Summary
Ferroportin (FPN1), the sole characterized mammalian iron exporter, has an iron responsive element (IRE) in its 5'UTR, which ensures that its translation is repressed by iron regulatory proteins in iron-deficient conditions to maintain cellular iron content. However, here we demonstrate that duodenal epithelial and erythroid precursor cells utilize an alternative upstream promoter to express a FPN1 transcript, FPN1B, which lacks the IRE and is not repressed in iron-deficient conditions. The FPN1B transcript encodes ferroportin with an identical open reading frame, and contributes significantly to ferroportin protein expression in erythroid precursors, and likely also in the duodenum of iron-starved animals. The identification of FPN1B reveals how FPN1 expression can bypass IRP-dependent repression in intestinal iron uptake, even when cells throughout the body are iron-deficient. In erythroid precursor cells, we hypothesize that FPN1B expression enhances real-time sensing of systemic iron status and facilitates restriction of erythropoiesis in response to low systemic iron.
Introduction
Iron is an essential element for living organisms, as it is required for activity of molecules involved in a series of crucial physiological events such as oxygen transport, mitochondrial respiration and DNA synthesis. However, because of its pro-oxidant properties, excess iron can potentially damage DNA, lipids and proteins, leading to cell death and tissue dysfunction. As mammals do not have a dedicated iron secretory pathway, organisms regulate body iron stores largely by regulating intestinal iron absorption, in part through the hepcidin-ferroportin interaction (Nemeth and Ganz, 2006; Darshan and Anderson, 2007; Wrighting and Andrews, 2008).
FPN1 (Donovan, et al., 2000), also known as IREG1 (McKie, et al., 2000), or MTP1 (Abboud and Haile, 2000)), is the sole iron exporter protein identified thus far (Donovan, et al., 2005). It is highly expressed in numerous cells, including two types that are important in systemic iron homeostasis, the duodenal enterocytes where FPN1 is responsible for transporting dietary iron across the basolateral membrane into the blood stream (for iron uptake) (Abboud and Haile, 2000; Donovan, et al., 2000; McKie, et al., 2000), and the reticuloendothelial macrophages where FPN1 exports iron from the cytosol into the blood stream (for iron recycling) (Yang, et al., 2002; Knutson, et al., 2003; Delaby, et al., 2005). In humans, some mutations in the coding region of FPN1 lead to autosomal dominant iron overload (Pietrangelo, 2004). In mice, targeted deletion of FPN1 causes embryonic lethality due to the inability to transfer iron from mother to FPN1 deficient embryos, while conditional knockout of FPN1 in the duodenum causes a significant anemia in mice, emphasizing its importance in iron absorption (Donovan, et al., 2005).
Ferroportin expression is regulated at several levels, including presumed transcriptional regulation in the duodenal mucosa and macrophages (McKie, et al., 2000; Knutson, et al., 2003), translational regulation by the IRE/IRP regulatory system (Abboud and Haile, 2000; McKie, et al., 2000; Liu, et al., 2002; Lymboussaki, et al., 2003), and post-translational regulation by the action of hepcidin, a small peptide hormone secreted by hepatocytes in response to increased iron abundance (Park, et al., 2001; Pigeon, et al., 2001). Circulating in the blood stream, hepcidin can bind to FPN1, which results in the internalization of FPN1 and degradation of FPN1 in lysosomes. Reduced export of iron from duodenal enterocytes and macrophages contributes to systemic iron deficiency (Nemeth, et al., 2004). Conversely, in iron-depleted animals, hepcidin expression is suppressed, FPN1 degradation is reduced, and more iron is therefore absorbed to satisfy systemic iron demands. Thus, by feedback regulation, hepcidin can interact with FPN1 to prevent systemic iron overload. Most types of hemochromatosis result from dysfunction of the hepcidin/FPN1 interaction, providing compelling evidence for the importance of the hepcidin-ferroportin interactions in regulation of systemic iron homeostasis (Nemeth and Ganz, 2006; Pietrangelo, 2006; Darshan and Anderson, 2007; Wrighting and Andrews, 2008).
Similar to numerous transcripts that encode iron metabolism proteins, FPN1 mRNA has a IRE in its 5'untranslated region (UTR), and its expression is translationally regulated by iron regulatory proteins (IRP1 and IRP2) (Abboud and Haile, 2000; McKie, et al., 2000; Liu, et al., 2002; Lymboussaki, et al., 2003). Other iron metabolism genes that contain IREs include L- and H-ferritin, an erythroid isoform of aminolevulinate synthase (e-ALAS) and mitochondrial aconitase, all of which contain 5'IREs that mediate translational repression, as well as the transferrin receptor1 (TfR1) and one isoform of divalent metal transporter (DMT1), both of which contain IREs in the 3'UTR that likely affect mRNA stability (Rouault, 2006; Wallander, et al., 2006). IRE-binding activities of IRPs are regulated by intracellular iron status, and are activated by iron deficiency. Binding by IRPs enables cells to replenish cytosolic iron by increasing iron uptake through TfR1 and DMT1, while simultaneously decreasing iron sequestration in ferritin, and suppressing iron export through FPN1.
FPN1 protein levels have been reported to be decreased in the duodenum of high-iron diet animals and increased in the duodenum of low-iron diet animals (Abboud and Haile, 2000; McKie, et al., 2000). However, in iron-deficient animals, activation of IRPs by iron deficiency would be expected to repress translation of FPN1, which raises the question of how enterocytes circumvent repression by IRE/IRP machinery to produce FPN1 protein. The IRE in FPN1 transcript was previously shown to be functional in reporter and competition assays (Abboud and Haile, 2000; McKie, et al., 2000; Liu, et al., 2002; Lymboussaki, et al., 2003). Moreover, in mice lacking intestinal expression of IRPs, FPN1 protein levels were significantly increased, whereas FPN1 mRNA levels remained constant, providing evidence that FPN1 was regulated by IRE/IRP machinery in vivo (Galy, et al., 2008). The IRE/IRP machinery is thought to be highly functional in duodenal enterocytes, as protein levels of L- and H-ferritin, two genes that contain 5'IREs, were dramatically decreased in the duodenum of mice maintained on a low-iron diet although their mRNA levels remained constant (McKie, et al., 1996; Pountney, et al., 1999). As FPN1 expression should also be similarly translationally repressed by IRPs, we considered the possibility that a non-IRE containing FPN1 transcript that could evade repression by the IRE/IRP system in iron deficiency might be expressed in duodenum to allow active export of iron into the blood stream to meet systemic iron demands.
In this work, we performed 5'-rapid amplification of cDNA end assays (RACE) on mouse cDNA libraries and identified a non-IRE containing transcript of FPN1 generated from an alternative upstream promoter. Characterization of its expression profiles and regulation by iron in vivo and in vitro, suggested that this non-IRE containing ferroportin transcript plays an important physiological role in two distinct cell types: in duodenal mucosal cells, where it likely facilitates intestinal iron uptake in iron-deficient animals, and also in erythroid precursors, where it drives iron export activity and perhaps renders erythroid precursors more responsive to changes in systemic iron status.
Results
FPN1 protein increased in the duodenum of iron-deficient mice
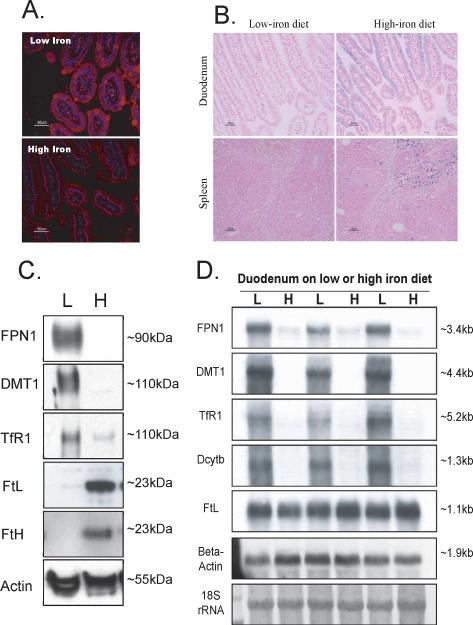
To evaluate expression levels of FPN1 in mice maintained on a low-iron diet, immunofluorescence experiments were performed on duodenal sections of mice maintained on a low-iron diet, and increased expression of FPN1 was detected throughout the duodenal epithelial cells, compared with mice maintained on high-iron diet (Figure 1A). As a confirmation of the iron status of these mice, Perls' Prussian blue staining on the duodenal and splenic sections showed more iron in tissues from animals on a high-iron diet compared to a low-iron diet (Figure 1B). Western blots confirmed that there was strong expression of FPN1 in the duodenum of low-iron diet mice and little if any in the duodenum of high-iron diet mice (Figure 1C). The increased expression of FPN1 in low-iron diet mice was consistent with previous reports, in which increased expression of FPN1 protein (Abboud and Haile, 2000; McKie, et al., 2000) and mRNA (McKie, et al., 2000) was reported in duodenal samples of iron-deficient animals.
Figure 1.
Expression of FPN1 increased in the duodenum of mice maintained on low-iron diet. (A) FPN1 expression (red) was detected with a polyclonal anti-FPN1 antibody and visualized by immunofluorescence in the duodenal sections of low- and high-iron diet mice. The nucleus was counterstained with DAPI (blue). (B) Prussian blue staining revealed more iron (blue) in the intestinal mucosa and spleen of high-iron diet mice than that of low-iron diet mice, counterstained with fast red for nucleus (red) and cytoplasm (pink). (C) Western blots revealed high expression of FPN1 in the lysate from duodenum of mice on a low-iron diet (40μg per lane). Levels of DMT1 and TfR1 increased on the low-iron diet, whereas FtL and FtH decreased. (D) Northern blots revealed that mRNA expression of FPN1, DMT1, TfR1 and Dcytb increased in duodenum of mice on a low-iron diet, whereas FtL mRNA did not change.
For comparison, expression levels of other IRE-containing genes in the intestinal mucosa were compared in animals on a low- vs. high-iron diet, and increased expression of DMT1 and TfR1, and decreased expression of FtL and FtH proteins were observed in animals on a low-iron diet (Figure 1C). At the mRNA level, transcripts encoding components of the iron uptake pathway, including FPN1, DMT1, TfR1 and Dcytb were significantly increased (Figure 1D), suggesting that, in addition to the well-known stabilization of the TfR1 mRNA, the transcriptional regulation of these genes is also important. Comparing the expression of ferritin mRNA levels with their protein levels, duodenal regulation of ferritin appeared to be almost entirely translational, as mRNA levels did not change, whereas protein levels increased significantly, indicating the IRE/IRP machinery was highly activated (Figure 1C, 1D) (McKie, et al., 1996; Pountney, et al., 1999). The mRNA level of FPN1 was about 5 fold elevated on the low-iron diet. However, because the FPN1 transcript contains a functional 5'IRE in its mRNA (Abboud and Haile, 2000; McKie, et al., 2000; Liu, et al., 2002; Lymboussaki, et al., 2003), its translation was expected to be repressed, similar to FtL and FtH, both of which are encoded by transcripts that contain 5' IREs as well. Therefore, the strikingly high expression of FPN1 protein on the low-iron diet (Figure 1C, top panel) was inconsistent with the expectation that translational repression would be mediated by the 5'IRE in the FPN1 mRNA. To better understand FPN1 expression in the duodenum, we undertook to carefully analyze the FPN1 transcript.
A transcript of FPN1 that lacked an IRE was identified by RACE
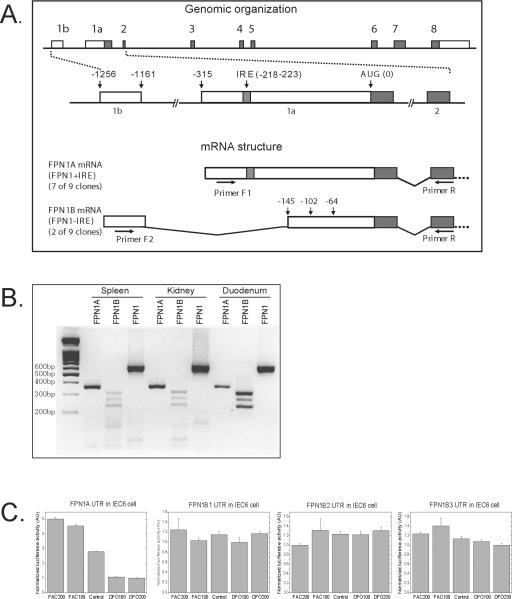
To fully characterize the 5'-end of the FPN1 transcript, we performed 5'-RACE assays with a Marathon-Ready mouse cDNA library. Although seven of the nine clones sequenced were almost identical to the FPN1 mRNA sequence provided by the “Entrez Gene” (NM_016917), two of them contained a novel 96 bp sequence in the 5'end of the transcript. By alignment with the genomic DNA, we found that this 96 bp sequence was from a new exon (exon1b) located 1161 bp 5' of the AUG (initiator methionine of the open reading frame), and 845 bp 5' of the previously annotated exon1 (exon1a) (Figure 2A). In the newly identified isoform, exon1b was connected by alternative splicing to a potential splice acceptor site within exon1a, which was 3' of the IRE, generating a transcript that lacked the IRE. Hereafter, we refer to the transcript that lacked the IRE as FPN1B, and the transcript that contained the IRE as FPN1A. FPN1B consists of 9 exons, including exon 1b, a truncated form of exon 1a, and the remaining 7 exons, whereas FPN1A consists of 8 exons, extending from exon1a to exon 8 (Figure 2A).
Figure 2.
A transcript of FPN1 that lacked an IRE was identified in mouse. (A) Schematic representation of genomic (top) and transcript structure (bottom) of FPN1. FPN1 has a total of nine exons (boxes 1b, 1a, 2 through 8, introns depicted as lines), and the IRE and AUG codon are located in exon 1a. By alternative usage of exon 1a or 1b, two FPN1 transcripts are generated. The FPN1A transcript extends from exon1a, and includes exons 2 to 8, producing a transcript that contains an IRE in its 5'UTR. The FPN1B transcript commences with exon 1b and includes a portion of exon 1a that is 3' of the IRE (from nucleotide -145, -102 or -64 depending on different acceptor sites), as well as exons 2 to 8, producing a transcript that does not have an IRE. Primers F1, F2 and R, as indicated on the graphic were used for PCR amplification of FPN1A and FPN1B. (B) PCR amplification of FPN1A (lanes 2, 5 and 8) and FPN1B (lanes 3, 6 and 9) using cDNA generated from spleen, kidney and duodenum revealed three bands which corresponded to the usage of three distinct acceptor sites within exon 1a of FPN1B. FPN1 was amplified by a pair of primers located within the ORF (lane 4, 7 and 10). The expected sizes for each of the bands were 596bp (FPN1), 356bp (FPN1A), and 312bp, 269bp, 231bp (FPN1B). PCR products were separated on a 2% agarose gel. (C) Expression of the FPN1B reporter construct was not repressed in iron starvation conditions. The 5'UTRs of FPN1A and FPN1B were cloned into the pGL3 luciferase vector, and then 1 μg of those vectors with 0.1 μg of renilla luciferase vector were transfected into IEC6 cells. One day later those cells were treated with either 200 μM, 100 μM FAC, 100 μM, or 200 μM DFO for 24h. The luciferase activity was measured by a dual luciferase activity assay. FPN1B1, B2 and B3 represent the longest, middle and shortest transcripts, respectively. Data are presented as mean ± SD.
To confirm the existence of FPN1B, we designed three primers, including one that was specific for the 5'-end of FPN1A (F1), one that was specific for exon 1b (F2), and a reverse primer specific for exon 2 (Primer R), a region that was shared by both transcripts (Figure 2A). With primers F1/R, we got a band of 356 bp as expected in cDNA from spleen, kidney and duodenum. However, with primers F2/R, we got three distinct bands in all of the three tissues tested (Figure 2B). Sequencing of each band showed that the largest band was derived from a transcript in which exon 1b connected to a nucleotide of exon 1a that was 145 bp 5' of the AUG start codon (-145), as was initially observed in the RACE assays. The two lower bands were generated by splicing from exon 1b to nucleotides at positions -102 and -64 of exon 1a, respectively (Figure 2A). Thus, there were actually three FPN1B transcripts, which we have deposited in GenBank: FPN1B1, accession number FJ207421, intron acceptor site -145; FPN1B2, accession number FJ207422, acceptor site -102; and FPN1B3, accession number FJ207423, acceptor site -64. For each of the three splice acceptor sites, the intron ends with “AG”, a dinucleotide sequence typical of intron acceptor sites. However, according to genome sequence compiled in the “Entrez Gene”, the genomic sequence for the intron acceptor site of FPN1B2 ends with “AC”, and only 2 of 12 sequences in the “Evidence Viewer” are “AG”, whereas the others are “AC”, indicating that a single nucleotide polymorphism exists in this site and FPN1B2 may not be expressed in all mouse strains.
The 5'UTR of FPN1B did not mediate translational repression in iron starved cells
To evaluate the regulation of the 5'UTR of the FPN1B transcript, we cloned the 5'UTRs of FPN1A and FPN1B into a pGL3 vector, in which a SV40 promoter drives transcription to produce a luciferase chimeric mRNA with the FPN1A or FPN1B 5'UTR as its 5'UTR, making it possible to check the translational regulation of FPN1A or FPN1B 5'UTR in vivo. In the rat intestinal epithelial cell line, IEC6, the expression of the FPN1A/luciferase construct decreased with iron starvation to 20% of the level in iron-replete cells (Figure 2C). In contrast, there was no change of the luciferase activity among the three FPN1B isoforms expressed in IEC6 cells subjected to different iron conditions. Similar results were observed in CACO2 and 293 cells (Supplemental Figure 1 A and B). Thus, none of the three identified 5'UTRs of FPN1B was regulated by iron.
FPN1B was specifically expressed in duodenum
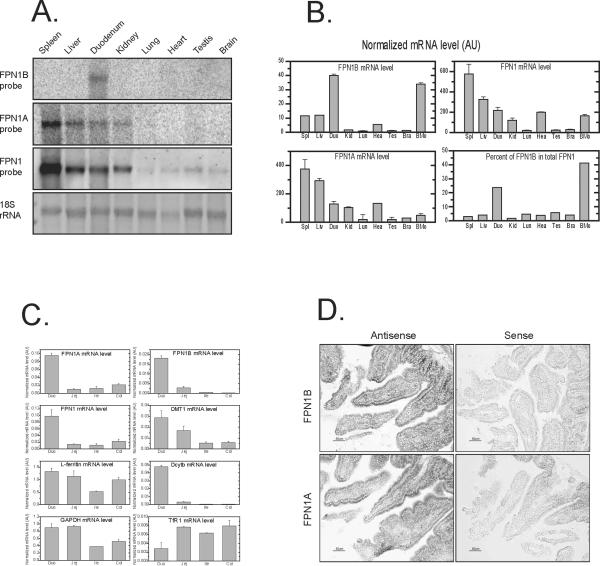
To characterize the expression profile of FPN1A and FPN1B in different tissues, we performed northern blots with transcript-specific probes. With the FPN1B probe, a band of about 3.4 kb was specifically identified in duodenum (Figure 3A). In contrast, the FPN1A band of about 3.4 kb was highly expressed in spleen, liver, duodenum and kidney. With a probe covering the coding region common to both transcripts, expression levels mirrored those for FPN1A. Because the difference in size between FPN1B and FPN1A is very small compared to the full-length transcript, changing by 74~154 bp out of 3400 bp total, we could not distinguish the FPN1B from FPN1A bands in northern blots performed using the common probe.
Figure 3.
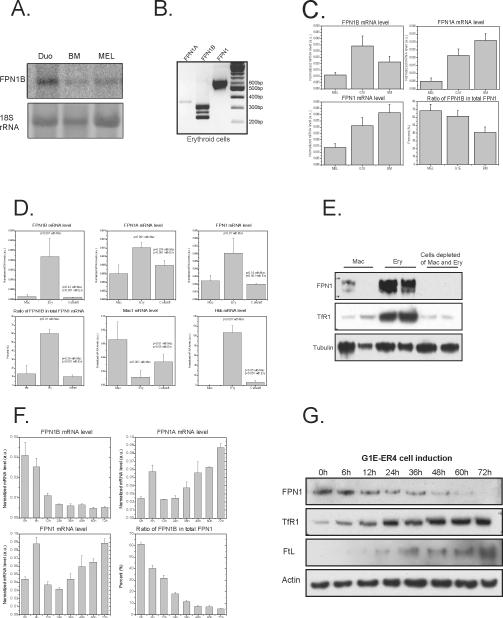
The FPN1B transcript was highly expressed in duodenum and bone marrow. (A) Total RNA of multiple tissues was probed with FPN1B or 1A specific probes in northern blots. (B) qRT-PCR was used to check the expression of FPN1B and FPN1A in cDNA of mouse tissues. The results revealed that FPN1B was specifically expressed in duodenum and bone marrow. Data are presented as mean ± SD. (C) qRT-PCR was used to check the expression of FPN1B, FPN1A, total FPN1, DMT1, Dcytb, TfR1 and FtL in cDNA from different parts of intestinal tract. The expression level was normalized to beta-actin. Data are presented as mean ± SD. (D) In situ hybridization to check the expression of the FPN1B and 1A transcripts revealed that both were expressed in duodenal mucosa cells.
To quantitatively compare the abundance of these two transcripts in different tissues, quantitative real-time PCR (qRT-PCR) was performed and results were consistent with northern blot results (Figure 3B). FPN1B was highly expressed in duodenum. Interestingly, FPN1B was also significantly expressed in bone marrow. FPN1B accounted for 25% of total FPN1 mRNA in duodenum and about 40% in bone marrow (Figure 3B). In contrast, FPN1B accounted for less than 6% of total FPN1 in other tissues. In different parts of the intestinal tract, FPN1B was expressed mainly in duodenum, as was FPN1A and Dcytb (Figure 3C). DMT1 expression was highest in duodenum, whereas TfR1 expression was lowest in duodenum among four parts of intestinal tract, and the expression of ferritin was comparable to GAPDH expression levels (Figure 3C). In situ hybridization studies revealed that both FPN1A and 1B transcripts were expressed strongly in enterocytes (Figure 3D).
Duodenal expression of FPN1B increased in animals on a low-iron diet
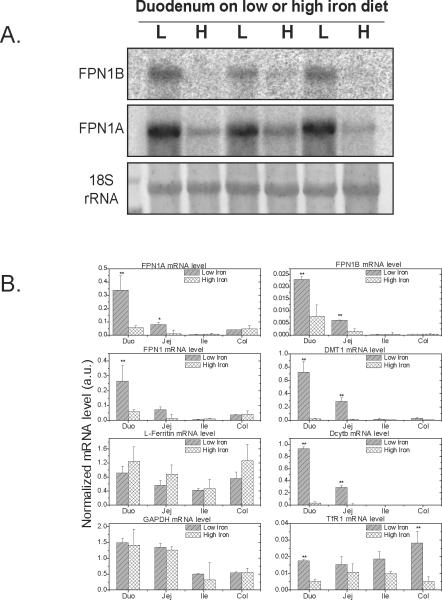
Because FPN1 protein levels increased in the duodenum of animals on a low-iron diet (Figure 1A, C), we evaluated FPN1B and FPN1A mRNA levels in northern blots of duodenal RNA from animals on low- vs. high-iron diets, and we found that both FPN1B and FPN1A mRNA levels significantly increased on the low-iron diet, similar to levels of total FPN1 mRNA (Figure 1D, 4A). The expression of FPN1B and FPN1A in four parts of intestinal tract on low- vs. high-iron diet were also studied by qRT-PCR. For FPN1B, mRNA levels increased three-fold in the duodenum and jejunum on the low-iron diet and FPN1A mRNA levels increased more than five-fold in duodenum and jejunum on the low-iron diet, whereas there was no change in either FPN1A or FPN1B levels in ileum and colon on the low-iron diet (Figure 4B). Transcript levels of DMT1 and Dcytb were dramatically increased on the low-iron diet in duodenum and jejunum, and TfR1 levels increased moderately, but ferritin and GADPH mRNA levels were unchanged in the intestinal tract from animals on a low-iron diet.
Figure 4.
Expression of the FPN1B transcript increased in the duodenum of mice maintained on a low-iron diet. (A) Northern blot and (B) qRT-PCR were used to check the expression of FPN1B and other iron genes in different parts of intestinal tract. The results revealed that FPN1B expression increased on a low-iron diet treatment. Data are presented as mean ± SD. * P<0.05 and ** P<0.01 comparing with corresponding high-iron diet.
FPN1B was specifically expressed in erythroid cells
To compare the FPN1B transcript in bone marrow and duodenum, we performed northern blots with the FPN1B specific probe on RNA from bone marrow and MEL (mouse erythroleukemia cell) cells (Figure 5A). In those two samples, we detected a 3.4 kb transcript similar to the FPN1B transcript in duodenum, and PCR results showed that all of the three sub-transcripts of FPN1B were expressed in erythroid cells of mouse bone marrow (Figure 5B), and in erythroid precursor cell lines, MEL cells and G1E cells (Supplemental Figure 2). We also checked the expression of FPN1B in other cell lines, including MSIE cells, MODE-k cells, RAW264.7 cells, and J774a.1 cells, but we did not detect FPN1B expression in non-erythroid cell lines (Results not shown). Quantification of FPN1B expression with qRT-PCR showed that FPN1B accounted for >60% of total FPN1 transcript in MEL and G1E cells in comparison with 40% in bone marrow (Figure 5C). MEL cells have some characteristics of BFU-E and CFU-E cells, and G1E cells are considered to represent proerythroblast cells that also have characteristics of BFU-E cells (Anguita, et al., 2004). The high expression of FPN1B in MEL and G1E cells revealed that FPN1B is expressed highly in erythroid precursor cells. To determine which cell type in bone marrow was responsible for high expression of FPN1B, we sorted bone marrow cells using antibodies to either macrophages (CD11b) or differentiating erythroid cells (Ter119). Quantification of ferroportin 1B and 1A transcripts revealed that almost all of the FPN1B was in cells of the erythroid lineage, where it accounted for >60% of FPN1 mRNA levels. In contrast, FPN1B transcript levels were 10-fold lower in macrophages, and comprised less than 15% of total FPN1 mRNA (Figure 5D). At the protein level, there was more than five times as much ferroportin protein in erythroid cells as in macrophages or other bone marrow cells (Figure 5E), and TfR1 mRNA and protein levels were high, as expected, in erythroid cells (Figure 5E and Supplemental Figure 3). Comparison of the expression of FPN1 transcripts with protein levels revealed that FPN1B mRNA levels correlated well with ferroportin protein levels in erythroid cells.
Figure 5.
FPN1B was specifically expressed in erythroid precursor cells and decreased with cell differentiation. (A) Northern blot showed that FPN1B expressed in bone marrow and MEL cells was identical in size to the approximately 3.4 kb duodenal FPN1B transcript. (B) PCR experiments verified that all of the three FPN1B sub-transcripts were expressed in mouse Ter119-positive erythroid cells. (C) qRT-PCR to check the expression of FPN1B, FPN1A and FPN1 mRNA expression in MEL cells, G1E cells and mouse bone marrow, normalized with actin, revealed high FPN1B expression in these two erythroid precursor cell lines. Data are presented as mean ± SD. (D) qRT-PCR results showed that FPN1B was specifically expressed in Ter119-positive erythroid cells (Ery) rather than CD11b-positive macrophages (Mac) or cells depleted of erythroid cells and macrophages. Mac1 (macrophage antigen alpha) and Hbb (Hemoglobin beta chain) were used as controls to verify the isolation. The expression levels of these genes were normalized with actin. Data are presented as mean ± SD. (E) Western blot showed that ferroportin was highly expressed in erythroid cells. TfR1 was used as a positive control for erythroid cells, and alpha-tubulin was used as a loading control. (F) FPN1B expression significantly decreased during erythroid cell differentiation. qRT-PCR was used to check the gene expression in G1E-ER4 cell at 6h, 12h, 24h, 36h, 48h, 60h and 72h after the addition of 10nM of estradiol to induce cell differentiation. The expression levels of these genes were normalized with actin. Data are presented as mean ± SD. (G) Western blot revealed that ferroportin protein decreased with differentiation of G1E-ER4 cells after addition of estradiol, whereas TfR1 and FtL expression increased with differentiation.
FPN1B accounts for most ferroportin expression in erythroid cells
To better characterize the role of FPN1B in erythroid cells, we obtained G1E-ER4 cells, an erythroid precursor cell line that expresses GATA-1 conditionally under control of estrogen receptor ligand binding domain, and we induced synchronous erythroid maturation for up to 72 hours by estradiol treatment (Weiss, et al., 1997; Rylski, et al., 2003; Welch, et al., 2004). At 0, 6, 12, 24, 36, 48, 60 and 72 hours after induction, we quantified levels of FPN1A and 1B transcripts (Figure 5F), and we discovered that FPN1B accounted for most FPN1 transcripts at early stages of differentiation. At late stages of differentiation when hemoglobin mRNA levels were high (Supplemental Figure 4), levels of FPN1A increased, but FPN1 protein levels did not increase along with FPN1A transcript levels. FPN1 protein levels reflected the levels of FPN1B transcript (Figure 5 F, G), suggesting that FPN1A expression was translationally repressed by IRPs in the differentiating erythroid cells. Consistent with previous reports, we found that TfR1 and FtL in both mRNA and protein levels were significantly increased during erythroid differentiation (Welch, et al., 2004), while there was no significant change of IRP1 or IRP2 expression (Figure 5G, Supplemental Figure 4). These results indicated that FPN1B is the major source of encoded FPN1 in erythroid precursors.
The FPN1B promoter has gut and erythroid-specific transcriptional regulation elements
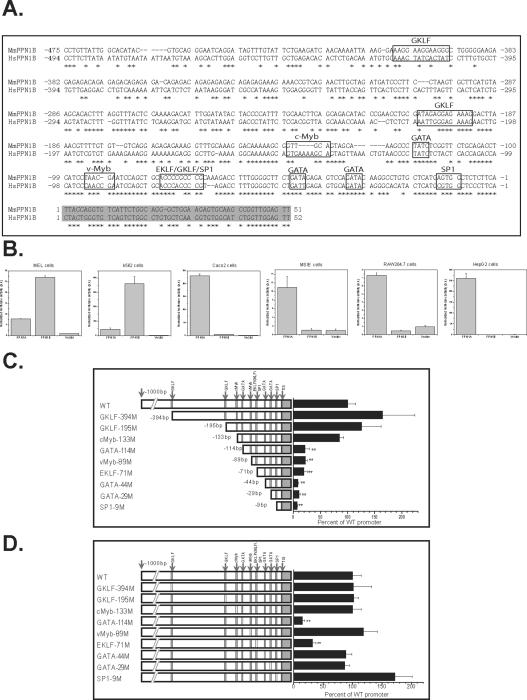
To investigate how FPN1B expression is regulated, we studied its promoter and the potential transcriptional elements. A CpG island was annotated in “GeneBuilder” and “Map Viewer” in the FPN1B promoter region. CpG islands are found in the promoters of about 56% human genes (Antequera and Bird, 1993). Sequence alignment of FPN1B promoter showed high conservation between mouse and human sequences in the 200bp region 5' of the transcription initiation site, which contained conserved consensus binding sites for several transcription factors, including GATA, EKLF and GKLF (Figure 6A). FPN1B and FPN1A promoter constructs were transfected into mammalian cell lines such as MEL cells, K562 cells, CACO2 cells, MSIE cells, RAW264.7 cells and HepG2 cells. Though the FPN1A promoter construct displayed promoter activity in all of the cell lines tested, the FPN1B promoter was active only in MEL and K562 cells, consistent with its endogenous pattern of mRNA expression among cell lines (Figure 6B, 5C). Serial deletion of the promoter region showed that the major elements of the FPN1B promoter were located within the 133bp region 5' of the transcriptional initiation site (Figure 6C, Supplemental Figure 5A). To characterize which transcriptional elements were essential for its promoter activity, each of the putative transcriptional elements in the FPN1B promoter was mutagenized. In MEL and K562 cells, mutation of GATA-114 site, EKLF/GKLF/SP1-71 (members of the KLF family share similar binding sites (Kaczynski, et al., 2003)) site, and the GATA-29 site resulted in significant loss of promoter activity, which indicated that these sites were important in FPN1B promoter activity (Figure 6D, Supplemental Figure 5B), and that GATA1, GATA2, EKLF, GKLF and SP1 all probably regulated FPN1B expression. GATA1, GATA2 and EKLF are erythroid-specific transcriptional factors (Miller and Bieker, 1993; Weiss and Orkin, 1995), whereas GKLF is specifically expressed in gastrointestinal tract (Shields, et al., 1996). The regulation of FPN1B by these transcription factors is consistent with its specific expression in duodenum and erythroid cells.
Figure 6.
The FPN1B promoter was regulated by putative GATA and KLF transcriptional elements. (A) Alignment of promoter region of mouse and human FPN1B. Sequences in black boxes indicated the consensus transcription factor binding sites, the sequence in gray indicated the new exon1, “*” indicated conserved sequences between human and mouse. (B) The FPN1B promoter construct displayed high activity in MEL and K562 cells, whereas FPN1A promoter displayed promoter activity in all of the cells tested. Empty pGL4-luciferase vector was used as a control. Data are presented as mean ± SD. (C) Serial deletion assays in MEL cells showed that the FPN1B promoter was located within 133bp region before the transcription initiation site. (D) Site-specific mutations in MEL cells showed that a GATA-114 site, and a EKLF/GKLF/SP1-71 site were essential for FPN1B promoter activity. The left panels showed the full sequence inserted in the pGL4 luciferase vector, from 1000bp before to 50bp after the transcription initiation site (TIS), the small black boxes indicated by arrows indicate the putative transcription factor binding sites, and white boxes indicate sites at which a consensus transcription factor binding site was mutagenized. The wide type (WT) construct was taken as 100% promoter activity. Data are presented as mean ± SD.
Discussion
Here, we have reported the identification of a ferroportin transcript, FPN1B, which lacks an IRE and is not subject to repression by the IRE/IRP system in iron-deficient conditions. Although FPN1B was generated from an alternative promoter, it shared the same open reading frame as FPN1A. FPN1B was specifically expressed in two locations: in the duodenum, especially in enterocytes, and in erythroid precursor cells. The discovery of FPN1B in duodenal epithelial cells helps to resolve the problem of how enterocytes can continue to export iron into the circulation when they are iron-depleted. The discovery that the FPN1B transcript encodes most of the ferroportin protein expressed in erythroid precursors adds an important new dimension to mechanisms of how erythroid precursor cells sense systemic iron levels and how their differentiation is attuned to iron status changes during erythropoiesis.
Discovery of a second ferroportin promoter and transcript fits well with the recent discovery that many nonproductive transcriptional initiation events occur in the vicinity of established promoters, some of which may produce mRNAs with altered translation efficiency, allowing transcripts to evolve to meet specific physiological needs (Buratowski, 2008). Alternative transcripts of FPN1 have been previously been observed in human erythroid cells (Cianetti, et al., 2005), where one of the major transcripts, referred to as FPNIIB, is homologous to the mouse FPN1B that we reported here. Along with the conservation between human and mouse in the promoter sequence and putative transcription factor binding sites, this human transcript suggests that FPN1B is conserved between human and mouse, and likely also in other mammals. Although the FPN1B transcript has not been previously observed in wild type mice, it was observed in polycythaemia (Pcm) mice (Mok, et al., 2004a), which were generated by radiation mutagenesis. Pcm mice contain a 58 bp deletion in the FPN1A promoter, which alters the transcription start site and eliminates the IRE in the 5'UTR. Among 10 FPN1 clones sequenced for Pcm mice, 4 clones included portions of exon 1b reported here, including one that was identical to FPN1B and three others that started from the middle of exon1b. The remaining clones initiated from exon1a, but the transcription start sites were 3' of the IRE region (Mok, et al., 2004a). Pcm mice suffered from semidominant spleen regression, which resulted from apoptotic cell death attributed to over-expression of FPN1 and decreased iron levels in this tissue (Mok, et al., 2004b). The phenotypes of Pcm mice also included a transient hypochromic and microcytic anemia that spontaneously corrected in young adult mice, where the correction correlated with increased hepcidin expression (Mok, et al., 2004a), suggesting that the conversion from FPN1B to FPN1A expression is essential to erythroid cells, especially in the early stages of life when hepcidin levels are not high enough to repress ferroportin expression (Figure 5F, G). Although the complete knockout of FPN1 is embryonic lethal, Pcm mice with a partial deletion of exon1a of FPN1 have a mild phenotype, suggesting that expression of FPN1B can compensate well for the loss of FPN1A in most physiological functions.
Analysis of the expression of FPN1B in erythroid lineage cells has enabled us to discern which FPN1 transcript encodes FPN1 protein by comparing amounts of transcript and protein at different stages of synchronously induced differentiation. The observation that levels of FPN1 protein correlate with FPN1B transcript levels in sorted erythroid cells and in differentiating G1E-ER4 cells, but not with levels of FPN1A transcript, suggests that FPN1B is functional and encodes most of the ferroportin protein generated, whereas FPN1A is repressed and has negligible contribution to ferroportin production. Similarly, even though FPN1B transcript accounts for only 20% of the total FPN1 mRNA in the duodenum, it may contribute significantly to enterocytic ferroportin production.
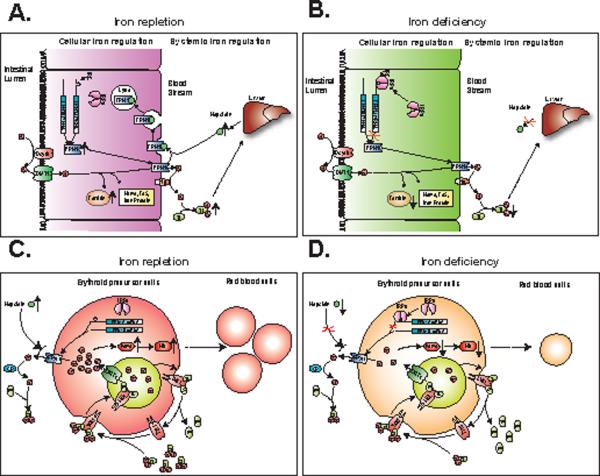
Enterocytes differ from other cells, because enterocytes must provide iron to satisfy systemic iron demands regardless of whether enterocytes themselves are iron-depleted. Whereas FPN1A can be repressed by IRE/IRP machinery, FPN1B is not regulated by the IRE/IRP system and is therefore likely to be important in enterocytic iron transport, especially when cells are iron-deficient (Figure 3, 4). Our results offer a clear rationale for why FPN1B is significantly expressed in duodenum (Figure 3A, B) and allow us to introduce an important modification to the existing model of duodenal iron uptake, which expands the hepcidin-ferroportin interaction model to explain not only how iron overload is prevented but also how duodenal iron absorption can continue in iron-deficient animals. In iron-replete conditions (Figure 7A), both FPN1A and FPN1B transcripts are translated into FPN1 protein, which trafficks to the basolateral membrane to transport iron into the circulation. Once the body senses that iron stores are high, the liver produces hepcidin, which causes ferroportin degradation and blocks iron absorption (Nemeth, et al., 2004). In iron-deficient conditions (Figure 7B), hepcidin expression decreases and the systemic iron-uptake blockage created by degradation of FPN1 is eliminated. However, iron deficiency activates the IRE/IRP system, which then represses FPN1A translation. During iron deficiency, expression of FPN1B increases markedly (as does FPN1A) and its ability to be translated into protein, regardless of IRP repressor activity likely enables it to generate enough FPN1 to satisfy systemic iron demands.
Figure 7.
A model for the physiological roles of the FPN1B transcript in the duodenal enterocytes and erythroid precursor cells. In duodenal enterocytes, FPN1 transports iron across the basolateral membrane to bind to transferrin in the blood stream, facilitated by the ferroxidase, hephaestin (Hp). (A) In iron-replete conditions, the liver senses body iron stores as reflected by transferrin saturation levels, and hepatocytes secrete high amounts of the iron-regulatory hormone hepcidin. In the duodenum, hepcidin is thought to bind to FPN1, and to thereby induce FPN1 internalization and degradation. Intracellularly, IRPs are inactivated in iron-replete animals, and both FPN1A and FPN1B can freely be translated into FPN1 protein. (B) In iron-deficient conditions, the liver ceases to produce hepcidin in response to the low levels of circulating iron. Therefore the hepcidin dependent degradation of FPN1 is eliminated. However, in iron-depleted cells, IRPs bind to the 5'IRE of FPN1A and repress the synthesis of FPN1 protein. Nevertheless, as levels of FPN1B transcript increase in iron deficiency, translation of FPN1B is not repressed by IRPs, and continued FPN1 expression allows sufficient iron export to satisfy the systemic iron demands of iron-deficient animals. In erythroid precursor cells, the Tf/TfR1 cycle actively transports iron into cells, and FPN1 generated mainly from FPN1B exports iron to Tf, facilitated by ceruloplasmin (CP). By active uptake and export of iron, erythroid precursor cells can precisely sense systemic iron levels and can determine whether erythropoietic expansion is warranted. (C) In iron-replete conditions, the Tf/TfR1 cycle transports more iron into the intracellular iron pool, and increased hepcidin expression likely blocks FPN1 from exporting iron out of cells, which increases intracellular iron levels and enhances red cell production and heme synthesis. (D) In iron-depleted conditions, because of decreased iron uptake from the Tf/TfR1 cycle and increased iron export from FPN1, diminished iron availability results in microcytic anemia characterized by decreased production of red blood cells and heme.
In erythroid precursor cells, FPN1B mRNA levels drive ferroportin protein expression to levels higher than those in macrophages, which are known for high ferroportin expression (Figure 5 D, E). The FPN1B promoter is regulated by GATA and EKLF transcription factors, and GATA1 and EKLF contribute to regulation of most erythroid-specific genes, including hemoglobin and TfR1, suggesting that iron homeostasis and red blood cell production are coordinately regulated in erythroid precursor cells. Although the promoter of FPN1A is ubiquitously expressed, the FPN1B promoter is specifically expressed in duodenum and erythroid cells. The regulation of the FPN1B promoter by GATA and EKLF factors and its down-regulation during erythroid cell differentiation suggest that erythroid cells need FPN1B to evade IRP/IRE repression and to export iron in erythroid precursor cells during the critical time period when cells commit to proliferation and differentiation. However, once the cells begin to produce hemoglobin, FPN1B expression diminishes, and FPN1A predominates, which allows erythroid cells to limit iron export through the IRE/IRP system, and to efficiently manufacture heme without developing microcytic anemia, as was observed in Pcm mice that almost do not express FPN1A.
Here, we propose that expression of FPN1B in erythroid precursor cells enhances both sensing of serum iron status and sensitivity to systemic iron status signals. By actively taking up (through TfR1) and exporting (by ferroportin translated from FPN1B) iron, erythroid precursor cells are rendered more sensitive to subtle shifts in systemic iron balance, and these shifts may ultimately determine whether the pluripotent cells commit to expansion and differentiation. For example, CFU-E cells will suffer developmental arrest when transferrin saturations subtly decrease, whereas other cell types such as lymphoid cells and megakaryocytes continue to develop (Kimura, et al., 1986; Goodnough, et al., 2000). In addition, the unattenuated expression of ferroportin in erythroid cells provides a possibility that the potential systemic iron regulator, hepcidin, is able to regulate the erythroid cell iron status and production, as suggested by the phenotype of the Pcm mice. By expressing FPN1B, the erythroid cells can express an iron exporter that is not regulated by intracellular iron, but is subject to regulation by systemic iron levels that are reflected by hepcidin expression. When systemic iron levels are high, the Tf/TfR1 cycle will absorb more iron into cells and increased hepcidin will block ferroportin from exporting iron out of erythroid cells, leading to increased hemoglobin and red cell production (Figure 7C). A model for the physiological role of FPN1B in erythroid cells is presented in Figure 7C and D. The potential cross-talk between hepcidin and ferroportin and their physiological roles in erythroid cells warrants further investigation.
In summary, our results demonstrate that FPN1B is specifically expressed in duodenal enterocytes and in erythroid precursor cells. In the duodenum, FPN1B could enable enterocytes to evade the repression by IRP/IRE machinery to export iron into the circulation of iron-deficient animals to fulfill systemic demands, whereas in erythroid precursors, expression of FPN1B likely enhances real-time sensing of systemic iron status when the decision to proliferate and differentiate is undertaken by precursor cells.
Experimental Procedures
Animals
All animals used in the procedures were approved by the institutional review board of animal care committee. For details, see the Supplementary Materials and Methods section.
Immunohistochemistry and Prussian blue staining
Immunohistochemistry were performed with paraffin embedded tissue sections (5μm), with Map23 rabbit anti-mouse FPN1 antibody at 1:100 dilution (raised by peptide 165-181 of mouse FPN1 in rabbit) (Wu, et al., 2004). Prussian blue staining was performed as described previously (LaVaute, et al., 2001). For details, see the Supplementary Materials and Methods section.
Rapid amplification of cDNA end assay (RACE) and promoter analysis
RACE assays were performed with the Marathon-Ready mouse cDNA library derived from spleen (Clontech Laboratories, Inc.) according to the manufacturer's protocol using primers shown in supplemental Table1. For details, see the Supplementary Materials and Methods section.
Cell culture and G1E cell induction
IEC-6 cells, CACO-2 cells, HEK293 cells, HepG2 cells, RAW264.7 cells and K562 cells were obtained from ATCC. Mouse small intestinal epithelium (MSIE) cells were kindly provided by Dr. Robert Whitehead. G1E and G1E-ER4 cells were gift from Dr. Mitchell Weiss. G1E cells were erythroid precursor cells from GATA-1-null mice. G1E-ER4 cells were G1E cells that stably expressed a conditional GATA-1 under control of estrogen receptor ligand binding domain, undergoing synchronous erythroid maturation upon estradiol treatment (Weiss, et al., 1997; Rylski, et al., 2003; Welch, et al., 2004). For details, see the Supplementary Materials and Methods section.
Plasmid preparation and dual-luciferase reporter assay
The 5'UTRs of FPN1A and FPN1B were cloned and inserted into the pGL3 control vector (Promega, Madison, WI). The promoter region of FPN1B and FPN1A were cloned into pGL4 luciferase reporter vector (Promega, Madison, WI). For details, see the Supplementary Materials and Methods section.
Bone marrow erythroid cells and macrophages isolation
Bone marrow erythroid cells (Ter119 positive) and macrophages (CD11b positive) were isolated from the femurs of 4~6 months old adult wide type mice with Dynabead® biotin binder (Invitrogen) according to the manufacturer's protocol. For details, see the Supplementary Materials and Methods section.
Western blot
Western blot was performed to detect the expression of FPN1, DMT1, TfR1, beta-actin, L- and H-ferritin. For details, see the Supplementary Materials and Methods section.
Northern blot
Northern blots were performed with the DIG Northern Starter Kit (Roche Applied Science), according to manufacturer's protocol with two minor modifications. For details, see the Supplementary Materials and Methods section.
In Situ Hybridization (ISH)
ISH was performed using digoxigenin (DIG)-labeled cRNA probes and alkaline phosphatase (AP) detection as described (Berger and Hediger, 1998). The specific UTR parts of FPN1A (170bp) and FPN1B (96bp) were cloned into the pSPT18 vector (Roche Diagnostics) (Primers shown in supplemental Table1), and used as template to prepare antisense or sense probe, respectively.
Quantitative real time PCR and reverse transcriptase PCR
The total RNA was prepared with TRIzol® reagent (Invitrogen); cDNA was prepared with High-Capacity cDNA Reverse Transcription Kits (Applied Biosystems, Foster City, CA). For details, see the Supplementary Materials and Methods section.
Statistical analysis
Results were expressed as mean ± SD, and all statistical analyses in this study were done with the Student's t-test.
Supplementary Material
Acknowledgements
We want to thank Dr. Mitchell Weiss for providing us G1E and G1E-ER4 cells, and Dr. Robert Whitehead for providing us MSIE and YAMC cells. This work was supported by the Intramural program of the National Institute of Child Health and Human Development.
Grant Support This work was supported by the intramural program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abboud S, Haile DJ. A novel mammalian iron-regulated protein involved in intracellular iron metabolism. J Biol Chem. 2000;275:19906–19912. doi: 10.1074/jbc.M000713200. [DOI] [PubMed] [Google Scholar]
- Anguita E, Hughes J, Heyworth C, Blobel GA, Wood WG, Higgs DR. Globin gene activation during haemopoiesis is driven by protein complexes nucleated by GATA-1 and GATA-2. Embo J. 2004;23:2841–2852. doi: 10.1038/sj.emboj.7600274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antequera F, Bird A. Number of CpG islands and genes in human and mouse. Proc Natl Acad Sci U S A. 1993;90:11995–11999. doi: 10.1073/pnas.90.24.11995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger UV, Hediger MA. Comparative analysis of glutamate transporter expression in rat brain using differential double in situ hybridization. Anat Embryol (Berl) 1998;198:13–30. doi: 10.1007/s004290050161. [DOI] [PubMed] [Google Scholar]
- Buratowski S. Transcription. Gene expression--where to start? Science. 2008;322:1804–1805. doi: 10.1126/science.1168805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cianetti L, Segnalini P, Calzolari A, Morsilli O, Felicetti F, Ramoni C, Gabbianelli M, Testa U, Sposi NM. Expression of alternative transcripts of ferroportin-1 during human erythroid differentiation. Haematologica. 2005;90:1595–1606. [PubMed] [Google Scholar]
- Darshan D, Anderson GJ. Liver-gut axis in the regulation of iron homeostasis. World J Gastroenterol. 2007;13:4737–4745. doi: 10.3748/wjg.v13.i35.4737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaby C, Pilard N, Goncalves AS, Beaumont C, Canonne-Hergaux F. Presence of the iron exporter ferroportin at the plasma membrane of macrophages is enhanced by iron loading and down-regulated by hepcidin. Blood. 2005;106:3979–3984. doi: 10.1182/blood-2005-06-2398. [DOI] [PubMed] [Google Scholar]
- Donovan A, Brownlie A, Zhou Y, Shepard J, Pratt SJ, Moynihan J, Paw BH, Drejer A, Barut B, Zapata A, et al. Positional cloning of zebrafish ferroportin1 identifies a conserved vertebrate iron exporter. Nature. 2000;403:776–781. doi: 10.1038/35001596. [DOI] [PubMed] [Google Scholar]
- Donovan A, Lima CA, Pinkus JL, Pinkus GS, Zon LI, Robine S, Andrews NC. The iron exporter ferroportin/Slc40a1 is essential for iron homeostasis. Cell Metab. 2005;1:191–200. doi: 10.1016/j.cmet.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Galy B, Ferring-Appel D, Kaden S, Grone HJ, Hentze MW. Iron regulatory proteins are essential for intestinal function and control key iron absorption molecules in the duodenum. Cell Metab. 2008;7:79–85. doi: 10.1016/j.cmet.2007.10.006. [DOI] [PubMed] [Google Scholar]
- Goodnough LT, Skikne B, Brugnara C. Erythropoietin, iron, and erythropoiesis. Blood. 2000;96:823–833. [PubMed] [Google Scholar]
- Kaczynski J, Cook T, Urrutia R. Sp1- and Kruppel-like transcription factors. Genome Biol. 2003;4:206. doi: 10.1186/gb-2003-4-2-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura H, Finch CA, Adamson JW. Hematopoiesis in the rat: quantitation of hematopoietic progenitors and the response to iron deficiency anemia. J Cell Physiol. 1986;126:298–306. doi: 10.1002/jcp.1041260221. [DOI] [PubMed] [Google Scholar]
- Knutson MD, Vafa MR, Haile DJ, Wessling-Resnick M. Iron loading and erythrophagocytosis increase ferroportin 1 (FPN1) expression in J774 macrophages. Blood. 2003;102:4191–4197. doi: 10.1182/blood-2003-04-1250. [DOI] [PubMed] [Google Scholar]
- LaVaute T, Smith S, Cooperman S, Iwai K, Land W, Meyron-Holtz E, Drake SK, Miller G, Abu-Asab M, Tsokos M, et al. Targeted deletion of the gene encoding iron regulatory protein-2 causes misregulation of iron metabolism and neurodegenerative disease in mice. Nat Genet. 2001;27:209–214. doi: 10.1038/84859. [DOI] [PubMed] [Google Scholar]
- Liu XB, Hill P, Haile DJ. Role of the ferroportin iron-responsive element in iron and nitric oxide dependent gene regulation. Blood Cells Mol Dis. 2002;29:315–326. doi: 10.1006/bcmd.2002.0572. [DOI] [PubMed] [Google Scholar]
- Lymboussaki A, Pignatti E, Montosi G, Garuti C, Haile DJ, Pietrangelo A. The role of the iron responsive element in the control of ferroportin1/IREG1/MTP1 gene expression. J Hepatol. 2003;39:710–715. doi: 10.1016/s0168-8278(03)00408-2. [DOI] [PubMed] [Google Scholar]
- McKie AT, Marciani P, Rolfs A, Brennan K, Wehr K, Barrow D, Miret S, Bomford A, Peters TJ, Farzaneh F, et al. A novel duodenal iron-regulated transporter, IREG1, implicated in the basolateral transfer of iron to the circulation. Mol Cell. 2000;5:299–309. doi: 10.1016/s1097-2765(00)80425-6. [DOI] [PubMed] [Google Scholar]
- McKie AT, Raja KB, Peters TJ, Farzaneh F, Simpson RJ. Expression of genes involved in iron metabolism in mouse intestine. Am J Physiol. 1996;271:G772–779. doi: 10.1152/ajpgi.1996.271.5.G772. [DOI] [PubMed] [Google Scholar]
- Miller IJ, Bieker JJ. A novel, erythroid cell-specific murine transcription factor that binds to the CACCC element and is related to the Kruppel family of nuclear proteins. Mol Cell Biol. 1993;13:2776–2786. doi: 10.1128/mcb.13.5.2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok H, Jelinek J, Pai S, Cattanach BM, Prchal JT, Youssoufian H, Schumacher A. Disruption of ferroportin 1 regulation causes dynamic alterations in iron homeostasis and erythropoiesis in polycythaemia mice. Development. 2004a;131:1859–1868. doi: 10.1242/dev.01081. [DOI] [PubMed] [Google Scholar]
- Mok H, Mendoza M, Prchal JT, Balogh P, Schumacher A. Dysregulation of ferroportin 1 interferes with spleen organogenesis in polycythaemia mice. Development. 2004b;131:4871–4881. doi: 10.1242/dev.01342. [DOI] [PubMed] [Google Scholar]
- Nemeth E, Ganz T. Regulation of iron metabolism by hepcidin. Annu Rev Nutr. 2006;26:323–342. doi: 10.1146/annurev.nutr.26.061505.111303. [DOI] [PubMed] [Google Scholar]
- Nemeth E, Tuttle MS, Powelson J, Vaughn MB, Donovan A, Ward DM, Ganz T, Kaplan J. Hepcidin regulates cellular iron efflux by binding to ferroportin and inducing its internalization. Science. 2004;306:2090–2093. doi: 10.1126/science.1104742. [DOI] [PubMed] [Google Scholar]
- Park CH, Valore EV, Waring AJ, Ganz T. Hepcidin, a urinary antimicrobial peptide synthesized in the liver. J Biol Chem. 2001;276:7806–7810. doi: 10.1074/jbc.M008922200. [DOI] [PubMed] [Google Scholar]
- Pietrangelo A. The ferroportin disease. Blood Cells Mol Dis. 2004;32:131–138. doi: 10.1016/j.bcmd.2003.08.003. [DOI] [PubMed] [Google Scholar]
- Pietrangelo A. Hereditary hemochromatosis. Annu Rev Nutr. 2006;26:251–270. doi: 10.1146/annurev.nutr.26.061505.111226. [DOI] [PubMed] [Google Scholar]
- Pigeon C, Ilyin G, Courselaud B, Leroyer P, Turlin B, Brissot P, Loreal O. A new mouse liver-specific gene, encoding a protein homologous to human antimicrobial peptide hepcidin, is overexpressed during iron overload. J Biol Chem. 2001;276:7811–7819. doi: 10.1074/jbc.M008923200. [DOI] [PubMed] [Google Scholar]
- Pountney DJ, Konijn AM, McKie AT, Peters TJ, Raja KB, Salisbury JR, Simpson RJ. Iron proteins of duodenal enterocytes isolated from mice with genetically and experimentally altered iron metabolism. Br J Haematol. 1999;105:1066–1073. doi: 10.1046/j.1365-2141.1999.01441.x. [DOI] [PubMed] [Google Scholar]
- Rouault TA. The role of iron regulatory proteins in mammalian iron homeostasis and disease. Nat Chem Biol. 2006;2:406–414. doi: 10.1038/nchembio807. [DOI] [PubMed] [Google Scholar]
- Rylski M, Welch JJ, Chen YY, Letting DL, Diehl JA, Chodosh LA, Blobel GA, Weiss MJ. GATA-1-mediated proliferation arrest during erythroid maturation. Mol Cell Biol. 2003;23:5031–5042. doi: 10.1128/MCB.23.14.5031-5042.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shields JM, Christy RJ, Yang VW. Identification and characterization of a gene encoding a gut-enriched Kruppel-like factor expressed during growth arrest. J Biol Chem. 1996;271:20009–20017. doi: 10.1074/jbc.271.33.20009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallander ML, Leibold EA, Eisenstein RS. Molecular control of vertebrate iron homeostasis by iron regulatory proteins. Biochim Biophys Acta. 2006;1763:668–689. doi: 10.1016/j.bbamcr.2006.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss MJ, Orkin SH. GATA transcription factors: key regulators of hematopoiesis. Exp Hematol. 1995;23:99–107. [PubMed] [Google Scholar]
- Weiss MJ, Yu C, Orkin SH. Erythroid-cell-specific properties of transcription factor GATA-1 revealed by phenotypic rescue of a gene-targeted cell line. Mol Cell Biol. 1997;17:1642–1651. doi: 10.1128/mcb.17.3.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch JJ, Watts JA, Vakoc CR, Yao Y, Wang H, Hardison RC, Blobel GA, Chodosh LA, Weiss MJ. Global regulation of erythroid gene expression by transcription factor GATA-1. Blood. 2004;104:3136–3147. doi: 10.1182/blood-2004-04-1603. [DOI] [PubMed] [Google Scholar]
- Wrighting DM, Andrews NC. Iron homeostasis and erythropoiesis. Curr Top Dev Biol. 2008;82:141–167. doi: 10.1016/S0070-2153(07)00006-3. [DOI] [PubMed] [Google Scholar]
- Wu LJ, Leenders AG, Cooperman S, Meyron-Holtz E, Smith S, Land W, Tsai RY, Berger UV, Sheng ZH, Rouault TA. Expression of the iron transporter ferroportin in synaptic vesicles and the blood-brain barrier. Brain Res. 2004;1001:108–117. doi: 10.1016/j.brainres.2003.10.066. [DOI] [PubMed] [Google Scholar]
- Yang F, Liu XB, Quinones M, Melby PC, Ghio A, Haile DJ. Regulation of reticuloendothelial iron transporter MTP1 (Slc11a3) by inflammation. J Biol Chem. 2002;277:39786–39791. doi: 10.1074/jbc.M201485200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.