Abstract
Context:
Twin studies indicate that the timing of pubertal onset is under strong genetic control. However, genes controlling pubertal timing in the general population have not yet been identified.
Objective:
To facilitate the identification of genes influencing the timing of pubertal growth and maturation, we conducted linkage mapping of constitutional delay of growth and puberty (CDGP), an extreme variant of normal pubertal timing, in extended families.
Participants and methods:
Fifty-two families multiply affected with CDGP were genotyped with 383 multiallelic markers. CDGP was defined based on growth charts (the age at onset of growth spurt, peak height velocity, or attaining adult height taking place at least 1.5 SD later than average). Chromosomal regions co-segregating with CDGP were identified with parametric affected only linkage analysis using CDGP as a dichotomized trait.
Results:
The genome-wide scan detected linkage of CDGP to a region on chromosome 2p13-2q13. The two-point HLOD-score was 1.62 (α 0.27) and the corresponding multipoint HLOD 2.54 (α 0.31). Fine-mapping the region at 1 cM resolution increased the multipoint HLOD-score to 4.44 (α 0.41). The linkage became weaker if also family members diagnosed with CDGP without growth data were included in the analyses.
Conclusions:
The pericentromeric region of chromosome 2 harbors a gene predisposing to pubertal delay in multiply affected pedigrees. Our data suggest that this locus may be a component of the internal clock controlling the timing of the onset of puberty.
Keywords: puberty timing, genes, linkage mapping
Introduction
Significant concordance in the timing of puberty between monozygotic twins (1, 2) and similarities in pubertal timing between family members (3-5) indicate that the onset of puberty is genetically determined. Furthermore, the hypothalamic neuroendocrine network initiating puberty seems to be influenced by a complex interplay of signals from peripheral tissues (e.g. body energy stores) (6) and environment (e.g. climate and light) (7, 8). However, the exact mechanisms of these impacts and the precise mechanisms controlling the onset of puberty are still poorly characterized.
The continuous distribution of the timing of puberty in the general population (9) suggests that multiple regulatory genes are involved, with possibly relatively small effect of a single gene. In support of genetic heterogeneity, two recent studies mapping genes influencing the continuous trait age at menarche both reported significant LOD-scores, but none of the identified genomic regions overlapped between the studies (10, 11). However, even though the regulation of the timing of puberty may be complex at the population level, the genetic makeup of individuals at the extreme ends of the normal spectrum is likely to represent a more homogenous genetic structure than the overall population. Furthermore, at the tails of the distribution, genetic variants with a strong impact on the trait may be expected. Supporting oligogenic inheritance, we and others have shown that constitutional delay of growth and puberty (CDGP), an extreme variant of normal pubertal timing strongly clusters in families and often segregates in an autosomal dominant pattern (4, 5). Therefore, in the present study, we set out to identify genes conferring susceptibility to pubertal delay by linkage mapping in extended pedigrees multiply affected with CDGP. We defined CDGP based on objective evidence of delayed pubertal growth spurt, thus avoiding the potential confounding ambiguity introduced by subjective recall of the phenotype. Furthermore, similar criteria for delayed central onset of puberty could be applied in both genders. Our family collection was maximized for a homogenous genetic composition and can be anticipated to be enriched for high-impact alleles that are amenable to linkage mapping.
Materials and methods
Identification of probands
Patients referred during 1982 through 2004 to specialist pediatric care due to delayed puberty were identified from the hospital discharge records of Helsinki, Kuopio, Tampere and Turku University Hospitals and two municipal hospitals in southern Finland. Patients fulfilling the diagnostic criteria for CDGP, defined as 2 SD later than average pubertal development (Tanner genital stage II, testis volume of more than 3 ml, beyond the age 13.5 yr in boys and Tanner breast stage II beyond the age 13.0 yr in girls) (12, 13) were included. Medical history, clinical examination, or routine laboratory tests failed to reveal any signs of chronic illnesses accounting for the delayed puberty, and hypogonadotropic hypogonadism, if suspected, was excluded by GnRH testing and by clinical follow-up ensuring spontaneous pubertal development. Pubertal growth spurt in probands was more than 2 SD later than average; age at acceleration of pubertal growth (take-off) beyond 13.8 and 12.2 yr, and age at peak height velocity (phv) later than 15.6 and 13.7 yr, in males and females, respectively (14).
Ascertainment of family members
Families of the CDGP patients were invited by a letter to participate. Information about medical history and pubertal timing was obtained by structured interviews, and a blood sample was drawn for DNA-extraction. Archived height measurement records were collected to draw growth charts. Whenever possible, the timing of puberty was assessed based on timing of pubertal growth spurt examined by three pediatricians in the group (KW, TL, LD). The criteria for CDGP in probands' family members were one of the three following: 1) age at take-off or 2) phv occurring 1.5 SD beyond the mean, i.e. age at take-off exceeding 12.9 and 11.3 yr, or age at phv exceeding 14.8 and 12.8 yr in males and females, or 3) age at attaining adult height more than 18 or 16 yr, in males and females, respectively (14). If growth charts were unavailable, the timing of puberty was based on interviews. In that case the criteria for CDGP were: recalling having undergone pubertal development or attained adult height more than 1.5 yr later than their peers (both sexes), or menarche after 14 yr. Those family members with chronic illnesses and other factors possibly affecting growth and development in childhood were excluded. Only families of Finnish origin were included.
Using growth chart based CDGP criteria we were able to identify 52 multiply affected families that were informative for linkage, encompassing a total of 410 subjects (213 males, 197 females). Of these, 179 individuals (97 males, 82 females) were classified as affected. The largest family contained 9 affected pedigree members. Of all families, 28 consisted of affected sibling pairs. These affected individuals, diagnosed based on growth chart information, were used in all primary linkage analyses. As an attempt to maximize the linkage information, we also run a sub-analysis, which included 56 additional affected subjects who had been classified as affected based on interviews alone. Thirty of these belonged to the 52 multiply affected pedigrees identified by growth chart criteria. Of the subjects, 26 classified as affected by interview belonged to 10 new multiply affected families.
All participants and/or the parents/guardians gave their written informed consent. The study protocol was approved by the Ethics Committee for Pediatrics, Adolescent Medicine and Psychiatry, Hospital District of Helsinki and Uusimaa and was extended to encompass also the sample collections at Kuopio, Tampere, and Turku University Hospitals. The study was conducted in accordance with the guidelines of The Declaration of Helsinki.
Genotyping and marker maps
Genome-wide linkage mapping was carried out using ABI PRISM Linkage mapping Set MD10 (Applied Biosystems). Standard PCR-protocols were used for the amplification of fragments using 10 ng of genomic DNA as a template. The fluorescently labeled PCR products were separated using ABI3730 (Applied Biosystems) automated electrophoresis system and the genotype calls were made using GeneMapper3.7 software. All allele calls were verified by two independent reviewers and any discrepancies were resolved. Genotypes were retrieved for 383 markers spanning all autosomes and the X-chromosome. The program GRR (Graphical Representation of Relationships) (15), which calculates the average identical-by-state allele-sharing between all pairs in a data-set, was used to screen for inconsistencies in familial relationships. Genotypes were checked for violation of Mendelian segregation of alleles using PEDCHECK (16).
Marker locations were based on DeCode map distances. In case a marker had not been placed on the DeCode map, estimates of their genetic locations were based on linear interpolation by using the physical location and the genetic locations of the immediately flanking deCODE markers in the University of California Santa Cruz (UCSC) database using Cartographer (https://apps.bioinfo.helsinki.fi/software/cartographer.aspx). Fine-mapping markers were selected from the UCSC database. Markers were selected one every 1 million bases preferentially picking markers with heterozygosity > 0.7.
Statistical analysis
Linkage analysis using pubertal delay as a dichotomized trait was carried out using the program Merlin (17). Since the expressivity of potential alleles co-segregating with CDGP is not known, the linkage analyses were run using the affected only approach, in which all unaffected family members were scored having unknown phenotype. Three dominant models were tested. The penetrances for affected heterozygotes and homozygotes were kept at 0.9 and unaffected in all models but the disease allele frequency was set to 0.01, 0.001 and 0.0001 in models 1, 2 and 3 respectively. The reported HLOD-scores are all obtained with model 3. Merlin was also used to reconstruct likely haplotypes segregating in the pedigrees corresponding to the most likely pattern of gene flow.
Results
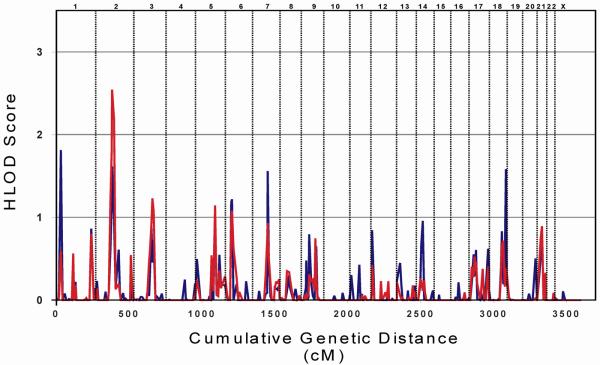
The genome was screened with 383 microsatellite markers in 52 families to identify chromosomal regions co-segregating with CDGP as diagnosed from growth charts. The average marker success rate was 98.0%, and the average information content 0.87. A graphic representation of all HLOD-scores obtained with parametric linkage analysis and a summary of loci yielding HLOD-scores above 1.5 are shown in Figure 1 and Table 1. The highest HLOD-score obtained with two-point analysis was detected at marker locus D1S2697 (HLOD 1.82; α 0.45). In addition, two-point analyses identified three other loci with HLOD-scores exceeding 1.5; on chromosome 2 (D2S2216, HLOD 1.62; α 0.27) and chromosomes 7 and 18 (Figure 1, Table 1). Of these four loci, only the locus on chromosome 2 was supported by multipoint analysis, which at marker locus D2S2216 resulted in an increase of the HLOD-score to 2.54 (α 0.31). Further support for linkage to CDGP was obtained from the neighboring loci D2S2333 and D2S160 yielding HLOD-scores of 1.79 and 2.18, respectively (Table 1). Multipoint analysis did not detect any other HLOD-scores exceeding 1.5.
FIG. 1.
HLOD-scores obtained from two- and multipoint linkage analyses of growth chart-based constitutional delay of growth and puberty. The results of the two-point analyses are represented by the blue line and the multipoint analyses by the red line.
TABLE 1.
Two-point and multipoint HLOD-scores exceeding 1.5 in the genome-wide linkage analysis of constitutional delay of growth and puberty.
| Locus | Position (cM) |
Two-point analysis | Multipoint analysis | ||||
|---|---|---|---|---|---|---|---|
|
|
|||||||
| HLOD | α | Information | HLOD | α | Information | ||
| D1S2697 | 32.0 | 1.82 | 0.45 | 0.55 | 0.59 | 0.22 | 0.78 |
| D2S2333 | 108.8 | 1.14 | 0.22 | 0.71 | 1.79 | 0.24 | 0.92 |
| D2S2216 | 111.5 | 1.62 | 0.27 | 0.67 | 2.54 | 0.31 | 0.91 |
| D2S160 | 124.9 | 0.97 | 0.20 | 0.67 | 2.18 | 0.26 | 0.86 |
| D7S657 | 103.5 | 1.56 | 0.31 | 0.74 | 0.93 | 0.19 | 0.92 |
| D18S462 | 116.4 | 1.58 | 0.47 | 0.60 | 0.39 | 0.19 | 0.88 |
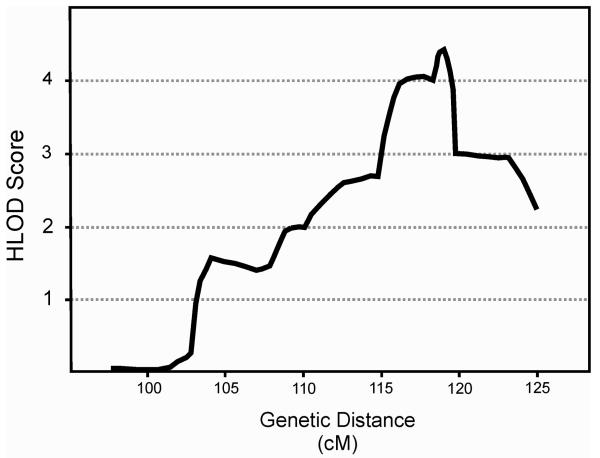
To further test whether the initial linkage signal on chromosome 2 would represent true co-segregation of the chromosomal region with CDGP or would be a false positive result associated only with the single marker D2S2216, we fine-mapped the region on chromosome 2p13-2q13. All family members were genotyped for 25 additional microsatellite markers covering the region at 0.97 cM intermarker distance increasing the overall multipoint information content to 0.97. Adding these markers to the linkage analysis clearly supported the initial linkage finding (Table 2, Figure 2). The highest two-point LOD-score was obtained at marker locus D2S2364 (HLOD 3.70; α 0.62). In addition, there were 6 loci yielding HLOD-scores exceeding 2. The highest multipoint LOD-score in the region (HLOD 4.44; α 0.41) was obtained at 119.0 cM (Figure 2). Adding individuals (n=56) with only interview-based phenotypes available did not improve the linkage signal (data not shown).
TABLE 2.
Two-point HLOD-scores obtained from linkage analysis of constitutional delay of growth and puberty exceeding 2, and corresponding multipoint HLOD-scores in the fine mapping analysis of chromosome 2p13-2q13.
| Locus | Position (cM) |
Two-point analysis | Multipoint analysis | ||||
|---|---|---|---|---|---|---|---|
|
|
|||||||
| HLOD | α | Information | HLOD | α | Information | ||
| D2S388 | 110.1 | 2.15 | 0.36 | 0.62 | 1.99 | 0.26 | 0.98 |
| D2S2159 | 112.6 | 2.12 | 0.33 | 0.73 | 2.61 | 0.33 | 0.99 |
| D2S2209 | 114.8 | 2.77 | 0.44 | 0.71 | 2.70 | 0.34 | 0.99 |
| D2S2264 | 116.1 | 2.17 | 0.41 | 0.71 | 3.95 | 0.41 | 0.99 |
| D2S2364 | 118.3 | 3.70 | 0.62 | 0.57 | 4.02 | 0.39 | 0.97 |
| D2S2229 | 119.0 | 3.38 | 0.43 | 0.73 | 4.44 | 0.41 | 0.99 |
| D2S293 | 119.8 | 2.87 | 0.40 | 0.76 | 3.01 | 0.37 | 0.99 |
FIG. 2.
Multipoint HLOD-scores obtained by linkage analyses of constitutional delay of growth and puberty after adding 25 microsatellite markers to the region on chromosome 2p13-2q13. The linkage analysis was run sampling information at each marker locus and at 3 positions between the markers.
Finally, we aimed at testing the hypothesis, whether stratifying the families based on place of origin would reduce the allelic heterogeneity. Most of our families were ascertained from southern and western Finland, but 7 families were living in the central and eastern parts of the country. The geographical origin of these families was not traced further back than to the 1940's, but according to previous works on monogenic disorders in Finland, this should suffice to trace the ancestry of disease-carrying alleles with a reasonable precision (18). Running these 7 families alone in the linkage analysis showed that they contributed to a substantial proportion of the linkage signal at chromosome 2. Multipoint analysis of the 7 families for all markers on chromosome 2p13-2q13 yielded a HLOD-score of 3.46 (α 0.75).
We then went ahead and scrutinized the segregating haplotypes in the 5 families contributing to the linkage signal. Based on two crossing-over events in the family yielding the second highest LOD-score, the minimal shared segment was spanning 6 Mb between markers D2S2209 and D2S293. None of the 5 families shared a haplotype pattern spanning more than 2 markers (Table 3). At marker locus D2S373 the allele 237 was co-segregating with delayed puberty in half of the families from eastern Finland. However, the frequency of that allele was very high also in families not showing evidence for linkage to chromosome 2q11-2q12, i.e. 0.5 as estimated from 167 founder chromosomes of the 30 families yielding the lowest LOD-scores at that locus. Nevertheless, the two families with the highest LOD-scores in Table 3 have almost similar looking haplotypes at marker loci D2S373, D2S2364 and D2S2229. The two haplotypes co-segregating with pubertal delay in each family, are distinguished only by 1 repeat unit at marker D2S2364, and could therefore theoretically represent the same ancestral chromosome with a mutation at marker locus D2S2364. However, the alleles present on these haplotypes were rather common with allele frequencies at the level of 0.35 – 0.45. The 273-[101/99]-181 haplotype was also present on 9% of the founder chromosomes of the 30 families yielding the smallest LOD-scores at chromosome 2q11-2q12.
TABLE 3.
Haplotypes on chromosome 2q11-2q12 co-segregating with constitutional delay of growth and puberty in 5 families from central and eastern Finland. LOD-score is obtained by multipoint analysis at marker position D2S2229. Family_5 shared 2 alleles throughout the region.
| Family | D2S2209 | D2S2264 | D2S373 | D2S2364 | D2S2229 | D2S293 | LOD- score |
|---|---|---|---|---|---|---|---|
| Family_1 | 190 | 251 | 237 | 101 | 181 | 178 | 2.07 |
| Family_2 | X | 255 | 237 | 99 | 181 | X | 1.77 |
| Family_3 | 198 | 233 | 223 | 99 | 191 | 176 | 0.29 |
| Family_4 | 192 | 251 | 237 | 95 | 181 | 174 | 0.29 |
| Family_5/1 | 198 | 243 | 223 | 95 | 205 | 174 | 0.16 |
| Family_5/2 | 202 | 253 | 245 | 99 | 189 | 182 |
Discussion
We here report the results of the first genome-wide search for genes conferring susceptibility to constitutional delay of growth and puberty (CDGP). By utilizing the linkage mapping approach, we successfully mapped a locus co-segregating with CDGP to the pericentromeric region on chromosome 2.
Linkage analysis is, indeed, a powerful tool to identify rare high-impact alleles co-segregating with specific traits in extended families. However, it is sensitive to misspecification of the phenotype. Therefore, we took several measures to avoid misclassification of the affectation status (i.e. delayed puberty). First of all, we chose to use a unique resource of archived height measurement records, which enabled objective assessment of the phenotype (i.e. timing of puberty) also in adults who had passed puberty already a long time ago. Furthermore, we attempted to exclude both under-nutrition and chronic illnesses by interviews, since both of them influence linear growth and pubertal maturation (19, 20). We also sought to exclude hypogonadotropic hypogonadism as it is characterized by later than average development and growth spurt during pubertal years, although in contrast to CDGP, spontaneous maturation is usually absent (21). The criterion for defining delayed puberty onset for some subjects was the age at attaining adult height. This phenotype could potentially also be caused either by estrogen receptor defects (22) or aromatase deficiency (23), which both will result in a prolonged growth period affecting the age at attaining adult height, because estrogen is the main regulator of epiphyseal closure (24). These conditions are, however, extremely rare and we only occasionally used the age at attaining adult height as the only criterion for CDGP, i.e. only when the growth measurements were too infrequent to allow estimation of the timing of acceleration or peak height velocity of pubertal linear growth.
The chromosomal region linked to delayed pubertal growth spurt in the present study did not coincide with the results of two previous studies mapping genes affecting another puberty timing phenotype, age at menarche (10, 11). Neither did it overlap with the genomic regions implicated in a genome-wide screen of idiopathic short stature (25), a phenotype also affecting longitudinal growth and partly overlapping with CDGP. These divergent results are likely to be reflections of phenotypic and genetic heterogeneity, as well as different study designs. After the activation of the hypothalamic-pituitary-gonadal (HPG) -axis, secondary sexual characteristics appear usually in a certain sequence with breast tissue growth and pubertal acceleration of linear growth being the first markers in girls (13, 14). In contrast, menarche is a late phenomenon in female puberty indicating that pubertal maturation is almost completed (13). Furthermore, there may be only a moderate correlation between age at menarche and timing of breast development (r = 0.37) (26). Pubertal growth spurt is regulated mainly by sex steroids, specifically estrogen, in both genders (27) and therefore reflects the full activation of the HPG-axis. In boys, pubertal growth spurt occurs later along somatic maturation than in girls (12, 14), but probably because estrogens in boys are derived from androgens by aromatization (28). Therefore, the timing of pubertal growth spurt may serve as a marker for central onset of puberty similarly in both genders.
In addition to using different markers for puberty onset, our study differs from the previous ones also by study design. First of all, as a consequence of studying age at menarche, only females were included in the previous scans (10, 11), while our analysis encompassed both genders. Secondly, the genome-wide screens of age at menarche both used the phenotype as a quantitative trait in the analysis, thus mapping genes affecting the whole distribution of pubertal timing present in the population. In contrast, our study population was selected from the extreme end of the continuous distribution of pubertal timing, i.e. probands with clinically verified CDGP (puberty taking place more that 2 SD later than average) and their relatives with pubertal growth spurt taking place more than 1.5 SD later than average, representing less than 10% of the general population. Individuals at the tail of a genetically determined continuous distribution might either be carrying a single allele with a strong impact on the phenotype, or alternatively multiple low- or medium-impact susceptibility alleles. By selecting extended families segregating CDGP in an autosomal dominant fashion, our study was clearly biased to detect rare alleles with a large impact on the phenotype. At present, when majority of the genetic structure underlying any complex trait still remains unidentified, it is not known how much overlap can be expected in the genetic variation of a complex continuous trait versus a complex discrete phenotype, such as pubertal timing versus CDGP. As an example, recent genome-wide association studies, ideally suited for detecting low-impact alleles, have successfully identified more than 40 loci associated with adult height (29-32). Still, majority of the genetic background of adult stature remains unaccounted for. Typically, these identified genetic regions don't overlap with height associated chromosomal regions identified by linkage mapping or with loci causing extreme height phenotypes, but HMGA2 representing the strongest identified association signal (29), also has alleles with large impact on height, causing short length in mice (33) and extremely tall stature in humans (34). Not finding any overlap between the loci implicated in the previous genome-wide scans of age at menarche and our own study might thus be a consequence of different mapping approaches targeting distinct milestone events of pubertal maturation. Regarding the differences between implicated chromosomal regions in the current study and the study on idiopathic short stature (25), it is likely that the majority of the individuals included in the idiopathic short stature study represent a completely distinct phenotype compared to CDGP studied by us. Finally, the lack of overlap of implicated chromosomal regions between the menarche scans further point to genetic heterogeneity of that phenotype.
Our initial hypothesis was that a selected study sample of multiply affected extended CDGP families would be enriched for high-impact alleles co-segregating with the phenotype. This turned out to be the case but, nevertheless, genetic heterogeneity was present also in our genetically fairly homogenous study population. Only forty percent of all studied families contributed to the significant linkage signal on chromosome 2p13-2q13. Interestingly, we found evidence for geographical stratification as majority of the linkage signal was accounted for by a minority of the families originating from central and eastern Finland. These regions represent the more recently settled parts of the country, to which the population spread from the early settlement region of the southern and western coastlines starting from the 16th century. Therefore, based on previous molecular characterizations of Finnish Mendelian disease genes, rather long genetic distances of up to 10 cM can be expected to flank the disease gene provided that the mutation arose on one of the founder chromosomes of the sub-settlement (18). In order to primarily verify the initial linkage signal we genotyped the linked region at 1 cM resolution. Even though the fine-mapping results supported linkage, we did not find evidence for linked families sharing chromosomal segments identical-by-descent. Therefore, the mutation co-segregating with CDGP might represent a common rather old genetic variant, which due to major bottle-necks and genetic drift has been enriched in the subpopulation of central and eastern Finland. This finding also implies that the detection of linkage disequilibrium surrounding the mutation is likely to require a rather dense map. Mapping genes in population isolates, such as the Finnish population, has several advantages including a more uniform genetic background (18), but because of the unique population structure the pertinence of a mapped locus to other populations is not automatically evident. Even though it is quite likely that the mapped allele co-segregating with CDGP is present also in other European populations, perhaps at a different frequency, it is nevertheless possible that it could represent a population specific mutation that has occurred on an ancestral chromosome a few thousand years ago.
As we couldn't restrict the linked region by linkage disequilibrium mapping at 1 cM resolution, we face the challenge of tracing the genetic variant conferring susceptibility to pubertal delay in a large genomic region of some 6-20 Mb. None of the genes in this region have previously shown involvement in HPG-axis functions, nor have they been implicated in the phenotypically related condition of hypogonadotropic hypogonadism. Nevertheless, there are a number of genes in the region with a potential connection to pubertal maturation, a few examples of which will be discussed below. One example is GPR45 belonging to the family of G-protein coupled receptors, which are widely expressed in the central nervous system. Mutations in other G-protein coupled receptor genes, such as GNRHR (35), KISS1R (36), and PROKR2 (37) are known to cause hypogonadotropic hypogonadism. Another example is POU3F3, also predominantly expressed in the brain and encoding for member of the homeobox family of POU domain transcription factors. Because inactivating mutations of another member of the POU family genes have been reported to cause combined pituitary hormone deficiency (38), one might speculate that POU3F3 could be a factor in development of the HPG-axis. Taken together, given the complexity of the phenotype and the large range of potentially involved pathways, in addition to the large size of the identified chromosomal region pinpointing the gene involved, identification of the causative variant and the evaluation of the impact of the identified locus at the population level must await follow-up studies.
The underlying genetic factors explaining the physiological variation of pubertal onset have hitherto remained elusive. However, the importance of understanding the mechanisms regulating the onset of pubertal growth and maturation is emphasized by the possibility of global advancement in pubertal timing and the evidence that early pubertal development may have long lasting implications (39, 40). In order to maximize the chances of identifying genetic variants influencing the onset of puberty, we chose to study delayed puberty representing the extreme tail of the normal distribution of pubertal timing. By locating a novel gene locus predisposing to CDGP, we have now taken one step toward unraveling the basic molecular mechanisms regulating the timing of the onset of puberty.
Acknowledgements
We are grateful to all the families for participating in the study. We are also to thank Dr. Hanna-Liisa Lenko and Dr. Jyrki Lähde for their help with initiating the family collection at Tampere University Hospital, and Mr. Kyösti Sutinen for skilful help with data management. The study was supported by the European Union: Puberty onset – influence of nutritional, environmental and endogenous regulators (PIONEER) –project; The Academy of Finland; The Program for Molecular Medicine of the Faculty of Medicine, University of Helsinki; The Sigrid Juselius Foundation; The Foundation for Pediatric Research, Finland; The Finnish Medical Foundation; The Paulo Foundation and The Päivikki and Sakari Sohlberg Foundation.
Grants: European Union: Puberty onset – influence of nutritional, environmental and endogenous regulators (PIONEER)- project (Contract No. 513991); The Academy of Finland: 120315 (to E.W.) and Center of Excellence in Complex Disease Genetics (to E.W., A.P.); The Program for Molecular Medicine of the Faculty of Medicine, University of Helsinki and The Sigrid Juselius Foundation (to A.P.); The Foundation for Pediatric Research, Finland (to K.W., T.L.); The Finnish Medical Foundation (to T.L.); The Paulo Foundation (to E.W. and K.W.) and The Päivikki and Sakari Sohlberg Foundation (to K.W.).
Abbreviations
- CDGP
constitutional delay of growth and puberty
- GNRHR
gonadotropin-releasing hormone receptor
- GPR45
G protein-coupled receptor 45
- HPG-axis
hypothalamic-pituitary-gonadal-axis
- KISS1R
KISS1 receptor
- PROKR2
prokineticin receptor 2
Footnotes
Disclosure information: The authors have nothing to disclose.
References
- 1.Sharma JC. The genetic contribution to pubertal growth and development studied by longitudinal growth data on twins. Ann Hum Biol. 1983;10:163–171. doi: 10.1080/03014468300006301. [DOI] [PubMed] [Google Scholar]
- 2.Kaprio J, Rimpelä A, Winter T, Viken RJ, Rimpelä M, Rose RJ. Common genetic influences on BMI and age at menarche. Hum Biol. 1995;67:739–753. [PubMed] [Google Scholar]
- 3.de Vries L, Kauschansky A, Shohat M, Phillip M. Familial central precocious puberty suggests autosomal dominant inheritance. J Clin Endocrinol Metab. 2004;89:1794–1800. doi: 10.1210/jc.2003-030361. [DOI] [PubMed] [Google Scholar]
- 4.Sedlmeyer IL, Hirschhorn JN, Palmert MR. Pedigree analysis of constitutional delay of growth and maturation: determination of familial aggregation and inheritance patterns. J Clin Endocrinol Metab. 2002;87:5581–5586. doi: 10.1210/jc.2002-020862. [DOI] [PubMed] [Google Scholar]
- 5.Wehkalampi K, Widén E, Laine T, Palotie A, Dunkel L. Patterns of inheritance of constitutional delay of growth and puberty in families of adolescent girls and boys referred to specialist pediatric care. J Clin Endocrinol Metab. 2008;93:723–728. doi: 10.1210/jc.2007-1786. [DOI] [PubMed] [Google Scholar]
- 6.Gamba M, Pralong FP. Control of GnRH neuronal activity by metabolic factors: the role of leptin and insulin. Mol Cell Endocrinol. 2006;254-255:133–139. doi: 10.1016/j.mce.2006.04.023. [DOI] [PubMed] [Google Scholar]
- 7.Albright DL, Voda AM, Smolensky MH, Hsi BP, Decker M. Seasonal characteristics of and age at menarche. Chronobiol Int. 1990;7:251–258. doi: 10.3109/07420529009056983. [DOI] [PubMed] [Google Scholar]
- 8.Magee K, Basinska J, Quarrington B, Stancer HC. Blindness and menarche. Life Sci. 1970;9:7–12. doi: 10.1016/0024-3205(70)90003-2. [DOI] [PubMed] [Google Scholar]
- 9.Tanner JM. Growth at Adolescence. 2nd ed. Blackwell; Oxford UK: 1962. [Google Scholar]
- 10.Guo Y, Shen H, Xiao P, Xiong DH, Yang TL, Guo YF, Long JR, Recker RR, Deng HW. Genomewide linkage scan for quantitative trait loci underlying variation in age at menarche. J Clin Endocrinol Metabolism. 2006;91:1009–1014. doi: 10.1210/jc.2005-2179. [DOI] [PubMed] [Google Scholar]
- 11.Rothenbuhler A, Fradin D, Heath S, Lefevre H, Bouvattier C, Lathrop M, Bougnères P. Weight-adjusted genome scan analysis for mapping quantitative trait loci for menarchal age. J Clin Endocrinol Metabolism. 2006;91:3534–3537. doi: 10.1210/jc.2006-0150. [DOI] [PubMed] [Google Scholar]
- 12.Marshall WA, Tanner JM. Variations in the pattern of pubertal changes in boys. Arch Dis Child. 1970;45:13–23. doi: 10.1136/adc.45.239.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marshall WA, Tanner JM. Variations in pattern of pubertal changes in girls. Arch Dis Child. 1969;44:291–303. doi: 10.1136/adc.44.235.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tanner JM, Whitehouse RH, Marubini E, Resele LF. The adolescent growth spurt of boys and girls of the Harpenden growth study. Ann Hum Biol. 1976;3:109–126. doi: 10.1080/03014467600001231. [DOI] [PubMed] [Google Scholar]
- 15.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. GRR: graphical representation of relationship errors. Bioinformatics. 2001;17:742–743. doi: 10.1093/bioinformatics/17.8.742. [DOI] [PubMed] [Google Scholar]
- 16.O'Connell JR, Weeks DE. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet. 1998;63:259–266. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin-rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 18.Peltonen L, Palotie A, Lange K. Use of population isolates for mapping complex traits. Nat Rev Genet. 2000;1:182–190. doi: 10.1038/35042049. [DOI] [PubMed] [Google Scholar]
- 19.Kulin HE, Bwibo N, Mutie D, Santner SJ. The effect of chronic childhood malnutrition on pubertal growth and development. Am J Clin Nutr. 1982;36:527–536. doi: 10.1093/ajcn/36.3.527. [DOI] [PubMed] [Google Scholar]
- 20.Pozo J, Argente J. Delayed puberty in chronic illness. Best Pract Res Clin Endocrinol Metab. 2002;16:73–90. doi: 10.1053/beem.2002.0182. [DOI] [PubMed] [Google Scholar]
- 21.Copeland KC, Paunier L, Sizonenko PC. The secretion of adrenal androgens and growth patterns of patients with hypogonadotropic hypogonadism and idiopathic delayed puberty. J Pediatr. 1977;91:985–990. doi: 10.1016/s0022-3476(77)80912-8. [DOI] [PubMed] [Google Scholar]
- 22.Smith EP, Boyd J, Frank GR, Takahashi H, Cohen RM, Specker B, Williams TC, Lubahn DB, Korach KS. Estrogen resistance caused by a mutation in the estrogen-receptor gene in a man. N Engl J Med. 1994;331:1056–1061. doi: 10.1056/NEJM199410203311604. [DOI] [PubMed] [Google Scholar]
- 23.Morishima A, Grumbach MM, Simpson ER, Fisher C, Qin K. Aromatase deficiency in male and female siblings caused by a novel mutation and the physiological role of estrogens. J Clin Endocrinol Metab. 1995;80:3689–3698. doi: 10.1210/jcem.80.12.8530621. [DOI] [PubMed] [Google Scholar]
- 24.Rochira V, Balestrieri A, Faustini-Fustini M, Carani C. Role of estrogen on bone in the human male: insights from the natural models of congenital estrogen deficiency. Mol Cell Endocrinol. 2001;178:215–220. doi: 10.1016/s0303-7207(01)00446-4. [DOI] [PubMed] [Google Scholar]
- 25.Dempfle A, Wudy SA, Saar K, Hagemann S, Friedel S, Scherag A, Berthold LD, Alzen G, Gortner L, Blum WF, Hinney A, Nurnberg P, Schafer H, Hebebrand J. Evidence for involvement of the vitamin D receptor gene in idiopathic short stature via a genome-wide linkage study and subsequent association studies. Hum Mol Genet. 2006;15:2772–2783. doi: 10.1093/hmg/ddl218. [DOI] [PubMed] [Google Scholar]
- 26.Biro FM, Huang B, Crawford PB, Lucky AW, Striegel-Moore R, Barton BA, Daniels S. Pubertal correlates in black and white girls. J Pediatr. 2006/2;148:234–240. doi: 10.1016/j.jpeds.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 27.Moll GW, Jr, Rosenfield RL, Fang VS. Administration of low-dose estrogen rapidly and directly stimulates growth hormone production. Am J Dis Child. 1986;140:124–127. doi: 10.1001/archpedi.1986.02140160042027. [DOI] [PubMed] [Google Scholar]
- 28.Veldhuis JD, Metzger DL, Martha PM, Jr, Mauras N, Kerrigan JR, Keenan B, Rogol AD, Pincus SM. Estrogen and testosterone, but not a nonaromatizable androgen, direct network integration of the hypothalamo-somatotrope (growth hormone)-insulin-like growth factor I axis in the human: evidence from pubertal pathophysiology and sex-steroid hormone replacement. J Clin Endocrinol Metab. 1997;82:3414–3420. doi: 10.1210/jcem.82.10.4317. [DOI] [PubMed] [Google Scholar]
- 29.Weedon MN, Lettre G, Freathy RM, Lindgren CM, Voight BF, Perry JR, Elliott KS, Hackett R, Guiducci C, Shields B, Zeggini E, Lango H, Lyssenko V, Timpson NJ, Burtt NP, Rayner NW, Saxena R, Ardlie K, Tobias JH, Ness AR, Ring SM, Palmer CN, Morris AD, Peltonen L, Salomaa V, Diabetes Genetics Initiative, Wellcome Trust Case Control Consortium. Davey Smith G, Groop LC, Hattersley AT, McCarthy MI, Hirschhorn JN, Frayling TM. A common variant of HMGA2 is associated with adult and childhood height in the general population. Nat Genet. 2007;39:1245–1250. doi: 10.1038/ng2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weedon MN, Lango H, Lindgren CM, Wallace C, Evans DM, Mangino M, Freathy RM, Perry JR, Stevens S, Hall AS, Samani NJ, Shields B, Prokopenko I, Farrall M, Dominiczak A, Diabetes Genetics Initiative, Wellcome Trust Case Control Consortium. Johnson T, Bergmann S, Beckmann JS, Vollenweider P, Waterworth DM, Mooser V, Palmer CN, Morris AD, Ouwehand WH, Cambridge GEM Consortium. Zhao JH, Li S, Loos RJ, Barroso I, Deloukas P, Sandhu MS, Wheeler E, Soranzo N, Inouye M, Wareham NJ, Caulfield M, Munroe PB, Hattersley AT, McCarthy MI, Frayling TM. Genome-wide association analysis identifies 20 loci that influence adult height. Nat Genet. 2008;40:575–583. doi: 10.1038/ng.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lettre G, Jackson AU, Gieger C, Schumacher FR, Berndt SI, Sanna S, Eyheramendy S, Voight BF, Butler JL, Guiducci C, Illig T, Hackett R, Heid IM, Jacobs KB, Lyssenko V, Uda M, Diabetes Genetics Initiative, FUSION, KORA, Prostate, Lung Colorectal and Ovarian Cancer Screening Trial, Nurses' Health Study. Sardi NIA, Boehnke M, Chanock SJ, Groop LC, Hu FB, Isomaa B, Kraft P, Peltonen L, Salomaa V, Schlessinger D, Hunter DJ, Hayes RB, Abecasis GR, Wichmann HE, Mohlke KL, Hirschhorn JN. Identification of ten loci associated with height highlights new biological pathways in human growth. Nat Genet. 2008;40:584–591. doi: 10.1038/ng.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gudbjartsson DF, Walters GB, Thorleifsson G, Stefansson H, Halldorsson BV, Zusmanovich P, Sulem P, Thorlacius S, Gylfason A, Steinberg S, Helgadottir A, Ingason A, Steinthorsdottir V, Olafsdottir EJ, Olafsdottir GH, Jonsson T, Borch-Johnsen K, Hansen T, Andersen G, Jorgensen T, Pedersen O, Aben KK, Witjes JA, Swinkels DW, den Heijer M, Franke B, Verbeek AL, Becker DM, Yanek LR, Becker LC, Tryggvadottir L, Rafnar T, Gulcher J, Kiemeney LA, Kong A, Thorsteinsdottir U, Stefansson K. Many sequence variants affecting diversity of adult human height. Nat Genet. 2008;40:609–615. doi: 10.1038/ng.122. [DOI] [PubMed] [Google Scholar]
- 33.Zhou X, Benson KF, Ashar HR, Chada K. Mutation responsible for the mouse pygmy phenotype in the developmentally regulated factor HMGI-C. Nature. 1995;376:771–774. doi: 10.1038/376771a0. [DOI] [PubMed] [Google Scholar]
- 34.Ligon AH, Moore SD, Parisi MA, Mealiffe ME, Harris DJ, Ferguson HL, Quade BJ, Morton CC. Constitutional rearrangement of the architectural factor HMGA2: a novel human phenotype including overgrowth and lipomas. Am J Hum Genet. 2005;76:340–348. doi: 10.1086/427565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.de Roux N, Young J, Misrahi M, Genet R, Chanson P, Schaison G, Milgrom E. A family with hypogonadotropic hypogonadism and mutations in the gonadotropin-releasing hormone receptor. N Engl J Med. 1997;337:1597–1602. doi: 10.1056/NEJM199711273372205. [DOI] [PubMed] [Google Scholar]
- 36.Seminara SB, Messager S, Chatzidaki EE, Thresher RR, Acierno JS, Jr, Shagoury JK, Bo-Abbas Y, Kuohung W, Schwinof KM, Hendrick AG, Zahn D, Dixon J, Kaiser UB, Slaugenhaupt SA, Gusella JF, O'Rahilly S, Carlton MB, Crowley WF, Jr, Aparicio SA, Colledge WH. The GPR54 gene as a regulator of puberty. N Engl J Med. 2003;349:1614–1627. doi: 10.1056/NEJMoa035322. [DOI] [PubMed] [Google Scholar]
- 37.Dodé C, Teixeira L, Levilliers J, Fouveaut C, Bouchard P, Kottler ML, Lespinasse J, Lienhardt-Roussie A, Mathieu M, Moerman A, Morgan G, Murat A, Toublanc JE, Wolczynski S, Delpech M, Petit C, Young J, Hardelin JP. Kallmann syndrome: mutations in the genes encoding prokineticin-2 and prokineticin receptor-2. PLoS Genetics. 2006;2:e175. doi: 10.1371/journal.pgen.0020175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Quentien MH, Barlier A, Franc JL, Pellegrini I, Brue T, Enjalbert A. Pituitary transcription factors: From congenital deficiencies to gene therapy. J Neuroendocrinol. 2006;18:633–642. doi: 10.1111/j.1365-2826.2006.01461.x. [DOI] [PubMed] [Google Scholar]
- 39.Rockhill B, Moorman PG, Newman B. Age at menarche, time to regular cycling, and breast cancer (North Carolina, United States) Cancer Causes Control. 1998;9:447–453. doi: 10.1023/a:1008832004211. [DOI] [PubMed] [Google Scholar]
- 40.Hulanicka B, Lipowicz A, Koziel S, Kowalisko A. Relationship between early puberty and the risk of hypertension/overweight at age 50: evidence for a modified barker hypothesis among polish youth. Econ Hum Biol. 2007;5:48–60. doi: 10.1016/j.ehb.2006.12.001. [DOI] [PubMed] [Google Scholar]