Abstract
Pumilio controls a number of processes in eukaryotes, including the translational repression of hunchback (hb) mRNA in early Drosophila embryos. The Pumilio Puf domain binds to a pair of 32 nucleotide (nt) Nanos response elements (NRE1 and NRE2) within the 3′ untranslated region of hb mRNA. Despite the elucidation of structures of human Pumilio Puf domain in complex with hb RNA elements, the nature of hb mRNA recognition remains unclear. In particular, the site that mediates regulation in vivo is significantly larger than the 8–10-nt RNA elements bound to single Puf molecules in crystal structures. Here we present biophysical and biochemical data that partially resolve the paradox. We show that each NRE is composed of two binding sites (Box A and Box B) and that two Puf domains can co-occupy a single NRE. The Puf domains have a higher affinity for the 3′ Box B site than the 5′ Box A site; binding to the intact NRE appears to be cooperative (at least in some experiments). We suggest that the 2 Pumilio:1 NRE complex is the functional regulatory unit in vivo.
Keywords: 3′ UTRs, Puf domain, Pumilio, hunchback, mRNA translation
INTRODUCTION
Pumilio (Pum) is a sequence-specific RNA binding protein that recognizes elements in the 3′ untranslated regions (3′ UTRs) of target transcripts to repress translation (Wharton and Aggarwal 2006). Pum has a number of defined targets, but perhaps the best characterized is hunchback (hb) mRNA in the developing Drosophila embryo (Lehmann and Nusslein-Volhard 1987; Murata and Wharton 1995). Repression of hb is nucleated by Pum binding to a pair of 32-nucleotide (nt) Nanos response elements (NREs) within the 3′ UTR (Murata and Wharton 1995). The binding of Pum to these NREs (NRE1 and NRE2), each comprised of a 5′-proximal Box A sequence (GUUGU) and a 3′-proximal Box B sequence (AUUGUA) (Fig. 1; Wharton and Struhl 1991; Zamore et al. 1997; Wharton et al. 1998), provides a platform for recruitment of the essential trans-acting cofactors, Nanos (Nos) and Brain Tumor (Brat) (Sonoda and Wharton 1999, 2001). The repression complex thus integrates the sequence specificity of Pum and the positional information of Nos to provide the correct spatiotemporal control of hb translation.
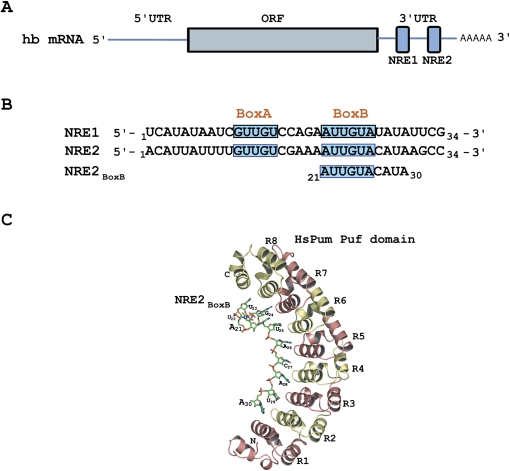
FIGURE 1.
Structure of hb mRNA. (A) Two NREs are located within the 3′ UTR of the hb mRNA. (B) Each NRE contains two Pum binding sites; Box A at the 5′end and Box B at the 3′ end. (C) Crystal structure of human Pum Puf domain (HsPum) bound to a fragment of NRE2 shows 8–10 nt contacting the concave surface of the protein (Wang et al. 2002).
The Pum RNA binding “Puf” domain (named after the founding family members Pumilio in Drosophila and FBF in Caenorhabditis elegans) is now listed among the common RNA binding folds such as the RRM, dsRBD, KH domain, and Y-box, among others (Wickens et al. 2002). However, despite the elucidation of the structure of the Puf domain, both alone and in complex with RNA elements (Edwards et al. 2001; Wang et al. 2001, 2002; Gupta et al. 2008; Miller et al. 2008), the precise nature of recognition of the hb NRE by Pum remains elusive. In particular, the site that mediates regulation in vivo is significantly larger than the 8- to 10-nt RNA elements bound to single Puf molecules in crystal structures (Fig. 1; Wang et al. 2002; Gupta et al. 2008; Miller et al. 2008). Transgenic experiments in flies reveal that the shortest element that confers translational repression by Pum is 32 nt (Wharton and Struhl 1991). Additionally, in vitro binding experiments suggest that an RNA sequence of at least 28 nt is required to recapitulate high affinity binding (Wharton et al. 1998). Thus, while specificity for binding an 8–10-nt RNA element is contained within a single Pum molecule (Wang et al. 2002), there appear to be additional requirements for in vitro binding and in vivo activity.
Here we present biophysical and biochemical data that suggest that each NRE has two Pum binding sites. We show that each site is recognized by a single Puf domain and that a single NRE can be co-occupied by two Puf domains. The Puf domains appear to bind cooperatively to the NRE, with a higher affinity for the 3′ Box B sequence than for the 5′ Box A sequence. We suggest that this 2 Pum:1 NRE complex is the functional unit that confers regulation in vivo.
RESULTS AND DISCUSSION
In the experiments described below, we primarily studied the human (Hs) Pum Puf domain, which is generally more soluble and stable than the Drosophila (Dm) protein. To further characterize the binding of HsPum to NRE1 and NRE2, we used fluorescence anisotropy (FA). For these binding experiments, we prepared five fluorescein (Fl)-labeled 34-nt RNA fragments:
NRE1wt (5′-Fl-UCAUAUAAUCGUUGUCCAGAAUUGUAUAUAUUCG-3′);
NRE2wt (5′-Fl-ACAUUAUUUUGUUGCGAAAAUUGUACAUAAGCC-3′);
NRE1MutA (5′-Fl-UCAUAUAAUCUAGGGCCAGAAUUGUAUAUAUUCG-3′);
NRE1MutB (5′-Fl-UCAUAUAAUCGUUGUCCAGAACGGGAUAUAUUCG-3′); and
NRE1MutA+B (5′-Fl-UCAUAUAAUCUAGGGCCAGAACGGGAUAUAUUCG-3′).
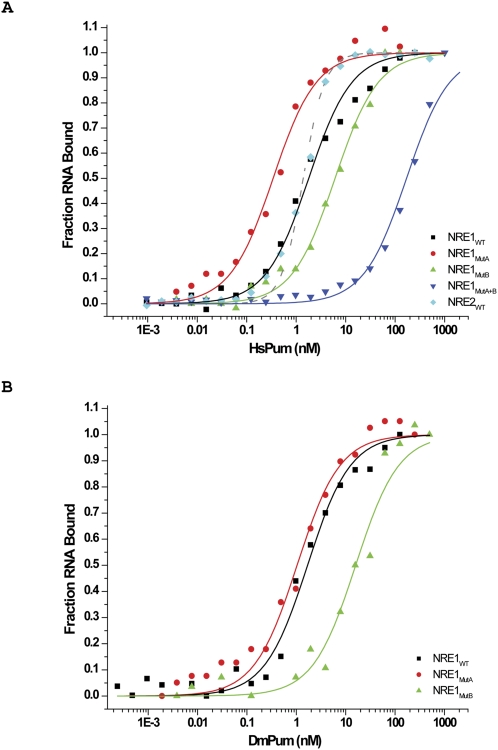
Briefly, NRE1wt and NRE2wt encompass the wild-type (wt) NRE1 and wt NRE2 sequences, respectively; NRE1MutA carries mutations in the Box A sequence; NRE1MutB carries mutations in the Box B sequence; and NRE1MutA+B carries mutations in both Box A and Box B sequences. As shown in Figure 2A, HsPum binds NRE1wt, NRE1MutA, and NRE1MutB with Kds of 1.8 nM ± 0.16 nM, 0.34 ± 0.03 nM, and 6.1 ± 0.5 nM, respectively. The double mutant NRE1MutA+B binds with much lower affinity, namely, with a Kd of 180 ± 9.8 nM. Together, these data show that the hb NRE1 is composed of two HsPum binding sites: a strong site that encompasses the Box B sequence and a weaker site that encompasses the Box A sequence. Also, HsPum appears to bind the two sequences independently in these experiments (although not in gel shift experiments, as described below), as the Kd for NRE1wt is between that for NRE1MutA and NRE1MutB (e.g., Kd for 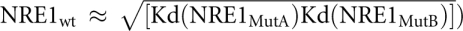 . DmPum yields broadly similar results (Fig. 2B); on NRE1MutA+B, the data were too noisy to be fit with an isotherm, probably because DmPum has a tendency to aggregate at the high concentrations required for the weak binding to NRE1MutA+B. HsPum binds cooperatively to NRE2wt. As shown in Figure 2A, the binding isotherm in this case is narrowly sigmoidal and is fit best with a two-site model. The overall affinity (Kd of 1.9 ± 0.2 nM) is, however, similar to that for NRE1wt.
. DmPum yields broadly similar results (Fig. 2B); on NRE1MutA+B, the data were too noisy to be fit with an isotherm, probably because DmPum has a tendency to aggregate at the high concentrations required for the weak binding to NRE1MutA+B. HsPum binds cooperatively to NRE2wt. As shown in Figure 2A, the binding isotherm in this case is narrowly sigmoidal and is fit best with a two-site model. The overall affinity (Kd of 1.9 ± 0.2 nM) is, however, similar to that for NRE1wt.
FIGURE 2.
Fluorescence anisotropy. (A) Binding curves for human Pum Puf domain (HsPum) with 5′-fluoroscein (5′-Fl)-labeled NRE1wt, NRE2wt, NRE1MutA, NRE1MutB, and NRE1MutA+B RNAs. (B) Binding curves for Drosophila Pum Puf domain (DmPum) with 5′-Fl NRE1wt, NRE1MutA, and NRE1MutB. On NRE1MutA+B, the data were too noisy to be fit with an isotherm, probably because DmPum has a tendency to aggregate at the high concentrations required for the weak binding to NRE1MutA+B. The experiments were performed as described in Materials and Methods. The errors were determined from the fit of the isotherms to the binding equations.
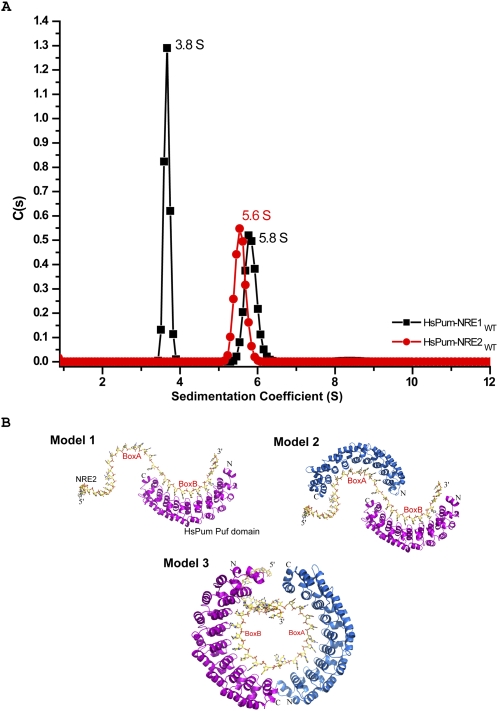
In order to fully characterize the stoichiometry of HsPum binding to a single NRE, we subjected the Pum-NRE complexes to analytical ultracentrifugation (AU) using absorption optics in sedimentation velocity experiments. We labeled the RNA molecules with Fl at the 5′ end so that only signal from RNA or RNA–protein complexes would be observed in the sedimentation profiles, thereby simplifying data analysis. Scans were collected at 1-min intervals at 495 nm (corresponding to the absorption wavelength of Fl) and were analyzed using the program SEDFIT (Schuck 2000; Schuck et al. 2002). As shown in Figure 3A, the HsPum–NRE1 complex sediments at 3.8 ± 0.4S and 5.8 ± 0.5S. From analysis of the data with programs SEDFIT (Schuck 2000; Schuck et al. 2002) and SEDPHAT (Vistica et al. 2004), the calculated mass for the first species (∼3.8S) is ∼50 kDa, for the second species (∼5.8S), ∼94 kDa. These masses correspond closely to the theoretical molecular masses of 1HsPum:1NRE (∼51.4 kDa) and 2HsPum:1NRE (∼91.4 kDa) complexes. Together, the results suggest a dynamic monomer–dimer equilibrium when HsPum binds NRE1. In contrast, the HsPum–NRE2 complex sediments as a single species at 5.6 ± 0.3S, close to the “dimer” S value obtained for the HsPum–NRE1 complex. For further analysis, we used the program HYDROPRO (Garcia De La Torre et al. 2000) to calculate theoretical sedimentation coefficients from atomic models of HsPum–NRE complexes. In particular, we built three models using the atomic coordinates from the crystal structure of HsPum Puf bound to a fragment (Box B) of hb NRE2 (Protein Data Bank [PDB] identification no. 1M8Y) (Wang et al. 2002). In model 1, we built one Pum molecule bound to the high affinity Box B site; in model 2, two Pum molecules bound to Box A and Box B on an “extended” NRE; and in model 3, two Pum molecules bound to Box A and Box B on a more compact “circular” NRE conformation (Fig. 3B). In models 2 and 3, the two Pum molecules were modeled in a head-to-tail orientation, with Puf repeats 8, 7, and 6 contacting the UGU sequences in Boxes A and B (Fig. 1). From HYDROPRO (Garcia De La Torre et al. 2000), the calculated sedimentation coefficients for models 1, 2, and 3 are 3.4S, 4.9S, and 5.3S, respectively. Bearing in mind the relatively small difference in S value between models 2 and 3, these calculations seem to suggest that the dimer species observed with NRE1 (5.8 ± 0.5S) and NRE2 (5.6 ± 0.3S) may be arranged more akin to the circular arrangement (5.3S) in model 3 than to the extended arrangement in model 2 (4.9S). The results from HYDRPRO for model 1 (3.4S) are also in close agreement for monomer species observed with NRE1 (3.8S). Taken together, the FA and AU data show that each NRE binds two Puf domains and that, at least in the case of NRE2, binding is cooperative.
FIGURE 3.
Analytical ultracentrifugation. (A) Sedimentation velocity analysis showing a plot of c(s) (continuous sedimentation coefficient) distribution versus sedimentation coefficient of human Pum Puf domain-NRE1 and -NRE2 complexes. The sedimentation coefficients have been corrected to standard conditions and are reported in units of Svedbergs, where 1 S = 1 × 10−13 sec. (B) Models of Pum-NRE2 complex. Model 1, one Pum molecule bound to the Box B site; model 2, two Pum molecules modeled head-to-tail on an “extended” NRE2; and model 3, two Pum molecules modeled head-to-tail on “circular” NRE2.
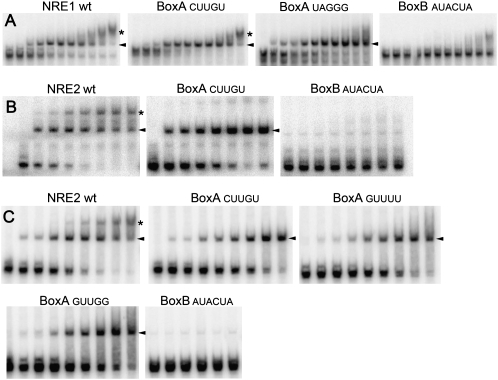
To further test the co-occupancy of two Pum molecules on a single NRE, we performed three sets of gel mobility shift experiments. First, we observe the formation of two complexes upon incubation of the HsPum Puf domain with RNA containing the wt hb NRE1 sequence (Fig. 4A). A single nucleotide substitution in Box A (GUUGU) has very little effect on binding; however, multiple substitutions in Box A (e.g., as in the FA experiment of Fig. 1) eliminate binding of the second molecule of HsPum. Substitutions in the 3′ Box B UGU essentially abolish complex formation. Second, we asked whether HsPum binds in a similar manner to hb NRE2, for which analysis of activity in vivo of various mutant sites is much better characterized (Wharton et al. 1998). As shown in Figure 4B, HsPum binds in a very similar manner to each NRE; the only significant difference we observed is that a single G-to-C substitution in Box A abolishes binding of the second Puf domain to NRE2, whereas the analogous substitution in NRE1 has essentially no effect on binding. Third, we asked whether binding of HsPum mirrors binding of the natural regulator of hb in vivo, DmPum. As shown in Figure 4C, single nucleotide substitutions in and around the Box A UGU abolish binding of a second molecule of DmPum, and substitutions in the Box B UGU abolish binding altogether.
FIGURE 4.
Gel mobility shift experiments. (A) Binding reactions with HsPum (0, 0.0062, 0.0125, 0.025, 0.05, 0.10, 0.21, 0.41, 0.83, 1.65, and 3.3 μM) and RNAs bearing the indicated NRE1 sites. (B) Binding reactions with HsPum (0, 0.05, 0.10, 0.21, 0.41, 0.83, 1.65, and 3.3 μM) and RNAs bearing the indicated NRE2 sites. (C) Binding reactions with DmPum (0, 0.025, 0.05, 0.10, 0.21, 0.41, 0.83, 1.65, and 3.3 μM) and RNAs bearing the indicated NRE2 sites. Reactions with the Box B AUACUA mutant contained 0, 0.025, 0.08, 0.26, 0.52, 1.03 μM DmPum. In all three panels, the presumptive 1 Pum:1 NRE and 2 Pum:1 NRE complexes are indicated with arrowheads and asterisks, respectively.
In summary, the data presented above show that each hb NRE is composed of two Pum binding sites, and that two Puf domains can co-occupy a single NRE. Binding of the HsPum and DmPum proteins appears to be essentially identical, perhaps not surprisingly given the structural similarity of the two Puf domains (Edwards et al. 2001; Wang et al. 2001), and that all of the key residues that interact with RNA are identical in the two proteins (Wang et al. 2002). The only significant discrepancy between the FA and gel shift experiments is that binding appears to be cooperative in gel shift experiments on both NRE1 and NRE2 (e.g., binding of Pum to the low affinity Box A sequence is dependent on binding to a wt Box B element); in contrast, two Pum molecules bind independently to NRE1 in FA experiments. While we do not understand the basis for this apparent discrepancy, we note that the two assays employ very different reaction conditions and time courses and that cooperative binding in vivo could easily be modified by portions of the protein outside the RBD. Nevertheless, all the data in Figures 2–4 support the main conclusion of this work: namely, that the two Puf domains co-occupy a single NRE, the functional unit in vivo.
The data we present here contrast with a previously published report that a single molecule of Pum binds to a hb NRE (Zamore et al. 1999). The finding that two Pum molecules can co-occupy a single NRE partially resolves a paradox: whereas mutations that simultaneously disrupt Pum binding in vitro and regulation in vivo are spread over 21 nt of hb NRE2 (Wharton and Struhl 1991; Wharton et al. 1998), cocrystal structures of Pum with fragments of hb NREs show that a single Puf domain contacts only 8–10 nt of RNA (Wang et al. 2002). The extended mutational footprint of Pum is too large to be accounted by a single Pum molecule but can be accounted by one Pum molecule binding to a “strong” Box B sequence at the 3′ end of the NRE and another molecule binding to a “weaker” Box A sequence at the 5′ end. While we are uncertain of the role of cooperative binding in vivo, our experiments, taken with other data (Wharton et al. 1998), suggest that co-occupancy of both Box A and Box B is essential for NRE activity. Consistent with this conclusion, measurement of the concentration of Pum in the early embryo (>40 nM) (Zamore et al. 1999) suggests that both Boxes are occupied in vivo.
In addition to its role in the translational repression of maternal hb mRNA, Pum also represses translation of maternal CycB mRNA in primordial germ cells (PGCs) in Drosophila (Lin and Spradling 1997; Asaoka-Taguchi et al. 1999). As with the hb NRE, mutations that disrupt binding of Pum to CycB NRE are distributed over a relatively large region (23 nt), and gel mobility shift experiments indicate two protein–RNA complexes (Kadyrova et al. 2007). However, the distance between UGU containing Boxes A and B in CycB NRE is longer than in hb NREs, resulting in a different “dimer” topography when Pum binds to these sites. This may explain why the Brat cofactor is recruited to hb but not to CycB (Kadyrova et al. 2007). Pum is also thought to regulate a number of other biological processes, including learning and memory (Dubnau et al. 2003; Menon et al. 2004; Ye et al. 2004). Pum may adopt other arrangements on the relevant mRNAs, though many of the relevant target mRNAs remain to be definitively identified (Wharton and Aggarwal 2006). Pum could even function as a monomer on some of the sites, as appears to be the case for Pum homologs in yeast and C. elegans (Olivas and Parker 2000; Gerber et al. 2004; Opperman et al. 2005). The ability to bind as one, two or more molecules expands both the repertoire of sites that Pum can target in vivo and the combinatorial interactions with potential cofactors.
MATERIALS AND METHODS
Protein purification
Expression of HsPum Puf (amino acids 828–1176) and DmPum Puf (amino acids 1092–1411) domains was achieved in E. coli BL21 (DE3) pLysS cells from a pET19b vector (Novagen) modified to contain a Tev protease recognition site for removal of the N-terminal 10× His tag. Cells were grown in LB medium with ampicillin (100 μg/mL) at 37°C to an OD600 of 0.8 and then induced overnight at 18°C with 0.4 mM IPTG. Cells were harvested by centrifugation, resuspended in buffer A (25 mM Tris, 500 mM NaCl, 10% glycerol at pH 7.9) with 1 mM PMSF, and sonicated in the presence of DNase I (1 U/mL). The insoluble fraction was removed by centrifugation, and the soluble fraction was filtered through 0.22-μm membranes prior to loading onto a 5 mL Ni2+-charged chelating column (Pharmacia). The column was washed with buffer A; buffer A plus 10, 20, 30, and 50 mM imidazole; and then eluted with buffer A plus 250 mM imidazole. The His-tag was removed by digestion with Tev protease overnight at 4°C. Monomeric protein was then purified by gel filtration by a Superdex75 16/60 column (Pharmacia) washed with 25 mM Tris (pH 7.4), 150 mM NaCl, 2 mM DTT, and 5% glycerol.
Fluorescence anisotropy
The 5′-Fl–labeled RNAs were synthesized and deprotected as recommended by the manufacturer (Dharmacon Research). Fluorescence emission intensities were collected on a Panvera Beacon 2000 fluorescence polarization system (at 20°C), and the anisotropy values calculated as previously described (Lone et al. 2007). Each reaction sample (total volume of 200 μL) consisted of 1 nM 5′Fl labeled RNA and increasing concentrations of the protein (from 0.0 to 500 nM) in binding buffer (10 mM HEPES at pH 7.5, 50 mM KCl, 1 mM DTT, 1 mM EDTA, 0.1 mg/mL BSA). Samples were equilibrated at room temperature for >30 min before the FA values were measured. For each data point, eight anisotropy values were recorded and averaged. Anisotropy values were referenced against a blank buffer at the beginning of each experiment to account for background correction. Anisotropy values were normalized by first subtracting the anisotropy value with no protein added and then dividing by the maximum anisotropy value for a particular RNA series. Anisotropy values were then plotted versus protein concentration and the data for NRE1 complexes fitted by nonlinear least squares regression, using Origin 7 (OriginLab), to the following quadratic equation:
where θ is the fraction of RNA bound, Ro is the total concentration of RNA, Po is the total protein concentration, and Kd is the dissociation constant.
The NRE2wt binding isotherm was fit to the equation below for cooperative two-site binding:
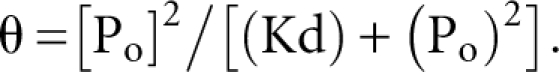 |
Analytical ultracentrifugation
The sedimentation velocity experiments were performed at 20°C in a Beckman XL-I Analytical Ultracentrifuge using an An60Ti rotor. Samples were prepared by mixing 10 μM of Fl labeled NRE1/NRE2 with 3 molar excess of HsPum in the buffer containing 10 mM HEPES (pH 7.5), and 50 mM KCl. Samples were centrifuged at 13,200 rpm for 10 min before loading onto aluminum double sector cell. The concentration profiles were recorded using the absorption scanning optics at a wavelength of 495 nm for Fl-labeled RNA. The experiments were carried out in aluminum double sector cells at 45,000 rpm. Data were analyzed according to the c(s) distribution function of the Lamm equation solutions calculated with the program SEDFIT (Schuck 2000; Schuck et al. 2002). The method also yields an apparent weight-average frictional ratio (f/f0), and the two values were used to calculate the molecular weights. The program SEDNTERP (http://www.rasmb.bbri.org) was used to calculate the partial specific volume (v), the density of the solution (ρ), and the viscosity (η).
Gel mobility shift experiments
Synthetic 32P-labeled RNA was prepared using HindIII- or EcoRI-linearized derivatives of plasmid T4425 which encode wt hb NRE1 embedded in hb 3′ UTR and Bluescript vector sequences (cuagCAUAUAAUCGUUGUCCAGAAUUGUAUAUAUUCGUggauc), a G10C Box A mutant (as in wt, but CUUGU), a multiply mutant Box A site (as in wt, but UAGGG), and a Box B mutant (as in wt, but AUACUA). (Vector-encoded nucleotides at the cloning sites are lower case, Box A and Box B nucleotides are bold, and substitutions are underlined.) Plasmids encoding wt hb NRE2 (cuaguAUUAUUUUGUUGUCGAAAAUUGUACAUAAGCCcuag, a G9C Box A mutant (as in wt, but CUUGU), a G12U Box A mutant (as in wt, but GUUUU), a U13G Box A mutant (as in wt, but GUUGG), and a UG21AC Box B mutant (as in wt, but AUACUA) were also used. The resulting transcripts are 112 nt long. Experiments were performed essentially as described (Murata and Wharton 1995) with the following modifications: Each 10 μL reaction contained purified protein, reaction buffer [10 mM HEPES at pH 7.4, 20 mM KCl, 1 mM DTT, 0.2 mg/mL heparin, 0.05 mg/mL poly(U), 5% glycerol], and heat-denatured radiolabeled RNA (10,000 cpm). Note that the FA reactions in Figure 2 are performed under different conditions and with significantly shorter RNAs, presumably accounting (at least in part) for the apparent difference in Kd for binding to the same sequences in the two experiments.
ACKNOWLEDGMENTS
We thank S. Pyle for performing some of the experiments of Figure 4. This work was supported by NIH grant GM062947 to A.K.A and R.P.W.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1327609.
REFERENCES
- Asaoka-Taguchi M., Yamada M., Nakamura A., Hanyu K., Kobayashi S. Maternal Pumilio acts together with Nanos in germline development in Drosophila embryos. Nat. Cell Biol. 1999;1:431–437. doi: 10.1038/15666. [DOI] [PubMed] [Google Scholar]
- Dubnau J., Chiang A.S., Grady L., Barditch J., Gossweiler S., McNeil J., Smith P., Buldoc F., Scott R., Certa U., et al. The staufen/pumilio pathway is involved in Drosophila long-term memory. Curr. Biol. 2003;13:286–296. doi: 10.1016/s0960-9822(03)00064-2. [DOI] [PubMed] [Google Scholar]
- Edwards T.A., Pyle S.E., Wharton R.P., Aggarwal A.K. Structure of Pumilio reveals similarity between RNA and peptide binding motifs. Cell. 2001;105:281–289. doi: 10.1016/s0092-8674(01)00318-x. [DOI] [PubMed] [Google Scholar]
- Garcia De La Torre J., Huertas M.L., Carrasco B. Calculation of hydrodynamic properties of globular proteins from their atomic-level structure. Biophys. J. 2000;78:719–730. doi: 10.1016/S0006-3495(00)76630-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber A.P., Herschlag D., Brown P.O. Extensive association of functionally and cytotopically related mRNAs with Puf family RNA-binding proteins in yeast. PLoS Biol. 2004;2:E79. doi: 10.1371/journal.pbio.0020079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta Y.K., Nair D.T., Wharton R.P., Aggarwal A.K. Structures of human Pumilio with noncognate RNAs reveal molecular mechanisms for binding promiscuity. Structure. 2008;16:549–557. doi: 10.1016/j.str.2008.01.006. [DOI] [PubMed] [Google Scholar]
- Kadyrova L.Y., Habara Y., Lee T.H., Wharton R.P. Translational control of maternal cyclin B mRNA by Nanos in the Drosophila germline. Development. 2007;134:1519–1527. doi: 10.1242/dev.002212. [DOI] [PubMed] [Google Scholar]
- Lehmann R., Nusslein-Volhard C. Involvement of the pumilio gene in the transport of an abdominal signal in the Drosophila embryo. Nature. 1987;329:167–170. [Google Scholar]
- Lin H., Spradling A.C. A novel group of pumilio mutations affects the asymmetric division of germline stem cells in the Drosophila ovary. Development. 1997;124:2463–2476. doi: 10.1242/dev.124.12.2463. [DOI] [PubMed] [Google Scholar]
- Lone S., Townson S.A., Uljon S.N., Johnson R.E., Brahma A., Nair D.T., Prakash S., Prakash L., Aggarwal A.K. Human DNA polymerase κ encircles DNA: Implications for mismatch extension and lesion bypass. Mol. Cell. 2007;25:601–614. doi: 10.1016/j.molcel.2007.01.018. [DOI] [PubMed] [Google Scholar]
- Menon K.P., Sanyal S., Habara Y., Sanchez R., Wharton R.P., Ramaswami M., Zinn K. The translational repressor Pumilio regulates presynaptic morphology and controls post-synaptic accumulation of translation factor eIF-4E. Neuron. 2004;44:663–676. doi: 10.1016/j.neuron.2004.10.028. [DOI] [PubMed] [Google Scholar]
- Miller M.T., Higgin J.J., Hall T.M. Basis of altered RNA-binding specificity by PUF proteins revealed by crystal structures of yeast Puf4p. Nat. Struct. Mol. Biol. 2008;15:397–402. doi: 10.1038/nsmb.1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata Y., Wharton R.P. Binding of Pumilio to maternal hunchback mRNA is required for posterior patterning in Drosophila embryos. Cell. 1995;80:747–756. doi: 10.1016/0092-8674(95)90353-4. [DOI] [PubMed] [Google Scholar]
- Olivas W., Parker R. The Puf3 protein is a transcript-specific regulator of mRNA degradation in yeast. EMBO J. 2000;19:6602–6611. doi: 10.1093/emboj/19.23.6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opperman L., Hook B., DeFino M., Bernstein D.S., Wickens M. A single spacer nucleotide determines the specificities of two mRNA regulatory proteins. Nat. Struct. Mol. Biol. 2005;12:945–951. doi: 10.1038/nsmb1010. [DOI] [PubMed] [Google Scholar]
- Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and Lamm equation modeling. Biophys. J. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuck P., Perugini M.A., Gonzales N.R., Howlett G.J., Schubert D. Size-distribution analysis of proteins by analytical ultracentrifugation: Strategies and application to model systems. Biophys. J. 2002;82:1096–1111. doi: 10.1016/S0006-3495(02)75469-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda J., Wharton R.P. Recruitment of Nanos to hunchback mRNA by Pumilio. Genes & Dev. 1999;13:2704–2712. doi: 10.1101/gad.13.20.2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda J., Wharton R.P. Drosophila brain tumor is a translational repressor. Genes & Dev. 2001;15:762–773. doi: 10.1101/gad.870801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vistica J., Dam J., Balbo A., Yikilmaz E., Mariuzza R.A., Rouault T.A., Schuck P. Sedimentation equilibrium analysis of protein interactions with global implicit mass conservation constraints and systematic noise decomposition. Anal. Biochem. 2004;326:234–256. doi: 10.1016/j.ab.2003.12.014. [DOI] [PubMed] [Google Scholar]
- Wang X., Zamore P.D., Hall T.M. Crystal structure of a Pumilio homology domain. Mol. Cell. 2001;7:855–865. doi: 10.1016/s1097-2765(01)00229-5. [DOI] [PubMed] [Google Scholar]
- Wang X., McLachlan J., Zamore P.D., Hall T.M. Modular recognition of RNA by a human Pumilio-homology domain. Cell. 2002;110:501–512. doi: 10.1016/s0092-8674(02)00873-5. [DOI] [PubMed] [Google Scholar]
- Wharton R.P., Aggarwal A.K. mRNA regulation by Puf domain proteins. Sci. STKE. 2006;2006:pe37. doi: 10.1126/stke.3542006pe37. [DOI] [PubMed] [Google Scholar]
- Wharton R.P., Struhl G. RNA regulatory elements mediate control of Drosophila body pattern by the posterior morphogen Nanos. Cell. 1991;67:955–967. doi: 10.1016/0092-8674(91)90368-9. [DOI] [PubMed] [Google Scholar]
- Wharton R.P., Sonoda J., Lee T., Patterson M., Murata Y. The Pumilio RNA-binding domain is also a translational regulator. Mol. Cell. 1998;1:863–872. doi: 10.1016/s1097-2765(00)80085-4. [DOI] [PubMed] [Google Scholar]
- Wickens M., Bernstein D.S., Kimble J., Parker R. A PUF family portrait: 3′ UTR regulation as a way of life. Trends Genet. 2002;18:150–157. doi: 10.1016/s0168-9525(01)02616-6. [DOI] [PubMed] [Google Scholar]
- Ye B., Petritsch C., Clark I.E., Gavis E.R., Jan L.Y., Jan Y.N. Nanos and Pumilio are essential for dendrite morphogenesis in Drosophila peripheral neurons. Curr. Biol. 2004;14:314–321. doi: 10.1016/j.cub.2004.01.052. [DOI] [PubMed] [Google Scholar]
- Zamore P.D., Williamson J.R., Lehmann R. The Pumilio protein binds RNA through a conserved domain that defines a new class of RNA-binding proteins. RNA. 1997;3:1421–1433. [PMC free article] [PubMed] [Google Scholar]
- Zamore P.D., Bartel D.P., Lehmann R., Williamson J.R. The PUMILIO–RNA interaction: A single RNA-binding domain monomer recognizes a bipartite target sequence. Biochemistry. 1999;38:596–604. doi: 10.1021/bi982264s. [DOI] [PubMed] [Google Scholar]