Abstract
Autoimmune polyendocrinopathy–candidiasis–ectodermal dystrophy (APECED) is a monogenic autoimmune disease caused by mutations in the autoimmune regulator (AIRE) gene. AIRE functions as a transcriptional regulator, and it has a central role in the development of immunological tolerance. AIRE regulates the expression of ectopic antigens in epithelial cells of the thymic medulla and has been shown to participate in the development of peripheral tolerance. However, the mechanism of action of AIRE has remained elusive. To further investigate the role of AIRE in host immune functions, we studied the properties and transcript profiles in in vitro monocyte-differentiated dendritic cells (moDCs) obtained from APECED patients and healthy controls. AIRE-deficient monocytes showed typical DC morphology and expressed DC marker proteins cluster of differentiation 86 and human leukocyte antigen class II. APECED patient-derived moDCs were functionally impaired: the transcriptional response of cytokine genes to pathogens was drastically reduced. Interestingly, some changes were observable already at the immature DC stage. Pathway analyses of transcript profiles revealed that the expression of the components of the host cell signaling pathways involved in cell–cell signalling, innate immune responses, and cytokine activity were reduced in APECED moDCs. Our observations support a role for AIRE in peripheral tolerance and are the first ones to show that AIRE has a critical role in DC responses to microbial stimuli in humans.
Keywords: AIRE, APS1, Dendritic cell, Transcript profile
Introduction
Autoimmune polyendocrinopathy–candidiasis–ectodermal dystrophy (APECED) is a monogenic disease characterized mainly by a triad of symptoms including hypoparathyroidism, Addison's disease, and chronic mucocutaneous candidiasis. Additionally, a number of other symptoms and target organs may be involved [1]. APECED is also called autoimmune polyglandular syndrome type I (APS1). APECED/APS1 is caused by different types of inactivating mutations in the autoimmune regulator (AIRE) gene [2].
AIRE protein is mainly expressed in thymic medullary epithelial cells (MECs), which have a pivotal role in the development of central tolerance. AIRE is believed to be involved in the expression of self-antigens to developing T cells [3-5]. However, AIRE is also restrictively expressed in the peripheral monocyte/dendritic cell lineage [6-9]. The expression of AIRE has been shown to be increased during the differentiation process of human monocyte to dendritic cells (moDCs) [10]. Moreover, recent reports propose that AIRE is required for normal development and function of dendritic cells [10, 11]. Furthermore, it has been shown that the tolerogenic activity of AIRE not only takes place centrally, i.e., in the thymus, but that AIRE is also involved in the regulation of peripheral tolerance, at the level of T cell–peripheral DC interaction [11]. This correlates with the findings that regulatory T cells in APECED patients showed differences from their normal counterparts [12]. The molecular mechanism of AIRE-dependent regulation in DCs has been suggested to be different from that seen in medullary epithelial cells [8]. However, the functions of AIRE in DCs are presently not known. The high prevalence of chronic mucocutaneous Candida albicans infections (CMC) in the APECED patients is a potential sign of compromised peripheral tolerance and/or innate immunity linked with the expression of defective AIRE protein. Curiously, CMC does not occur in Aire-deficient mice. At present, the molecular basis of the increased susceptibility for CMC in APECED patients is not known.
Dendritic cells exist in two states: immature DCs (iDC) and mature DCs (mDC). The critical step in regulating tolerance and immunity resides in the maturation of iDC to mDC. Normally, when no “danger signals” are present, DCs remain immature and induce immunological tolerance, i.e., T cells to become anergic, apoptotic, or regulatory T cells (Tregs). However, upon stimulation with microbes, DCs induce immunity by priming T cells and by stimulating effector T cell responses [13]. If iDC do not function, T cells do not get silenced or induced to become Tregs and, thus, tolerance to “self” is disrupted. If mDCs are impaired, proper effector T cell responses are not generated and immunity will not be activated.
Dendritic cells thus play an important role as an interphase between innate and adaptive immune responses. Upon contact with microbes or their genetic material, DCs undergo a maturation process characterized by upregulation of expression of cell surface adhesion molecules and major histocompatibility complex (MHC) class II. Activated DCs migrate to local lymph nodes, produce cytokines, and present antigens to naïve T cells to initiate adaptive immune responses. As a whole, DCs regulate the magnitude and quality of the immune response. At present, the specific role of DCs in coordinating central and peripheral tolerance is under active investigation. It is becoming increasingly evident that a failure in DC functions maintaining tolerance can lead to autoimmune and/or inflammatory diseases [14, 15].
In order to analyze the role of AIRE in DC biology, we compared the molecular and functional properties of monocyte-derived DCs (moDC) generated from APECED patients carrying the most common human AIRE mutation (R257X) to those from healthy controls. Our results indicate that these APECED moDCs show low basal cytokines expression levels and an impaired ability to mature in response to microbial stimuli. Genome-wide transcript profiles of APECED moDCs revealed anomalies in critical immunological pathways under both unstimulated and C. albicans-stimulated conditions. Our data indicate that AIRE has an important role in DC functions in humans with potential implications for a role in the maintenance of peripheral tolerance regulated by dendritic cells.
Materials and methods
Patients and samples
The diagnostic criteria for APECED was the presence of at least two of the following symptoms: hypoparathyroidism, primary adrenocortical failure, and chronic mucocutaneous candidiasis [16]. Patients with APECED were systematically screened for mutations in the coding region of AIRE as described in Björses et al. [17]. This study included six female APECED patients between the ages of 26 and 51 years old and all patients carried the homozygote FinnMajor mutation (R257X) in the AIRE gene. Genomic DNA was extracted from 10–20-ml blood samples, according to standard procedures [18]. As controls, we used buffy coats from voluntary blood donors obtained from the Finnish Red Cross Transfusion Service (Helsinki, Finland). The controls were sex- and age-matched with the APECED patients. All of the samples were drawn in accordance with the Helsinki Declaration, and the study was approved by the Helsinki University Hospital Ethics Committee and the Ethics Committee of Red Cross Transfusion Service.
Cell culture
Peripheral blood mononuclear cells (PBMC) were isolated from 70–100 ml of ethylenediaminetetraacetic acid blood over Ficoll–Paque gradient (Amersham Biosciences, Chalfont, UK). PBMCs were plated at a cell density of 10×106 cells/ml onto six-well, 24-well, or 96-well plates. Monocytes were allowed to adhere for 2 h at 37°C, after which nonadherent cells were removed by washing with phosphate-buffered saline. Monocytes were differentiated into moDCs in RPMI supplemented with 2 mM L-glutamine, 20 mM HEPES, 10% fetal calf serum, and containing 20 ng/ml interleukin (IL)-4 (R&D systems) and 10 ng/ml granulocyte–macrophage colony-stimulating factor (BioSource International), as described [19]. Fresh medium was added every 2 days. Plasmacytoid and myeloid DCs were isolated directly from PBMC with BDCA-4- or BDCA-1-conjugated magnetic beads, respectively (Miltenyi Biotec, Gladbach, Germany) using the method described previously [20].
moDC stimulations and infections
In vitro differentiated moDCs were infected with Sendai virus (Cantell strain, multiplicity of infection (MOI) of five) as previously described [21] or stimulated with live C. albicans (MOI of one) or Escherichia coli lipopolysaccharide (purified LPS from E. coli, HB101, Sigma, St. Louis, MO, USA) at a concentration of 100 ng/ml. Cells for isolation of total cellular RNA were collected at 24 h after stimulation or infection.
Fluorescence-activated cell sorting
Paraformaldehyde-fixed DCs were stained with flourescein isothiocyanate-conjugated anti-cluster of differentiation (CD) 86, anti-MHC class II-DR, isotype controls (all from Caltag Laboratories, Burlingame, CA, USA), anti-CD1a, or DC-specific ICAM-grabbing nonintegrin (DC-SIGN; both from BD Biosciences, San Jose, CA, USA) antibodies for 30 min at +4°C. After staining, the cells were washed and analyzed with FACScan flow cytometric device using CellQuest software (Becton Dickinson, Franklin Lakes, NJ, USA).
Cytokine assays
Supernatants from unstimulated, Sendai virus-infected, or LPS-/C. albicans-stimulated DCs were collected at 24 h after stimulation and stored at −20°C until analyzed. Human Th1/Th2 10plex Kit II (Bender Medsystems GmbH, Vienna, Austria), containing a panel of ten cytokines, was used. Assays were performed according to the manufacturer's recommendation and analyzed using the FACScan instrument and FlowCytomix Pro 1.0 Software.
Affymetrix arrays and analysis
Expression arrays containing nearly 50,000 RNA transcripts and variants of human genes or open reading frames were used and microarray analyses were performed according to the Affymetrix standard protocol (Affymetrix, Santa Clara, CA, USA). Each chip was processed individually, i.e., using RNA from one patient/control per chip (no pooling). In brief, total RNA was extracted from DCs using the RNeasy Mini kit (Qiagen, Venlo, The Netherlands). Quantity of RNA was determined with spectrophotometer and the quality was controlled using the Agilent Bioanalyzer 2100 (Agilent Technologies, Palo Alto, CA, USA). One microgram of total cellular RNA was treated according to the conventional Affymetrix eukaryotic RNA labeling protocols (Affymetrix) and then used to prepare biotinylated cRNA with the Enzo Bioarray High Yield RNA transcript labeling kit (Enzo, Life Sciences, Farmingdale, NY, USA). Fifteen micrograms of biotin-labeled cRNA were fragmented according to the Affymetrix eukaryotic sample protocol and hybridized to the U133 Plus 2.0 chip (Affymetrix). Hybridization, staining, and washing were performed using the Affymetrix Fluidics Station 450 and Hybridization Oven 640 under standard conditions. Also scanning was performed according to the Affymetrix protocol.
The raw data were first processed with the Affymetrix Microarray Suite 5 (Affymetrix) software in order to create lists of good quality genes. The initial selection procedure utilized Affymetrix expression level flags (present, marginal, absent). Genes that were flagged either present or marginally expressed in all replicates were considered expressed and genes that were flagged absent in all replicates were considered absent. Genes with inconsistent flagging were not included in the analysis. Furthermore, a cross gene error model was devised with the GeneSpring GX 7.3.1 (Agilent Technologies) software in order to calculate the lowest reliable expression value for each set of chips and to filter away the genes with unreliably low intensities. The raw data were then reprocessed with GeneSpring GX 7.3.1 using the GC robust multi-array average (GC-RMA) algorithm [22] with default parameters. The preprocessed intensities that had a magnitude lower than 0.01 were set to 0.01. The intensities of individual genes were normalized by dividing by the median intensity of a set of appropriate replicates. The resulting data were log2-transformed and geometrical means were calculated to give one expression value for a set of replicates. Differentially expressed genes were discovered by filtering the data using the following parameters: fold-change≥2.0 and p-value≤0.05.
The potential regulation of sets of genes with biological associations (pathways), which may not be defined by individual, statistically significant gene expression changes were also analyzed basically as described by Kiialainen and coworkers [23] (Saharinen, unpublished). The gene sets were derived from the Gene Ontology (GO) Consortium [24] as represented in Ensembl database (release 46 [25]). The GO-directed acyclic tree was iterated from the annotated GO class to the root of the tree, following all possible routes, which were added to the annotations of the given gene. For the detection of regulation in the gene sets, an iterative hypergeometric distribution p-value calculation was used [23]. For visualization, a p-value cut-off of ≤0.0001 was chosen for the pathways that were included in the pathway trees. False discovery rates were also calculated and were ≤9×10−5 for every group.
The stimulated/nonstimulated and over two-fold expression level changes for the genes were studied manually. We checked the annotated genes from PubMed and Online Mendelian Inheritance in Man. Genes outside these criteria were studied if their functions were associated with those in the studied analyses (lists in supplementary data).
Relative quantitative RT-PCR (TaqMan)
Quantitative reverse transcriptase polymerase chain reaction (Q-RT-PCR) was performed with ABI PRISM 5700 device using TaqMan gene expression assays (Applied Biosystems, Foster City, CA, USA). The 0.2 μg, 0.5 μg, or 1 μg of RNA was used as starting material for reverse transcriptase using TaqMan Reverse Transcriptase kit (Applied Biosystems). cDNA samples (50 ng in 1 μl) were amplified using a TaqMan Universal PCR MasterMix and the commercial Gene Expression Assay with primers and probes for: CXCL8 (IL-8) Hs_00174103_m1, tumor necrosis factor (TNF)-alpha Hs_00174128_m1, IL-12p35 Hs_00168405_m1, IL-12p40 Hs_002333688_m1, interferon (IFN)-beta Hs_00277188_s1, IL-29 Hs_00601677_g1, CXCL10 Hs_00171042_m1 AIRE Hs_00230829_m1, DC-SIGN Hs_01036031_m1IL-21R Hs_00222310_m1, suppressor of cytokine signaling 1 (SOCS1) Hs_00705164_s1, wingless-type MMTV integration site family, member 5A (WNT5A) Hs_00180103_m1, toll-like receptor 2 (TLR2) Hs_00152932_m1, or Hs_01872448_s1, Fc-epsilon R1A_2 Hs00758599_m1, CD1a Hs_00381754_g1, and IRF4 Hs_00180031_m1 from Applied Biosystems. Expression values were normalized with cyclophilin (for AIRE) or β-actin (for all other genes) as endogenous controls. Changes in gene expression levels were calculated according to TaqMan's comparative delta (CT) method. Values of 2−ΔCt and standard errors were calculated and these values are shown in the figures. Significance was calculated using Student's t-test. Standard curves were prepared using an RNA specimen collected from Sendai virus infected DCs (9 h time point) of a healthy donor.
Results
AIRE is expressed in human DCs and its expression remains constant during microbial stimulation
Previous findings have indicated that AIRE is expressed at low levels in monocyte-derived DCs [9, 10]. First, we wanted to compare the expression of AIRE in moDCs generated from six patients and six controls as well as in the plasmacytoid DC (BDCA 4+) and myeloid DC (CD1c+, BDCA 1) subsets isolated directly from the blood of three healthy control individuals. The expression analysis of AIRE mRNA was performed using quantitative, real-time PCR (Q-RT-PCR), using cyclophilin expression as an endogenous control [3]. The expression of AIRE mRNA was relatively low and practically equal in all three DC populations studied (data not shown). We also studied whether the expression level of AIRE mRNA would change during stimulation of moDCs with different microbes or their components. As analyzed by Q-RT-PCR, the expression of AIRE mRNA did not change significantly in response to Sendai virus infection or stimulation with C. albicans (data not shown).
Monocytes from APECED patients differentiate normally to immature moDCs
Next, we studied whether monocytes isolated from peripheral blood of APECED patients can differentiate normally and show typical features of moDCs. Microscopical examination showed that the in vitro differentiated moDCs from APECED patients and controls looked very similar. In addition, infection of cells with Sendai virus or stimulation with C. albicans revealed that both patient-derived and control moDCs produced cellular processes typical for differentiated moDCs (data not shown). To obtain further evidence that the AIRE-deficient moDCs had differentiated normally, the surface marker expression and cytokine production of the moDCs were analyzed by flow cytometry (fluorescence-activated cell sorting, FACS) and by Q-RT-PCR.
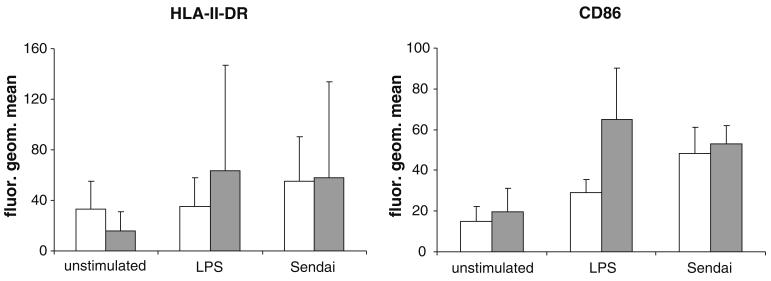
The basal and microbe-induced expression of human leukocyte antigen (HLA) class II and CD86 was analyzed by flow cytometry. The mean fluorescent intensity of HLA-II-DR of APECED moDCs was slightly lower than the corresponding levels of the controls. However, upon stimulation with E. coli-derived lipopolysaccharide or infection with Sendai virus, HLA-II-DR seemed to be expressed at higher levels in patient moDCs as compared to control cells (Fig. 1). Similarly, although expressed at the similar level prior to stimulation, CD86 steady-state expression levels were somewhat higher in stimulated patient DCs. However, there was a lot of individual variation in the expression of HLA class II and CD86 and the differences between the patient and control groups were not statistically significant.
Fig. 1.
Expression of HLA II DR and CD86 show that monocytes from APECED patients are able to differentiate into immature DCs in vitro. Purified monocytes obtained from APECED patients (n=3) and control individuals (n=6) were differentiated in vitro into moDCs. Cells were unstimulated (left), stimulated with E. coli lipopolysaccha-ride (LPS; 100 ng/ml), or infected with Sendai virus (MOI 5). Patient (gray bars) and control (white bars) moDCs both express cell-surface HLA class II and CD86. The mean (+/−1 standard deviation unit) fluorescence intensities are shown. Although patient moDCs express higher amounts of the markers especially upon LPS stimulation, differences between the groups are not statistically significant
APECED patient moDCs express typical DC specific genes
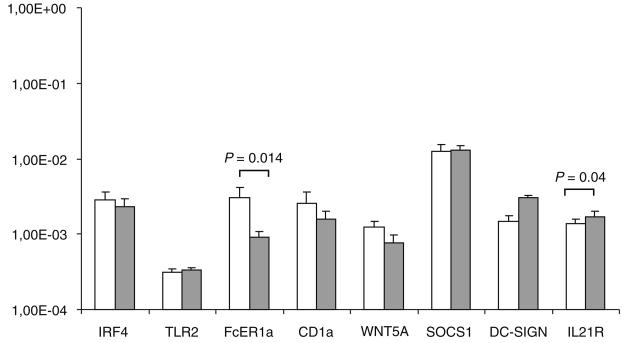
Recently, it was identified that human monocyte-derived DCs express a typical pattern of genes that is characteristic of differentiated immature DCs [26]. The transcript levels of moDC-specific genes (CD1α, DC-SIGN, IL-21 receptor, IRF4, FcεR1a, SOCS1, WNT5A, and TLR2) were analyzed by Q-RT-PCR in order to study whether there would be significant differences in the basal expression of these genes in patient and control moDCs (Fig. 2). Relative expression levels of different mRNAs were calculated using the delta CT method from two to six patients (depending on the marker) and from six controls and the expression levels are related to β-actin mRNA, which was used as a housekeeping gene. APECED moDCs showed somewhat higher expression of DC-SIGN (not significant) and IL-21R (p=0.04) and a lower expression of FcεR1a (p=0.014) as compared to cells obtained from control individuals. For the other marker genes studied, the differences between patient and control moDCs were small and not significant (Fig. 2).
Fig. 2.
Expression of DC-specific genes in differentiated control individual and APECED patient DCs. Monocytes were differentiated into moDCs for 6 days and total cellular RNA was isolated. Expression of dendritic cell-specific genes was measured using QRT-PCR from moDCs derived from patients (n=3–6; gray bars) and controls (n=6; white bars). Relative expression of mRNAs is related to the expression of β-actin mRNA. Standard errors are shown and significance was calculated using Student's t-test
AIRE-deficient moDCs show impaired expression of inflammatory cytokine genes in response to microbial stimulation
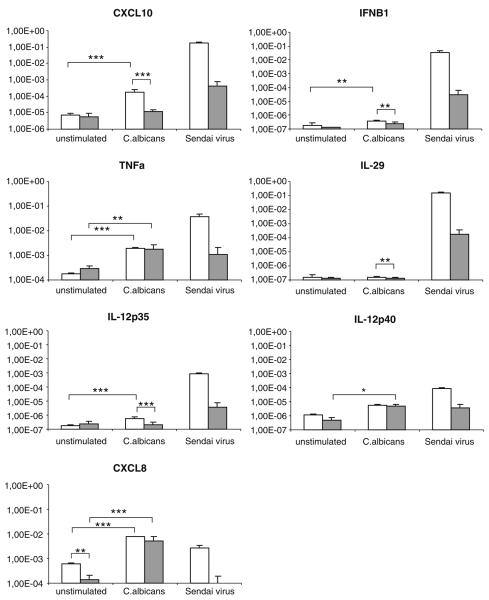
In order to seek a possible functional defect in patient DCs, we characterized microbe-induced cytokine gene expression in APECED moDCs using live C. albicans and Sendai virus, which have been widely used as microbial stimuli in DC research. Previous to stimulation, the expression of most cytokine and chemokine mRNAs appeared to be similar in APECED and control DCs (Fig. 3), which further suggests that patient DCs undergo a normal monocytes–DC differentiation. Only CXCL8 transcript levels were lower in patients vs. controls (Fig. 3; p≤0.01).
Fig. 3.
APECED patient derived moDCs respond poorly to microbial stimulus. MoDCs differentiated from APECED patients or control individuals were stimulated with C. albicans (MOI 1) or infected with Sendai virus (MOI 5); cells were collected at 24 h after microbial stimulation and total cellular RNA was isolated. Expression of cytokine genes relevant for moDCs were studied using QRT-PCR of unstimulated, Sendai virus infected, or C. albicans-stimulated moDCs from patients (gray bars) and from control individuals (white bars). Means of triplicate specimen and their standard deviations are shown using the delta CT method (controls n=6, patients n=2–5). The expression of cytokine mRNA is shown in relation to β-actin mRNA. Standard errors are shown and significance was calculated using Student's t-test
To compare the expression of cytokine mRNAs in response to microbial stimuli, we first used Q-RT-PCR analysis. All cytokine mRNA levels were compared to β-actin mRNA levels, which enables the comparison between patient and control samples. Control moDCs showed clear induction of TNF-α, IFN-β, IFN-λ1 (IL-29), IL-12p35, IL-12p40, CXCL8 (IL-8), and CXCL10 (IP-10) transcript levels in response to Sendai virus infection (p≤0.01; Fig. 3). However, although APECED moDC responses remained at levels at least a hundred fold lower, the differences were not statistically significant (p>0.05) in our study sample. C. albicans was a relatively poor inducer of IL-29, which was practically not induced by this pathogen in patients or controls. Unlike in the case of Sendai virus infection, TNF-α and CXCL8 transcript levels were comparable in APECED control moDCs in response to C. albicans stimulation, whereas CXCL10 gene was induced more weakly in APECED moDCs (p≤0.001). Of the examined genes, CXCL8 and TNFα were upregulated significantly in both APECED and control moDCs in response to C. albicans in our study sample, whereas in control cells, additionally IL-12p35, IFN-β, and CXCL10 mRNA expression was stimulated significantly (p≤0.01 or p≤0.001) by C. albicans (Fig. 3).
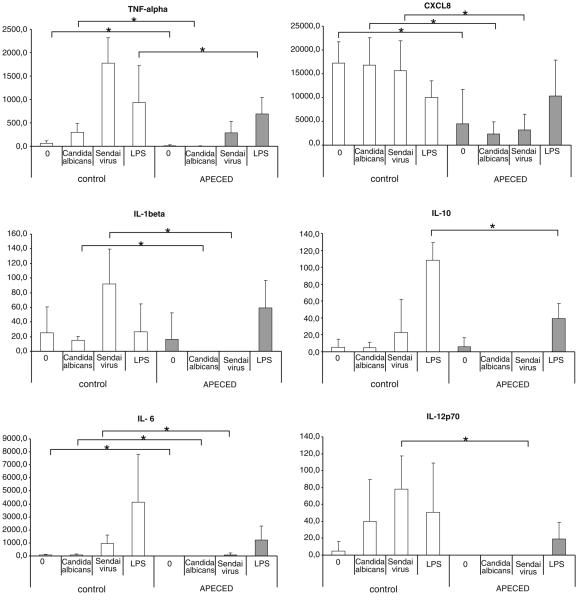
APECED patient moDCs show reduced cytokine production in response to microbial stimuli
To further analyze cytokine gene expression at the polypeptide level, we measured cytokine concentrations in cell culture supernatants of moDCs that were infected with Sendai virus or stimulated with C. albicans or LPS for 24 h. This was performed using a FACS-based multiplex bead assay measuring TNF-α, IL-1β, IL-2, IL-4, IL-5, IL-6, CXCL8, IL-10, IL-12p10, and IFN-γ. As shown in Fig. 4, the basal expression of TNF-α, IL-1β, IL-6, CXCL8, and IL-12 in unstimulated cells was lower in APECED moDCs. Upon stimulation with C. albicans, the production of TNF-α, IL-1β, IL-6, CXCL8, IL-10, and IL-12 was clearly higher in controls and the differences were statistically significant in the case of TNF-α, IL-1β, IL-6, and CXCL8. Similarly, there was a clear difference in the ability of APECED moDCs and control moDCs to produce cytokines in response to Sendai virus infection or LPS stimulation (Fig. 4). It was of interest that Sendai virus infection induced the highest levels of TNF-α, IL-1β, and IL-12, while LPS was the best inducer of IL-6 and IL-10 of the stimuli used.
Fig. 4.
Monocyte-derived DCs of APECED patients produce low amounts of cytokines. MoDCs differentiated from APECED patients or control individuals were stimulated with LPS (100 ng/ml), C. albicans (MOI 1), or infected with Sendai virus (MOI 5) and cell culture supernatants were collected at 24 h after microbial stimulation and cytokine levels were determined by flow cytometric device using Human Th1/Th2 10plex Kit II containing a panel of ten cytokines. The data for TNF-α, IL-1β, IL-6, CXCL8, IL-10, and IL-12 levels in control individuals (n=6, white bars) and APECED patients (n=2–6, gray bars) are presented. The levels of other cytokines tested, IL-2, IL-4, IL-5, and IFN-γ remained undetectable. The statistical significances between the groups were calculated using Students t-test (*p<0.05)
Expression profiles of AIRE-deficient DCs show aberrations in immunologically important pathways
In order to study differences in the genome-wide transcript patterns in APECED and control moDCs, we performed Affymetrix expression array analyses using total cellular RNAs from three APECED patient-derived and three healthy donor-derived moDCs. RNAs from the moDCs were hybridized onto human Affymetrix chips U133 Plus 2.0 individually. Additionally, monocyte-derived DCs from the same three patients and healthy controls were stimulated with C. albicans for 24 h and the cells were collected; RNA was isolated and hybridized onto individual Affymetrix chips. After the preprocessing and conditioning of the raw expression data (see “Materials and methods” for details), lists of individual genes that were over two-fold up- or downregulated (p<0.05) in APECED moDCs prior to and after stimulation were obtained (supplementary data). Of these, the ones belonging to the most consistently and differentially expressed Gene Ontology categories (see below) are highlighted in the volcano plots shown in Fig. 5a,b. Some genes, such as CXCL8, IL-10, and TNF-α, were down-regulated at both protein (cytokine assay, Fig. 4) and at mRNA level in Q-RT-PCR (Fig. 3) as well as/or in Affymetrix array analyses (Fig. 5 and supplementary data).
Fig. 5.
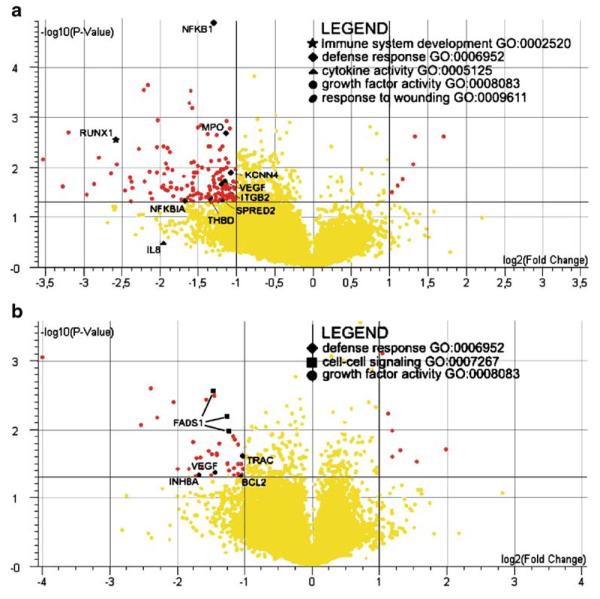
APECED patient moDCs show differential expression of genes compared to control moDCs. Volcano plots from Affymetrix arrays of a nonstimulated and b C. albicans-stimulated patient derived DCs vs. controls. Log2-transformed fold changes of gene expression are plotted as a function of the log of the p-values from the statistical test for differential expression (p-value cut-off 0.05. fold change cut-off two times). Genes belonging to Gene Ontology categories (e.g., GO:0002520) differentially expressed in the pathway analysis (see Fig. 6) are highlighted with different symbols as described in the legend table
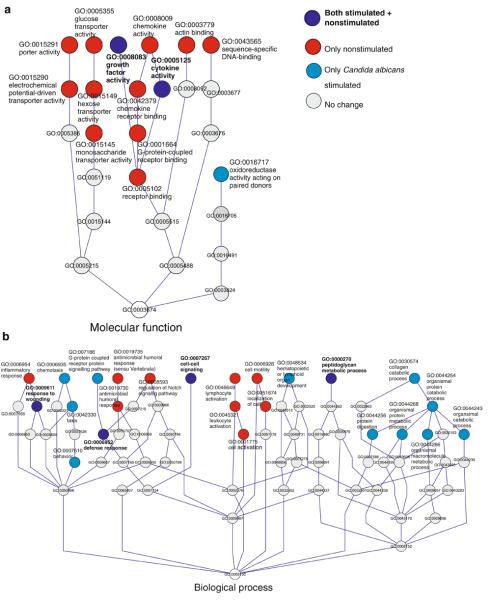
In addition to changes seen in individual genes, an in-house developed tool for pathway analysis (Saharinen, unpublished) was used to study possible changes in patient moDCs in the functional groups of genes or their regulatory pathways using the Gene Ontology category classification. These analyses were done in order to gain more insight into the possible, important groups of genes, or pathways modestly downregulated in the patients. Such changes do not come up at analysis on the single gene level. Several immunologically important pathways were downregulated. In nonstimulated APECED, moDCs downregulated pathways included cell–cell signaling, cytokine and chemokine activity, host defense and inflammatory response, lymphocyte activity, and cell motility. In C. albicans-stimulated APECED moDCs, the pathways described above, as well as those involved in chemotaxis and G-protein regulated signaling pathway, were clearly downregulated (Fig. 6a,b). Several pathways, such as cell–cell signaling, defense response, and cytokine activity (see also Fig. 7) were downregulated in both experimental settings. The implications of the observed changes for the DC functions remain to be elaborated.
Fig. 6.
Schematic representation of the a molecular function and b biological process Gene Ontology classes, in which the most significant gene sets differentially expressed in nonstimulated and C. albicans-stimulated APECED patient moDCs are enriched according to the pathway analysis. Colored circles denote regulated gene sets (pathways) and gray circles represent nonregulated pathways, connecting nodes leading to the root of the tree (cut-off p≤0.0001). Several immunologically relevant molecular functions and biological processes were downregulated in both unstimulated and stimulated patient cells indicating important differences in patient moDCs compared to control moDCs. A larger number of pathways were downregulated in nonstimulated patient moDCs than C. albicans-stimulated moDCs (red vs. light blue circles) suggesting differences in patient moDCs perceptible already at the unstimulated state
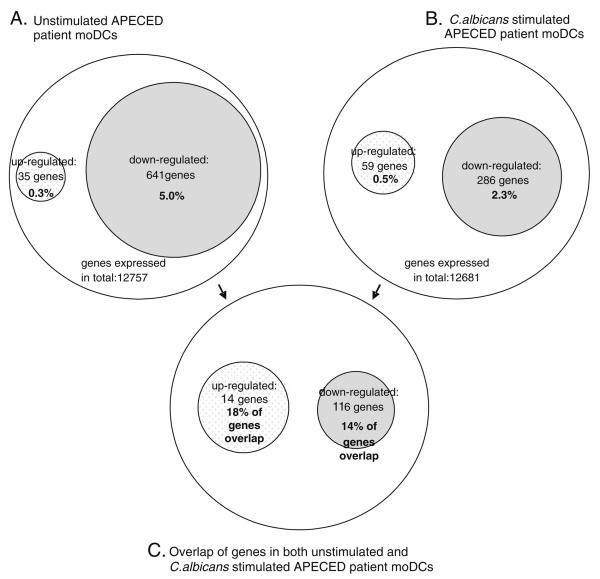
Fig. 7.
Differential expression in different experimental settings. Expression level changes from Affymetrix array analyses of unstimulated and in C. albicans-stimulated patient moDCs compared to control moDCs are summarized in this figure. Genes exhibiting more that two-fold up- or downregulation were considered differentially expressed. A, B The number and percentages of differentially expressed genes are given in corresponding experimental settings. Overall, in APECED patients, the prevailing mode of differential expression was downregulation. This is consistent with known role for AIRE as transcriptional activator. C The number and percentage of genes in the intersection between the gene sets differentially expressed in different experimental settings. The percentage is calculated from upregulated genes in C minus upregulated genes in A+B minus the genes upregulated in both given e.g., [14/(35+59−14)]×100%. Although fewer genes were differentially expressed in the stimulated cells, a large fraction of these genes were also differentially expressed in the nonstimulated cells
Discussion
Although the function of AIRE in thymic medullary epithelial cells and in central tolerance is reasonably well established, the role of AIRE in the periphery remains to be elucidated. Increasing evidence indicates that AIRE has a role in the peripheral immune system and that this role may be different from that of its involvement in the regulation of self-antigen presentation in the thymus [9-11]. In the present study, we carried out a comparison of the genome-wide expression profiles of monocyte-derived DCs from APECED patients and healthy controls, which revealed clear differences in the expression of several immune responses regulating genes and pathways, most importantly those involved in cytokine production and cell–cell signaling. Furthermore, we present evidence that AIRE-deficient monocytes differentiate apparently normally to immature moDCs although certain DC-specific genes were expressed differentially between the patient and control DCs. However, upon stimulation with different microbes or their components, APECED DCs showed clearly impaired expression of inflammatory cytokine genes.
APECED monocytes showed typical changes in morphology while differentiating in vitro into immature moDCs. Analysis of mRNA levels of selected genes by Q-RT-PCR revealed that the expression of DC-specific molecular markers, such as CD86, and HLA-II-DR in moDCs of APECED patients, was similar to that seen in the moDCs of healthy controls. Intriguingly, the mRNA level of DC-SIGN was weakly, though not significantly upregulated in AIRE-deficient moDCs. DC-SIGN, a pattern recognition, adhesion, and pathogen uptake receptor [27], functions as one of the major receptors for C. albicans enhancing its internalization [28]. Interestingly, the expression of several other genes such as CD205, CD206, Dectin-1, and TLR2, which are involved in cellular responses to C. albicans [29, 30], was similar in APECED and control cells and their expression was not changed during Candida stimulation. At present, the reason why APECED patients are prone to candidiasis is still unknown.
Analysis of the gene expression profile revealed that a number of functionally important genes were downregulated in APECED moDCs. Among them were, for example, nuclear factor-kappa B1 (NFKB1), important for DC activation and maturation [31, 32], CD58 (lymphocyte function-associated antigen 3, LFA-3), an MHC antigen-mediating cell adhesion and T cell activation [33, 34], and integrin beta 2 (ITGB2; also called CD18 or LFA-1), also important in the interactions of DCs with target cells [34, 35] (discussed below). The downregulation of such genes as these may partly contribute to the observed aberrations in the response to stimuli by APECED and control cells. Expression of typical APECED-related autoantigen genes was not significantly altered. The basal and C. albicans induced expression of type I (IFN-b) and type III (IL-29/IL-λ1) interferons remained at very low levels in AIRE-deficient moDCs (Fig. 3 and array data). However, in Sendai virus-infected DCs, the expression of IFN mRNAs was at least 100-fold higher in normal cells. Similarly, the expression of most cytokine genes tested was impaired in APECED DCs.
Interestingly, although the transcript levels of some DC-specific genes was weakly elevated, the cytokine production was decreased in APECED moDCs, especially in response to microbial stimulus. Different types of stimuli were used and all of the three stimuli used, live C. albicans, LPS, and Sendai virus, yielded dramatically reduced levels of DC cytokine production, implying that patient moDCs respond weakly to microbial stimuli. As cytokine synthesis is connected with the maturation of DCs [19], this may indicate that APECED patients have some impairment in DC maturation processes. However, an important observation was that certain differences in the gene expression profiles of moDCs existed already at the iDC stage of APECED patients. Gene expression levels of FcεR1a and IL21R (by Q-RT-PCR) or CD1a and CD58 (by array analysis) as well as pathway analyses (discussed below) showed reduced expression of several genes and gene sets, e.g., those involved in cytokine activity, host defense response, and cell–cell signaling already in iDCs. This may suggest that the tolerogenic capacity of the iDCs would be lower in APECED patients.
Importantly, using Affymetrix arrays, we were able to show that the transcript profiles of APECED patients carrying the same homozygote mutation differ in many respects from those of healthy donors. The expression of a number of genes involved in DC immunology was reduced in APECED moDCs. Interestingly, the percentage of overlapping genes or percentage of the same genes found over- or underexpressed in both unstimulated and in C. albicans-stimulated patient moDCs was 14% of the down-regulated and 18% of the upregulated genes (Fig. 7). Using a threshold of two-fold differential expression, the total number of upregulated genes was only 0.2% (25 genes) in nonstimulated and 0.4% (52 genes) in C. albicans-stimulated patient moDCs, whereas the numbers of down-regulated genes were drastically higher—5.9% (735 genes) in nonstimulated and 2.5% (314 genes) in C. albicans-stimulated APECED moDCs (Fig. 7). For example, the expression of TNF-α, CXCL2, CXCL8, IL-15, IL-18, and transcription factor WNT5B was lower in APECED moDCs (data not shown). The ability for antigen uptake of APECED moDCs is likely to be normal since DEC-205 and other uptake receptors were expressed at the level comparable to controls. The antigen presenting capacity of APECED moDCs appeared normal, since the expression of HLA class II genes was even weakly upregulated in APECED moDCs both at mRNA and protein level.
In the pathway analysis using the array data, the expression of several immunologically relevant functional categories was flagged. These pathway results fit in well with previous findings on the role of AIRE in the maturation of DCs [10] and their role in peripheral activation of T cells [11]. Considering the low cytokine production levels by the APECED moDCs, it is not surprising that the pathways regulating cell–cell signaling and cytokine activity would be downregulated. A decrease in DC–T cell communication could, in turn, lead to diminished lymphocyte activation and subsequently decreased activation of other target cells and, thus, an impaired T cell response. However, since APECED patients are not known to be more prone to infections other than those caused by C. albicans, these findings are slightly puzzling. Our results indirectly invoke a hypothesis that APECED patients would be susceptible to other infections apart from C. albicans. An example of such would be the herpes simplex virus 1 infections, which can remain undiagnosed in APECED patients due to the fact that they can be hidden by the chronic candidiasis [36]. Nonetheless, while our findings suggest a somewhat impaired functional capacity of APECED moDCs, this does not result in increased overall susceptibility to most microbial pathogens.
We also analyzed the highest 200 up- or downregulated genes from Johnnidis et al. [45] arrays performed on thymic medullary epithelial cells from Aire-deficient and wt mice by comparing both their individual differentially expressed genes manually and by running their genes with our pathway analysis program. Not a single homologous gene was found to be similarly up- or downregulated in the mouse arrays and in our DC analysis. However, when the mouse and human array data were compared by pathway analysis method, we found that both species shared some differentially regulated functional categories. In Aire-knockout mouse, MECs and human AIRE-deficient DCs cytokine activity, inflammatory response, G-protein-coupled receptor binding, chemotaxis, taxis, chemokine activity, and chemokine receptor binding were downregulated. Interestingly, in Aire-deficient mice pathways involved in T cell activation and proliferation as well as MHC class II, biosynthesis were upregulated, whereas only few pathways were found to be upregulated in APECED moDCs (mRNA processing, antigen processing and presentation, MHC protein complex). These differences in gene expression profiles may partly be due to the species and cell type-specific differences but possibly also due to the apparently different role of AIRE in central vs. peripheral tolerance in human and mouse.
There are a few other studies where the gene expression patterns in AIRE/Aire-deficient and control cells have been analyzed. Sillanpää et al. [10] compared the expression profiles of AIRE-positive and AIRE-negative monocytic U937 cell lines. AIRE-regulated expression of tissue-specific antigens that was seen in the thymus was not detected in the U937 cells. This suggests that the role of AIRE in the periphery may be different from that in the thymus. Array data presented by Anderson et al. [46] on medullary epithelial cells from Aire-deficient mice showed alterations in the expression of several genes involved in antigen processing and presentation. These cells also showed less effective antigen presenting capacity. However, it was not clear whether the antigen presentation of splenic dendritic cells from Aire-deficient mice was less effective as compared to wild type mice. In contrast, Ramsey et al. [11] found that peripheral dendritic cells from Aire-negative mice were more effective activators of naïve T cells and, furthermore, they reported that the amount of dendritic cells in the spleen, lymph nodes, and blood of these mice was increased. Evidence from Aire-deficient mice and APECED patients suggested that decreased expression of the vascular cell adhesion molecule-1 (VCAM-1) was responsible for the altered dendritic cell function. In our study, VCAM-1 was upregulated in controls after stimulation with C. albicans, but no changes in VCAM-1 expression levels could be seen in APECED patients compared to controls.
Our findings of impaired DC functions are in agreement with the findings of Kekäläinen et al. [12] who suggested that the defect in AIRE-deficient regulatory T cells in humans may be due to impaired DCs-stimulated Treg development. A defect in both DCs and Tregs could implicate a possible defect in the interplay between these two cell types. Alternatively, a defect in DC functions could lead to impaired peripheral immune responses. We also found downregulation of ITGB2 and CD58 (LFA-3) genes, which are involved in DC–Treg interplay [33, 35, 37-39]. DC maturation and DC–Treg interactions are crucial for immune response and tolerance [40].
Functional defects in DCs, especially defects in DC maturation, have been shown to be involved in other common autoimmune diseases such as type 1 diabetes [41] (also lower IL-12 in diabetic children) [42], systemic lupus erythematosus [43], and arthritis [44]. Findings concerning the monogenic disease APECED and its causative gene AIRE have indicated that well-functioning peripheral and central tolerance are important in preventing autoimmunity in humans.
To conclude, in the present study, we show that moDCs of APECED patients show impaired cytokine production in response to microbial stimuli as well as the expression of genes involved in immune regulatory pathways is reduced. These observations emphasize the need for further functional studies related to peripheral tolerance and how it is regulated in APECED patients. Revealing the functions of AIRE will help us to understand the fundamental molecular mechanisms controlling the development of immune tolerance.
Supplementary Material
Acknowledgments
We thank Drs. T. Petteri Arstila and Jaakko Perheentupa for the help in obtaining the patient samples. Sampo Sammalisto and Samuli Ripatti are acknowledged for helping with the statistical analysis, and T. Petteri Arstila and Seppo Meri are thanked for critical reading of the manuscript. Lea Puhakka and Anne Vikman are thanked for the diligent technical assistance. Pamela Österlund and the other members of the Julkunen group are warmly thanked for help in their laboratory.
Funding This study was supported by the Magnus Ehrnrooth foundation, Helsinki Biomedical Graduate School in (HBGS), EU FP6 program project EURAPS, the Center of Excellence in Disease Genetics of the Academy of Finland, Academy of Finland (grant 206282), and The Lilly Foundation (Finland).
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s00109-008-0374-7) contains supplementary material, which is available to authorized users.
Conflict of interest statement The authors have no conflicting financial interest, connections, or other conflicting situations regarding this manuscript.
References
- 1.Perheentupa J. Autoimmune polyendocrinopathy–candidiasis–ectodermal dystrophy. J Clin Endocrinol Metab. 2006;91:2843–2850. doi: 10.1210/jc.2005-2611. [DOI] [PubMed] [Google Scholar]
- 2.Halonen M, Kangas H, Ruppell T, Ilmarinen T, Ollila J, Kolmer M, Vihinen M, Palvimo J, Saarela J, Ulmanen I, et al. APECED-causing mutations in AIRE reveal the functional domains of the protein. Human Mutat. 2004;23:245–257. doi: 10.1002/humu.20003. [DOI] [PubMed] [Google Scholar]
- 3.Anderson MS, Venanzi ES, Klein L, Chen Z, Berzins SP, Turley SJ, Von Boehmer H, Bronson R, Dierich A, Benoist C, et al. Projection of an immunological self shadow within the thymus by the AIRE protein. Science. 2002;298:1395–1401. doi: 10.1126/science.1075958. [DOI] [PubMed] [Google Scholar]
- 4.Liston A, Gray DH, Lesage S, Fletcher AL, Wilson J, Webster KE, Scott HS, Boyd RL, Peltonen L, Goodnow CC. Gene dosage-limiting role of Aire in thymic expression, clonal deletion, and organ-specific autoimmunity. J Exp Med. 2004;200:1015–1026. doi: 10.1084/jem.20040581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng MH, Shum AK, Anderson MS. What's new in the Aire? Trends Immunol. 2007;28:321–327. doi: 10.1016/j.it.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 6.Heino M, Peterson P, Kudoh J, Nagamine K, Lagerstedt A, Ovod V, Ranki A, Rantala I, Nieminen M, Tuukkanen J, et al. Autoimmune regulator is expressed in the cells regulating immune tolerance in thymus medulla. Biochem Biophys Res Commun. 1999;257:821–825. doi: 10.1006/bbrc.1999.0308. [DOI] [PubMed] [Google Scholar]
- 7.Kogawa K, Kudoh J, Nagafuchi S, Ohga S, Katsuta H, Ishibashi H, Harada M, Hara T, Shimizu N. Distinct clinical phenotype and immunoreactivity in Japanese siblings with autoimmune polyglandular syndrome type 1 (APS-1) associated with compound heterozygous novel AIRE gene mutations. Clin Immunol. 2002;103:277–283. doi: 10.1006/clim.2002.5208. [DOI] [PubMed] [Google Scholar]
- 8.Zheng X, Yin L, Liu Y, Zheng P. Expression of tissue-specific autoantigens in the hematopoietic cells leads to activation-induced cell death of autoreactive T cells in the secondary lymphoid organs. Eur J Immunol. 2004;34:3126–3134. doi: 10.1002/eji.200425177. [DOI] [PubMed] [Google Scholar]
- 9.Kogawa K, Nagafuchi S, Katsuta H, Kudoh J, Tamiya S, Sakai Y, Shimizu N, Harada M. Expression of AIRE gene in peripheral monocyte/dendritic cell lineage. Immunol Lett. 2002;80:195–198. doi: 10.1016/s0165-2478(01)00314-5. [DOI] [PubMed] [Google Scholar]
- 10.Sillanpää N, Magureanu CG, Murumagi A, Reinikainen A, West A, Manninen A, Lahti M, Ranki A, Saksela K, Krohn K, et al. Autoimmune regulator induced changes in the gene expression profile of human monocytes–dendritic cell-lineage. Mol Immunol. 2004;41:1185–1198. doi: 10.1016/j.molimm.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 11.Ramsey C, Hassler S, Marits P, Kampe O, Surh CD, Peltonen L, Winqvist O. Increased antigen presenting cell-mediated T cell activation in mice and patients without the autoimmune regulator. Eur J Immunol. 2006;36:305–317. doi: 10.1002/eji.200535240. [DOI] [PubMed] [Google Scholar]
- 12.Kekäläinen E, Tuovinen H, Joensuu J, Gylling M, Franssila R, Pontynen N, Talvensaari K, Perheentupa J, Miettinen A, Arstila TP. A defect of regulatory T cells in patients with autoimmune polyendocrinopathy–candidiasis–ectodermal dystrophy. J Immunol. 2007;178:1208–1215. doi: 10.4049/jimmunol.178.2.1208. [DOI] [PubMed] [Google Scholar]
- 13.Mahnke K, Schmitt E, Bonifaz L, Enk AH, Jonuleit H. Immature, but not inactive: the tolerogenic function of immature dendritic cells. Immunol Cell Biol. 2002;80:477–483. doi: 10.1046/j.1440-1711.2002.01115.x. [DOI] [PubMed] [Google Scholar]
- 14.Steinman RM, Nussenzweig MC. Avoiding horror autotoxicus: the importance of dendritic cells in peripheral T cell tolerance. Proc Natl Acad Sci USA. 2002;99:351–358. doi: 10.1073/pnas.231606698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Manuel SL, Rahman S, Wigdahl B, Khan ZK, Jain P. Dendritic cells in autoimmune diseases and neuroinflammatory disorders. Front Biosci. 2007;12:4315–335. doi: 10.2741/2390. [DOI] [PubMed] [Google Scholar]
- 16.Ahonen P, Myllarniemi S, Sipila I, Perheentupa J. Clinical variation of autoimmune polyendocrinopathy–candidiasis–ectodermal dystrophy (APECED) in a series of 68 patients. N Engl J Med. 1990;322:1829–1836. doi: 10.1056/NEJM199006283222601. [DOI] [PubMed] [Google Scholar]
- 17.Björses P, Halonen M, Palvimo JJ, Kolmer M, Aaltonen J, Ellonen P, Perheentupa J, Ulmanen I, Peltonen L. Mutations in the AIRE gene: effects on sub-cellular location and transactivation function of the APECED protein. Am J Hum Genet. 2000;66:378–392. doi: 10.1086/302765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vandenplas S, Wiid I, Grobler-Rabie A, Brebner K, Ricketts M, Wallis G, Bester A, Boyd C, Mathew C. Blot hybridisation analysis of genomic DNA. J Med Genet. 1984;21:164–172. doi: 10.1136/jmg.21.3.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Veckman V, Miettinen M, Pirhonen J, Siren J, Matikainen S, Julkunen I. Streptococcus pyogenes and Lactobacillus rhamnosus differentially induce maturation and production of Th1-type cytokines and chemokines in human monocyte-derived dendritic cells. J Leukoc Biol. 2004;75:764–771. doi: 10.1189/jlb.1003461. [DOI] [PubMed] [Google Scholar]
- 20.Veckman V, Julkunen I. Streptococcus pyogenes activates human plasmacytoid and myeloid dendritic cells. J Leukoc Biol. 2007;83:296–304. doi: 10.1189/jlb.0707457. [DOI] [PubMed] [Google Scholar]
- 21.Osterlund P, Veckman V, Siren J, Klucher KM, Hiscott J, Matikainen S, Julkunen I. Gene expression and antiviral activity of alpha/beta interferons and interleukin-29 in virus-infected human myeloid dendritic cells. J Virol. 2005;79:9608–9617. doi: 10.1128/JVI.79.15.9608-9617.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu ZJ, Irizarry RA, Gentleman R, Martinez-Murillo F, Spencer F. A model-based background adjustment for oligonucleotide expression arrays. J Am Stat Assoc. 2004;99:909–917. [Google Scholar]
- 23.Kiialainen A, Veckman V, Saharinen J, Paloneva J, Gentile M, Hakola P, Hemelsoet D, Ridha B, Kopra O, Julkunen I, et al. Transcript profiles of dendritic cells of PLOSL patients link demyelinating CNS disorders with abnormalities in pathways of actin bundling and immune response. J Mol Med. 2007;85:971–983. doi: 10.1007/s00109-007-0191-4. [DOI] [PubMed] [Google Scholar]
- 24.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene Ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hubbard TJ, Aken BL, Beal K, Ballester B, Caccamo M, Chen Y, Clarke L, Coates G, Cunningham F, Cutts T, et al. Ensembl 2007. Nucleic Acids Res. 2007;35:D610–D617. doi: 10.1093/nar/gkl996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lehtonen A, Ahlfors H, Veckman V, Miettinen M, Lahesmaa R, Julkunen I. Gene expression profiling during differentiation of human monocytes to macrophages or dendritic cells. J Leukoc Biol. 2007;82:710–720. doi: 10.1189/jlb.0307194. [DOI] [PubMed] [Google Scholar]
- 27.Geijtenbeek TB, Torensma R, van Vliet SJ, van Duijnhoven GC, Adema GJ, van Kooyk Y, Figdor CG. Identification of DC-SIGN, a novel dendritic cell-specific ICAM-3 receptor that supports primary immune responses. Cell. 2000;100:575–585. doi: 10.1016/s0092-8674(00)80693-5. [DOI] [PubMed] [Google Scholar]
- 28.Cambi A, Gijzen K, de Vries JM, Torensma R, Joosten B, Adema GJ, Netea MG, Kullberg BJ, Romani L, Figdor CG. The C-type lectin DC-SIGN (CD209) is an antigen-uptake receptor for Candida albicans on dendritic cells. Eur J Immunol. 2003;33:532–538. doi: 10.1002/immu.200310029. [DOI] [PubMed] [Google Scholar]
- 29.Goodridge HS, Simmons RM, Underhill DM. Dectin-1 stimulation by Candida albicans yeast or zymosan triggers NFAT activation in macrophages and dendritic cells. J Immunol. 2007;178:3107–3115. doi: 10.4049/jimmunol.178.5.3107. [DOI] [PubMed] [Google Scholar]
- 30.Donini M, Zenaro E, Tamassia N, Dusi S. NADPH oxidase of human dendritic cells: role in Candida albicans killing and regulation by interferons, dectin-1 and CD206. Eur J Immunol. 2007;37:1194–1203. doi: 10.1002/eji.200636532. [DOI] [PubMed] [Google Scholar]
- 31.Kieran M, Blank V, Logeat F, Vandekerckhove J, Lottspeich F, Le Bail O, Urban MB, Kourilsky P, Baeuerle PA, Israel A. The DNA binding subunit of NF-kappa B is identical to factor KBF1 and homologous to the rel oncogene product. Cell. 1990;62:1007–1018. doi: 10.1016/0092-8674(90)90275-j. [DOI] [PubMed] [Google Scholar]
- 32.Chaussabel D, Semnani RT, McDowell MA, Sacks D, Sher A, Nutman TB. Unique gene expression profiles of human macrophages and dendritic cells to phylogenetically distinct parasites. Blood. 2003;102:672–681. doi: 10.1182/blood-2002-10-3232. [DOI] [PubMed] [Google Scholar]
- 33.Suciu-Foca N, Manavalan JS, Scotto L, Kim-Schulze S, Galluzzo S, Naiyer AJ, Fan J, Vlad G, Cortesini R. Molecular characterization of allospecific T suppressor and tolerogenic dendritic cells: review. Int Immunopharmacol. 2005;5:7–11. doi: 10.1016/j.intimp.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 34.Sanchez-Madrid F, Krensky AM, Ware CF, Robbins E, Strominger JL, Burakoff SJ, Springer TA. Three distinct antigens associated with human T-lymphocyte-mediated cytolysis: LFA-1, LFA-2, and LFA-3. Proc Natl Acad Sci USA. 1982;79:7489–7493. doi: 10.1073/pnas.79.23.7489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Das H, Sugita M, Brenner MB. Mechanisms of Vdelta1 gammadelta T cell activation by microbial components. J Immunol. 2004;172:6578–6586. doi: 10.4049/jimmunol.172.11.6578. [DOI] [PubMed] [Google Scholar]
- 36.Nagafuchi S, Umene K, Yamanaka F, Oohashi S, Shindo M, Kurisaki H, Kudoh J, Shimizu N, Hara T, Harada M. Recurrent herpes simplex virus infection in a patient with autoimmune polyendocrinopathy–candidiasis–ectodermal dystrophy associated with L29P and IVS9-1G>C compound heterozygous autoimmune regulator gene mutations. J Intern Med. 2007;261:605–610. doi: 10.1111/j.1365-2796.2007.01786.x. [DOI] [PubMed] [Google Scholar]
- 37.Varga G, Balkow S, Wild MK, Stadtbaeumer A, Krummen M, Rothoeft T, Higuchi T, Beissert S, Wethmar K, Scharffetter-Kochanek K, et al. Active MAC-1 (CD11b/CD18) on DCs inhibits full T-cell activation. Blood. 2007;109:661–669. doi: 10.1182/blood-2005-12-023044. [DOI] [PubMed] [Google Scholar]
- 38.Grossman WJ, Verbsky JW, Barchet W, Colonna M, Atkinson JP, Ley TJ. Human T regulatory cells can use the perforin pathway to cause autologous target cell death. Immunity. 2004;21:589–601. doi: 10.1016/j.immuni.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 39.Wethmar K, Helmus Y, Luhn K, Jones C, Laskowska A, Varga G, Grabbe S, Lyck R, Engelhardt B, Bixel MG, et al. Migration of immature mouse DC across resting endothelium is mediated by ICAM-2 but independent of beta2-integrins and murine DC-SIGN homologues. Eur J Immunol. 2006;36:2781–2794. doi: 10.1002/eji.200526311. [DOI] [PubMed] [Google Scholar]
- 40.Hubert P, Jacobs N, Caberg JH, Boniver J, Delvenne P. The cross-talk between dendritic and regulatory T cells: good or evil? J Leukoc Biol. 2007;82:781–794. doi: 10.1189/jlb.1106694. [DOI] [PubMed] [Google Scholar]
- 41.Angelini F, Del Duca E, Piccinini S, Pacciani V, Rossi P, Manca Bitti ML. Altered phenotype and function of dendritic cells in children with type 1 diabetes. Clin Exp Immunol. 2005;142:341–346. doi: 10.1111/j.1365-2249.2005.02916.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Trucco M, Giannoukakis N. Immunoregulatory dendritic cells to prevent and reverse new-onset type 1 diabetes mellitus. Expert Opin Biol Ther. 2007;7:951–963. doi: 10.1517/14712598.7.7.951. [DOI] [PubMed] [Google Scholar]
- 43.Monrad S, Kaplan MJ. Dendritic cells and the immunopathogenesis of systemic lupus erythematosus. Immunol Res. 2007;37:135–145. doi: 10.1007/BF02685895. [DOI] [PubMed] [Google Scholar]
- 44.Laborde EA, Vanzulli S, Beigier-Bompadre M, Isturiz MA, Ruggiero RA, Fourcade MG, Catalan Pellet AC, Sozzani S, Vulcano M. Immune complexes inhibit differentiation, maturation, and function of human monocyte-derived dendritic cells. J Immunol. 2007;179:673–681. doi: 10.4049/jimmunol.179.1.673. [DOI] [PubMed] [Google Scholar]
- 45.Johnnidis JB, Venanzi ES, Taxamn DJ, Ting JP, Benoist CO, Mathis DJ. Chromosomal clustering of genes controlled by the aire transcription factor. Proc Natl Acad Sci USA. 2005;102(20):7233–7238. doi: 10.1073/pnas.0502670102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Anderson MS, Venanzi ES, Chen Z, Berzins SP, Benoist C, Mathis D. The cellular mechanism of aire control of T cell tolerance. Immunity. 2005;23(2):227–239. doi: 10.1016/j.immuni.2005.07.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.