Abstract
Areal BMD (aBMD) and areal bone size (ABS) are biologically correlated traits and are each important determinants of bone strength and risk of fractures. Studies showed that aBMD and ABS are genetically correlated, indicating that they may share some common genetic factors, which, however, are largely unknown. To study the genetic factors influencing both aBMD and ABS, bivariate whole genome linkage analyses were conducted for aBMD-ABS at the femoral neck (FN), lumbar spine (LS), and ultradistal (UD)-forearm in a large sample of 451 white pedigrees made up of 4498 individuals. We detected significant linkage on chromosome Xq27 (LOD = 4.89) for LS aBMD-ABS. In addition, we detected suggestive linkages at 20q11 (LOD = 3.65) and Xp11 (LOD = 2.96) for FN aBMD-ABS; at 12p11 (LOD = 3.39) and 17q21 (LOD = 2.94) for LS aBMD-ABS; and at 5q23 (LOD = 3.54), 7p15 (LOD = 3.45), Xq27 (LOD = 2.93), and 12p11 (LOD = 2.92) for UD-forearm aBMD-ABS. Subsequent discrimination analyses indicated that quantitative trait loci (QTLs) at 12p11 and 17q21 may have pleiotropic effects on aBMD and ABS. This study identified several genomic regions that may contain QTLs important for both aBMD and ABS. Further endeavors are necessary to follow these regions to eventually pinpoint the genetic variants affecting bone strength and risk of fractures.
Key words: BMD, bone structure, bone size, whole genome linkage scan, bivariate
INTRODUCTION
Osteoporosis is a significant public health problem that is responsible for >2 million fractures and direct medical costs of $17 billion in 2005 for the United States.(1) BMD provides a useful evaluation of material property of bone and is the most prominent risk factor of osteoporotic fractures,(2) but not the only one. From a biomechanical viewpoint, the fracture risk depends on structural features of the bone as well (e.g., bone size, shape, and architecture), which may significantly influence stress or stress distribution of the applied force.(3,4) Many studies have shown that areal bone size (ABS), which is derived from the projection area of the interested region of a specific bone under the X-ray beam of DXA, is an independent determinant of bone strength and a major risk factor of fractures.(5–8)
Although BMD and ABS reflect different aspects of bone composition and structure, they are biologically closely correlated. In ossification process, osteoid formation determines initial bone size and provides the matrix for subsequent mineralization and bone maturation. Studies also showed that BMD and bone size change in a synergistic manner when adapting to the mechanical load in bone development and turnover.(9,10) In accordance with their biological correlation, studies showed that BMD and ABS measurements have significant genetic correlations.(11) During the past decade, univariate linkage scans have identified a number of genomic regions, respectively, important for BMD and ABS(12); however, the shared genetic factors underlying these two important osteoporosis-related phenotypes are still largely unknown.
Bivariate linkage analysis provides a formal way to identify genomic regions harboring quantitative trait locus (QTLs) influencing two correlated traits. By incorporating correlation information, bivariate linkage analysis can improve the statistical power considerably and facilitate the identification of QTLs whose effects are too small to be detected by univariate linkage analyses.(13,14) An additional strength of bivariate linkage analysis is its ability to differentiate pleiotropic effects of a single locus influencing two correlated traits from coincident linkage of tightly clustered loci each influencing different trait.(15)
Given the strong genetic correlation between BMD and ABS but the lack of studies to show the common genetic effects underlying this correlation, in this study, we aimed to fill the gap by performing a bivariate whole genome linkage study for aBMD-ABS pairs at the femoral neck (FN), lumbar spine (LS), and ultradistal (UD)-forearm in the same sample used in our earlier univariate linkage scans for aBMD and ABS.(16,17)
MATERIALS AND METHODS
Subjects
The study was approved by institutional review boards of Creighton University and University of Missouri-Kansas City. All subjects signed informed-consent documents before entering the study. All the study subjects were whites of European origin and were recruited from the vicinity of Creighton University. The sampling scheme and exclusion criteria have been detailed previously.(18) Briefly, individuals with chronic diseases and conditions that might affect bone mass, structure, or metabolism were excluded. The study sample contains a total of 4498 subjects from 451 pedigrees. The pedigrees vary in size from 4 to 416 individuals, with a mean (SD) of 11.6 (28.5). This large sample size provides an exceedingly large number of relative pairs (>150,000) informative for linkage analyses. The basic characteristics of the study subjects are summarized in Table 1.
Table 1.
Basic Characteristics of the Study Subjects
| Total (n = 4498) | Female (n = 2682) | Male (n = 1816) | |
| Height (m) | 1.70 ± 0.10 | 1.64 ± 0.07 | 1.78 ± 0.07* |
| Weight (kg) | 78.5 ± 18.2 | 71.3 ± 16.0 | 89.4 ± 15.8* |
| Age (yr) | 47.7 ± 16.0 | 47.6 ± 16.0 | 48.0 ± 16.1 |
| Areal BMD (g/cm2) | |||
| Femoral neck | 0.826 ± 0.147 | 0.797 ± 0.143 | 0.867 ± 0.144* |
| Lumbar spine | 1.036 ± 0.162 | 1.011 ± 0.163 | 1.072 ± 0.153* |
| Ultradistal-forearm | 0.467 ± 0.085 | 0.430 ± 0.070 | 0.521 ± 0.076* |
| Areal bone size (cm2) | |||
| Femoral neck | 16.193 ± 1.835 | 5.042 ± 0.396 | 5.912 ± 0.490* |
| Lumbar spine | 63.384 ± 8.341 | 58.588 ± 5.843 | 70.375 ± 6.475* |
| Ultradistal-forearm | 3.892 ± 0.553 | 3.573 ± 0.373 | 4.341 ± 0.441* |
Values are means ± SD of the raw data without adjustment for covariates.
* Significant difference exists between males and females (p < 0.05).
Measurements
Areal BMD (aBMD; g/cm2) and ABS (cm2) were measured by Hologic 1000, 2000+, or 4500 DXA machines (Hologic, Bedford, MA, USA). The skeletal sites measured include the FN (the narrowest portion of the FN), LS 1–4, and the UD-forearm region (including UD regions of the ulna and radius). All measurements used posteroanterior projection. ABS was derived from the projection area of these regions under the X-ray beam of the DXA machines. All machines were calibrated daily, and the long-term precision was monitored with external phantoms. aBMD measures obtained from different machines were transformed into compatible ones using the formula proposed by Genant et al.(19) and the algorithm that we developed in-house and used extensively. The measurement precision, as reflected by CVs for FN, LS, and UD-forearm, were 1.9%, 0.9%, and 2.3% for aBMD and 2.1%, 1.1%, and 2.9% for ABS, respectively. Members of the same pedigree were usually measured on the same type of machine, which ensured minimum or no effect on our linkage analyses because of phenotype measurements taken by different machines. Weight (kg) and height (m) were measured at the same visit as the DXA measurement.
Genotyping
For each subject, DNA was extracted from peripheral blood using the Puregene DNA isolation kit (Gentra Systems, Minneapolis, MN, USA). A total of 4126 subjects of the entire sample were successfully genotyped for 410 microsatellite markers (including 392 markers for autosomes and 18 markers for X chromosome) from the Marshfield map data Set 14 by Marshfield Center for Medical Genetics (Marshfield, WI, USA). These markers had an average population heterozygosity of 0.75 and were spaced on an average of 8.9 cM apart. Pedcheck(20) was used to ensure that the genotype data conformed to a Mendelian inheritance pattern at all the marker loci. RELPAIR(21) was run to confirm the relatedness for each subject against the claimed relationship. In addition, we used MERLIN(22) to detect genotyping errors through unlikely recombination (e.g., double recombination) in our sample. The genotyping error rate was shown to be on a very low level of ∼0.3%.
Statistical analysis
We adopted variance component analysis method implemented in SOLAR (sequential oligogenic linkage analysis routines)(23) to conduct the bivariate whole genome linkage scans for aBMD-ABS pairs at the FN, LS, and UD-forearm. In the framework of variance component analysis, the phenotypic variance is dissected into components attributable to different resource, including major QTLs, residual genetic factors, environmental factors, covariates, etc. LOD score was computed to test the linkage by comparing the maximum likelihood of the model in which the genetic variance attributable to the major QTL under scrutiny is estimated to that in which the major QTL effect is constrained to 0. The bivariate test statistic (2 × Ln10 × LOD) follows an asymptotic mixture of 1/4χ 0 2 :1/2χ 1 2 :1/4χ 2 2.(24) Multipoint LOD scores were calculated for chromosomes 1 through 22. Two-point LOD scores were computed for the X chromosome, because SOLAR cannot handle multipoint linkage analysis for the X chromosome. Some other software, such as GENEHUNTER and MERLIN, may have options of multipoint linkage analysis for the X chromosome, but they cannot handle large pedigrees as used in this study. Because of 2 degrees of freedom involved, the LOD score in bivariate linkage analysis is not directly comparable to the classical LOD score of linkage analysis. Adopting the p value matching method described elsewhere,(15,25) the threshold for “suggestive” linkage is LOD score of 2.37 (p = 1.7 × 10−3) and 3.85 (p = 4.9 × 10−5) for “significant” linkage. We further adjusted the threshold to account for the multiple testing problem caused by joint analyses for three skeletal sites, yielding the LOD scores of 2.83 (p = 5.7 × 10−4) for “suggestive” linkage and 4.32 (p = 1.6 × 10−6) for “significant” linkage.
When bivariate linkage was found, we used a likelihood-based test(15) to differentiate pleiotropy from coincident linkage. The likelihood for the fitted model in which rhoq, a measure of shared genetic effects due to the major QTL, was compared with the likelihood of a model in which rhoq was constrained to 1 (complete pleiotropy), and the likelihood of a model in which rhoq was constrained to 0 (complete coincident linkage). Twice the difference between the likelihoods follows a 1/2χ 0 2 :1/2χ 1 2 mixture distribution under the null hypothesis of complete pleiotropy. For the test of coincident linkage, twice the difference follows a χ 1 2 distribution. When p value is less than a specific threshold, we statistically reject the corresponding null hypothesis. The test was conducted only at the location showing at least suggestive linkage in bivariate analysis. The pleiotropic test is currently not applicable for X chromosome in present version of SOLAR.
Before linkage analyses, aBMD and ABS measurements were adjusted for covariates including age, sex, age-by-sex interaction, height, and weight. A Box-Cox transformation was applied to ensure both traits followed normal distributions. Finally, data for both traits were standardized to the N(0, 1) distribution such that phenotypic distributions of both traits were transformed into comparable.
We further used six bioinformatics tools (i.e., DGP,(26) Endeavour,(27) GeneSeeker,(28) Prioritizer,(29) PROSPECTR,(30) and SUSPECTS(31)) to identify promising candidate genes in the linkage regions. These bioinformatics tools may search and/or predict candidate genes based on multiple lines of evidence, such as sequence, expression, phenotype, functional annotation, protein interactions, pathways, and literature mining. Following previously reported methods,(32,33) all genes pinpointed by GeneSeeker or DGP were considered as “suggested,” whereas only top-ranked 25 genes were considered as “suggested” for the other four methods. Genes that were suggested by at least three applications were considered as promising candidate genes.
RESULTS
The genotyping error rate was ∼0.3%. About 4.8% of the subjects do not conform to the claimed relatedness according to RELPAIR. These markers and subjects were excluded from further linkage analyses. Table 2 presents the heritability estimates for aBMD and ABS, as well as the genetic and environmental correlations between them. It was shown that genetic correlations of aBMD and ABS were substantial at the FN and LS but relatively modest at the UD-forearm, although still significant (p < 0.01).
Table 2.
Heritability and Correlations of the Studied Phenotype Pairs
| Skeletal sites |
Heritability
|
ρG (SE) | ρE (SE) | ρP | |
| aBMD | ABS | ||||
| Femoral neck | 0.61 | 0.40 | −0.31 (0.04) | 0.001 (0.05) | −0.15 |
| Lumbar spine | 0.61 | 0.69 | 0.46 (0.03) | 0.24 (0.04) | 0.38 |
| Ultradistal-forearm | 0.48 | 0.66 | −0.12 (0.04) | −0.21 (0.04) | −0.16 |
ρG, genetic correlation; ρE, environmental correlation; ρP, phenotypic correlation.
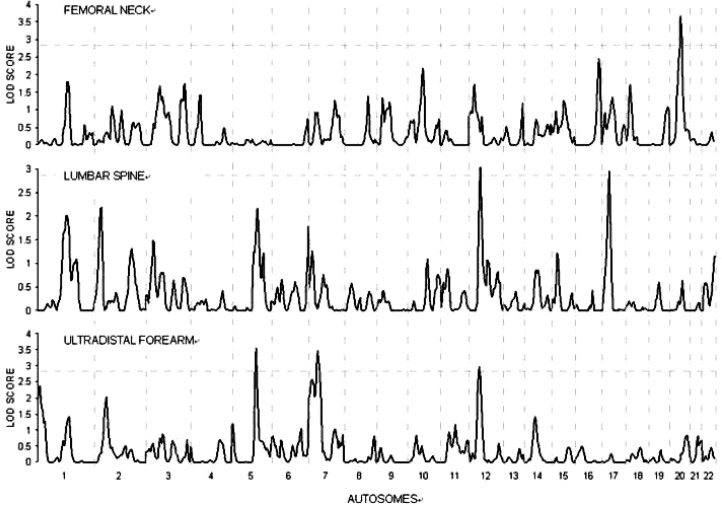
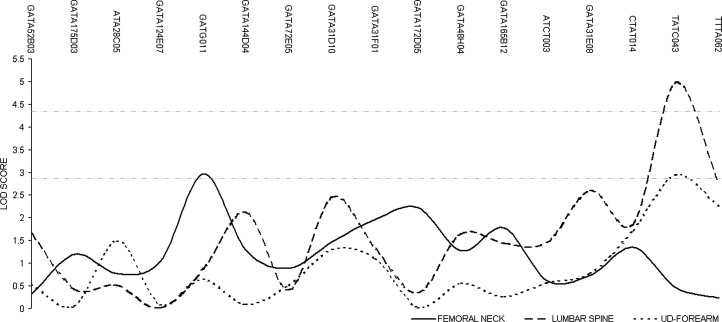
Applying LOD scores of 2.83 and 4.32 as the thresholds for “suggestive” and “significant” linkages, respectively, we identified significant linkage for LS aBMD-ABS at Xq27 (LOD = 4.89). We also detected suggestive linkages for FN aBMD-ABS at 20q11 (LOD = 3.65) and Xp11 (LOD = 2.96), for LS aBMD-ABS at 12p11 (LOD = 3.39) and 17q21 (LOD = 2.94), and for UD-forearm aBMD-ABS at 5q23 (LOD = 3.54), 7p15 (LOD = 3.45), Xq27 (LOD = 2.93), and 12p11 (LOD = 2.92). The results are plotted in Figs. 1 and 2.
FIG. 1.
Results of bivariate multipoint linkage scans on autosomes. The vertical lines are the borders of chromosomes. The horizontal dash-dotted line indicates the suggestive threshold (LOD = 2.83). The top, middle, and the bottom charts summary the results of bivariate linkage scans for aBMD-ABS at femoral neck, lumbar spine, and UD-forearm, respectively.
FIG. 2.
Results of bivariate two-point linkage scans on X chromosome. The two dash-dotted horizontal lines indicate the suggestive threshold (LOD = 2.83) and the significant threshold (LOD = 4.32), respectively.
Table 3 summarizes the results of bivariate linkage analysis and subsequent discrimination tests of pleiotropy versus coincident linkage. The probabilities of pleiotropy and coincident linkage are denoted by p 1 and p 0, respectively. Using the threshold of p < 0.10 for rejection of the corresponding null hypothesis, we detected significant pleiotropic effects at 12p11 (p 1 = 0.13, p 0 = 0.07) and 17q21 (p 1 = 0.50, p 0 = 0.02) for LS aBMD-ABS, coincident linkage at 20q11 (p 1 = 0.07, p 0 = 019) for FN aBMD-ABS, and at 7p15 (p 1 = 0.001, p 0 = 0.17) for UD-forearm aBMD-ABS. For ease of comparison of the results with classical LOD scores, we also present the equivalent LOD scores. We did not detect any significant linkage signals in sex-stratified bivariate linkage analyses.
Table 3.
Results of the Bivariate Linkage Analysis
| Skeletal sites | LOD | LODE* | p | p1† | p0† | Location | Nearest marker |
| Femoral neck | 3.65‡ | 3.11 | 7.64 E−05 | 0.07 | 0.19 | 20q11.23 (57) | GATA42A03 |
| 2.96 | 2.46 | 3.79 E−04 | — | — | Xp11.4 (58) | GATG011 | |
| Lumbar spine | 4.89§ | 4.30 | 4.25 E−06 | — | — | Xq27.3 (166) | TATC043 |
| 3.39 | 2.86 | 1.41 E−04 | 0.13 | 0.07 | 12p11.23 (39) | GATA6C01 | |
| 2.94 | 2.44 | 3.98 E−04 | 0.50 | 0.02 | 17q21.2 (64) | GATA25A04 | |
| Ultradistal-forearm | 3.54‡§ | 3.01 | 9.76 E−05 | 0.001 | 0.002 | 5q23.1 (127) | GATA62A04 |
| 3.45§ | 2.92 | 1.22 E−04 | 0.001 | 0.17 | 7p15.1 (35) | GATA41G07 | |
| 2.93§ | 2.43 | 4.08 E−04 | — | — | Xq27.3 (166) | TATC043 | |
| 2.92 | 2.42 | 4.19 E−04 | 0.13 | 0.47 | 12p11.23 (39) | GATA6C01 |
* LODE indicates the equivalent LOD scores comparable to traditional univariate ones.
† p 1 and p 0 indicate the probabilities of complete pleiotropy and complete coincident linkage, respectively. The concordant p 1 and p 0 are shown in bold italics, which means one of them is <0.1 and the other is >0.1.
‡ The locus linked to bone size in our previous univariate linkage study.(17)
§ The locus linked to BMD in our previous univariate linkage study.(16)
—, pleiotropic tests on X chromosome are not supported by the current version of SOLAR.
A total of 280, 272, 279, 500, 211, 575, and 399 genes were suggested by at least one tool for the loci 5q23, 7p15, 12p11, 17q21, 20q11, Xp11, and Xq27, respectively. For ease of presentation, we only list in Table 4 the most promising candidate genes, the genes that were suggested by at least three bioinformatics tools.
Table 4.
Most Promising Candidate Genes Suggested by Bioinformatics Tools
| Gene symbol* | Linkage signal in this study | DGP | Endeavour | GeneSeeker | Prioritizer | PROSPECTR | SUSPECTS | Total† |
| COL1A1 | LOD = 2.94 for LS at 17q21 | ● | ● | ● | ● | ● | ● | 6 |
| LBP | LOD = 3.65 for FN at 20q11 | ● | ● | ● | ● | ● | 5 | |
| CCR7 | LOD = 2.94 for LS at 17q21 | ● | ● | ● | ● | ● | 5 | |
| HSD17B4 | LOD = 3.54 for UD-forearm at 5q23 | ● | ● | ● | ● | 4 | ||
| KRT13 | LOD = 2.94 for LS at 17q21 | ● | ● | ● | ● | 4 | ||
| KRT15 | LOD = 2.94 for LS at 17q21 | ● | ● | ● | ● | 4 | ||
| KRT19 | LOD = 2.94 for LS at 17q21 | ● | ● | ● | ● | 4 | ||
| KRT10 | LOD = 2.94 for LS at 17q21 | ● | ● | ● | ● | 4 | ||
| FKBP10 | LOD = 2.94 for LS at 17q21 | ● | ● | ● | ● | 4 | ||
| ATP6V0A1 | LOD = 2.94 for LS at 17q21 | ● | ● | ● | ● | 4 | ||
| IGFBP4 | LOD = 2.94 for LS at 17q21 | ● | ● | ● | ● | 4 | ||
| KRT17 | LOD = 2.94 for LS at 17q21 | ● | ● | ● | ● | 4 | ||
| FMR1 | LOD = 2.93 for UD-forearm at Xq27 | ● | ● | ● | 3 | |||
| USP9X | LOD = 2.96 for FN at Xp11 | ● | ● | ● | 3 | |||
| SEMA6A | LOD = 3.54 for UD-forearm at 5q23 | ● | ● | ● | 3 | |||
| EPB41L1 | LOD = 3.65 for FN at 20q11 | ● | ● | ● | 3 | |||
| NNAT | LOD = 3.65 for FN at 20q11 | ● | ● | ● | 3 | |||
| CTNNBL1 | LOD = 3.65 for FN at 20q11 | ● | ● | ● | 3 | |||
| TGIF2 | LOD = 3.65 for FN at 20q11 | ● | ● | ● | 3 | |||
| RBL1 | LOD = 3.65 for FN at 20q11 | ● | ● | ● | 3 | |||
| SLA2 | LOD = 3.65 for FN at 20q11 | ● | ● | ● | 3 | |||
| NDRG3 | LOD = 3.65 for FN at 20q11 | ● | ● | ● | 3 | |||
| GHRH | LOD = 3.65 for FN 20q11 | ● | ● | ● | 3 | |||
| SCAND1 | LOD = 3.65 for FN 20q11 | ● | ● | ● | 3 | |||
| KRT16 | LOD = 2.94 for LS at 17q21 | ● | ● | ● | 3 | |||
| ACLY | LOD = 2.94 for LS at 17q21 | ● | ● | ● | 3 | |||
| KCNH4 | LOD = 2.94 for LS at 17q21 | ● | ● | ● | 3 | |||
| CNTNAP1 | LOD = 2.94 for LS at 17q21 | ● | ● | ● | 3 | |||
| HCRT | LOD = 2.94 for LS at 17q21 | ● | ● | ● | 3 | |||
| KRT14 | LOD = 2.94 for LS at 17 q21 | ● | ● | ● | 3 | |||
| KRT35 | LOD = 2.94 for LS at 17q21 | ● | ● | ● | 3 | |||
| NAGLU | LOD = 2.94 for LS at 17q21 | ● | ● | ● | 3 | |||
| ARNTL2 | LOD = 3.39 for LS at 12p11 | ● | ● | ● | 3 | |||
| MED21 | LOD = 3.39 for LS at 12p11 | ● | ● | ● | 3 |
Solid circles in this table indicate the corresponding genes were suggested by this method. All genes pinpointed by Geneseeker or DGP were considered “suggested,” whereas only the top-ranked 25 genes were considered “suggested” for the other four methods. Here we only listed the most interesting candidate genes that were suggested by at least three tools.
* Gene symbol according to HUGO Gene Nomenclature Committee.
† The times of the corresponding gene getting suggested by these six software applications.
DISCUSSION
This is the first bivariate linkage study to search for QTLs important for both aBMD and ABS at three important skeletal sites. In this study, the most significant linkage was found at Xq27 for LS aBMD-ABS (LOD = 4.89). This region also showed suggestive linkage to UD-forearm aBMD-ABS (LOD = 2.93). Linkage of Xq27 to aBMD was repeatedly observed in previous studies (e.g., for UD-forearm aBMD [LOD = 2.78,(16) LOD = 4.30(34)] and hip aBMD [LOD = 2.57]).(34) Potential candidate genes in the vicinity of this region include biglycan (BGN) and interleukin-1 receptor-associated kinase 1 (IRAK1). Bgn (homologous to human Xq27)-deficient mice was reported to exhibit an osteoporosis-like phenotype at the femora.(35) Ishida et al.(36) detected significant association of haplotypes in the IRAK1 gene with low radial aBMD in two independent populations.
Chromosome 12p11 achieved suggestive linkage for aBMD-ABS at the LS (LOD = 3.39) and UD-forearm (LOD = 2.92). The subsequent pleiotropic test showed this region may contain a QTL that has pleiotropic effects on LS aBMD-ABS. To our knowledge, this is the first study showing the importance of this region to bone. Previous studies failed to detect that linkage of either BMD or bone size to 12p11 may be partially caused by the limited power of univariate linkage analysis. At 12p11, low-density lipoprotein receptor-related protein 6 (LRP6) is a candidate gene that has been associated with vertebral body size and fracture risk.(37) LRP6 plays a broad role in the transduction of Wnt signals, which actively involve in osteoblast and chondrocyte differentiation.(38) In ringelschwanz mutant mice, LRP6 was shown to be necessary for proper osteogenesis.(39)
In this study, 20q11 achieved a LOD score of 3.65 for FN aBMD-ABS. Our earlier univariate linkage scan also detected the linkage of hip ABS to this region (LOD = 2.18).(17) Consistently, Beamer et al.(40) found femoral volumetric BMD (vBMD; LOD = 3.14) of mice was linked to a homologous region of human chromosome 20q11. In this region, growth differentiation factor 5 (GDF5), also known as cartilage-derived morphogenetic protein 1 (CDMP1), is an important candidate gene. The protein product of GDF5 is a member of the bone morphogenetic protein (BMP) family, which plays an important role in skeletal development and metabolism.(41)
Suggestive linkage at 17q21 was observed for LS aBMD-ABS (LOD = 2.94), with potential pleiotropic effects. Our results are in accordance with previously reported linkages at 17q21 to aBMD(42,43) and femur head width.(44) As a strong candidate gene in this region, collagen, type I, α 1 (COL1A1) was associated with bone-related phenotypes in multiple studies.(12) In particular, our previous studies showed that the COL1A1 gene is important for both LS aBMD (p = 0.027)(45) and UD-forearm ABS,(46) partially in agreement with the observed bivariate linkage of 17q21 to LS aBMD-ABS in this study. In addition, chondroadherin (CHAD), homeobox B cluster (HOXB@), and sclerosteosis (SOST) are among the promising candidate genes for this region. CHAD has been reported to promote attachment of osteoblastic cells to solid-state substrates and to bind chondrocytes through their integrin α2β1 receptors.(47) The importance of HOXB (homeobox B cluster) for regulation of skeletal patterning has been shown in numerous animal systems.(48) The SOST gene polymorphisms were associated with aBMD in elderly whites.(49)
We found suggestive linkage at 5q23 for UD-forearm aBMD-ABS. Interestingly, 5q23 was linked to UD-forearm aBMD (LOD = 3.39)(16) and LS ABS(17) (LOD = 1.78) in our previous univariate linkage studies using the same sample, further supporting the existence of a QTL with dual effects on aBMD and ABS in this region. Interleukin 4 (IL4) is a potential candidate gene for this region, which was associated with human bone resorption and aBMD.(50) Lysyl oxidase (LOX) is another interesting gene in this region. Hong et al.(51) showed that regulation of lysyl oxidase activity plays a key role in the control of collagen deposition by osteoblast cultures.
The importance of chromosome 7p15, linked to UD-forearm aBMD-ABS in this study, was also suggested in an earlier univariate linkage scan for hip ABS (LOD = 2.53)(52) and cortical thickness at the FN (LOD = 1.86).(53) Potential candidate genes at 7p15 include interleukin 6 (IL6),(54) neuropeptide Y (NPY),(55) and homeobox A cluster (HOXA@),(56) a gene cluster homologous to foregoing HOXB@. HOX genes are important transcriptional regulator of embryonic development in development of skeletal structure on the anterior–posterior axis.(57) In this study, we concurrently detected linkage of UD-forearm aBMD-ABS to HOXA@ locus (7p15) and linkage of LS aBMD-ABS to HOXB@ locus (17q21). This is consistent with the observation that HOX genes express and function in a position-specific manner.(58) We also obtained a LOD score of 2.96 at Xp11 for FN aBMD-ABS. Bone morphogenetic protein 15 (BMP15) is a promising candidate gene in this region. In Table 5, we provide a brief summary of previous linkage studies for bone phenotypes at the loci detected in this study.
Table 5.
Brief Summary of Previous Linkage Studies for Bone Phenotypes at the Loci Detected in This Study
|
Current results for aBMD-ABS
|
Previous linkage evidence
|
||||
| Region | LODE* | Sites | Phenotypes | LOD or p values | References |
| 20q11 | 3.11 | FN | FN vBMD in mice | LOD = 3.14 | (40) |
| Peak whole body aBMD in mice | p < 0.0020 | (43) | |||
| UD ABS | LOD = 2.24 | (52) | |||
| Whole body vBMD in mice | LOD = 6.6 | (61) | |||
| Xp11 | 2.46 | FN | Hip aBMD | LOD = 2.15 | (34) |
| Cross-sectional area at FN | LOD = 2.23 | (53) | |||
| Cortical thickness at FN | LOD = 2.38 | ||||
| Cortical thickness at FN | LOD = 3.45 | (62) | |||
| Xq27 | 4.30 | LS | UD aBMD | LOD = 2.78 | (16) |
| 2.43 | UD | UD aBMD | LOD = 4.30 | (34) | |
| Hip aBMD | LOD = 2.57 | ||||
| 17q21 | 2.44 | LS | LS aBMD in mice | p < 0.0001 | (42) |
| Femur head width | LOD = 3.6 | (44) | |||
| Peak bone mass in mice | LOD = 10.8 | (63) | |||
| 5q23 | 3.01 | UD | UD aBMD | LOD = 3.39 | (16) |
| UD aBMD in females | LOD = 2.82 | ||||
| 7p15 | 2.92 | UD | Hip ABS | LOD = 2.53 | (52) |
| Cortical thickness at FN in females | LOD = 1.86 | (53) | |||
| LS aBMD | LOD = 2.15 | (64) | |||
* LODE indicates the equivalent LOD scores comparable to traditional univariate ones.
FN, femoral neck; LS, lumbar spine; UD, ultradistal forearm; aBMD, areal BMD; vBMD, volumetric BMD.
This study has several strengths. First, compared with traditional univariate analyses, bivariate linkage analyses use more information and considerably improve the power to detect QTLs with modest effects on correlated traits.(14,59) This power advantage in this study is reflected by (1) our previous univariate whole genome linkage scans for aBMD(16) and ABS(17) failed to disclose the common QTLs important to both traits and (2) the higher LOD scores achieved at 5q23, 7p15, and Xq27 for UD-forearm aBMD-ABS in this study compared with those achieved in univariate linkage analysis for UD-forearm aBMD in the same sample.(16) Second, bivariate linkage analysis can improve precision of parameter estimation, including QTL position and effect size,(59,60) which may greatly facilitate subsequent fine mapping and functional studies. Third, genes are sometimes tightly clustered. When a specific genomic region is shown to harbor QTLs affecting multiple phenotypes, it is still important to differentiate pleiotropic effects (i.e., a single locus influencing both traits) from coincident linkage (i.e., separate tightly clustered loci each influencing a single trait). The bivariate linkage analysis adopted in this study is able to fulfill this purpose in a high power.(15) Fourth, given the close biological correlation between BMD and ABS, knowledge about genes with dual effects on BMD and ABS may provide additional clues to our understanding on bone metabolism. Linkages observed in univariate linkage scans but not in this study imply that these regions may harbor QTLs affecting either aBMD or ABS, but not both.
In practice, promising candidate genes can usually be selected from the linkage regions. However, precise and reliable inference of candidate genes remains a challenge in the field. Recent advancement in using bioinformatics tools to prioritize causative genes for diabetes and obesity(32,33) exemplified the usefulness of computational biology methods in gene discovery. In this study, we adopted six bioinformatics tools to search or predict the candidate genes in the linkage regions. These genes deserve further studies to testify their potential roles in determination of BMD and ABS.
In summary, this study, for the first time, identified several genomic regions that may contain QTLs influencing both aBMD and ABS. Further follow-up studies for these regions are needed to eventually pinpoint the genes contributing to bone strength and risk of osteoporotic fractures.
ACKNOWLEDGMENTS
Investigators of this work were partially supported by grants from the NIH (R01 AR050496, K01 AR02170-01, R01 AR45349-01, R01 GM60402-01A1, R21 AG027110-01A1, and R01 AG026564-01A2) and the Dickson/Missouri endowment fund. The study benefited from grants from the National Science Foundation of China (30570875), Huo YingDong Education Foundation, Xi’an Jiaotong University, and the Ministry of Education of China. XGL was partially supported by Doctoral Foundation of Xi’an Jiaotong University (DFXJTU2004-11). The authors thank Feiyan Deng for helpful comments and revision on the final manuscript.
Footnotes
The authors state that they have no conflicts of interest.
REFERENCES
- 1.Burge R, Dawson-Hughes B, Solomon DH, Wong JB, King A, Tosteson A. Incidence and economic burden of osteoporosis-related fractures in the United States, 2005-2025. J Bone Miner Res. 2007;22:465–475. doi: 10.1359/jbmr.061113. [DOI] [PubMed] [Google Scholar]
- 2.Cummings SR, Kelsey JL, Nevitt MC, O’Dowd KJ. Epidemiology of osteoporosis and osteoporotic fractures. Epidemiol Rev. 1985;7:178–208. doi: 10.1093/oxfordjournals.epirev.a036281. [DOI] [PubMed] [Google Scholar]
- 3.Currey JD. The mechanical adaptations of bones. In: Currey JD, editor. The Mechanical Properties of Materials and the Structure of Bone. Princeton, NJ, USA: Princeton University Press; 1984. pp. 3–37. [Google Scholar]
- 4.Seeman E. Invited review: Pathogenesis of osteoporosis. J Appl Physiol. 2003;95:2142–2151. doi: 10.1152/japplphysiol.00564.2003. [DOI] [PubMed] [Google Scholar]
- 5.Seeman E, Duan Y, Fong C, Edmonds J. Fracture site-specific deficits in bone size and volumetric density in men with spine or hip fractures. J Bone Miner Res. 2001;16:120–127. doi: 10.1359/jbmr.2001.16.1.120. [DOI] [PubMed] [Google Scholar]
- 6.Duan Y, Parfitt A, Seeman E. Vertebral bone mass, size, and volumetric density in women with spinal fractures. J Bone Miner Res. 1999;14:1796–1802. doi: 10.1359/jbmr.1999.14.10.1796. [DOI] [PubMed] [Google Scholar]
- 7.Vega E, Ghiringhelli G, Mautalen C, Rey Valzacchi G, Scaglia H, Zylberstein C. Bone mineral density and bone size in men with primary osteoporosis and vertebral fractures. Calcif Tissue Int. 1998;62:465–469. doi: 10.1007/s002239900462. [DOI] [PubMed] [Google Scholar]
- 8.Deng HW, Xu FH, Davies KM, Heaney R, Recker RR. Differences in bone mineral density, bone mineral content, and bone areal size in fracturing and non-fracturing women, and their interrelationships at the spine and hip. J Bone Miner Metab. 2002;20:358–366. doi: 10.1007/s007740200052. [DOI] [PubMed] [Google Scholar]
- 9.Beck TJ, Oreskovic TL, Stone KL, Ruff CB, Ensrud K, Nevitt MC, Genant HK, Cummings SR. Structural adaptation to changing skeletal load in the progression toward hip fragility: The study of osteoporotic fractures. J Bone Miner Res. 2001;16:1108–1119. doi: 10.1359/jbmr.2001.16.6.1108. [DOI] [PubMed] [Google Scholar]
- 10.Lorentzon M, Mellstrom D, Ohlsson C. Association of amount of physical activity with cortical bone size and trabecular volumetric BMD in young adult men: The GOOD study. J Bone Miner Res. 2005;20:1936–1943. doi: 10.1359/JBMR.050709. [DOI] [PubMed] [Google Scholar]
- 11.Wang YB, Lei SF, Dvornyk V, Sun X, Jiang DK, Li MX, Deng HW. The genetic, environmental and phenotypic correlations of bone phenotypes at the spine and hip in Chinese. Ann Hum Biol. 2006;33:500–509. doi: 10.1080/03014460600814135. [DOI] [PubMed] [Google Scholar]
- 12.Liu YJ, Shen H, Xiao P, Xiong DH, Li LH, Recker RR, Deng HW. Molecular genetic studies of gene identification for osteoporosis: A 2004 update. J Bone Miner Res. 2006;21:1511–1535. doi: 10.1359/JBMR.051002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marlow AJ, Fisher SE, Francks C, MacPhie IL, Cherny SS, Richardson AJ, Talcott JB, Stein JF, Monaco AP, Cardon LR. Use of multivariate linkage analysis for dissection of a complex cognitive trait. Am J Hum Genet. 2003;72:561–570. doi: 10.1086/368201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Allison DB, Thiel B, St Jean P, Elston RC, Infante MC, Schork NJ. Multiple phenotype modeling in gene-mapping studies of quantitative traits: Power advantages. Am J Hum Genet. 1998;63:1190–1201. doi: 10.1086/302038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Almasy L, Dyer TD, Blangero J. Bivariate quantitative trait linkage analysis: Pleiotropy versus co-incident linkages. Genet Epidemiol. 1997;14:953–958. doi: 10.1002/(SICI)1098-2272(1997)14:6<953::AID-GEPI65>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 16.Xiao P, Shen H, Guo YF, Xiong DH, Liu YZ, Liu YJ, Zhao LJ, Long JR, Guo Y, Recker RR, Deng HW. Genomic regions identified for BMD in a large sample including epistatic interactions and gender-specific effects. J Bone Miner Res. 2006;21:1536–1544. doi: 10.1359/jbmr.060717. [DOI] [PubMed] [Google Scholar]
- 17.Shen H, Long JR, Xiong DH, Guo YF, Xiao P, Liu YZ, Zhao LJ, Liu YJ, Deng HY, Li JL, Recker RR, Deng HW. A genomewide scan for quantitative trait loci underlying areal bone size variation in 451 Caucasian families. J Med Genet. 2006;43:873–880. doi: 10.1136/jmg.2006.041251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deng HW, Deng H, Liu YJ, Liu YZ, Xu FH, Shen H, Conway T, Li JL, Huang QY, Davies KM, Recker RR. A genomewide linkage scan for quantitative-trait loci for obesity phenotypes. Am J Hum Genet. 2002;70:1138–1151. doi: 10.1086/339934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Genant HK, Grampp S, Gluer CC, Faulkner KG, Jergas M, Engelke K, Hagiwara S, Van Kuijk C. Universal standardization for dual x-ray absorptiometry: Patient and phantom cross-calibration results. J Bone Miner Res. 1994;9:1503–1514. doi: 10.1002/jbmr.5650091002. [DOI] [PubMed] [Google Scholar]
- 20.O’Connell JR, Weeks DE. PedCheck: A program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet. 1998;63:259–266. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Epstein MP, Duren WL, Boehnke M. Improved inference of relationship for pairs of individuals. Am J Hum Genet. 2000;67:1219–1231. doi: 10.1016/s0002-9297(07)62952-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin–rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 23.Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62:1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Self SGLK-Y. Asymptotic properties of maximum likelihood estimators and likelihood ratio tests under nonstandard conditions. J Am Stat Assoc. 1987;82:605–610. [Google Scholar]
- 25.Turner ST, Kardia SL, Boerwinkle E, Andrade Md M. Multivariate linkage analysis of blood pressure and body mass index. Genet Epidemiol. 2004;27:64–73. doi: 10.1002/gepi.20002. [DOI] [PubMed] [Google Scholar]
- 26.Lopez-Bigas N, Ouzounis CA. Genome-wide identification of genes likely to be involved in human genetic disease. Nucleic Acids Res. 2004;32:3108–3114. doi: 10.1093/nar/gkh605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aerts S, Lambrechts D, Maity S, Van Loo P, Coessens B, De Smet F, Tranchevent LC, De Moor B, Marynen P, Hassan B, Carmeliet P, Moreau Y. Gene prioritization through genomic data fusion. Nat Biotechnol. 2006;24:537–544. doi: 10.1038/nbt1203. [DOI] [PubMed] [Google Scholar]
- 28.van Driel MA, Cuelenaere K, Kemmeren PP, Leunissen JA, Brunner HG, Vriend G. GeneSeeker: Extraction and integration of human disease-related information from web-based genetic databases. Nucleic Acids Res. 2005;33:W758–W761. doi: 10.1093/nar/gki435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Franke L, Bakel H, Fokkens L, de Jong ED, Egmont-Petersen M, Wijmenga C. Reconstruction of a functional human gene network, with an application for prioritizing positional candidate genes. Am J Hum Genet. 2006;78:1011–1025. doi: 10.1086/504300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Adie EA, Adams RR, Evans KL, Porteous DJ, Pickard BS. Speeding disease gene discovery by sequence based candidate prioritization. BMC Bioinformatics. 2005;6:55. doi: 10.1186/1471-2105-6-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adie EA, Adams RR, Evans KL, Porteous DJ, Pickard BS. SUSPECTS: Enabling fast and effective prioritization of positional candidates. Bioinformatics. 2006;22:773–774. doi: 10.1093/bioinformatics/btk031. [DOI] [PubMed] [Google Scholar]
- 32.Tiffin N, Adie E, Turner F, Brunner HG, van Driel MA, Oti M, Lopez-Bigas N, Ouzounis C, Perez-Iratxeta C, Andrade-Navarro MA, Adeyemo A, Patti ME, Semple CA, Hide W. Computational disease gene identification: A concert of methods prioritizes type 2 diabetes and obesity candidate genes. Nucleic Acids Res. 2006;34:3067–3081. doi: 10.1093/nar/gkl381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Elbers CC, Onland-Moret NC, Franke L, Niehoff AG, van der Schouw YT, Wijmenga C. A strategy to search for common obesity and type 2 diabetes genes. Trends Endocrinol Metab. 2007;18:19–26. doi: 10.1016/j.tem.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 34.Shen H, Zhang YY, Long JR, Xu FH, Liu YZ, Xiao P, Zhao LJ, Xiong DH, Liu YJ, Dvornyk V, Rocha-Sanchez S, Liu PY, Li JL, Conway T, Davies KM, Recker RR, Deng HW. A genome-wide linkage scan for bone mineral density in an extended sample: Evidence for linkage on 11q23 and Xq27. J Med Genet. 2004;41:743–751. doi: 10.1136/jmg.2004.020396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu T, Bianco P, Fisher LW, Longenecker G, Smith E, Goldstein S, Bonadio J, Boskey A, Heegaard AM, Sommer B, Satomura K, Dominguez P, Zhao C, Kulkarni AB, Robey PG, Young MF. Targeted disruption of the biglycan gene leads to an osteoporosis-like phenotype in mice. Nat Genet. 1998;20:78–82. doi: 10.1038/1746. [DOI] [PubMed] [Google Scholar]
- 36.Ishida R, Emi M, Ezura Y, Iwasaki H, Yoshida H, Suzuki T, Hosoi T, Inoue S, Shiraki M, Ito H, Orimo H. Association of a haplotype (196Phe/532Ser) in the interleukin-1-receptor-associated kinase (IRAK1) gene with low radial bone mineral density in two independent populations. J Bone Miner Res. 2003;18:419–423. doi: 10.1359/jbmr.2003.18.3.419. [DOI] [PubMed] [Google Scholar]
- 37.van Meurs JB, Rivadeneira F, Jhamai M, Hugens W, Hofman A, van Leeuwen JP, Pols HA, Uitterlinden AG. Common genetic variation of the low-density lipoprotein receptor-related protein 5 and 6 genes determines fracture risk in elderly white men. J Bone Miner Res. 2006;21:141–150. doi: 10.1359/JBMR.050904. [DOI] [PubMed] [Google Scholar]
- 38.Johnson ML, Harnish K, Nusse R, Van Hul W. LRP5 and Wnt signaling: A union made for bone. J Bone Miner Res. 2004;19:1749–1757. doi: 10.1359/JBMR.040816. [DOI] [PubMed] [Google Scholar]
- 39.Kokubu C, Heinzmann U, Kokubu T, Sakai N, Kubota T, Kawai M, Wahl MB, Galceran J, Grosschedl R, Ozono K, Imai K. Skeletal defects in ringelschwanz mutant mice reveal that Lrp6 is required for proper somitogenesis and osteogenesis. Development. 2004;131:5469–5480. doi: 10.1242/dev.01405. [DOI] [PubMed] [Google Scholar]
- 40.Beamer WG, Shultz KL, Donahue LR, Churchill GA, Sen S, Wergedal JR, Baylink DJ, Rosen CJ. Quantitative trait loci for femoral and lumbar vertebral bone mineral density in C57BL/6J and C3H/HeJ inbred strains of mice. J Bone Miner Res. 2001;16:1195–1206. doi: 10.1359/jbmr.2001.16.7.1195. [DOI] [PubMed] [Google Scholar]
- 41.Erlacher L, McCartney J, Piek E, ten Dijke P, Yanagishita M, Oppermann H, Luyten FP. Cartilage-derived morphogenetic proteins and osteogenic protein-1 differentially regulate osteogenesis. J Bone Miner Res. 1998;13:383–392. doi: 10.1359/jbmr.1998.13.3.383. [DOI] [PubMed] [Google Scholar]
- 42.Benes H, Weinstein RS, Zheng W, Thaden JJ, Jilka RL, Manolagas SC, Shmookler Reis RJ. Chromosomal mapping of osteopenia-associated quantitative trait loci using closely related mouse strains. J Bone Miner Res. 2000;15:626–633. doi: 10.1359/jbmr.2000.15.4.626. [DOI] [PubMed] [Google Scholar]
- 43.Klein RF, Mitchell SR, Phillips TJ, Belknap JK, Orwoll ES. Quantitative trait loci affecting peak bone mineral density in mice. J Bone Miner Res. 1998;13:1648–1656. doi: 10.1359/jbmr.1998.13.11.1648. [DOI] [PubMed] [Google Scholar]
- 44.Koller DL, Liu G, Econs MJ, Hui SL, Morin PA, Joslyn G, Rodriguez LA, Conneally PM, Christian JC, Johnston CC, Jr, Foroud T, Peacock M. Genome screen for quantitative trait loci underlying normal variation in femoral structure. J Bone Miner Res. 2001;16:985–991. doi: 10.1359/jbmr.2001.16.6.985. [DOI] [PubMed] [Google Scholar]
- 45.Long JR, Liu PY, Lu Y, Dvornyk V, Xiong DH, Zhao LJ, Deng HW. Tests of linkage and/or association of TGF-beta1 and COL1A1 genes with bone mass. Osteoporos Int. 2005;16:86–92. doi: 10.1007/s00198-004-1650-1. [DOI] [PubMed] [Google Scholar]
- 46.Long JR, Liu PY, Lu Y, Xiong DH, Zhao LJ, Zhang YY, Elze L, Recker RR, Deng HW. Association between COL1A1 gene polymorphisms and bone size in Caucasians. Eur J Hum Genet. 2004;12:383–388. doi: 10.1038/sj.ejhg.5201152. [DOI] [PubMed] [Google Scholar]
- 47.Camper L, Heinegard D, Lundgren-Akerlund E. Integrin alpha2beta1 is a receptor for the cartilage matrix protein chondroadherin. J Cell Biol. 1997;138:1159–1167. doi: 10.1083/jcb.138.5.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gehring WJ. New Haven, CT, USA: Yale University Press; 1998. Master Control Genes in Development and Evolution: The Homeobox Story. [Google Scholar]
- 49.Uitterlinden AG, Arp PP, Paeper BW, Charmley P, Proll S, Rivadeneira F, Fang Y, van Meurs JB, Britschgi TB, Latham JA, Schatzman RC, Pols HA, Brunkow ME. Polymorphisms in the sclerosteosis/van Buchem disease gene (SOST) region are associated with bone-mineral density in elderly whites. Am J Hum Genet. 2004;75:1032–1045. doi: 10.1086/426458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saidenberg-Kermanac’h N, Bessis N, Lemeiter D, de Vernejoul MC, Boissier MC, Cohen-Solal M. Interleukin-4 cellular gene therapy and osteoprotegerin decrease inflammation-associated bone resorption in collagen-induced arthritis. J Clin Immunol. 2004;24:370–378. doi: 10.1023/B:JOCI.0000029116.12371.bf. [DOI] [PubMed] [Google Scholar]
- 51.Hong HH, Pischon N, Santana RB, Palamakumbura AH, Chase HB, Gantz D, Guo Y, Uzel MI, Ma D, Trackman PC. A role for lysyl oxidase regulation in the control of normal collagen deposition in differentiating osteoblast cultures. J Cell Physiol. 2004;200:53–62. doi: 10.1002/jcp.10476. [DOI] [PubMed] [Google Scholar]
- 52.Huang QY, Xu FH, Shen H, Deng HY, Conway T, Liu YJ, Liu YZ, Li JL, Li MX, Davies KM, Recker RR, Deng HW. Genome scan for QTLs underlying bone size variation at 10 refined skeletal sites: Genetic heterogeneity and the significance of phenotype refinement. Physiol Genomics. 2004;17:326–331. doi: 10.1152/physiolgenomics.00161.2002. [DOI] [PubMed] [Google Scholar]
- 53.Xiong DH, Shen H, Xiao P, Guo YF, Long JR, Zhao LJ, Liu YZ, Deng HY, Li JL, Recker RR, Deng HW. Genome-wide scan identified QTLs underlying femoral neck cross-sectional geometry that are novel studied risk factors of osteoporosis. J Bone Miner Res. 2006;21:424–437. doi: 10.1359/JBMR.051202. [DOI] [PubMed] [Google Scholar]
- 54.Chung HW, Seo JS, Hur SE, Kim HL, Kim JY, Jung JH, Kim LH, Park BL, Shin HD. Association of interleukin-6 promoter variant with bone mineral density in pre-menopausal women. J Hum Genet. 2003;48:243–248. doi: 10.1007/s10038-003-0020-8. [DOI] [PubMed] [Google Scholar]
- 55.Heikkinen AM, Niskanen LK, Salmi JA, Koulu M, Pesonen U, Uusitupa MI, Komulainen MH, Tuppurainen MT, Kroger H, Jurvelin J, Saarikoski S. Leucine7 to proline7 polymorphism in prepro-NPY gene and femoral neck bone mineral density in postmenopausal women. Bone. 2004;35:589–594. doi: 10.1016/j.bone.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 56.Hassan MQ, Tare R, Lee SH, Mandeville M, Weiner B, Montecino M, van Wijnen AJ, Stein JL, Stein GS, Lian JB. HOXA10 controls osteoblastogenesis by directly activating bone regulatory and phenotypic genes. Mol Cell Biol. 2007;27:3337–3352. doi: 10.1128/MCB.01544-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wellik DM, Capecchi MR. Hox10 and Hox11 genes are required to globally pattern the mammalian skeleton. Science. 2003;301:363–367. doi: 10.1126/science.1085672. [DOI] [PubMed] [Google Scholar]
- 58.Favier B, Dolle P. Developmental functions of mammalian Hox genes. Mol Hum Reprod. 1997;3:115–131. doi: 10.1093/molehr/3.2.115. [DOI] [PubMed] [Google Scholar]
- 59.Williams JT, Van Eerdewegh P, Almasy L, Blangero J. Joint multipoint linkage analysis of multivariate qualitative and quantitative traits. I. Likelihood formulation and simulation results. Am J Hum Genet. 1999;65:1134–1147. doi: 10.1086/302570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jiang C, Zeng ZB. Multiple trait analysis of genetic mapping for quantitative trait loci. Genetics. 1995;140:1111–1127. doi: 10.1093/genetics/140.3.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Masinde GL, Li X, Gu W, Wergedal J, Mohan S, Baylink DJ. Quantitative trait loci for bone density in mice: The genes determining total skeletal density and femur density show little overlap in F2 mice. Calcif Tissue Int. 2002;71:421–428. doi: 10.1007/s00223-001-1113-z. [DOI] [PubMed] [Google Scholar]
- 62.Shen H, Long JR, Xiong DH, Liu YJ, Liu YZ, Xiao P, Zhao LJ, Dvornyk V, Zhang YY, Rocha-Sanchez S, Liu PY, Li JL, Deng HW. Mapping quantitative trait loci for cross-sectional geometry at the femoral neck. J Bone Miner Res. 2005;20:1973–1982. doi: 10.1359/JBMR.050715. [DOI] [PubMed] [Google Scholar]
- 63.Shimizu M, Higuchi K, Bennett B, Xia C, Tsuboyama T, Kasai S, Chiba T, Fujisawa H, Kogishi K, Kitado H, Kimoto M, Takeda N, Matsushita M, Okumura H, Serikawa T, Nakamura T, Johnson TE, Hosokawa M. Identification of peak bone mass QTL in a spontaneously osteoporotic mouse strain. Mamm Genome. 1999;10:81–87. doi: 10.1007/s003359900949. [DOI] [PubMed] [Google Scholar]
- 64.Devoto M, Spotila LD, Stabley DL, Wharton GN, Rydbeck H, Korkko J, Kosich R, Prockop D, Tenenhouse A, Sol-Church K. Univariate and bivariate variance component linkage analysis of a whole-genome scan for loci contributing to bone mineral density. Eur J Hum Genet. 2005;13:781–788. doi: 10.1038/sj.ejhg.5201411. [DOI] [PubMed] [Google Scholar]