Abstract
Ovarian cancer patients treated with cisplatin-based chemotherapy often develop acquired cisplatin resistance and, consequently, cancer recurrence. We have previously reported that annexin A11 is associated with cisplatin resistance and related to tumor recurrence in ovarian cancer patients. In this study, we used small interfering RNA to suppress annexin A11 expression in ovarian cancer cells followed by various in vitro assays. We showed that knockdown of annexin A11 expression reduced cell proliferation and colony formation ability of ovarian cancer cells. Epigenetic silencing of annexin A11 conferred cisplatin resistance to ovarian cancer cells. Through a comprehensive time course study of cisplatin response in ovarian cancer cells with/without suppression of annexin A11 expression using whole-genome oligonucleotide microarrays, we identified a set of differentially expressed genes associated with annexin A11 expression and some patterns of gene expressions in response to cisplatin exposure. These identified genes/patterns were further validated by real-time polymerase chain reaction and immunoblot analysis. Many of them such as HMOX1, TGFBI, LY6D, S100P, EIF4EBP2, DHRS2, and PCSK9 have been involved in apoptosis, cell cycling/proliferation, cell adhesion/migration, transcription regulation, and signal transduction. In addition, immunohistochemistry analyses indicated that annexin A11 immunointensity inversely correlated with HMOX1 immunoreactivity in 142 ovarian cancer patients. In contrast to annexin A11, HMOX1 immunoreactivity positively correlated with in vitro cisplatin resistance in ovarian cancers. Collectively, annexin A11 is directly involved in cell proliferation and cisplatin resistance of ovarian cancer. Manipulation of annexin A11 and its associated genes may represent a novel therapeutic strategy in human ovarian cancers.
Introduction
Ovarian cancer is the fifth leading cause of cancer death among U.S. women and has the highest mortality rate of all gynecologic cancers [1]. Up to 80% of patients whose conditions have been diagnosed at advanced stages of ovarian cancer die within 5 years [1,2]. Many of them, after undergoing debulking surgery, initially respond to chemotherapy yet later relapse with recurrent tumors that are refractory to the original treatment, eventually succumbing to the disease [3,4]. Acquisition of cisplatin resistance during chemotherapy is highly related to cancer mortality and remains a major clinical challenge. Cisplatin is a cytotoxic compound that causes apoptosis through DNA damage by the formation of interstrand or intrastrand adducts. The response to cisplatin is a complex and multifactorial process that leads to the activation of several pathways organized in a large network and transmitting proapoptotic or antiapoptotic signals [5,6]. The major mechanisms of resistance that have been identified so far involve reduced drug uptake, increased drug efflux, increased repair of platinum-DNA adducts, increased tolerance of DNA damage, and increased levels of intracellular thiols such as glutathione and metallothionein [5–8]. Although many putative cisplatin resistance mechanisms have been identified and characterized in vitro, their relevance to clinical resistance has been difficult to prove. The mechanisms underlying clinical cisplatin resistance remain poorly understood because no single over-arching mechanism predominates even within the same histological subtype of ovarian cancer. A better understanding of the molecular mechanisms of drug resistance is fundamental to reduce cancer mortality in ovarian cancer patients.
Annexin A11 is a member of the annexin superfamily of structurally related Ca2+-dependent phospholipid-binding proteins. Despite their structural similarities, annexins have diverse functions including cell division, apoptosis, Ca2+ signaling, growth regulation, and secretory function [9–11]. Annexin A11 contains a conserved structural element, four tandem annexin repeats, in which the Ca2+-binding sites are located; a unique N-terminal domain rich in glycine, proline, and tyrosine residues involved in binding to calcyclin (S100A6) and the apoptosis-linked protein ALG2 [12,13]. Previous studies have suggested that annexin A11 may play a role in cellular DNA synthesis and in cell proliferation as well as in membrane trafficking events such as exocytosis [14–18]. Several members of the annexin superfamily had been demonstrated to be involved in drug resistance in a variety of human cancers [19–22]. Different drugs may have different effects on the expression of certain proteins. Recently, we have shown that annexin A11 was downregulated in cisplatin-resistant ovarian cancer cells compared with their parental cells; expressions of annexin A11 were significantly lower in recurrent tumors than those in the primary ovarian cancers; a lower expression of annexin A11 was significantly associated with earlier recurrence of ovarian cancers; and annexin A11 immunoreactivity inversely correlated with in vitro cisplatin resistance in ovarian cancers [23].
To further elucidate the molecular mechanism underlying the observed association between annexin A11 and cisplatin resistance in ovarian cancer, we carried out the functional study using small interfering RNA (siRNA) followed by various in vitro assays. To identify potential downstream annexin A11-associated targets, a comprehensive time course study of cisplatin response in ovarian cancer cells with/without suppression of annexin A11 expression using whole-genome oligonucleotide microarrays was performed in this study.
Materials and Methods
Cell Lines and Culture
Two cisplatin-sensitive ovarian cancer cell lines, 2008 and HEY, which were kindly provided by Dr. S.B. Howell, were used in this study [23]. All parental cell lines were maintained in drug-free RPMI-1640 medium (Invitrogen, Carlsbad, CA) supplemented with 10% (v/v) heat-inactivated fetal bovine serum (Hyclone, Logan, UT) and 1% penicillin-streptomycin (Invitrogen) at 37°C in a humidified atmosphere containing 5% CO2. All transfected cell lines were cultured in the same growth medium without antibiotics.
siRNA Knockdown of Annexin A11 Gene Expression
All stealth RNA interference (RNAi) sequences were purchased from Invitrogen. The three stealth RNAi that targeted different annexin A11 sequences were as follows: A1, GGCCGUGGUGAAAUGUCUCAAGAAU; A2, CCUCCUGGACAUCAGAUCAGAGUAU; and A3, GGGAUUACCGGAAGAUUCUGCUGAA. The stealth RNAi-negative control duplex (medium GC) was used as a negative control. Transfection of annexin A11-specific siRNA and the negative control was performed using Lipofectamine 2000 (Invitrogen). The optimized dose and duration of RNAi silencing were experimentally determined. Briefly, cancer cells were seeded the day before siRNA transfection in either six-well plates or T25 flasks and were 30% to 50% confluent at the time of transfection. Stealth RNAi and Lipofectamine were diluted in Opti-MEM I Medium (Invitrogen), and 40 nM of the siRNA duplex was used in each transfection mixture. 2008 or HEY cells were transfected with one annexin A11-specific siRNA (A1 or A2 or A3) or a combination of three different siRNA at the equal amount (A1–3) or negative control for 2 to 3 days and were then harvested for the downstream experiments.
Cell Proliferation Assay
Cell Counting Kit-8 (CCK-8; Dojindo, Gaithersburg, MD) was used in cell proliferation assay. Briefly, 2008 and HEY cells were cultured in T25 flasks and transfected with annexin A11-specific siRNA (A1) or negative control for 3 days. Cells were then collected by trypsinization, counted by using a hemacytometer with trypan blue dye, and plated at 3000 viable cells per well into 96-well tissue culture plates in a final volume of 100 µl. Every 24 hours, a plate was subjected to assay by adding 10 µl of CCK-8 solution to each well, and the plate was further incubated for 4 hours at 37°C. The absorbance at 450 nm was measured with a microplate reader (EL 312e; Biotek Instruments, Winooski, VT). The experiment was performed in eight replicates.
Cell Colony Formation Assay
2008 and HEY cells were cultured in T25 flasks and transfected with A1 or negative control for 3 days. Cells were then collected, counted, and plated at 3000 viable cells per well into six-well plates. Six days after plating, cells were fixed with methanol and stained with 0.1% crystal violet, and colonies were counted under the light microscope. The experiment was performed in six replicates.
Cell Cytotoxicity Assay
2008 cells were cultured in T25 flasks and transfected with A1 or negative control for 3 days. Cells were then collected, counted, and plated at 3000 viable cells per well into 96-well plates in a final volume of 100 µl. After incubating overnight, cells were treated with various concentrations of cisplatin diluted in 100 µl of conditioned medium (the final concentrations of cisplatin were 0, 1.56, 3.13, 6.25, 12.5, 25, 50, and 100 µg/ml). After incubating for 72 hours, the plates were assayed by CCK-8 as above. The experiment was performed in four replicates.
Time Course Experiment of Annexin A11-Associated Cisplatin Response and Sample Preparations
2008 cells were cultured in T150 flasks and transfected with A1 or negative control for 2 days. Cells were then collected, counted, and placed into 100-mm dishes. After incubating overnight, transfected cells were at 50% confluence and treated with 10 µM cisplatin (Sigma-Aldrich, St. Louis, MO) for 0, 8, 16, and 24 hours and then harvested in two portions for both total RNA and total protein extractions at every single time point. The optimized dose and duration of cisplatin treatment were experimentally determined in a previous study [23]. Total RNA was isolated using TRIZOL reagent (Invitrogen) followed by RNeasy mini kit with DNase on-column digestion (Qiagen, Valencia, CA). RNA was quantified with NanoDrop ND-1000 followed by quality assessment with the 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA) according to manufacturer's protocol.
Agilent Whole-Genome Oligo Microarray
Total RNA was labeled using Agilent Low RNA Input Fluorescent Linear Amplification Kit (Agilent) following the manufacturer's instruction with minor modifications. Briefly, 0.4 µg of RNA was reverse-transcribed into cDNA by MMLV-RT using an oligo dT primer (System Biosciences, Mountain View, CA) that incorporated a T7 promoter sequence. The cDNA was then used as a template for in vitro transcription in the presence of T7 RNA polymerase and cyanine-3-labeled CTPs (Perkin Elmer Life Sciences, Boston, MA). RNA spike-in controls (Agilent) were added to RNA samples before amplification and labeling. The labeled cRNA was purified using the RNeasy mini kit (Qiagen). A total of 0.825 µg of each Cy3-labeled sample was used for hybridization on Agilent 4x 44K whole human genome microarray at 65°C for 17 hours in a hybridization oven with rotation. After hybridization, slides were washed and dried using stabilization and drying solution according to the Agilent microarray processing protocol. Slides were scanned using the Agilent Microarray Scanner controlled by Agilent Scan Control 7.0 software.
Microarray Data Analysis
Microarray data were extracted with Agilent Feature Extraction 9.5.3.1 software and imported into GeneSpring GX 10 (Agilent). Normalization was done with all intensities higher than 5 by cross-array quantile normalization in log2 scale. Data were then transformed back to original scale for the remaining analysis. Features with intensities smaller than 300 at all time points were excluded from the analysis, and the resulting data were used for principal component analysis using MATLAB version 7.5 software. To identify genes that were differentially expressed at different time points, genes that were either upregulated or downregulated more than two-fold at 8, 16, or 24 hours compared with 0 hour in both cell lines after cisplatin exposure were selected. To identify genes that were differentially expressed between transfected cells with or without annexin A11 silencing, genes with a fold up-regulation or down-regulation of at least two at every single time point were chosen. The identified genes were then clustered and the heat maps representing gene expression at different time points were generated using the Cluster and TreeView software. Gene ontology analysis was performed using the Ingenuity pathway analysis program.
Real-time Reverse Transcription-Polymerase Chain Reaction
Total RNA was isolated from the different cancer cell lines using TRIZOL (Invitrogen) according to the manufacturer's instructions. One microgram of total RNA was used to generate cDNA using the iScript cDNA Synthesis Kit (Bio-Rad, Hercules, CA). One microliter of the resulting cDNA was used in the subsequent polymerase chain reaction (PCR) in iQ SYBR Green Supermix (Bio-Rad), with the following cycles: 95°C for 3 minutes followed by 50 cycles at 95°C for 30 seconds, at 60°C for 30 seconds, and at 72°C for 55 seconds. An experiment consisted of triplicate amplification reactions for each gene product being analyzed. The GAPDH mRNA was used as an internal control for equal sampling of total RNA from one cell to another. The cycle threshold number (CT) was determined for each PCR using iQ5 Real-time PCR Detection System (Bio-Rad). The comparative CT method was used to calculate the relative abundance of a target transcript with regard to an internal control (GAPDH). Results are expressed as relative abundance of a specific mRNA between control and experimental sample (fold change, mean ± SD). Sequences and product sizes for all of genes are shown in Table W1.
Immunoblot Analysis
The denatured samples were electrophoresed on 4% to 15% gradient SDS-PAGE gels (Bio-Rad), electroblotted on nitrocellulose membranes (Bio-Rad), and probed with the respective antibodies against different targets. Both anti-annexin A11 (1:10,000) and anti-annexin A5 (1:2000) monoclonal antibodies were purchased from BD Biosciences (San Jose, CA). Rabbit anti-human HMOX1 polyclonal antibody (1:500) and mouse polyclonal anti-LY6D (1:500) were purchased from GenWay Biotech (San Diego, CA) and Novus Biologicals (Littleton, CO), respectively. The bound antibodies were visualized with horseradish peroxidase-conjugated secondary antibodies and enhanced chemiluminescence (Amersham, Pittsburgh, PA). Actin in the corresponding cell lysates was used as an additional control to show equal loading.
Tissue Microarrays and Immunohistochemistry
In accordance with the human subject research guidelines of institutional review board, formalin-fixed, paraffin-embedded tissues were obtained from the Department of Pathology at Johns Hopkins Hospital. These included 150 ovarian carcinoma tissues, which are 90 primary tumors, 52 first recurrent tumors, and 8 second or third recurrent tumors. Detailed clinicopathologic characteristics of the study cohort have been previously described [23]. All patients underwent primary debulking surgery followed by platinum/paclitaxel-based combined chemotherapy. Tissue microarrays were constructed to facilitate immunohistochemistry (IHC) using EnVision + System-HRP kit (Dako, Carpinteria, CA) with an anti-annexin A11 monoclonal antibody (1:200; BD Biosciences) [23] and an anti-HMOX1 polyclonal antibody (1:200; BioVision, Mountain View, CA). The IHC staining of the protein were scored semiquantitatively as described previously [23]. In vitro cisplatin responses of tumors were assessed by the extreme drug resistance (EDR) assay (Oncotech, Tustin, CA) and have been previously described [23].
Statistical Analysis
All of the statistical analyses were performed using the Statistica 6.1 (Statsoft, Tulsa, OK). Data were subjected to Student's unpaired t test or Fisher's exact test. Differences with P < .05 were considered statistically significant.
Results
Effect and Duration of Silencing of Annexin A11 Using siRNA
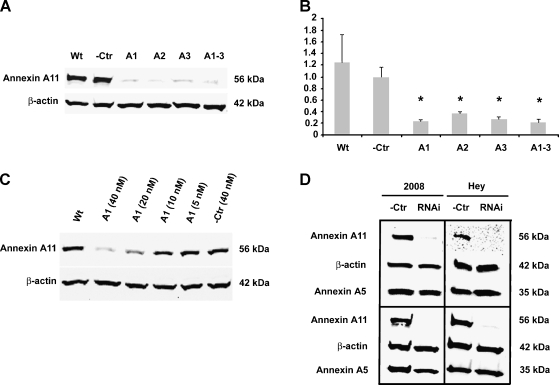
As shown in Figure 1, A and B, after 3 days of siRNA transfection at the concentration of 40 nM, annexin A11-specific siRNA, either applied individually (A1, A2, and A3) or in combination (A1–3), significantly decreased annexin A11 mRNA and protein expression levels in 2008 cells. Quantitative real-time PCR revealed that there were about three-fold to four-fold of down-regulation in annexin A11 mRNA expression levels in RNAi-treated cells (A1, A2, A3, and A1–3) compared with negative control cells (-Ctr, P < .05). Immunoblot analysis showed that there were only barely detectable annexin A11 protein expressions in RNAi-treated cells (A1, A2, A3, and A1–3) compared with annexin A11 strong expressions in negative control cells (-Ctr) and parental cells without treatment (Wt). Immunoblot analysis revealed a dose-dependent silencing effect of annexin A11 expression in RNAi (A1)-treated 2008 cells at the concentrations ranging from 5 to 40 nM (Figure 1C). In addition, our experimental data demonstrated that the effect of silencing of annexin A11 protein expressions in 2008 and HEY cells lasted at least for 10 or 7 days after 3 or 2 days of siRNA transfection at the concentration of 40 nM, respectively (Figure 1D).
Figure 1.
Knockdown of annexin A11 expression in ovarian cancer cells. (A and B) Effect of silencing of annexin A11 using different siRNA. 2008 cells were treated with one (A1 or A2 or A3) or a combination (A1-3) of three stealth RNA against annexin A11 or nonspecific sequence (-Ctr) at the concentration of 40 nM or without treatment (Wt) for 3 days. Immunoblot analysis (A) and real-time PCR (B) were performed to confirm the suppression of annexin A11 mRNA and protein expressions in the cells. β-Actin was taken as an additional control for equal sampling in immunoblot analysis (A). The relative mRNA expression level of each sample was normalized to GAPDH expression and compared with -Ctr sample. *P < .05 (B). (C) Dose-dependent silencing of annexin A11 by siRNA. 2008 cells were treated with RNAi (A1) at the indicated concentrations of 40, 20, 10, or 5 nM or -Ctr at the concentration of 40 nM or without treatment (Wt) for 3 days. Immunoblot analysis (C) was performed to check the annexin A11 expression levels in the cells. β-Actin was taken as an additional control for equal sampling. (D) Duration of silencing of annexin A11 using siRNA. 2008 or HEY cells were treated with RNAi (A1) or -Ctr at the concentration of 40 nM. Immunoblot analysis was performed to analyze the annexin A11 expression levels in these cells. Note that the level of annexin A11 protein was significantly decreased by day 3 (2008; left top), day 10 (2008; left bottom), day 2 (HEY; right top), or day 7 (HEY; right bottom), respectively. β-Actin or annexin A5 was taken as a loading control or off-target effect control.
Knockdown of Annexin A11 Reduced Cancer Cell Proliferation and Colony Formation Ability
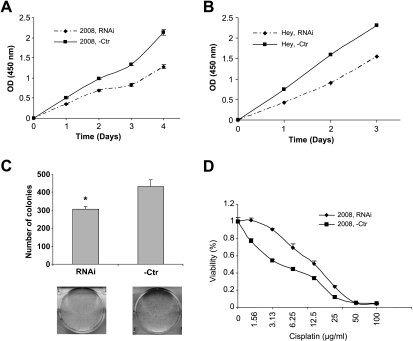
Cell growth and apoptosis are intimately related [24–28]. To determine the effect of annexin A11 on cell growth of ovarian cancer, we performed cell proliferation assay and cell colony formation assay after RNAi silencing of annexin A11 expression in 2008 and HEY cells. We observed a significantly (P < .05) slower rate of proliferation (40% or 34% decreased) of the annexin A11-specific siRNA transfectants compared with that of the negative control transfectants in both 2008 and HEY cells (Figure 2, A and B). Suppression of annexin A11 expression also greatly damaged 2008 cell colony formation abilities (P < .01; Figure 2C). HEY cells did not form countable colonies during their growth process. These data suggested that annexin A11 plays an important role in cell proliferation of ovarian cancer.
Figure 2.
Epigenetic silencing of annexin A11 reduces cell proliferation, colony formation ability, and confers cisplatin resistance to ovarian cancer cells. (A and B) Cell proliferation assay. 2008 (A) or HEY (B) cells were treated with RNAi (A1) or -Ctr at the concentration of 40 nM for 3 days, respectively, and then plated at 3000 viable cells per well into 96-well plates. Every 24 hours, one plate was subjected to assay by CCK-8 kit. The data in each time point are averaged values from eight replicates (P < .05). (C) Colony formation assay. 2008 cells were treated in same way as above for 3 days and then plated at 3000 viable cells per well into six-well plates. Six days after plating, cells were fixed with methanol and stained with 0.1% crystal violet and colonies were counted. The experiment was performed in six replicates (P < .01). (D) Cell cytotoxicity assay. 2008 cells were treated in same way as above for 3 days and then plated at 3000 viable cells per well into 96-well plates. After incubating overnight, cells were treated with various concentration of cisplatin diluted in 100 µl of conditioned medium (the final concentrations of cisplatin were 0, 1.56, 3.13, 6.25, 12.5, 25, 50, and 100 µg/ml). After continuous incubation for 72 hours, the plates were subjected to assay by CCK-8 kit. The experiment was performed in three replicates (P < .01). Three independent experiments were performed for each assay.
Epigenetic Silencing of Annexin A11 Conferred Chemoresistance to Ovarian Cancer Cells
Previously, we reported that annexin A11 is associated with cisplatin resistance and related to tumor recurrence in ovarian cancer patients [23]. To directly demonstrate the involvement of annexin A11 in cisplatin resistance of ovarian cancer cells, the cisplatin-sensitive 2008 cells were transfected with an annexin A11-specific siRNA or negative control followed by cell cytotoxicity assay. The sensitivities of the pair of cell lines to the cytotoxic effect of cisplatin were determined. Dose-response curves were plotted on a semilog scale as the percentage of the control cell number, which was obtained from the sample without drug exposure. The experimental data showed that epigenetic silencing of annexin A11 expression significantly enhanced cisplatin resistance in 2008 cells (P < .01; Figure 2D). IC50 in two cell lines are 42 and 16 µM, respectively, with a 2.6-fold increase in RNAi cells compared with control cells. These data are consistent with the previous observation of an association between annexin A11 and cisplatin resistance in ovarian cancer [23].
Dynamic Response of Gene Expression to Cisplatin Treatment and Annexin A11-Associated Gene Expression Alterations
To better understand the molecular mechanisms through which annexin A11 plays an important role in cell proliferation and drug resistance of ovarian cancer and to identify potential downstream annexin A11-associated targets, we performed time course profiling of gene expressions of both annexin A11-specific siRNA (R group) and negative control (N group) transfected ovarian cancer cells treated with cisplatin for different durations using the Agilent 44K whole genome oligo microarrays. Unsupervised analysis using principal component analysis indicated that cisplatin treatment has a major effect on global gene expression patterns of both cell lines at all time points (data not shown).
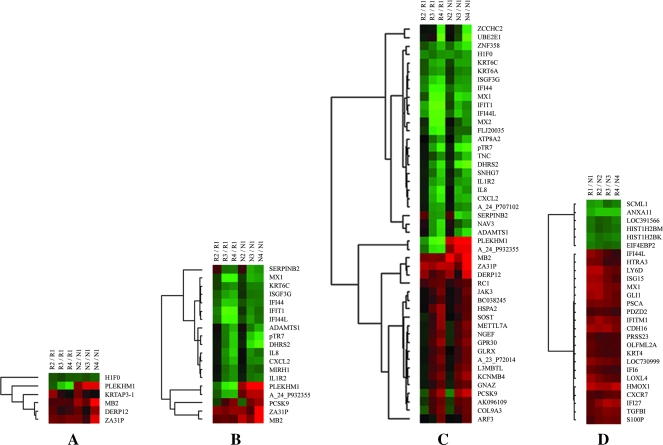
During the time course of cisplatin exposure, a total of 6 genes were either upregulated or downregulated at 8 hours after treatment with cisplatin, 19 after 16 hours and 47 after 24 hours in both groups of cells (Figure 3, A–C). Most genes that altered at 8 hours (5/6) maintained their alterations of gene expression at 16 and/or 24 hours, representing a set of genes that were earlier and lasting responders to cisplatin exposure (Figure 3A). H1F0, MB2, DERP12, and ZA31P showed consistent alterations (either up-regulation or down-regulation) of gene expression at all time points in both groups of cells. By 16 hours of cisplatin exposure, another major pattern of gene expression began to emerge. There were 15 genes that were altered at 16 hours but not at 8 hours and maintained their alterations at 24 hours. Among them, 12 genes including MX1, KRT6C, ISGF3G, IFI44, IFIT1, IFI44L, ADAMTS1, pTR7, DHRS2, IL8, CXCL2, and IL1R2 showed consistent down-regulations of gene expression at 16 and 24 hours in both groups of cells, which were organized into a major cluster (Figure 3B). SERPINB2 was increased at 8 hours but decreased at 16 and 24 hours in both groups of cells, whereas PCSK9 was decreased at 8 hours but increased at 16 and 24 hours in both groups of cells. By 24 hours, more genes were included into this major cluster of downregulated genes, and a new major cluster of upregulated genes including PCSK9 was also formed as shown in Figure 3C, suggesting the establishment of a large gene expression program in response to cisplatin exposure. Interestingly, among these genes, both PLEKHM1 and A_24_P932355 showed total different responses to cisplatin treatment in both groups of cells. They were decreased in R group cells at 8, 16, and 24 hours, whereas they increased in N group cells at 8, 16, and 24 hours. The above identified genes altered during the time course of cisplatin exposure include some genes involved in apoptosis (PCSK9, SERPINB2, MX1), cell cycle/cell proliferation (H1F0, IL8, ADAMTS1, ATP8A2, DHRS2, KRT6C), signal transduction (IL8, ZCCHC2, ARF3, JAK3, KCNMB4, NGEF, PLEKHM1, TNC, CXCL2, GNAZ, GPR30, MX1, SOST), transcription regulation (L3MBTL, ZNF358, ISGF3G), cell adhesion (IL8, TNC), cell motility/migration (IL8, S100P), metabolism (DHRS2, PCSK9, METTL7A, UBE2E1), immune response (IL8, CXCL2, IL1R2, IFIT1, ISGF3G, MX1, MX2), and nucleotide binding (ARF3, ATP8A2, GNAZ, H1F0, HSPA2, JAK3, MX1, MX2, NAV3, ZCCHC2, ZAF358).
Figure 3.
Dynamic response of gene expression to cisplatin treatment and annexin A11-associated gene expression alterations. (A–C) Hierarchical clustering of gene expression alterations. Hierarchical clustering of genes either upregulated or downregulated more than two-fold change at 8 (A), 16 (B), and 24 hours (C) compared with 0 hour in both RNAi (R groups) and control (N groups) cell lines are shown. (D) Hierarchical clustering of genes with a fold up-regulation or down-regulation of at least two (R vs N) at every single time point are also shown. R1–R4 or N1–N4 represents different time points at 0, 8, 16, or 24 hours in order, respectively, in R or N groups. Clustering was performed using the Cluster and TreeView software. Genes that were increased are shown in red, whereas genes that were decreased are indicated in blue.
A total of 26 genes were identified as annexin A11-associated genes (Figure 3D). Their expression levels were either increased (n = 21) or decreased (n = 5) after silencing of annexin A11. In this study, only genes with a fold up-regulation or down-regulation of at least 2 at every single time point were selected for validation. Table 1 lists these genes with averaged fold changes over all the time points, which were ordered accordingly. The identified annexin A11-associated genes include some genes involved in apoptosis/cell proliferation (HMOX1, MX1, GLI1, TGFBI, IFITM1, EIF4EBP2, HTRA3, IFI6, KRT4), DNA binding (HMOX1, MX1, HIST1H2BM, HIST1H2BK), signal transduction (HMOX1, GLI1, CXCR7, EIF4EBP2, IFITM1), transcription regulation (HMOX1, GLI1, SCML1), cell adhesion (TGFBI, CDH16, LY6D, PDZD2), and immune response (IFI27, IFI6, ISG15, MX1). Other genes were either only one gene in one category or without available annotation of gene ontology.
Table 1.
Genes Altered upon Expression of Annexin A11.
| Gene Symbol | Accession No. | Description | Fold Change |
| Up-regulation | |||
| HMOX1 | NM_002133 | Heme oxygenase (decycling) 1 | +5.13 |
| CDH16 | NM_004062 | Cadherin 16, KSP-cadherin | +4.24 |
| MX1 | NM_002462 | Myxovirus (influenza virus) resistance 1 | +4.14 |
| LY6D | NM_003695 | Lymphocyte antigen 6 complex, locus D | +4.08 |
| IFI27 | NM_005532 | Interferon, a-inducible protein 27 | +4.05 |
| GLI1 | NM_005269 | Glioma-associated oncogene homolog 1 (zinc finger protein) | +3.87 |
| IFITM1 | NM_003641 | Interferon-induced transmembrane protein 1 (9–27) | +3.72 |
| ISG15 | NM_005101 | ISG15 ubiquitin-like modifier | +3.62 |
| LOC730999 | XM_001131389 | Hypothetical protein LOC730999 | +3.30 |
| LOXL4 | NM_032211 | Lysyl oxidase-like 4 | +3.28 |
| PSCA | NM_005672 | Prostate stem cell antigen | +3.24 |
| TGFBI | NM_000358 | Transforming growth factor, β-induced, 68 kDa | +3.19 |
| IFI44L | NM_006820 | Interferon-induced protein 44-like | +2.98 |
| S100P | NM_005980 | S100 calcium binding protein P | +2.90 |
| HTRA3 | NM_053044 | HtrA serine peptidase 3 | +2.73 |
| CXCR7 | NM_020311 | Chemokine (C-X-C motif) receptor 7 | +2.56 |
| OLFML2A | NM_182487 | Olfactomedin-like 2A | +2.52 |
| IFI6 | NM_022873 | Interferon, α-inducible protein 6 | +2.48 |
| KRT4 | NM_002272 | Keratin 4 | +2.43 |
| PRSS23 | NM_007173 | Protease, serine, 23 | +2.36 |
| PDZD2 | NM_178140 | PDZ domain containing 2 | +2.36 |
| Down-regulation | |||
| HIST1H2BM | NM_003521 | Histone cluster 1, H2bm | -2.21 |
| LOC391566 | XR_018583 | Similar to histone H2B 291B | -2.27 |
| EIF4EBP2 | NM_004096 | Eukaryotic translation initiation factor 4E binding protein 2 | -2.30 |
| HIST1H2BK | NM_080593 | Histone cluster 1, H2bk | -2.49 |
| SCML1 | NM_006746 | Sex comb on midleg-like 1 | -2.63 |
| ANXA11 | NM_145869 | Annexin A11 | -3.43 |
Fold Change (R/N) is the averaged value over all time points.
The design of the time course gene expressions profiling study did not include replicates to allow for proper estimate of false discovery rate for results in Figure 3, A–C. However, for the results in Figure 3D, the identification of annexin A11-associated genes, using sample label permutation, the average number of genes with at least two-fold changes at every single time point was 5.8, indicating an estimated potential false discovery rate of 22%.
Validation of DNA Microarray Data Using Real-time PCR
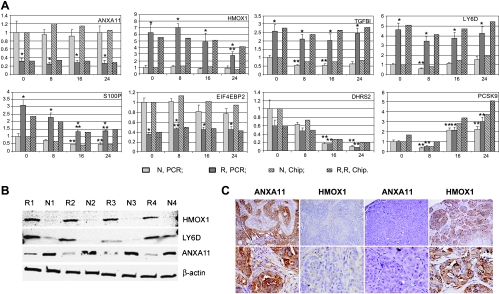
Before further effort to unravel the molecular pathways through which annexin A11 is involved in cell proliferation and drug resistance of ovarian cancer, the DNA microarray profiling data need to be validated using alternative platform. We performed real-time PCR assays to independently determine mRNA expression levels on a set of genes that were representative of the above gene ontology classes: HMOX1, PCSK9, and EIF4EBP2 for apoptosis; HMOX1, TGFBI, DHRS2, and EIF4EBP2 for cell proliferation; TGFBI and LY6D for cell adhesion; HMOX1 for transcription regulation; HMOX1 and EIF4EBP2 for signal transduction; DHRS2 and PCSK9 for metabolism; HMOX1 for DNA binding; and S100P for cell migration. As shown in Figure 4A and Table W2, the consistent up-regulation (HMOX1, TGFBI, LY6D, and S100P) and down-regulation (EIF4EBP2) of genes subjected to epigenetic silencing of annexin A11 (ANXA11) across all time points were well validated using real-time PCR. The down-regulation of DHRS2 that was one representative of the major cluster emerged at 16 and 24 hours after cisplatin exposure was verified in both groups of cells. PCSK9 that was identified as one of the major clusters of genes upregulated at the later time point(s) was also investigated using real-time PCR. Our results showed dynamic responsive patterns of gene expression that were extremely similar to the microarray data, which was decreased at 8 hours but increased at 16 and 24 hours in both groups of cells. Overall, the real-time PCR results agreed well with the microarray data and confirmed that epigenetic silencing of annexin A11 in ovarian cancer cells followed by cisplatin exposure led to significant changes in the expression of genes involved in apoptosis, cell cycling/proliferation, cell adhesion, cell migration, transcription regulation, and signal transduction.
Figure 4.
Validation of DNA microarray data and immunohistochemical analysis. (A) Validation of DNA microarray data by real-time PCR. The up-regulation (HMOX1, TGFBI, LY6D, and S100P) and down-regulation (EIF4EBP2) of genes associated with annexin A11 expression (ANXA11) and the dynamic response of gene expression to cisplatin treatment (DHRS2 and PCSK9) were validated using real-time PCR. N represents control cells and R represents RNAi cells. For each individual gene, the expression levels at different time points were normalized to the control sample (N, PCR, 0 h). In addition, the relative mRNA expression levels were normalized to GAPDH expression. Each gene was amplified in triplicate, and each experiment was performed three times. *P < .05, R versus N. **P < .05, either [R2 (or R3 or R4) vs R1] or [N2 (or N3 or N4) vs N1]. (B) Suppression of annexin A11 upregulated HMOX1 and LY6D protein expressions. Immunoblot analysis was performed to confirm the suppression of annexin A11 protein expressions in the cells. The up-regulations of HMOX1 and LY6D protein expression levels in R1, R2, R3, and R4 compared with N1, N2, N3, and N4 were demonstrated. β-Actin was taken as an additional control for equal sampling in immunoblot analysis. (C) Annexin A11 immunointensity inversely correlated with HMOX1 immunoreactivity in ovarian cancer patients. Two representative pairs of tissue sections (left two sections from a primary tumor with low EDR and right two sections from a first recurrent tumor with extreme EDR) stained with two different antibodies are shown. Both sections of each pair were from similar areas of the same specimen. Original magnifications: upper panel, x100; lower panel, x400.
Suppression of Annexin A11 Upregulated HMOX1 and LY6D Protein Expressions
According to our DNA microarray results, HMOX1 and LY6D were consistently increased by approximately 5.13- or 4.08-fold, respectively, in cells subjected to epigenetic silencing of annexin A11 across all time points.Using immunoblot analysis, the suppressions of annexin A11 protein expressions in the R group cells compared with those in the N group cells were confirmed. Immunoblot analysis showed that suppression of annexin A11 also upregulated HMOX1 and LY6D protein expression levels in R1, R2, R3, and R4 compared with N1, N2, N3, and N4, respectively (Figure 4B).
Annexin A11 Immunointensity Inversely Correlated with HMOX1 Immunoreactivity in Ovarian Cancer Patients
Owing to the extensive involvement of HMOX1 in different cellular processes including cell proliferation and apoptosis, we further evaluated the correlation of protein expressions between annexin A11 and HMOX1 in 150 ovarian carcinoma tissues with IHC staining. HMOX1 immunoreactivity was observed in the cytoplasm of tumor cells (Figure 4C) and significantly inversely correlated with annexin A11 immunointensity in 142 primary and first recurrent ovarian cancer patients (P = .04; Figure 4C and Table 2). This inverse correlation exists even more significantly in 52 first recurrent tumors (P = .01; Table 2). In addition, among 81 tumors for which the EDR results were available for analysis, HMOX1 immunoreactivity significantly positively correlated with in vitro cisplatin resistance (P = .04; Figure 4C). More specifically, there were approximately 60.6% of ovarian carcinomas with extreme and intermediate cisplatin resistance exhibited positive HMOX1 immunoreactivity, whereas only 37.5% of tumors with low cisplatin resistance showed positive HMOX1 immunoreactivity.
Table 2.
Annexin A11 Immunointensity Correlates Inversely with HMOX1 Immunoreactivity in Ovarian Cancer Patients.
| HMOX1 | ANXA11 | Total | |||
| Negative | Weak | Moderate | Strong | ||
| Negative | 5 (41.7%) | 19 (30.2%) | 23 (59%) | 13 (46.4%) | 60 |
| Positive | 7 (58.3%) | 44 (69.8%) | 16 (41%) | 15 (53.6%) | 82 |
| Total | 12 | 63 | 39 | 28 | 142 (P = .04) |
| Negative | 4 (50%) | 5 (16.7%) | 6 (66.7%) | 3 (60%) | 18 |
| Positive | 4 (50%) | 25 (83.3%) | 3 (33.3%) | 2 (40%) | 34 |
| Total | 8 | 30 | 9 | 5 | 52 (P = .01) |
There is an inverse correlation of protein expressions between annexin A11 and HMOX1 in 142 primary and first recurrent ovarian cancer patients (P = .04). This inverse correlation exists even more significantly in 52 first recurrent tumors (P = .01).
Discussion
In this study, we demonstrated that annexin A11 was directly involved in cell proliferation and cisplatin resistance of ovarian cancer. Specifically, using RNAi techniques, we showed that knockdown of annexin A11 expression reduced cell proliferation and colony formation ability of ovarian cancer cells. Furthermore, we showed that epigenetic silencing of annexin A11 conferred cisplatin resistance to ovarian cancer cells. We have previously shown that decreased expression of annexin A11 was characteristic for cisplatin-resistant ovarian cancer cells and may contribute to tumor recurrence in ovarian cancer patients [23]. The experimental results in this study are in agreement with our previous observation and further underscored the biological relevance of annexin A11 in the drug resistance of ovarian cancer.
Annexin A11 is a member of the annexin superfamily of Ca2+ and phospholipid-binding, membrane-associated proteins implicated in Ca2+ signal transduction processes associated with cell growth and differentiation [9–11]. Although diverse functions have been ascribed to annexins, there is no consensus about the role played by the annexin protein family as a whole [11]. The exact cellular functions of individual annexin members remain to be determined. Annexin A11 is ubiquitously expressed in a variety of tissues and cell types of eukaryotes, but its subcellular distribution varies considerably [14,17]. The nuclear localization of annexin A11 has been demonstrated to be cell type-specific and developmentally dependent [14]. Using recombinant human annexin A11-specific autoantibodies cloned by phage display, annexin A11 was found to be associated with the mitotic spindles and might play a role in cell division [17]. A combination of confocal and video time-lapse microscopy revealed that annexin A11 was required for midbody formation and completion of the terminal phase of cytokinesis [29]. A recent genome-wide association study identified ANXA11 as a new susceptibility locus for sarcoidosis and surmised that a depletion or dysfunction of annexin A11 may affect the apoptosis pathway in individuals with sarcoidosis and hence destroy the balance between apoptosis and survival of activated inflammatory cells [30]. In consistent with these observations, in this study, knockdown of annexin A11 expression resulted in a slower rate of cell growth in two ovarian cancer cell lines, 2008 and HEY, providing the first evidence that annexin A11 plays an important role in cell proliferation of ovarian cancer. In addition to the classic mechanisms, there are also several new molecular factors that have been linked to chemoresistance such as altered cell signaling pathways or presence of quiescent noncycling cells [22]. The cell cycle and apoptosis are intimately related, as evidenced by the central role of p53, both in cell cycle arrest and in the induction of apoptosis [25]. Conversely, this intimate relation was also demonstrated both in vitro and clinically; tumor cells that undergo a growth arrest or have a lower proliferation activity may be protected from apoptosis and may therefore be ultimately resistant to the cytotoxic agent [24,26]. Our results demonstrated that epigenetic silencing of annexin A11 expression reduced cell proliferation and conferred cisplatin resistance to ovarian cancer cells, suggesting the possibility that the observed association between annexin A11 and cisplatin resistance may be mediated through alterations in cell cycling/proliferation. The exact mechanism underlying this phenomenon and whether it is universal or tumor cell type-specific remains to be further investigated.
Although cancer cells with intrinsic or acquired cisplatin resistance have been extensively analyzed to identify genomic or proteomic markers involved in drug resistance, the exact timing of transcriptional response to cisplatin treatment remains unclear. This longitudinal analysis of both annexin A11-specific siRNA and negative control-transfected ovarian cancer cells for their response to cisplatin treatment allowed us to identify some patterns of gene expressions in response to cisplatin exposure. A set of genes altered at 8 hours and maintained their alterations of gene expression at 16 and 24 hours, representing earlier and lasting responders to cisplatin exposure. By 16 hours of cisplatin exposure, another major pattern of gene expression (down-regulation) began to emerge and maintained their alterations, with more genes included into this major cluster at 24 hours. Furthermore, the third major pattern of gene expression (up-regulation) was formed at 24 hours after initial downregulations of gene expressions at 8 hours of cisplatin exposure. These major patterns of gene expression suggested the establishment of a large gene expression program in response to cisplatin exposure. Many of these genes have been involved in apoptosis, cell cycle/proliferation, signal transduction, transcription regulation, cell adhesion, cell motility/migration, metabolism, and immune response. Tumor cells, in contrast to normal cells, respond to cisplatin exposure with transient gene expression to protect or repair their chromosomes. Some genes could serve as the master switch for turning on other genes in response to DNA-damaging agents and play a major role in cisplatin resistance. PLEKHM1 was previously reported to be involved in colon cancer cells' response to cisplatin exposure [31]. Interestingly, in this study, PLEKHM1 showed totally different responses to cisplatin treatment in both groups of cells.
In this study, we also identified a set of genes that are differentially expressed at all time points between two groups of cell lines, which represents the annexin A11-associated gene expression alterations.Many of these genes have been involved in apoptosis/cell proliferation, DNA binding, signal transduction, transcription regulation, and cell adhesion. Among them, the up-regulation of heme oxygenase 1 (HMOX1) or heat shock protein 32 (HSP32) seems particularly interesting because this inducible isoform of heme oxygenase has been shown to occur in various tumor tissues and contribute to tumor progression [32,33]. HMOX1 was reported to modulate different cellular functions including cytokine production, cell proliferation, and apoptosis and can exert unique cytoprotective effects [32–34]. It has previously been shown that HMOX1 attenuated the cisplatin-induced apoptosis of auditory cells [34] and that suppression of Nrf2-driven HMOX1 enhanced the chemosensitivity of lung cancer cells toward cisplatin [33]. In this study, HMOX1 immunoreactivity inversely correlated with annexin A11 immunointensity and positively correlated with in vitro cisplatin resistance in ovarian cancer patients, which suggested that HMOX1 may also collectively serve as a potential marker for ovarian cancer chemoresistance, and inhibition of intratumoral annexin A11-regulated HMOX1 activity may be a potential therapeutic strategy in human ovarian cancers. The extracellular matrix protein TGFBI induced microtubule stabilization and sensitized ovarian cancers to paclitaxel [35]. LY6D was reported to be a chemotherapy-induced antigen and has been used both as a therapeutic target and as a diagnostic marker for head and neck cancer [36–38]. S100P sensitizes ovarian cancer cells to carboplatin and paclitaxel in vitro [39]. IFITM1 was identified as a potent marker of cis-platinum response in esophageal cancer [40]. In this study, these annexin A11-associated genes were coordinately regulated to provide relatively different baselines in terms of gene expression and might be responsible for the observed phenotype changing of cancer cells.
In conclusion, this study shows that annexin A11 is directly involved in cell proliferation and cisplatin resistance of ovarian cancer. Through a time course study of cisplatin response in ovarian cancer cells with/without suppression of annexin A11 expression, we identified a set of differentially expressed genes associated with annexin A11 expression and patterns of gene expressions in response to cisplatin exposure. Many of them such as HMOX1, TGFBI, LY6D, S100P, EIF4EBP2, DHRS2, and PCSK9 have been involved in apoptosis, cell cycling/proliferation, cell adhesion/migration, transcription regulation, and signal transduction. HMOX1 immunoreactivity inversely correlated with annexin A11 immunointensity and positively correlated with in vitro cisplatin resistance in ovarian cancer patients. Further characterization of these genes may contribute to a better understanding of the molecular mechanism through which annexin A11 plays an important role in cell proliferation and drug resistance of ovarian cancer. Manipulation of annexin A11 and its associated genes may represent a novel therapeutic strategy in human ovarian cancers.
Supplementary Material
Abbreviations
- RNAi
RNA interference
- siRNA
small interfering RNA
- IHC
immunohistochemistry
- EDR
extreme drug resistance assay
Footnotes
This research was partially supported by a Department of Defense IDEA grant DAMD17-OC03-IDEA, a National Institutes of Health/National Cancer Institute Early Detection Research Network grant CA115102-01, and National Institutes of Health/National Cancer Institute grants 1P50 CA83639, CA129080, and CA103937.
This article refers to supplementary materials, which are designated by Tables W1 and W2 and are available online at www.neoplasia.com.
References
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, Thun MJ. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 2.Holschneider CH, Berek JS. Ovarian cancer: epidemiology, biology, and prognostic factors. Semin Surg Oncol. 2000;19:3–10. doi: 10.1002/1098-2388(200007/08)19:1<3::aid-ssu2>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 3.Bristow RE, Tomacruz RS, Armstrong DK, Trimble EL, Montz FJ. Survival effect of maximal cytoreductive surgery for advanced ovarian carcinoma during the platinum era: a meta-analysis. J Clin Oncol. 2002;20:1248–1259. doi: 10.1200/JCO.2002.20.5.1248. [DOI] [PubMed] [Google Scholar]
- 4.Tummala MK, McGuire WP. Recurrent ovarian cancer. Clin Adv Hematol Oncol. 2005;3:723–736. [PubMed] [Google Scholar]
- 5.Boulikas T, Vougiouka M. Cisplatin and platinum drugs at the molecular level. (Review) Oncol Rep. 2003;10:1663–1682. [PubMed] [Google Scholar]
- 6.Siddik ZH. Cisplatin: mode of cytotoxic action and molecular basis of resistance. Oncogene. 2003;22:7265–7279. doi: 10.1038/sj.onc.1206933. [DOI] [PubMed] [Google Scholar]
- 7.Fink D, Aebi S, Howell SB. The role of DNA mismatch repair in drug resistance. Clin Cancer Res. 1998;4:1–6. [PubMed] [Google Scholar]
- 8.Johnson SW, Laub PB, Beesley JS, Ozols RF, Hamilton TC. Increased platinum-DNA damage tolerance is associated with cisplatin resistance and cross-resistance to various chemotherapeutic agents in unrelated human ovarian cancer cell lines. Cancer Res. 1997;57:850–856. [PubMed] [Google Scholar]
- 9.Brichory FM, Misek DE, Yim AM, Krause MC, Giordano TJ, Beer DG, Hanash SM. An immune response manifested by the common occurrence of annexins I and II autoantibodies and high circulating levels of IL-6 in lung cancer. Proc Natl Acad Sci USA. 2001;98:9824–9829. doi: 10.1073/pnas.171320598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chetcuti A, Margan SH, Russell P, Mann S, Millar DS, Clark SJ, Rogers J, Handelsman DJ, Dong Q. Loss of annexin II heavy and light chains in prostate cancer and its precursors. Cancer Res. 2001;61:6331–6334. [PubMed] [Google Scholar]
- 11.Monastyrskaya K, Babiychuk EB, Hostettler A, Rescher U, Draeger A. Annexins as intracellular calcium sensors. Cell Calcium. 2007;41:207–219. doi: 10.1016/j.ceca.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 12.Fernandez MP, Jenkins NA, Gilbert DJ, Copeland NG, Morgan RO. Sequence and chromosomal localization of mouse annexin XI. Genomics. 1996;37:366–374. doi: 10.1006/geno.1996.0571. [DOI] [PubMed] [Google Scholar]
- 13.Furge LL, Chen K, Cohen S. Annexin VII and annexin XI are tyrosine phosphorylated in peroxovanadate-treated dogs and in platelet-derived growth factor-treated rat vascular smooth muscle cells. J Biol Chem. 1999;274:33504–33509. doi: 10.1074/jbc.274.47.33504. [DOI] [PubMed] [Google Scholar]
- 14.Mamiya N, Iino S, Mizutani A, Kobayashi S, Hidaka H. Development-related and cell-type specific nuclear localization of annexin XI: immunolocalization analysis in rat tissues. Biochem Biophys Res Commun. 1994;202:403–409. doi: 10.1006/bbrc.1994.1942. [DOI] [PubMed] [Google Scholar]
- 15.Misaki Y, Pruijn GJ, van der Kemp AW, van Venrooij WJ. The 56K autoantigen is identical to human annexin XI. J Biol Chem. 1994;269:4240–4246. [PubMed] [Google Scholar]
- 16.Jorgensen CS, Levantino G, Houen G, Jacobsen S, Halberg P, Ullman S, Khamashta MA, Asmussen K, Oxholm P, Jorgensen MK, et al. Determination of autoantibodies to annexin XI in systemic autoimmune diseases. Lupus. 2000;9:515–520. doi: 10.1177/096120330000900707. [DOI] [PubMed] [Google Scholar]
- 17.Farnaes L, Ditzel HJ. Dissecting the cellular functions of annexin XI using recombinant human annexin XI-specific autoantibodies cloned by phage display. J Biol Chem. 2003;278:33120–33126. doi: 10.1074/jbc.M210852200. [DOI] [PubMed] [Google Scholar]
- 18.Williams LH, McClive PJ, Van Den Bergen JA, Sinclair AH. Annexin XI co-localises with calcyclin in proliferating cells of the embryonic mouse testis. Dev Dyn. 2005;234:432–437. doi: 10.1002/dvdy.20548. [DOI] [PubMed] [Google Scholar]
- 19.Han EK, Tahir SK, Cherian SP, Collins N, Ng SC. Modulation of paclitaxel resistance by annexin IV in human cancer cell lines. Br J Cancer. 2000;83:83–88. doi: 10.1054/bjoc.2000.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Y, Serfass L, Roy MO, Wong J, Bonneau AM, Georges E. Annexin-I expression modulates drug resistance in tumor cells. Biochem Biophys Res Commun. 2004;314:565–570. doi: 10.1016/j.bbrc.2003.12.117. [DOI] [PubMed] [Google Scholar]
- 21.Le Moguen K, Lincet H, Deslandes E, Hubert-Roux M, Lange C, Poulain L, Gauduchon P, Baudin B. Comparative proteomic analysis of cisplatin sensitive IGROV1 ovarian carcinoma cell line and its resistant counterpart IGROV1-R10. Proteomics. 2006;6:5183–5192. doi: 10.1002/pmic.200500925. [DOI] [PubMed] [Google Scholar]
- 22.Stewart JJ, White JT, Yan X, Collins S, Drescher CW, Urban ND, Hood L, Lin B. Proteins associated with cisplatin resistance in ovarian cancer cells identified by quantitative proteomic technology and integrated with mRNA expression levels. Mol Cell Proteomics. 2006;5:433–443. doi: 10.1074/mcp.M500140-MCP200. [DOI] [PubMed] [Google Scholar]
- 23.Song J, Shih Ie M, Salani R, Chan DW, Zhang Z. Annexin XI is associated with cisplatin resistance and related to tumor recurrence in ovarian cancer patients. Clin Cancer Res. 2007;13:6842–6849. doi: 10.1158/1078-0432.CCR-07-0569. [DOI] [PubMed] [Google Scholar]
- 24.Dimanche-Boitrel MT, Pelletier H, Genne P, Petit JM, Le Grimellec C, Canal P, Ardiet C, Bastian G, Chauffert B. Confluence-dependent resistance in human colon cancer cells: role of reduced drug accumulation and low intrinsic chemosensitivity of resting cells. Int J Cancer. 1992;50:677–682. doi: 10.1002/ijc.2910500502. [DOI] [PubMed] [Google Scholar]
- 25.Kohn KW, Jackman J, O'Connor PM. Cell cycle control and cancer chemotherapy. J Cell Biochem. 1994;54:440–452. doi: 10.1002/jcb.240540411. [DOI] [PubMed] [Google Scholar]
- 26.Waldman T, Zhang Y, Dillehay L, Yu J, Kinzler K, Vogelstein B, Williams J. Cell-cycle arrest versus cell death in cancer therapy. Nat Med. 1997;3:1034–1036. doi: 10.1038/nm0997-1034. [DOI] [PubMed] [Google Scholar]
- 27.Itamochi H, Kigawa J, Akeshima R, Sato S, Kamazawa S, Takahashi M, Kanamori Y, Suzuki M, Ohwada M, Terakawa N. Mechanisms of cisplatin resistance in clear cell carcinoma of the ovary. Oncology. 2002;62:349–353. doi: 10.1159/000065067. [DOI] [PubMed] [Google Scholar]
- 28.Itamochi H, Kigawa J, Sugiyama T, Kikuchi Y, Suzuki M, Terakawa N. Low proliferation activity may be associated with chemoresistance in clear cell carcinoma of the ovary. Obstet Gynecol. 2002;100:281–287. doi: 10.1016/s0029-7844(02)02040-9. [DOI] [PubMed] [Google Scholar]
- 29.Tomas A, Futter C, Moss SE. Annexin 11 is required for midbody formation and completion of the terminal phase of cytokinesis. J Cell Biol. 2004;165:813–822. doi: 10.1083/jcb.200311054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hofmann S, Franke A, Fischer A, Jacobs G, Nothnagel M, Gaede KI, Schurmann M, Muller-Quernheim J, Krawczak M, Rosenstiel P, et al. Genome-wide association study identifies ANXA11 as a new susceptibility locus for sarcoidosis. Nat Genet. 2008;40:1103–1106. doi: 10.1038/ng.198. [DOI] [PubMed] [Google Scholar]
- 31.Duale N, Lindeman B, Komada M, Olsen AK, Andreassen A, Soderlund EJ, Brunborg G. Molecular portrait of cisplatin induced response in human testis cancer cell lines based on gene expression profiles. Mol Cancer. 2007;6:53. doi: 10.1186/1476-4598-6-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marinissen MJ, Tanos T, Bolos M, de Sagarra MR, Coso OA, Cuadrado A. Inhibition of heme oxygenase-1 interferes with the transforming activity of the Kaposi sarcoma herpesvirus-encoded G protein-coupled receptor. J Biol Chem. 2006;281:11332–11346. doi: 10.1074/jbc.M512199200. [DOI] [PubMed] [Google Scholar]
- 33.Kim HR, Kim S, Kim EJ, Park JH, Yang SH, Jeong ET, Park C, Youn MJ, So HS, Park R. Suppression of Nrf2-driven heme oxygenase-1 enhances the chemosensitivity of lung cancer A549 cells toward cisplatin. Lung Cancer. 2008;60:47–56. doi: 10.1016/j.lungcan.2007.09.021. [DOI] [PubMed] [Google Scholar]
- 34.Kim HJ, So HS, Lee JH, Lee JH, Park C, Park SY, Kim YH, Youn MJ, Kim SJ, Chung SY, et al. Heme oxygenase-1 attenuates the cisplatin-induced apoptosis of auditory cells via down-regulation of reactive oxygen species generation. Free Radic Biol Med. 2006;40:1810–1819. doi: 10.1016/j.freeradbiomed.2006.01.018. [DOI] [PubMed] [Google Scholar]
- 35.Ahmed AA, Mills AD, Ibrahim AE, Temple J, Blenkiron C, Vias M, Massie CE, Iyer NG, McGeoch A, Crawford R, et al. The extracellular matrix protein TGFBI induces microtubule stabilization and sensitizes ovarian cancers to paclitaxel. Cancer Cell. 2007;12:514–527. doi: 10.1016/j.ccr.2007.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.de Bree R, Roos JC, Plaizier MA, Quak JJ, van Kamp GJ, den Hollander W, Snow GB, van Dongen GA. Selection of monoclonal antibody E48 IgG or U36 IgG for adjuvant radioimmunotherapy in head and neck cancer patients. Br J Cancer. 1997;75:1049–1060. doi: 10.1038/bjc.1997.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nieuwenhuis EJ, Leemans CR, Kummer JA, Denkers F, Snow GB, Brakenhoff RH. Assessment and clinical significance of micrometastases in lymph nodes of head and neck cancer patients detected by E48 (Ly-6D) quantitative reverse transcription-polymerase chain reaction. Lab Invest. 2003;83:1233–1240. doi: 10.1097/01.lab.0000083532.46536.56. [DOI] [PubMed] [Google Scholar]
- 38.Rubinfeld B, Upadhyay A, Clark SL, Fong SE, Smith V, Koeppen H, Ross S, Polakis P. Identification and immunotherapeutic targeting of antigens induced by chemotherapy. Nat Biotechnol. 2006;24:205–209. doi: 10.1038/nbt1185. [DOI] [PubMed] [Google Scholar]
- 39.Wang Q, He Z, Gao J, Hu S, Huang M, Liu M, Zheng J, Tang H. S100P sensitizes ovarian cancer cells to carboplatin and paclitaxel in vitro. Cancer Lett. 2008;272:277–284. doi: 10.1016/j.canlet.2008.07.017. [DOI] [PubMed] [Google Scholar]
- 40.Fumoto S, Shimokuni T, Tanimoto K, Hiyama K, Otani K, Ohtaki M, Hihara J, Yoshida K, Hiyama E, Noguchi T, et al. Selection of a novel drug-response predictor in esophageal cancer: a novel screening method using microarray and identification of IFITM1 as a potent marker gene of CDDP response. Int J Oncol. 2008;32:413–423. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.