Scholar]
- 15.Gancedo JM. Yeast carbon catabolite repression. Microbiol. Mol. Biol. Rev. 1998;62:334–361. doi: 10.1128/mmbr.62.2.334-361.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carlson M. Glucose repression in yeast. Curr. Opin. Microbiol. 1999;2:202–207. doi: 10.1016/S1369-5274(99)80035-6. [DOI] [PubMed] [Google Scholar]
- 17.Honigberg SM, Lee RH. Snf1 kinase connects nutritional pathways controlling meiosis in Saccharomyces cerevisiae. Mol Cell Biol. 1998;18:4548–4555. doi: 10.1128/mcb.18.8.4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ashrafi K, Lin SS, Manchester JK, Gordon JI. Sip2p and its partner Snf1p kinase affect aging in S. cerevisiae. Genes Dev. 2000;14:1872–1885. [PMC free article] [PubMed] [Google Scholar]
- 19.Cullen PJ, Sprague GF., Jr Glucose depletion causes haploid invasive growth in yeast. Proc. Natl. Acad. Sci. USA. 2000;97:13619–13624. doi: 10.1073/pnas.240345197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuchin S, Vyas VK, Carlson M. Snf1 protein kinase and the repressors Nrg1 and Nrg2 regulate FLO11, haploid invasive growth, and diploid pseudohyphal differentiation. Mol. Cell. Biol. 2002;22:3994–4000. doi: 10.1128/MCB.22.12.3994-4000.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thompson-Jaeger S, Francois J, Gaughran JP, Tatchell K. Deletion of SNF1 affects the nutrient response of yeast and resembles mutations which activate the adenylate cyclase pathway. Genetics. 1991;129:697–706. doi: 10.1093/genetics/129.3.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alepuz PM, Cunningham KW, Estruch F. Glucose repression affects ion homeostasis in yeast through the regulation of the stress-activated ENA1 gene. Mol Microbiol. 1997;26:91–98. doi: 10.1046/j.1365-2958.1997.5531917.x. [DOI] [PubMed] [Google Scholar]
- 23.McCartney RR, Schmidt MC. Regulation of Snf1 kinase. Activation requires phosphorylation of threonine 210 by an upstream kinase as well as a distinct step mediated by the Snf4 subunit. J. Biol. Chem. 2001;276:36460–36466. doi: 10.1074/jbc.M104418200. [DOI] [PubMed] [Google Scholar]
- 24.Dubacq C, Chevalier A, Mann C. The protein kinase Snf1 is required for tolerance to the ribonucleotide reductase inhibitor hydroxyurea. Mol Cell Biol. 2004;24:2560–2572. doi: 10.1128/MCB.24.6.2560-2572.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Portillo F, Mulet JM, Serrano R. A role for the non-phosphorylated form of yeast Snf1: tolerance to toxic cations and activation of potassium transport. FEBS Lett. 2005;579:512–516. doi: 10.1016/j.febslet.2004.12.019. [DOI] [PubMed] [Google Scholar]
- 26.Hong SP, Carlson M. Regulation of Snf1 protein kinase in response to environmental stress. J Biol Chem. 2007 doi: 10.1074/jbc.M700146200. [DOI] [PubMed] [Google Scholar]
- 27.Woods A, Munday MR, Scott J, Yang X, Carlson M, Carling D. Yeast SNF1 is functionally related to mammalian AMP-activated protein kinase and regulates acetyl-CoA carboxylase in vivo. J. Biol. Chem. 1994;269:19509–19516. [PubMed] [Google Scholar]
- 28.Jiang R, Carlson M. Glucose regulates protein interactions within the yeast SNF1 protein kinase complex. Genes Dev. 1996;10:3105–3115. doi: 10.1101/gad.10.24.3105. [DOI] [PubMed] [Google Scholar]
- 29.Wilson WA, Hawley SA, Hardie DG. Glucose repression/derepression in budding yeast: SNF1 protein kinase is activated by phosphorylation under derepressing conditions, and this correlates with a high AMP:ATP ratio. Curr. Biol. 1996;6:1426–1434. doi: 10.1016/s0960-9822(96)00747-6. [DOI] [PubMed] [Google Scholar]
- 30.Hong S-P, Leiper FC, Woods A, Carling D, Carlson M. Activation of yeast Snf1 and mammalian AMP-activated protein kinase by upstream kinases. Proc Natl Acad Sci USA. 2003;100:8839–8843. doi: 10.1073/pnas.1533136100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nath N, McCartney RR, Schmidt MC. Yeast Pak1 kinase associates with and activates Snf1. Mol. Cell. Biol. 2003;23:3909–3917. doi: 10.1128/MCB.23.11.3909-3917.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sutherland CM, Hawley SA, McCartney RR, Leech A, Stark MJ, Schmidt MC, Hardie DG. Elm1p is one of three upstream kinases for the Saccharomyces cerevisiae SNF1 complex. Curr. Biol. 2003;13:1299–1305. doi: 10.1016/s0960-9822(03)00459-7. [DOI] [PubMed] [Google Scholar]
- 33.Tu J, Carlson M. REG1 binds to protein phosphatase type 1 and regulates glucose repression in Saccharomyces cerevisiae. EMBO J. 1995;14:5939–5946. doi: 10.1002/j.1460-2075.1995.tb00282.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sanz P, Alms GR, Haystead TAJ, Carlson M. Regulatory interactions between the Reg1-Glc7 protein phosphatase and the Snf1 protein kinase. Mol. Cell. Biol. 2000;20:1321–1328. doi: 10.1128/mcb.20.4.1321-1328.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vincent O, Townley R, Kuchin S, Carlson M. Subcellular localization of the Snf1 kinase is regulated by specific β subunits and a novel glucose signaling mechanism. Genes Dev. 2001;15:1104–1114. doi: 10.1101/gad.879301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hedbacker K, Townley R, Carlson M. cAMP-dependent protein kinase regulates the subcellular localization of Snf1-Sip1 protein kinase. Mol. Cell. Biol. 2004;24:1836–1843. doi: 10.1128/MCB.24.5.1836-1843.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hedbacker K, Hong SP, Carlson M. Pak1 protein kinase regulates activation and nuclear localization of Snf1-Gal83 protein kinase. Mol Cell Biol. 2004;24:8255–8263. doi: 10.1128/MCB.24.18.8255-8263.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Woods A, Johnstone SR, Dickerson K, Leiper FC, Fryer LG, Neumann D, Schlattner U, Wallimann T, Carlson M, Carling D. LKB1 Is the upstream kinase in the AMP-activated protein kinase dascade. Curr. Biol. 2003;13:2004–2008. doi: 10.1016/j.cub.2003.10.031. [DOI] [PubMed] [Google Scholar]
- 39.Hawley SA, Boudeau J, Reid JL, Mustard KJ, Udd L, Makela TP, Alessi DR, Hardie DG. Complexes between the LKB1 tumor suppressor, STRADalpha/beta and MO25alpha/beta are upstream kinases in the AMP-activated protein kinase cascade. J Biol. 2003;2:28. doi: 10.1186/1475-4924-2-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shaw RJ, Kosmatka M, Bardeesy N, Hurley RL, Witters LA, DePinho RA, Cantley LC. The tumor suppressor LKB1 kinase directly activates AMP-activated kinase and regulates apoptosis in response to energy stress. Proc Natl Acad Sci U S A. 2004;101:3329–3335. doi: 10.1073/pnas.0308061100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Woods A, Dickerson K, Heath R, Hong SP, Momcilovic M, Johnstone SR, Carlson M, Carling D. Ca(2+)/calmodulin-dependent protein kinase kinase-beta acts upstream of AMP-activated protein kinase in mammalian cells. Cell Metab. 2005;2:21–33. doi: 10.1016/j.cmet.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 42.Hawley SA, Pan DA, Mustard KJ, Ross L, Bain J, Edelman AM, Frenguelli BG, Hardie DG. Calmodulin-dependent protein kinase kinase-beta is an alternative upstream kinase for AMP-activated protein kinase. Cell Metab. 2005;2:9–19. doi: 10.1016/j.cmet.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 43.Hurley RL, Anderson KA, Franzone JM, Kemp BE, Means AR, Witters LA. The Ca2+/Calmodulin-dependent Protein Kinase Kinases Are AMP-activated Protein Kinase Kinases. J Biol Chem. 2005;280:29060–29066. doi: 10.1074/jbc.M503824200. [DOI] [PubMed] [Google Scholar]
- 44.Hong SP, Momcilovic M, Carlson M. Function of mammalian LKB1 and Ca2+/calmodulin-dependent protein kinase kinase alpha as Snf1-activating kinases in yeast. J Biol Chem. 2005;280:21804–21809. doi: 10.1074/jbc.M501887200. [DOI] [PubMed] [Google Scholar]
- 45.Momcilovic M, Hong SP, Carlson M. Mammalian TAK1 activates Snf1 protein kinase in yeast and phosphorylates AMP-activated protein kinase in vitro. J Biol Chem. 2006;281:25336–25343. doi: 10.1074/jbc.M604399200. [DOI] [PubMed] [Google Scholar]
- 46.Goffrini P, Ficarelli A, Donnini C, Lodi T, Puglisi PP, Ferrero I. FOG1 and FOG2 genes, required for the transcriptional activation of glucose-repressible genes of Kluyveromyces lactis, are homologous to GAL83 and SNF1 of Saccharomyces cerevisiae. Curr Genet. 1996;29:316–326. [PubMed] [Google Scholar]
- 47.Dong J, Dickson RC. Glucose represses the lactose-galactose regulon in Kluyveromyces lactis through a SNF1 and MIG1- dependent pathway that modulates galactokinase (GAL1) gene expression. Nucleic Acids Res. 1997;25:3657–3664. doi: 10.1093/nar/25.18.3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Petter R, Chang YC, Kwon-Chung KJ. A gene homologous to Saccharomyces cerevisiae SNF1 appears to be essential for the viability of Candida albicans. Infect Immun. 1997;65:4909–4917. doi: 10.1128/iai.65.12.4909-4917.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kanai T, Ogawa K, Ueda M, Tanaka A. Expression of the SNF1 gene from candida tropicalis is required for growth on various carbon sources, including glucose. Arch Microbiol. 1999;172:256–263. doi: 10.1007/s002030050768. [DOI] [PubMed] [Google Scholar]
- 50.Townley R, Shapiro L. Crystal Structures of the Adenylate Sensor from Fission Yeast AMP-Activated Protein Kinase. Science. 2007 doi: 10.1126/science.1137503. [DOI] [PubMed] [Google Scholar]
- 51.Celenza JL, Carlson M. Structure and expression of the SNF1 gene of Saccharomyces cerevisiae. Mol. Cell. Biol. 1984;4:54–60. doi: 10.1128/mcb.4.1.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Crute BE, Seefeld K, Gamble J, Kemp BE, Witters LA. Functional domains of the alpha1 catalytic subunit of the AMP-activated protein kinase. J Biol Chem. 1998;273:35347–35354. doi: 10.1074/jbc.273.52.35347. [DOI] [PubMed] [Google Scholar]
- 53.Leech A, Nath N, McCartney RR, Schmidt MC. Isolation of mutations in the catalytic domain of the snf1 kinase that render its activity independent of the snf4 subunit. Eukaryot Cell. 2003;2:265–273. doi: 10.1128/EC.2.2.265-273.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Estruch F, Treitel MA, Yang X, Carlson M. N-terminal mutations modulate yeast SNF1 protein kinase function. Genetics. 1992;132:639–650. doi: 10.1093/genetics/132.3.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jiang R, Carlson M. The Snf1 protein kinase and its activating subunit, Snf4, interact with distinct domains of the Sip1/Sip2/Gal83 component in the kinase complex. Mol. Cell. Biol. 1997;17:2099–2106. doi: 10.1128/mcb.17.4.2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ludin K, Jiang R, Carlson M. Glucose-regulated interaction of a regulatory subunit of protein phosphatase 1 with the Snf1 protein kinase in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 1998;95:6245–6250. doi: 10.1073/pnas.95.11.6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Elbing K, McCartney RR, Schmidt MC. Purification and characterization of the three Snf1-activating kinases of Saccharomyces cerevisiae. Biochem J. 2006 doi: 10.1042/BJ20051213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Elbing K, Rubenstein EM, McCartney RR, Schmidt MC. Subunits of the Snf1 kinase heterotrimer show interdependence for association and activity. J Biol Chem. 2006;281:26170–26180. doi: 10.1074/jbc.M603811200. [DOI] [PubMed] [Google Scholar]
- 59.Rudolph MJ, Amodeo GA, Bai Y, Tong L. Crystal structure of the protein kinase domain of yeast AMP-activated protein kinase Snf1. Biochem Biophys Res Commun. 2005;337:1224–1228. doi: 10.1016/j.bbrc.2005.09.181. [DOI] [PubMed] [Google Scholar]
- 60.Nayak V, Zhao K, Wyce A, Schwartz MF, Lo WS, Berger SL, Marmorstein R. Structure and dimerization of the kinase domain from yeast Snf1, a member of the Snf1/AMPK protein family. Structure. 2006;14:477–485. doi: 10.1016/j.str.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 61.Matsumoto K, Toh-e A, Oshima Y. Isolation and characterization of dominant mutations resistant to carbon catabolite repression of galactokinase synthesis in Saccharomyces cerevisiae. Mol Cell Biol. 1981;1:83–93. doi: 10.1128/mcb.1.2.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schmidt MC, McCartney RR. beta-subunits of Snf1 kinase are required for kinase function and substrate definition. EMBO J. 2000;19:4936–4943. doi: 10.1093/emboj/19.18.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wiatrowski HA, van Denderen BJW, Berkey CD, Kemp BE, Stapleton D, Carlson M. Mutations in the Gal83 glycogen-binding domain activate the Snf1/Gal83 kinase pathway by a glycogen-independent mechanism. Mol. Cell. Biol. 2004;24 doi: 10.1128/MCB.24.1.352-361.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vincent O, Carlson M. Gal83 mediates the interaction of the Snf1 kinase complex with the transcription activator Sip4. EMBO J. 1999;18:6672–6681. doi: 10.1093/emboj/18.23.6672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O'Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 66.Nath N, McCartney RR, Schmidt MC. Purification and characterization of Snf1 kinase complexes containing a defined Beta subunit composition. J Biol Chem. 2002;277:50403–50408. doi: 10.1074/jbc.M207058200. [DOI] [PubMed] [Google Scholar]
- 67.Lin SS, Manchester JK, Gordon JI. Sip2, an N-myristoylated beta subunit of Snf1 kinase, regulates aging in Saccharomyces cerevisiae by affecting cellular histone kinase activity, recombination at rDNA loci, and silencing. J Biol Chem. 2003;278:13390–13397. doi: 10.1074/jbc.M212818200. [DOI] [PubMed] [Google Scholar]
- 68.Vyas VK, Kuchin S, Berkey CD, Carlson M. Snf1 kinases with different β-subunit isoforms play distinct roles in regulating haploid invasive growth. Mol. Cell. Biol. 2003;23:1341–1348. doi: 10.1128/MCB.23.4.1341-1348.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lin SS, Manchester JK, Gordon JI. Enhanced gluconeogenesis and increased energy storage as hallmarks of aging in Saccharomyces cerevisiae. J. Biol. Chem. 2001;276:36000–36007. doi: 10.1074/jbc.M103509200. [DOI] [PubMed] [Google Scholar]
- 70.Kuchin S, Treich I, Carlson M. A regulatory shortcut between the Snf1 protein kinase and RNA polymerase II holoenzyme. Proc. Natl. Acad. Sci. USA. 2000;97:7916–7920. doi: 10.1073/pnas.140109897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.McCartney RR, Rubenstein EM, Schmidt MC. Snf1 kinase complexes with different beta subunits display stress-dependent preferences for the three Snf1-activating kinases. Curr Genet. 2005;47:335–344. doi: 10.1007/s00294-005-0576-2. [DOI] [PubMed] [Google Scholar]
- 72.Corvey C, Koetter P, Beckhaus T, Hack J, Hofmann S, Hampel M, Stein T, Karas M, Entian KD. Carbon Source-dependent assembly of the Snf1p kinase complex in Candida albicans. J Biol Chem. 2005;280:25323–25330. doi: 10.1074/jbc.M503719200. [DOI] [PubMed] [Google Scholar]
- 73.Neigeborn L, Carlson M. Genes affecting the regulation of SUC2 gene expression by glucose repression in Saccharomyces cerevisiae. Genetics. 1984;108:845–858. doi: 10.1093/genetics/108.4.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Entian K-D, Zimmermann FK. New genes involved in carbon catabolite repression and derepression in the yeast Saccharomyces cerevisiae. J. Bacteriol. 1982;151:1123–1128. doi: 10.1128/jb.151.3.1123-1128.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bateman A. The structure of a domain common to archaebacteria and the homocystinuria disease protein. Trends Biochem Sci. 1997;22:12–13. doi: 10.1016/s0968-0004(96)30046-7. [DOI] [PubMed] [Google Scholar]
- 76.Adams J, Chen ZP, Van Denderen BJ, Morton CJ, Parker MW, Witters LA, Stapleton D, Kemp BE. Intrasteric control of AMPK via the gamma1 subunit AMP allosteric regulatory site. Protein Sci. 2004;13:155–165. doi: 10.1110/ps.03340004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Scott JW, Hawley SA, Green KA, Anis M, Stewart G, Scullion GA, Norman DG, Hardie DG. CBS domains form energy-sensing modules whose binding of adenosine ligands is disrupted by disease mutations. J Clin Invest. 2004;113:274–284. doi: 10.1172/JCI19874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Scott JW, Ross FA, Liu JK, Hardie DG. Regulation of AMP-activated protein kinase by a pseudosubstrate sequence on the gamma subunit. EMBO J. 2007;26:806–815. doi: 10.1038/sj.emboj.7601542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jackson T, Kwon E, Chachulska AM, Hyman LE. Novel roles for elongin C in yeast. Biochim Biophys Acta. 2000;1491:161–176. doi: 10.1016/s0167-4781(00)00052-x. [DOI] [PubMed] [Google Scholar]
- 80.Hedbacker K, Carlson M. Regulation of the nucleocytoplasmic distribution of Snf1-Gal83 protein kinase. Eukaryot Cell. 2006;5:1950–1956. doi: 10.1128/EC.00256-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Orlova M, Kanter E, Krakovich D, Kuchin S. Nitrogen availability and TOR regulate the Snf1 protein kinase in Saccharomyces cerevisiae. Eukaryot Cell. 2006;5:1831–1837. doi: 10.1128/EC.00110-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kim MD, Hong SP, Carlson M. Role of Tos3, a Snf1 protein kinase kinase, during growth of Saccharomyces cerevisiae on nonfermentable carbon sources. Eukaryot Cell. 2005;4:861–866. doi: 10.1128/EC.4.5.861-866.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rubenstein EM, McCartney RR, Schmidt MC. Regulatory domains of Snf1-activating kinases determine pathway specificity. Eukaryot Cell. 2006;5:620–627. doi: 10.1128/EC.5.4.620-627.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bouquin N, Barral Y, Courbeyrette R, Blondel M, Snyder M, Mann C. Regulation of cytokinesis by the Elm1 protein kinase in Saccharomyces cerevisiae. J. Cell. Sci. 2000;113:1435–1445. doi: 10.1242/jcs.113.8.1435. [DOI] [PubMed] [Google Scholar]
- 85.Blacketer M, Koehler C, Coats S, Myers A, Madaule P. Regulation of dimorphism in Saccharomyces cerevisiae involvement of the novel protein kinase homolog Elm1p and protein phosphatase 2A. Mol. Cell. Biol. 1993;13:5567–5581. doi: 10.1128/mcb.13.9.5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sreenivasan A, Kellogg D. The Elm1 kinase functions in a mitotic signaling network in budding yeast. Mol. Cell. Biol. 1999;19:7983–7994. doi: 10.1128/mcb.19.12.7983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sreenivasan A, Bishop AC, Shokat KM, Kellogg DR. Specific inhibition of Elm1 kinase activity reveals functions required for early G1 events. Mol Cell Biol. 2003;23:6327–6337. doi: 10.1128/MCB.23.17.6327-6337.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tu J, Carlson M. The GLC7 type 1 protein phosphatase is required for glucose repression in Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:6789–6796. doi: 10.1128/mcb.14.10.6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Dombek KM, Voronkova V, Raney A, Young ET. Functional analysis of the yeast Glc7-binding protein Reg1 identifies a protein phosphatase type 1-binding motif as essential for repression of ADH2 expression. Mol. Cell. Biol. 1999;19:6029–6040. doi: 10.1128/mcb.19.9.6029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Young ET, Dombek KM, Tachibana C, Ideker T. Multiple pathways are co-regulated by the protein kinase Snf1 and the transcription factors Adr1 and Cat8. J Biol Chem. 2003;278:26146–26158. doi: 10.1074/jbc.M301981200. [DOI] [PubMed] [Google Scholar]
- 91.Treitel MA, Kuchin S, Carlson M. Snf1 protein kinase regulates phosphorylation of the Mig1 repressor in Saccharomyces cerevisiae. Mol. Cell. Biol. 1998;18:6273–6280. doi: 10.1128/mcb.18.11.6273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ostling J, Ronne H. Negative control of the Mig1p repressor by Snf1p-dependent phosphorylation in the absence of glucose. Eur J Biochem. 1998;252:162–168. doi: 10.1046/j.1432-1327.1998.2520162.x. [DOI] [PubMed] [Google Scholar]
- 93.Hedges D, Proft M, Entian K-D. CAT8, a new zinc cluster-encoding gene necessary for derepression of gluconeogenic enzymes in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 1995;15:1915–1922. doi: 10.1128/mcb.15.4.1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lesage P, Yang X, Carlson M. Yeast SNF1 protein kinase interacts with SIP4, a C6 zinc cluster transcriptional activator a new role for SNF1 in the glucose response. Mol. Cell. Biol. 1996;16:1921–1928. doi: 10.1128/mcb.16.5.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Young ET, Kacherovsky N, Van Riper K. Snf1 protein kinase regulates Adr1 binding to chromatin but not transcription activation. J Biol Chem. 2002;277:38095–38103. doi: 10.1074/jbc.M206158200. [DOI] [PubMed] [Google Scholar]
- 96.Lo W, Duggan SL, Emre NC, Belotserkovskya R, Lane WS, Shiekhattar R, Berger SL. Snf1--a histone kinase that works in concert with the histone acetyltransferase Gcn5 to regulate transcription. Science. 2001;293:1142–1146. doi: 10.1126/science.1062322. [DOI] [PubMed] [Google Scholar]
- 97.Shirra MK, Rogers SE, Alexander DE, Arndt KM. The Snf1 protein kinase and Sit4 protein phosphatase have opposing functions in regulating TATA-binding protein association with the Saccharomyces cerevisiae INO1 promoter. Genetics. 2005;169:1957–1972. doi: 10.1534/genetics.104.038075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Treitel MA, Carlson M. Repression by SSN6-TUP1 is directed by MIG1, a repressor/activator protein. Proc. Natl. Acad. Sci. USA. 1995;92:3132–3136. doi: 10.1073/pnas.92.8.3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tzamarias D, Struhl K. Distinct TPR motifs of Cyc8 are involved in recruiting the Cyc8-Tup1 corepressor complex to differentially regulated promoters. Genes Dev. 1995;9:821–831. doi: 10.1101/gad.9.7.821. [DOI] [PubMed] [Google Scholar]
- 100.Smith FC, Davies SP, Wilson WA, Carling D, Hardie DG. The SNF1 kinase complex from Saccharomyces cerevisiae phosphorylates the transcriptional repressor protein Mig1p in vitro at four sites within or near regulatory domain 1. FEBS Lett. 1999;453:219–223. doi: 10.1016/s0014-5793(99)00725-5. [DOI] [PubMed] [Google Scholar]
- 101.DeVit MJ, Johnston M. The nuclear exportin Msn5 is required for nuclear export of the Mig1 glucose repressor of Saccharomyces cerevisiae. Curr Biol. 1999;9:1231–1241. doi: 10.1016/s0960-9822(99)80503-x. [DOI] [PubMed] [Google Scholar]
- 102.Papamichos-Chronakis M, Conlan RS, Gounalaki N, Copf T, Tzamarias D. Hrs1/Med3 is a Cyc8-Tup1 corepressor target in the RNA polymerase Ii holoenzyme. J. Biol. Chem. 2000;275:8397–8403. doi: 10.1074/jbc.275.12.8397. [DOI] [PubMed] [Google Scholar]
- 103.Randez-Gil F, Bojunga N, Proft M, Entian K-D. Glucose derepression of gluconeogenic enzymes in Saccharomyces cerevisiae correlates with phosphorylation of the gene activator Cat8p. Mol. Cell. Biol. 1997;17:2502–2510. doi: 10.1128/mcb.17.5.2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Vincent O, Carlson M. Sip4, a Snf1 kinase-dependent transcriptional activator, binds to the carbon source-responsive element of gluconeogenic genes. EMBO J. 1998;17:7002–7008. doi: 10.1093/emboj/17.23.7002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Charbon G, Breunig KD, Wattiez R, Vandenhaute J, Noel-Georis I. Key role of Ser562/661 in Snf1-dependent regulation of Cat8p in Saccharomyces cerevisiae and Kluyveromyces lactis. Mol Cell Biol. 2004;24:4083–4091. doi: 10.1128/MCB.24.10.4083-4091.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tamai KT, Liu X, Silar P, Sosinowski T, Thiele DJ. Heat shock transcription factor activates yeast metallothionein gene expression in response to heat and glucose starvation via distinct signalling pathways. Mol Cell Biol. 1994;14:8155–8165. doi: 10.1128/mcb.14.12.8155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hahn JS, Thiele DJ. Activation of the Saccharomyces cerevisiae heat shock transcription factor under glucose starvation conditions by Snf1 protein kinase. J Biol Chem. 2004;279:5169–5176. doi: 10.1074/jbc.M311005200. [DOI] [PubMed] [Google Scholar]
- 108.Mayordomo I, Estruch F, Sanz P. Convergence of the target of rapamycin and the Snf1 protein kinase pathways in the regulation of the subcellular localization of Msn2, a transcriptional activator of STRE (Stress Response Element)-regulated genes. J Biol Chem. 2002;277:35650–35656. doi: 10.1074/jbc.M204198200. [DOI] [PubMed] [Google Scholar]
- 109.De Wever V, Reiter W, Ballarini A, Ammerer G, Brocard C. A dual role for PP1 in shaping the Msn2-dependent transcriptional response to glucose starvation. Embo J. 2005;24:4115–4123. doi: 10.1038/sj.emboj.7600871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bertram PG, Choi JH, Carvalho J, Chan TF, Ai W, Zheng XF. Convergence of TOR-nitrogen and Snf1-glucose signaling pathways onto Gln3. Mol Cell Biol. 2002;22:1246–1252. doi: 10.1128/MCB.22.4.1246-1252.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Shirra MK, Arndt KM. Evidence for the involvement of the Glc7-Reg1 phosphatase and the Snf1-Snf4 kinase in the regulation of INO1 transcription in Saccharomyces cerevisiae. Genetics. 1999;152:73–87. doi: 10.1093/genetics/152.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lo WS, Gamache ER, Henry KW, Yang D, Pillus L, Berger SL. Histone H3 phosphorylation can promote TBP recruitment through distinct promoter-specific mechanisms. Embo J. 2005;24:997–1008. doi: 10.1038/sj.emboj.7600577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Liu Y, Xu X, Singh-Rodriguez S, Zhao Y, Kuo MH. Histone H3 Ser10 phosphorylation-independent function of Snf1 and Reg1 proteins rescues a gcn5- mutant in HIS3 expression. Mol Cell Biol. 2005;25:10566–10579. doi: 10.1128/MCB.25.23.10566-10579.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hardy TA, Huang D, Roach PJ. Interactions between cAMP-dependent and SNF1 protein kinases in the control of glycogen accumulation in Saccharomyces cerevisiae. J. Biol. Chem. 1994;269:27907–27913. [PubMed] [Google Scholar]
- 115.Wang Z, Wilson WA, Fujino MA, Roach PJ. Antagonistic controls of autophagy and glycogen accumulation by Snf1p, the yeast homolog of AMP-activated protein kinase, and the cyclin-dependent kinase Pho85p. Mol Cell Biol. 2001;21:5742–5752. doi: 10.1128/MCB.21.17.5742-5752.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Shirra MK, Patton-Vogt J, Ulrich A, Liuta-Tehlivets O, Kohlwein SD, Henry SA, Arndt KM. Inhibition of acetyl coenzyme A carboxylase activity restores expression of the INO1 gene in a snf1 mutant strain of Saccharomyces cerevisiae. Mol Cell Biol. 2001;21:5710–5722. doi: 10.1128/MCB.21.17.5710-5722.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Francois J, Parrou JL. Reserve carbohydrates metabolism in the yeast Saccharomyces cerevisiae. FEMS Microbiology Reviews. 2001;25:124–145. doi: 10.1111/j.1574-6976.2001.tb00574.x. [DOI] [PubMed] [Google Scholar]
- 118.Cannon JF, Pringle JR, Fiechter A, Khalil M. Characterization of glycogen-deficient glc mutants of Saccharomyces cerevisiae. Genetics. 1994;136:485–503. doi: 10.1093/genetics/136.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Huang D, Farkas I, Roach PJ. Pho85p, a cyclin-dependent protein kinase, and the Snf1p protein kinase act antagonistically to control glycogen accumulation in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:4357–4365. doi: 10.1128/mcb.16.8.4357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Tomas-Cobos L, Sanz P. Active Snf1 protein kinase inhibits expression of the Saccharomyces cerevisiae HXT1 glucose transporter gene. Biochem J. 2002;368:657–663. doi: 10.1042/BJ20020984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kaniak A, Xue Z, Macool D, Kim JH, Johnston M. Regulatory network connecting two glucose signal transduction pathways in Saccharomyces cerevisiae. Eukaryot Cell. 2004;3:221–231. doi: 10.1128/EC.3.1.221-231.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wiedemuth C, Breunig KD. Role of Snf1p in regulation of intracellular sorting of the lactose and galactose transporter Lac12p in Kluyveromyces lactis. Eukaryot Cell. 2005;4:716–721. doi: 10.1128/EC.4.4.716-721.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ye T, Garcia-Salcedo R, Ramos J, Hohmann S. Gis4, a new component of the ion homeostasis system in the yeast Saccharomyces cerevisiae. Eukaryot Cell. 2006;5:1611–1621. doi: 10.1128/EC.00215-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Platara M, Ruiz A, Serrano R, Palomino A, Moreno F, Arino J. The transcriptional response of the yeast Na(+)-ATPase ENA1 gene to alkaline stress involves three main signaling pathways. J Biol Chem. 2006;281:36632–36642. doi: 10.1074/jbc.M606483200. [DOI] [PubMed] [Google Scholar]
- 125.Purnapatre K, Piccirillo S, Schneider BL, Honigberg SM. The CLN3/SWI6/CLN2 pathway and SNF1 act sequentially to regulate meiotic initiation in Saccharomyces cerevisiae. Genes Cells. 2002;7:675–691. doi: 10.1046/j.1365-2443.2002.00551.x. [DOI] [PubMed] [Google Scholar]
- 126.Palecek SP, Parikh AS, Huh JH, Kron SJ. Depression of Saccharomyces cerevisiae invasive growth on non-glucose carbon sources requires the Snf1 kinase. Mol Microbiol. 2002;45:453–469. doi: 10.1046/j.1365-2958.2002.03024.x. [DOI] [PubMed] [Google Scholar]
- 127.Xie M, Zhang D, Dyck JR, Li Y, Zhang H, Morishima M, Mann DL, Taffet GE, Baldini A, Khoury DS, Schneider MD. A pivotal role for endogenous TGF-beta-activated kinase-1 in the LKB1/AMP-activated protein kinase energy-sensor pathway. Proc Natl Acad Sci U S A. 2006;103:17378–17383. doi: 10.1073/pnas.0604708103. [DOI] [PMC free article] [PubMed] [Google Scholar]