Abstract
Proteins of the GW182 family are essential for miRNA-mediated gene silencing in animal cells; they interact with Argonaute proteins (AGOs) and are required for both the translational repression and mRNA degradation mediated by miRNAs. To gain insight into the role of the GW182–AGO1 interaction in silencing, we generated protein mutants that do not interact and tested them in complementation assays. We show that silencing of miRNA targets requires the N-terminal domain of GW182, which interacts with AGO1 through multiple glycine–tryptophan (GW)-repeats. Indeed, a GW182 mutant that does not interact with AGO1 cannot rescue silencing in cells depleted of endogenous GW182. Conversely, silencing is impaired by mutations in AGO1 that strongly reduce the interaction with GW182 but not with miRNAs. We further show that a GW182 mutant that does not localize to P-bodies but interacts with AGO1 rescues silencing in GW182-depleted cells, even though in these cells, AGO1 also fails to localize to P-bodies. Finally, we show that in addition to the N-terminal AGO1-binding domain, the middle and C-terminal regions of GW182 (referred to as the bipartite silencing domain) are essential for silencing. Together our results indicate that miRNA silencing in animal cells is mediated by AGO1 in complex with GW182, and that P-body localization is not required for silencing.
Keywords: Argonaute, GW182, GW-repeats, miRNAs, mRNA decay, P-bodies, TNRC6
INTRODUCTION
Proteins of the GW182 family interact with Argonaute proteins and are required for miRNA-mediated gene silencing in animal cells (for review, see Ding and Han 2007; Eulalio et al. 2007a, 2008a). Studies in Drosophila melanogaster showed that GW182 interacts with Argonaute-1 (AGO1, the Argonaute protein dedicated to the miRNA pathway in flies) (Behm-Ansmant et al. 2006a,b; Schneider et al. 2006); furthermore, depleting GW182 suppresses silencing of miRNA targets, irrespective of whether they are translationally repressed or directed to degradation (Rehwinkel et al. 2005; Behm-Ansmant et al. 2006a,b; Eulalio et al. 2008b). These and additional observations have shaped the model in which silencing by miRNAs is effected by a protein complex consisting minimally of AGO1 and GW182 (Eulalio et al. 2008a,b).
The essential role of GW182 proteins in the miRNA pathway has been more difficult to demonstrate in other multicellular organisms because they contain multiple paralogs with partially redundant functions. Indeed, there are three GW182 paralogs in vertebrates (known as TNRC6A, TNRC6B, and TNRC6C) and two in Caenorhabditis elegans (AIN-1 and AIN-2). Both AIN-1 and AIN-2 interact with C. elegans Argonaute proteins 1 and 2 (ALG-1 and ALG-2); moreover, co-depleting AIN-1 and AIN-2 suppresses silencing more efficiently than depleting each protein individually, indicating that AIN-1 and AIN-2 are functionally redundant (Ding et al. 2005; Zhang et al. 2007; Ding and Grosshans 2009). The idea that GW182 proteins are redundant in human cells is supported by the observation that TRNC6A, TNRC6B, and TNRC6C interact with all four human Argonaute proteins (AGO1–4), and with a common set of mRNA targets (Landthaler et al. 2008). Furthermore, depleting either TRNC6A or B partially relieves silencing mediated by siRNAs and miRNAs (Jakymiw et al. 2005; Liu et al. 2005a; Meister et al. 2005; Chu and Rana 2006).
D. melanogaster GW182 and the vertebrate TNRC6A–C proteins are characterized by multiple glycine–tryptophan (GW) repeats scattered within the N-terminal third of the proteins. Additional repeats can be present in variable numbers in the middle and C-terminal regions, which are predicted to be unstructured (Eystathioy et al. 2002; Behm-Ansmant et al. 2006a; for review, see Ding and Han 2007). The N-terminal and middle GW-repeat-containing regions are separated by an ubiquitin-associated (UBA)-like domain and a glutamine/asparagine-rich (Q/N-rich) region, whereas between the middle and C-terminal regions lies a conserved RNA recognition motif (RRM) (Eystathioy et al. 2002; Behm-Ansmant et al. 2006a; Eulalio et al. 2009a). The C. elegans AIN-1 and AIN-2 proteins are divergent members of the GW182 family. They share similarities with the N-terminal GW-repeats of insect GW182 and vertebrate TNRC6A–C proteins but lack all downstream sequences including the UBA-like and RRM domains, as well as the Q/N-rich, middle, and C-terminal unstructured regions (Ding et al. 2005; Behm-Ansmant et al. 2006a; Zhang et al. 2007).
In D. melanogaster GW182, the N-terminal GW-repeat-containing region is required for the interaction with AGO1, whereas P-body localization requires additional sequences comprising the UBA-like and the Q/N-rich domains (Behm-Ansmant et al. 2006a). How these and the additional domains affect the activity of GW182 proteins is less well understood. In this context, it is particularly intriguing that the C. elegans proteins function in the miRNA pathway, despite the fact that they consist exclusively of the N-terminal GW-repeat region.
To probe the role of GW182 in the miRNA pathway, we generated a series of deletion and point mutants and tested if in cells depleted of endogenous GW182, these mutants mediate silencing. We also tested if the mutant proteins could interact with both AGO1 and miRNAs, or localize to P-bodies. We show that for GW182 to function in the miRNA pathway, it must interact with AGO1; in contrast, it does not need to localize to P-bodies. In agreement with this, to mediate silencing, AGO1 requires the interaction with GW182. Finally, we show that in addition to the N-terminal AGO1-binding domain, the middle and C-terminal low-complexity regions of GW182 are essential for silencing. Together our results indicate that GW182 acts downstream from the step where miRNA loads onto AGO1; furthermore, its role in silencing is unrelated to its ability to promote AGO1 accumulation in P-bodies.
RESULTS
A GW-repeat embedded in motif I of GW182 provides a major binding site for AGO1
Previously, we showed that the N-terminal region of D. melanogaster GW182 (residues 1–539) is necessary and sufficient for the protein to interact with AGO1 (Behm-Ansmant et al. 2006a). This region is characterized by two highly conserved motifs (motifs I and II) and 12 glycine–tryptophan (GW) repeats (Fig. 1A). Notably, for a variety of proteins, GW-repeats were recently shown to determine the specificity for Argonaute proteins (El-Shami et al. 2007; Till et al. 2007). Moreover, Till et al. (2007) identified a GW-repeat-containing sequence in the N-terminal region of human TNRC6B (termed the AGO-hook, downstream from motif II), which is sufficient to mediate an interaction with human AGO2 in vitro. It has not been established, however, whether the AGO-hook is necessary for the AGO2 interaction in the context of full-length TNRC6B. Furthermore, the AGO-hook motif is not conserved in D. melanogaster GW182.
FIGURE 1.
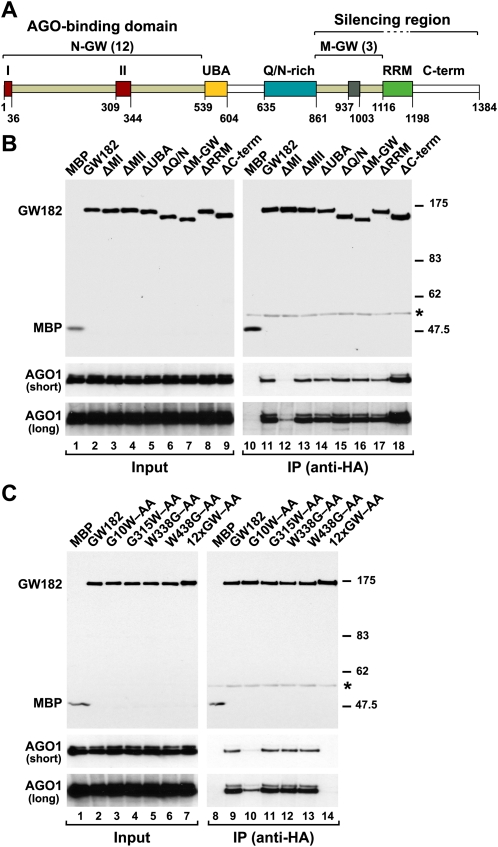
A GW-repeat in motif I of D. melanogaster GW182 provides a major binding site for AGO1. (A) Domain organization of GW182. N-GW and M-GW: N-terminal and middle GW-repeat-containing regions, respectively, with the number of repeats indicated in brackets; (UBA) ubiquitin associated-like domain; (Q/N-rich) region rich in glutamine (16.7%) and asparagine (14.5%); (RRM) RNA recognition motif; (C-term) C-terminal region. Red boxes I and II: two conserved motifs within the N-terminal GW-repeats. Gray box: conserved motif III in the middle region. The bipartite silencing region includes the M-GW and C-terminal regions but not the RRM, which is dispensable for silencing (Eulalio et al. 2009a). Amino acid positions at domain boundaries are indicated. (B,C) Lysates from S2 cells expressing HA-tagged versions of MBP, wild-type GW182, or GW182 mutants were immunoprecipitated using a monoclonal anti-HA antibody. Inputs (1.5%) and immunoprecipitates (30%) were analyzed by Western blotting using a polyclonal anti-HA antibody. Endogenous AGO1 was detected by Western blotting using anti-AGO1 antibodies (two different exposures are shown, short and long). (Asterisks) Indicate cross-reactivity with the immunoglobulin heavy chain by the secondary antibody (IP panels).
To generate GW182 mutants that do not interact with AGO1, we substituted N-terminal GW-repeats with alanines; we also deleted motifs I and II (see Supplemental Table 1 for a complete list of GW182 mutants). The interaction of GW182 mutants with AGO1 was tested in coimmunoprecipitation assays. To this end, HA-epitope-tagged wild-type and mutant GW182s were transiently expressed in D. melanogaster S2 cells and tested for the ability to coimmunoprecipitate endogenous AGO1. We found that deleting motif I (ΔMI), or substituting the single GWG-repeat in this motif with alanines (G10W–AA), strongly impaired the interaction with AGO1 (Fig. 1B, lane 12, C, lane 10). In contrast, deleting motif II did not prevent the interaction with AGO1 (Fig. 1B, ΔMII, lane 13). Similarly, the interaction between GW182 and AGO1 was not affected by substituting alanines for GW-repeats outside of motifs I and II (Fig. 1C, lane 13; Supplemental Table 1).
Only when all 12 N-terminal GW-repeats were substituted with alanines was the interaction between GW182 and AGO1 completely abolished (Fig. 1C, 12xGW–AA, lane 14). Together these results indicate that in the context of full-length D. melanogaster GW182, motif I is the major binding site for AGO1. The additional GW-repeats appear to contribute to the interaction by providing low-affinity binding sites. These results also indicate that the GW-repeats in the middle domain of D. melanogaster GW182 do not contribute to the interaction with AGO1. In agreement with this, deleting this middle region had no effect on the AGO1 interaction (Fig. 1B, ΔM-GW, lane 16).
GW182 interaction with AGO1 is essential for silencing
To investigate whether the GW182 mutants described above function in silencing, we used a complementation assay described before (Eulalio et al. 2009a). Briefly, in this assay, endogenous GW182 is depleted using dsRNAs that target the 5′- and 3′-UTR sequences of GW182 mRNA. This depletion inhibits miRNA-mediated silencing (Figs. 2, 3), although less efficiently than by dsRNAs targeting GW182 ORF. GW182 mutants are then tested for the ability to restore silencing in GW182-depleted cells. Mutant proteins are expressed from mRNAs including only the ORF of GW182; therefore, their expression is immune to the action of the dsRNAs.
FIGURE 2.
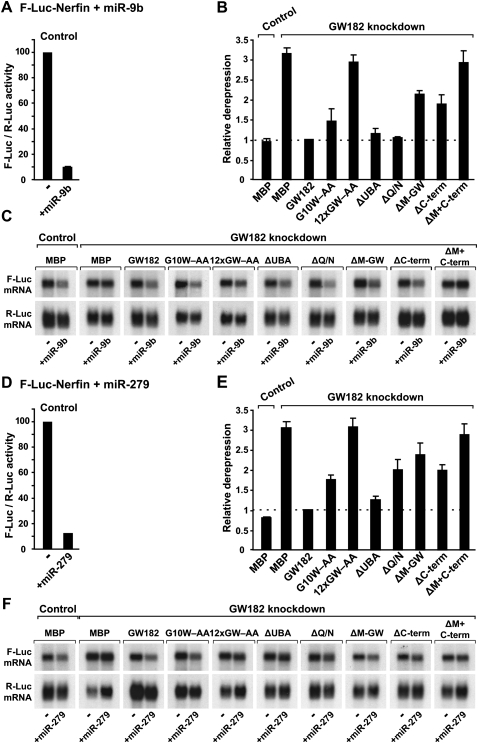
Role of GW182 domains in silencing. (A–F) S2 cells were treated with dsRNA targeting the 5′- and 3′-UTRs of GW182 mRNA. Control cells were treated with GFP dsRNA. These cells were subsequently transfected with a mixture of three plasmids: one expressing the F-Luc-Nerfin-1 reporter; another expressing miRNA primary transcripts (+miR-9b or miR-279) or the corresponding empty vector (−); and a third expressing Renilla luciferase (R-Luc). Plasmids (25 ng) encoding wild-type HA-GW182, HA-GW182 mutants, or HA-MBP were included in the transfection mixtures, as indicated. Firefly luciferase activities were normalized to those of the Renilla luciferase transfection control and set to 100 in cells transfected with the empty vector (i.e., in the absence of the miRNAs). (A,D) Normalized Firefly luciferase activities in the absence or presence of miRNAs in control cells (i.e., cells treated with GFP dsRNA and transfected with MBP). (B,E) Relative fold de-repression for each condition. Mean values ± standard deviations from three independent experiments are shown. (C,F) Northern blot analysis of representative RNA samples shown in B and E.
FIGURE 3.
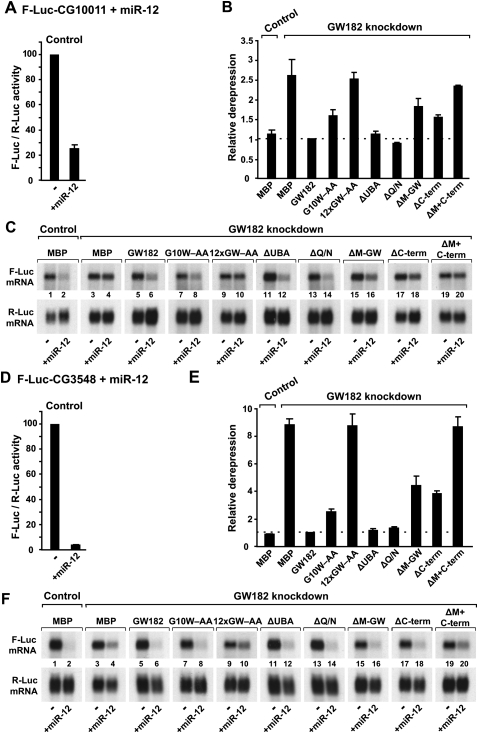
Role of GW182 domains in silencing. (A–F) S2 cells were treated with dsRNA targeting the 5′- and 3′-UTRs of GW182 mRNA. Control cells were treated with GFP dsRNA. These cells were subsequently transfected with a mixture of three plasmids: one expressing the indicated F-Luc reporters; another expressing miR-12 primary transcripts (+miR-12) or the corresponding empty vector (−); and a third expressing Renilla luciferase (R-Luc). Plasmids (25 ng) encoding wild-type HA-GW182, HA-GW182 mutants, or HA-MBP were included in the transfection mixtures, as indicated. Firefly luciferase activities were normalized to those of the Renilla luciferase transfection control and analyzed as described in Figure 2. (C,F) Northern blot analysis of representative RNA samples shown in B and E.
We monitored miRNA activity using previously characterized firefly luciferase reporters. These include the F-Luc-Nerfin-1 (silenced by miR-9b and miR-279) (Fig. 2) and the F-Luc-CG10011 and F-Luc-CG3548 reporters (silenced by miR-12) (Fig. 3). miR-9b and miR-279 silence the F-Luc-Nerfin-1 reporter mainly at the translational level (Fig. 2A,C,D,F), whereas miR-12 directs the CG10011 and CG3548 reporters to degradation (Fig. 3A,C,D,F; Behm-Ansmant et al. 2006a,b; Eulalio et al. 2007b, 2008b).
Depleting endogenous GW182 suppressed silencing of all reporters leading to a 2.5-fold to ninefold increase in firefly luciferase expression (Figs. 2B,E, 3B,E). A corresponding increase in mRNA levels was observed for the reporters that were degraded (Fig. 3C,F, cf. lane 4 and lane 2). Transfecting GW182-depleted cells with a plasmid expressing wild-type GW182, fully restored silencing (Figs. 2B,E, 3B,E). Silencing was restored over a broad range of expression plasmid transfected (from 10 ng to 1 μg) (data not shown).
Proteins that carried alanine substitutions of GW-repeats and yet retained the ability to interact with AGO1 were active in the complementation assay (Supplemental Table 1). The GW182 mutant (G10W–AA), which was impaired in AGO1 binding, was also impaired in the complementation assay when low amounts of expression plasmid were transfected (Figs. 2B,E, 3B,E). This mutant, however, complemented silencing of the F-Luc-Nerfin-1 reporter when higher amounts of expression plasmid were transfected (Supplemental Fig. 1E,F), indicating that the residual AGO1-binding activity of this mutant is sufficient to rescue silencing when it is overexpressed.
In contrast, the GW182 mutant in which all 12 N-terminal GW-repeats were substituted by alanines (12xGW–AA) failed to rescue silencing at all concentrations tested (Figs. 2B,E, 3B,E; Supplemental Fig. 1D–F). The expression levels of wild-type and mutant GW812 proteins were comparable (Fig. 1). Because the expression levels of AGO1 or of miRNAs are not affected in cells depleted of GW182 (Fig. 6A,B, below), these results provide compelling evidence that the interaction between GW182 and AGO1 is essential for silencing.
FIGURE 6.
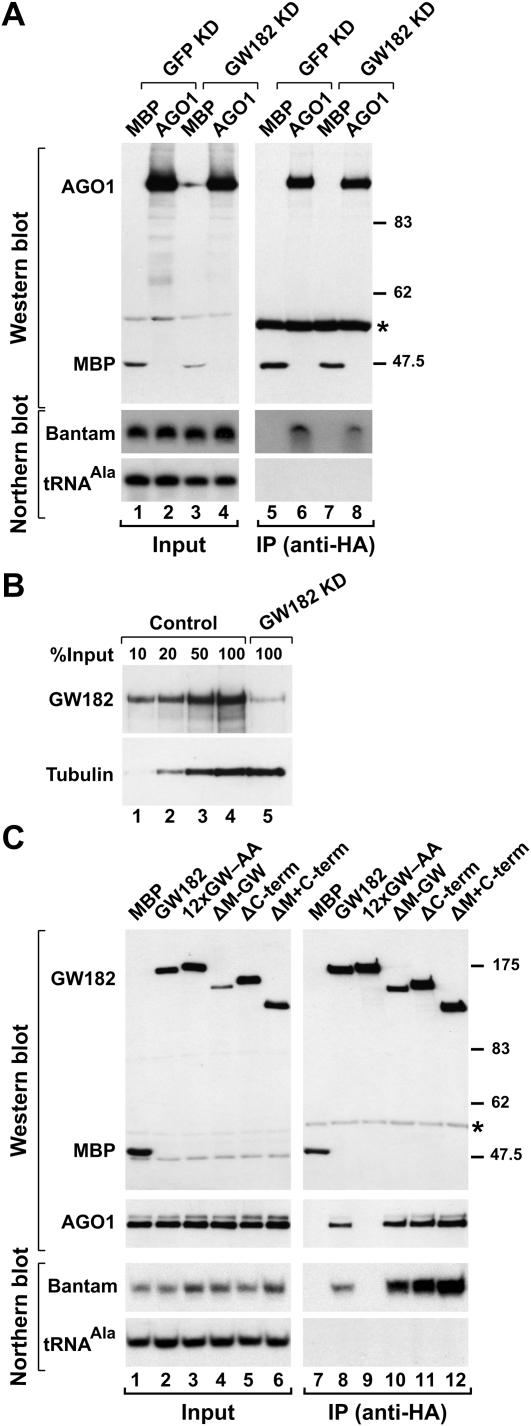
GW182 functions downstream from miRNA loading onto AGO1. (A) S2 cells were treated (Control) GFP or (GW182 KD) GW182 dsRNAs on days 0 and 4, and transfected on day 6 with plasmids expressing HA-MBP or HA-AGO1. Proteins were immunoprecipitated using a monoclonal anti-HA antibody. Inputs (1.5%) and immunoprecipitates (30%) were analyzed by Western blotting using a polyclonal anti-HA antibody. The association between HA-AGO1 and miRNAs was analyzed by Northern blotting. tRNAAla served as a loading control for the Northern blots. (B) The effectiveness of the GW182 depletion was analyzed by Western blotting using anti-GW182 antibodies. (Lanes 1–4) Dilutions of control cells (treated with GFP dsRNA) were loaded. Tubulin served as a loading control. (C) Lysates from S2 cells expressing HA-tagged versions of MBP, wild-type GW182, or GW182 mutants were immunoprecipitated using a monoclonal anti-HA antibody. Inputs (1.5%) and immunoprecipitates (30%) were analyzed by Northern blotting, as described in panel A. Endogenous AGO1 was detected by Western blotting using anti-AGO1 antibodies.
Uncoupling AGO1 interaction with miRNAs and GW182
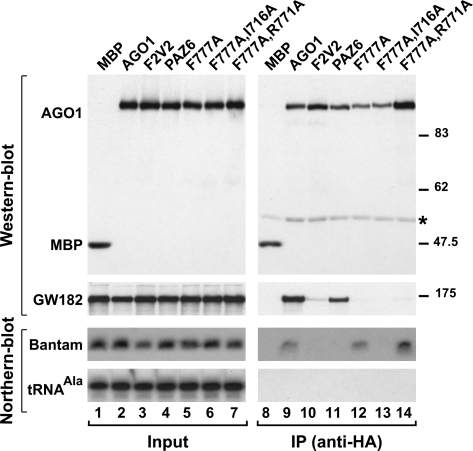
To confirm further the importance of the GW182–AGO1 interaction in miRNA-mediated gene silencing we sought to generate AGO1 mutants that no longer interact with GW182. Previous studies showed that mutations in the mid and PIWI domains of human AGO2 abolish GW182 binding (Till et al. 2007). These mutations may also interfere with miRNA binding, as some of the residues involved in the interaction with GW182 are located in the miRNA 5′-binding pocket (Till et al. 2007). Based on mutations described in a study by Till et al. (2007), we generated the corresponding mutations in D. melanogaster AGO1. To obtain AGO1 mutants that interact with GW182 but not with miRNAs, we mutated the PAZ domain (PAZ6) (Supplemental Table 2; Liu et al. 2005b). As a negative control in parallel, we analyzed an AGO1 mutant carrying valine substitutions of phenylalanines 594 (F594V) and 629 (F629V) (referred to as F2V2); this mutant is inactive in silencing because it cannot interact with either miRNAs or GW182 (Eulalio et al. 2008b).
HA-tagged AGO1 mutants were expressed in D. melanogaster S2 cells and tested for their ability to coimmunoprecipitate endogenous miRNAs and GW182. The presence of miRNAs and GW182 in the immunoprecpitates was detected by Northern and Western blotting, respectively. We found endogenous bantam and GW182 coimmunoprecipitated with wild-type HA-AGO1 but not the F2V2 mutant, as reported before (Fig. 4, lanes 9,10; Eulalio et al. 2008b). Similarly, AGO1 mutants carrying the substitutions R707A, Y749A, or K785A were also strongly impaired in miRNA and GW182 binding (Supplemental Fig. 2A; Supplemental Table 2). In fact, the only mutations that disrupted binding to miRNAs but not to GW182 were those located in the PAZ domain (Fig. 4, PAZ6, lane 11; Supplemental Fig. 2B; Supplemental Table 2).
FIGURE 4.
Mutations in AGO1 that uncouple miRNA and GW182 binding. (A) Lysates from S2 cells expressing HA-tagged versions of MBP, wild-type AGO1, or AGO1 mutants were immunoprecipitated using a monoclonal anti-HA antibody. Inputs (1.5%) and immunoprecipitates (30%) were analyzed by Western blotting using a polyclonal anti-HA antibody. Endogenous GW182 was detected using anti-GW182 antibodies. The association between HA-AGO1 and endogenous bantam was analyzed by Northern blotting. tRNAAla served as a loading control. (Asterisk) IgG cross-reactivity.
Importantly, we identified residues in AGO1 required for the interaction with GW182 but not with miRNAs. Specifically, alanine substitutions of AGO1 residue F777, or of F777 in combination with R771, abolished the interaction with GW182 but not with miRNAs (Fig. 4, lanes 12,14). Nevertheless, the proteins carrying these mutations localized to P-bodies, a process that depends on the interaction with GW182 (Fig. 7, below); thus, these mutants retained a residual binding affinity for GW182, despite being negative in coimmunoprecipitation assays.
FIGURE 7.
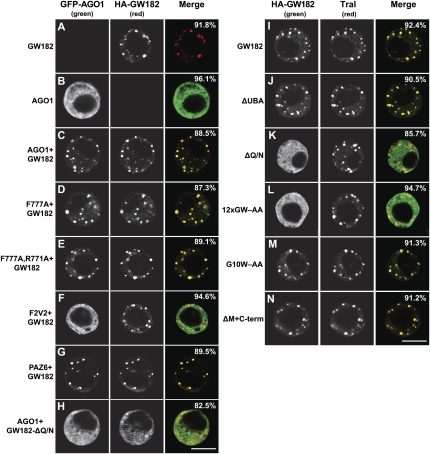
GW182 Q/N-rich region is required for P-body localization. (A–H) GFP-tagged AGO1 or mutants were expressed in S2 cells. In panels C–H, the effect of cotransfecting HA-GW182 or HA-GW182 mutants on the localization of AGO1 was examined. The merged images show the GFP signal in green, the HA signal in red. (I–N) Confocal fluorescent micrographs of fixed S2 cells expressing HA-tagged fusions of full-length GW182 or GW182 mutants. Cells were stained with affinity-purified anti-Tral antibodies. The merged images show the HA signal in green and the anti-Tral signal in red. In all panels, the fraction of cells stained identically to the representative panel was determined by scoring at least 100 cells in two independent transfections performed per protein. Scale bar, 5 μm.
We could not, however, identify additional mutations that completely abolished the interaction of AGO1 with GW182, without affecting miRNA binding (Supplemental Fig. 2; Supplemental Table 2). For instance, a protein carrying a double alanine substitution of residues F777 and I716 no longer interacted with GW182 and miRNAs (Fig. 4, lane 13). The I716A and R771A mutations, on their own, have only a slight effect on GW182 binding (Supplemental Fig. 2A; Supplemental Table 2). These results provide further evidence that the miRNA- and GW182-binding sites on Argonaute proteins are partially overlapping, as has been reported by Till et al. (2007). However, since these two activities can be uncoupled by specific mutations, the binding sites are not interdependent.
The silencing activity of AGO1 depends on the interaction with GW182
To investigate whether AGO1 mutants that are impaired in GW182 binding could still function in miRNA-mediated silencing, we performed complementation assays as described above; however, in this case, we depleted endogenous AGO1 using a specific siRNA (AGO1-siRNA). An siRNA targeting GFP was used as a negative control (GFP-siRNA). AGO1 depletion fully suppressed silencing of the F-Luc reporters as shown before (Fig. 5; Eulalio et al. 2008b). Silencing was restored by expressing an siRNA-resistant version of AGO1 (Fig. 5).
FIGURE 5.
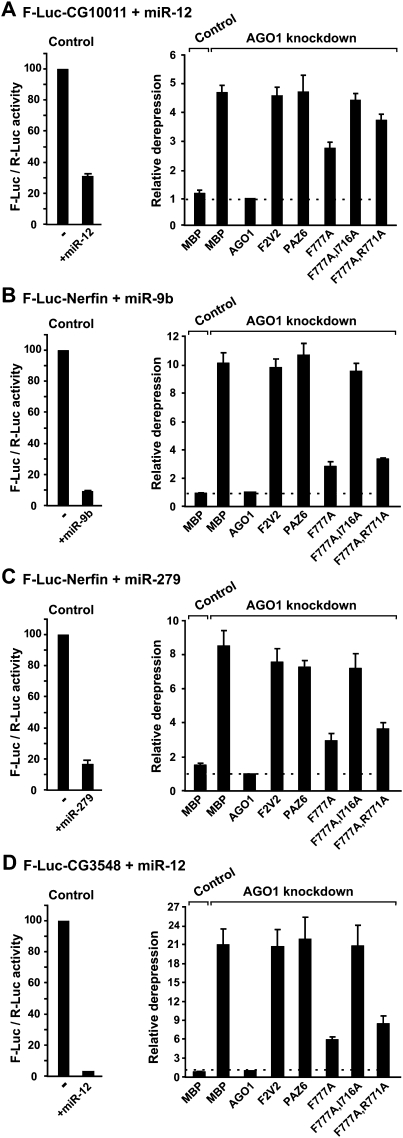
AGO1 mutants impaired in GW182-binding are also impaired in the complementation assay. (A–D) S2 cells were transfected with (Control) GFP-siRNA or (AGO1 knockdown) AGO1-siRNA plus a mixture of three plasmids: one expressing the indicated F-Luc reporters; another expressing miRNA primary transcripts (+miRNA) or the corresponding empty vector (−); and a third expressing Renilla luciferase (R-Luc). Plasmids encoding siRNA-resistant versions of wild-type or mutant HA-AGO1 proteins were included in the transfection mixtures, as indicated. Control cells were cotransfected with a plasmid encoding HA-MBP. Firefly luciferase activities were normalized to those of the Renilla luciferase transfection control and analyzed as described in Figure 2.
As expected, in AGO1-depleted cells, mutants that lost the ability to interact with both miRNAs and GW182 could not rescue silencing (Fig. 5, e.g., F2V2, F777A,I716A), as shown before for the F2V2 mutant (Eulalio et al. 2008b). In addition, the AGO1 PAZ6 mutant was also inactive in the complementation assay as expected; this mutant did not interact with miRNAs but retained the ability to interact with GW182 (Fig. 5, PAZ6). Remarkably, mutant F777A and the double mutant (Fig. 5, F777A,R771A), which interacted with miRNAs but were impaired in GW182 binding, were also impaired in the complementation assay. These results together with the results described above indicate that AGO1 requires the interaction with GW182 to mediate miRNA silencing.
GW182 acts downstream from miRNA loading onto AGO1
To define more precisely the step of silencing at which GW182 is required, we investigated whether GW182 was required for the interaction of AGO1 with miRNAs. To this end, HA-tagged AGO1 was expressed in control cells or cells depleted of GW182. The interaction between AGO1 and endogenous miRNAs was tested for by coimmunoprecipitation assays using anti-HA antibodies. We observed that HA-AGO1 coimmunoprecipitated endogenous bantam, both in control cells and in cells depleted of GW182 (Fig. 6A, lanes 6,8). The residual levels of GW182 in depleted cells were below 10% of those detected in control cells (Fig. 6B), yet AGO1 and bantam expression levels remained unaffected in these cells (Fig. 6A; data not shown), indicating that GW182 is not required for miRNA biogenesis.
We also tested the association of GW182 with miRNAs by coimmunoprecipitation followed by Northern blotting. We observed that wild-type GW182 coimmunoprecipitated bantam. In contrast, the GW182 mutant (12xGW–AA), which did not interact with AGO1, failed to associate with bantam (Fig. 6C, lane 9). Thus, AGO1 bridges the association of GW182 with miRNAs. Together these results indicate that GW182 acts downstream from miRNA loading onto AGO1.
AGO1 accumulation in P-bodies requires the interaction with GW182 but not with miRNAs
We previously showed that GFP-AGO1 spreads throughout the cytoplasm in transiently transfected S2 cells but accumulates in P-bodies when coexpressed with HA-GW182 (Fig. 7, cf. B and C; Behm-Ansmant et al. 2006a). As mentioned above, the AGO1 mutants (F777A and F777A, R771A), which did not interact with GW182 in coimmunoprecipitation assays, localized to P-bodies when coexpressed with HA-GW182 (Fig. 7D,E; Supplemental Table 2), suggesting that the residual GW182-binding activity of these mutants is sufficient for their accumulation in P-bodies. Mutations in AGO1 that abolished the interaction with GW182 and miRNAs prevented AGO1 accumulation in P-bodies (Fig. 7F; Supplemental Table 2). This was the case, for instance, for the F2V2 mutant (Fig. 7F); however, the AGO1 PAZ6 mutant that interacted with GW182 but not with miRNAs localized to P-bodies (Fig. 7G). These results suggest that AGO1 accumulation in P-bodies is mediated by GW182 but does not require AGO1 to bind miRNAs. Thus, the pool of AGO1 in P-bodies may not be necessarily active in silencing.
The role of GW182 in silencing is unrelated to P-body localization
It has been proposed that the role of GW182 in silencing is to direct AGO1 and miRNA targets to P-bodies where translational repression and mRNA decay could occur (Liu et al. 2005b; Pillai et al. 2005; Bhattacharyya et al. 2006). We therefore investigated the possibility that the essential role of GW182 in silencing is due to its ability to localize to P-bodies and to promote the accumulation of AGO1 in P-bodies.
In previous studies, we showed that a fragment of GW182 containing the N-terminal GW-repeats, the UBA-like domain, and the Q/N-rich region was necessary and sufficient for P-body localization, although none of these regions was sufficient on their own (Behm-Ansmant et al. 2006a). To define the P-body localization domain of GW182 more precisely, we deleted the UBA-like domain or the Q/N-rich region in the context of full-length GW182 (GW182-ΔUBA; GW182-ΔQ/N).
We found a GW182 protein lacking the UBA-like domain localized to cytoplasmic foci, as observed for wild-type GW182 (Fig. 7, cf. J and I). These foci correspond to endogenous P-bodies, as judged by the staining with an antibody recognizing Trailer Hitch (Tral) (Eulalio et al. 2007a). Furthermore, the GW182 mutant lacking the UBA-like domain interacted with AGO1 and was fully active in the complementation assay (Figs. 1B, 2, 3), indicating that the UBA-like domain is dispensable for the silencing activity of GW182.
In contrast, the protein lacking the Q/N-rich region spread throughout the cytoplasm and failed to localize to P-bodies (Fig. 7K). Consequently, although this mutant could interact with AGO1, it did not promote the accumulation of AGO1 in P-bodies (Figs. 1B, 7H). In the complementation assay, however, GW182-ΔQ/N restored silencing as efficiently as the wild-type protein for all but one of the reporters tested (Figs. 2, 3). The exception was the F-Luc-Nerfin-1 reporter, whose silencing by miR-279 was only partially restored (Fig. 2E). These results indicate that the Q/N-rich region is not essential for, but may contribute to silencing in a target-specific manner. More importantly, because GW182-ΔQ/N does not accumulate in P-bodies and fails to localize AGO1 to P-bodies, these results indicate that the role of GW182 in silencing can be uncoupled from its ability to localize to P-bodies. In agreement with this, we showed in previous studies that P-body integrity is not required for silencing (Eulalio et al. 2007c).
It is important to note that the Q/N-rich region is necessary but not sufficient for P-body localization because mutations in the N-terminal GW-repeats, which disrupted AGO1 interaction, also prevented the accumulation of GW182 in P-bodies (Fig. 7L). Thus, GW182 localization to P-bodies requires both the interaction with AGO1 and the Q/N-rich region.
The middle and C-terminal regions of GW182 are essential for silencing
In previous studies, we showed that the RRM domain of GW182 is not essential for, but contributes to silencing, in a miRNA target-specific manner (Eulalio et al. 2009a). The results described above indicated that the UBA-like domain and Q/N-rich region are also dispensable for silencing, although they may contribute to the repression of specific mRNAs. We therefore tested whether the middle and C-terminal regions contribute to GW182 silencing activity.
Deleting the middle region (M-GW), which contains three GW-repeats, did not affect the GW182 interaction with AGO1 or miRNAs (Figs. 1B, lane 16, 6C, lane 10); this is in agreement with the observation that these GW-repeats do not contribute to the AGO1 interaction. Unexpectedly, deleting the C-terminal region (C-term) slightly increased the association of GW182 with AGO1 and miRNAs (Figs. 1B, lane 18, 6C, lane 11). Nevertheless, despite their ability to interact with AGO1 and miRNAs, proteins lacking the middle or C-terminal regions were impaired in the complementation assay; thus, these regions contribute to silencing (Figs. 2, 3). Accordingly, a protein lacking both the middle and C-terminal regions (ΔM+C-term) did not restore silencing in cells depleted of endogenous GW182 (Figs. 2, 3), despite the fact that this protein coimmunoprecipitated AGO1 and miRNAs more efficiently than wild-type GW182 (Fig. 6C, lane 12; Supplemental Fig. 1). Furthermore, the GW182 mutant lacking both the middle and C-terminal regions localized to P-bodies (Fig. 7N), indicating lack of correlation between silencing activity and the ability to localize to P-bodies.
Within the middle region, a motif of about 60 residues shows a higher degree of conservation among GW182 proteins (Fig. 1A, gray box). However, deleting this motif alone did not affect GW182 silencing activity (Supplemental Fig. 3A–D, ΔMIII), indicating that the additional, less conserved sequences in the middle region are involved in silencing. In summary, our results indicate that in addition to the N-terminal AGO1-binding domain, the middle and C-terminal regions of GW182 are essential for silencing and may constitute a bipartite silencing domain.
DISCUSSION
Proteins of the GW182 family have been shown to play an essential role in the miRNA pathway in diverse organisms (Ding and Han 2007; Eulalio et al. 2008a); yet the step of silencing at which these proteins function remained to be established. Here we show that GW182 proteins act in the effector step of silencing, downstream from miRNA loading onto AGO1. Two domains in these proteins play an essential role in silencing: the N-terminal domain, which provides multiple binding sites for Argonaute proteins, and a bipartite silencing domain that is required for translational repression and decay of miRNA targets. We further show that impairing the interaction between AGO1 and GW182 inhibits silencing. Together, our results provide compelling evidence that miRNA silencing in animal cells is mediated by AGO1 in complex with GW182.
Role of GW182 in silencing
AGO1–GW182 complexes repress translation and/or direct miRNA targets to degradation. The mechanism of miRNA-mediated translational repression, in the absence of mRNA degradation, remains to be elucidated (Nilsen et al. 2007). In contrast, it is well established that degradation of miRNA targets (having partially complementary miRNA-binding sites) is mediated by deadenylation and decapping, followed by 5′ to 3′ decay of the mRNA body (Bagga et al. 2005; Rehwinkel et al. 2005; Wu and Belasco 2005; Behm-Ansmant et al. 2006a,b; Giraldez et al. 2006; Mishima et al. 2006; Wu et al. 2006; Eulalio et al. 2007b, 2008b, 2009b). This process requires GW182, AGO1, the CCR4-NOT1-CAF1 deadenylase complex, the decapping enzyme DCP2, and several decapping activators including DCP1, Ge-1, EDC3, RCK/Me31B, and HPat (Rehwinkel et al. 2005; Behm-Ansmant et al. 2006a; Chu and Rana 2006; Eulalio et al. 2007b, 2008b, 2009b). The precise mechanism by which these proteins are recruited to miRNA targets is unknown; but it is possible that decay factors are recruited only after the GW182–AGO1 complexes bind to the target or after the mRNP target undergoes conformational rearrangements.
The role of GW182 in silencing is not limited to promoting mRNA degradation. As shown in this and additional studies, GW182 proteins are also required for translational repression mediated by miRNAs (Liu et al. 2005a; Meister et al. 2005; Behm-Ansmant et al. 2006a; Chu and Rana 2006; Eulalio et al. 2008b). The GW182 silencing domain is also essential for this process, suggesting possible cross talk between this domain and the translational machinery. However, without clearly understanding the mechanisms of miRNA-mediated gene silencing, we cannot discern whether translational repression and/or mRNA decay cause silencing directly or are indirect consequences of a primary effect of miRNA–effector complexes on mRNP composition and/or conformation.
Regardless, it is surprising that the middle and C-terminal GW182 regions, although essential for silencing, are predicted to be unstructured and are not highly conserved within the GW182 protein family. Indeed, the silencing activity of human TNRC6A, B, and C also resides in the corresponding C-terminal silencing domains (Lazzaretti et al. 2009). Within the middle region, a motif of about 60 residues shows a higher degree of conservation among GW182 proteins (Fig. 1A, gray box); however, deleting this motif did not affect GW182 silencing activity, indicating that the additional, less conserved sequences within the middle region contribute to silencing. Although the role of the middle and C-terminal regions in silencing remains to be elucidated, it is possible that these regions provide sites for protein–protein interactions relevant for miRNA-mediated gene silencing. Furthermore, the high content of serine residues in these regions (17.1% and 26.6%, respectively) suggests that GW182 activity may be regulated by phosphorylation.
MATERIALS AND METHODS
DNA constructs
Luciferase reporters and plasmids for expression of miRNAs and HA-tagged proteins were described before (Eulalio et al. 2007b, 2008b). Renilla luciferase cloned between the KpnI and XhoI sites of vector pAc5.1A (Invitrogen) served as a transfection control. Mutants of D. melanogaster AGO1 or GW182 were generated by site-directed mutagenesis using the plasmid pAc5.1B-λN-HA-AGO1 (siRNA resistant) or pAc5.1B-λN-HA-GW182 as template, respectively, using the Quick change mutagenesis kit from Stratagene. Plasmid pAc5.1B-λNHA-AGO1 (siRNA resistant) carries mutations that prevent the interaction with AGO1-siRNA without changing the protein sequence.
RNA interference, luciferase assays, and Northern blots
RNA interference was performed as described before (Eulalio et al. 2007b, 2008b) with the exception that cells were depleted on days 0 and 4, transfected on day 7 and collected on day 10. Transfections of S2 cells were performed in 6-well plates, using Effectene transfection reagent (QIAGEN). The following siRNAs were used: GFP siRNA (5′-GCGACGUAAACGGCCACAAGUUCUU) and AGO1 siRNA-1 (5′-CGAAGGAGAUCAAGGGUUUUU). The siRNAs were transfected at a final concentration of 75 nM. For miRNA-mediated silencing assays, the transfection mixtures contained 0.1 μg of firefly luciferase reporter plasmid, 0.4 μg of the Renilla transfection control, and 0.5 μg of plasmids expressing miRNA primary transcripts or the corresponding vector without insert. When indicated, 0.025–1 μg of plasmids expressing recombinant proteins were cotransfected. Firefly and Renilla luciferase activities were measured 3 d (GW182 complementation assay) or 4 d (AGO1 complementation assay) after transfection using the Dual-Luciferase Reporter Assay System (Promega). Total RNA was isolated using TriFast (Peqlab Biotechnologies) and analyzed as described before (Eulalio et al. 2007b, 2008b).
Coimmunoprecipitations and Western blots
The interaction of AGO1 with endogenous miRNAs and GW182, or the interaction of GW182 with endogenous miRNAs and AGO1 was tested as described (Rehwinkel et al. 2006; Eulalio et al. 2008b). Antibodies to D. melanogaster GW182 were described before (Behm-Ansmant et al. 2006a). Antibodies to AGO1 (dilution 1:1000) were purchased from Abcam (catalog number ab5070). A monoclonal anti-HA antibody was purchased from Covance Research Products (catalog number MMS-101P). Polyclonal anti-HA and anti-tubulin antibodies were purchased from Sigma (catalog numbers H6908 and T6199, respectively). Bound primary antibodies were detected with alkaline-phosphatase-coupled secondary antibodies (Western-Star kit from Tropix).
Immunofluorescence
Immunostainings were performed as described by Eulalio et al. (2007c). Cells were stained with affinity-purified anti-Tral antibodies diluted 1:250 in PBS containing 1% BSA. Alexa-594-coupled goat anti-rat secondary antibody (Molecular probes, catalog number A11007) was used at a dilution of 1:1000. HA-tagged proteins were detected with a monoclonal anti-HA antibody (Covance Research Products, catalog number MMS-101P) diluted 1:1000 in PBS containing 1% BSA. Alexa Fluor 594–coupled or Alexa Fluor 488–coupled goat anti-mouse secondary antibody (Molecular probes, catalog numbers A11001 and A11005, respectively) was used at a dilution of 1:1000. Cells were mounted using Fluoromount-G (Southern Biotechnology Associates, Inc.). Images were acquired using a Leica TCS SP2 confocal microscope.
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
This study was supported by the Max Planck Society, by grants from the Deutsche Forschungsgemeinschaft (DFG, FOR855, and the Gottfried Wilhelm Leibniz Program awarded to E.I.), and by the Sixth Framework Programme of the European Commission through the SIROCCO Integrated Project LSHG-CT-2006-037900.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1605509.
REFERENCES
- Bagga S., Bracht J., Hunter S., Massirer K., Holtz J., Eachus R., Pasquinelli A.E. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–563. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- Behm-Ansmant I., Rehwinkel J., Doerks T., Stark A., Bork P., Izaurralde E. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes & Dev. 2006a;20:1885–1898. doi: 10.1101/gad.1424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behm-Ansmant I., Rehwinkel J., Izaurralde E. MicroRNAs silence gene expression by repressing protein expression and/or by promoting mRNA decay. Cold Spring Harb. Symp. Quant. Biol. 2006b;71:523–530. doi: 10.1101/sqb.2006.71.013. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya S.N., Habermacher R., Martine U., Closs E.I., Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- Chu C.Y., Rana T.M. Translation repression in human cells by microRNA-induced gene silencing requires RCK/p54. PLoS Biol. 2006;4:e210. doi: 10.1371/journal.pbio.0040210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X.C., Großhans H. Repression of C. elegans microRNA targets at the initiation level of translation requires GW182 proteins. EMBO J. 2009;28:213–222. doi: 10.1038/emboj.2008.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L., Han M. GW182 family proteins are crucial for microRNA-mediated gene silencing. Trends Cell Biol. 2007;17:411–416. doi: 10.1016/j.tcb.2007.06.003. [DOI] [PubMed] [Google Scholar]
- Ding L., Spencer A., Morita K., Han M. The developmental timing regulator AIN-1 interacts with miRISCs and may target the argonaute protein ALG-1 to cytoplasmic P bodies in C. elegans . Mol. Cell. 2005;19:437–447. doi: 10.1016/j.molcel.2005.07.013. [DOI] [PubMed] [Google Scholar]
- El-Shami M., Pontier D., Lahmy S., Braun L., Picart C., Vega D., Hakimi M.A., Jacobsen S.E., Cooke R., Lagrange T. Reiterated WG/GW motifs form functionally and evolutionarily conserved ARGONAUTE-binding platforms in RNAi-related components. Genes & Dev. 2007;21:2539–2544. doi: 10.1101/gad.451207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A., Behm-Ansmant I., Izaurralde E. P bodies: At the crossroads of post-transcriptional pathways. Nat. Rev. Mol. Cell Biol. 2007a;8:9–22. doi: 10.1038/nrm2080. [DOI] [PubMed] [Google Scholar]
- Eulalio A., Rehwinkel J., Stricker M., Huntzinger E., Yang S.F., Doerks T., Dorner S., Bork P., Boutros M., Izaurralde E. Target-specific requirements for enhancers of decapping in miRNA-mediated gene silencing. Genes & Dev. 2007b;21:2558–2570. doi: 10.1101/gad.443107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A., Behm-Ansmant I., Schweizer D., Izaurralde E. P-body formation is a consequence, not the cause of RNA-mediated gene silencing. Mol. Cell. Biol. 2007c;27:3970–3981. doi: 10.1128/MCB.00128-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A., Huntzinger E., Izaurralde E. Getting to the root of miRNA-mediated gene silencing. Cell. 2008a;132:9–14. doi: 10.1016/j.cell.2007.12.024. [DOI] [PubMed] [Google Scholar]
- Eulalio A., Huntzinger E., Izaurralde E. GW182 interaction with Argonaute is essential for miRNA-mediated translational repression and mRNA decay. Nat. Struct. Mol. Biol. 2008b;15:346–353. doi: 10.1038/nsmb.1405. [DOI] [PubMed] [Google Scholar]
- Eulalio A., Tritschler F., Buettner R., Weichenrieder O., Izaurralde E., Truffault V. The RRM domain in GW182 proteins contributes to miRNA-mediated gene silencing. Nucl. Acids Res. 2009a doi: 10.1093/nar/gkp173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A., Huntzinger E., Nishihara T., Rehwinkel J., Fauser M., Izaurralde E. Deadenylation is a widespread effect of miRNA regulation. RNA. 2009b;15:21–32. doi: 10.1261/rna.1399509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eystathioy T., Chan E.K., Tenenbaum S.A., Keene J.D., Griffith K., Fritzler M.J. A phosphorylated cytoplasmic autoantigen, GW182, associates with a unique population of human mRNAs within novel cytoplasmic speckles. Mol. Biol. Cell. 2002;13:1338–1351. doi: 10.1091/mbc.01-11-0544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldez A.J., Mishima Y., Rihel J., Grocock R.J., Van Dongen S., Inoue K., Enright A.J., Schier A.F. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- Jakymiw A., Lian S., Eystathioy T., Li S., Satoh M., Hamel J.C., Fritzler M.J., Chan E.K. Disruption of GW bodies impairs mammalian RNA interference. Nat. Cell Biol. 2005;7:1267–1274. doi: 10.1038/ncb1334. [DOI] [PubMed] [Google Scholar]
- Landthaler M., Gaidatzis D., Rothballer A., Chen P.Y., Soll S.J., Dinic L., Ojo T., Hafner M., Zavolan M., Tuschl T. Molecular characterization of human Argonaute-containing ribonucleoprotein complexes and their bound target mRNAs. RNA. 2008;14:2580–2596. doi: 10.1261/rna.1351608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzaretti D., Tournier I., Izaurralde E. The C-terminal domains of human TNRC6A, TNRC6B, and TNRC6C silence bound transcripts independently of the Argonaute proteins. RNA. 2009;15 doi: 10.1261/rna.1606309. (this issue). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Rivas F.V., Wohlschlegel J., Yates J.R., III, Parker R., Hannon G.J. A role for the P-body component GW182 in microRNA function. Nat. Cell Biol. 2005a;7:1261–1266. doi: 10.1038/ncb1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Valencia-Sanchez M.A., Hannon G.J., Parker R. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat. Cell Biol. 2005b;7:719–723. doi: 10.1038/ncb1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G., Landthaler M., Peters L., Chen P.Y., Urlaub H., Lührmann R., Tuschl T. Identification of novel Argonaute-associated proteins. Curr. Biol. 2005;15:2149–2155. doi: 10.1016/j.cub.2005.10.048. [DOI] [PubMed] [Google Scholar]
- Mishima Y., Giraldez A.J., Takeda Y., Fujiwara T., Sakamoto H., Schier A.F., Inoue K. Differential regulation of germline mRNAs in soma and germ cells by zebrafish miR-430. Curr. Biol. 2006;16:2135–2142. doi: 10.1016/j.cub.2006.08.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsen T.W. Mechanisms of microRNA-mediated gene regulation in animal cells. Trends Genet. 2007;23:243–249. doi: 10.1016/j.tig.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Pillai R.S., Bhattacharyya S.N., Artus C.G., Zoller T., Cougot N., Basyuk E., Bertrand E., Filipowicz W. Inhibition of translational initiation by let-7 microRNA in human cells. Science. 2005;309:1573–1576. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- Rehwinkel J., Behm-Ansmant I., Gatfield D., Izaurralde E. A crucial role for GW182 and the DCP1:DCP2 decapping complex in miRNA-mediated gene silencing. RNA. 2005;11:1640–1647. doi: 10.1261/rna.2191905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehwinkel J., Natalin P., Stark A., Brennecke J., Cohen S.M., Izaurralde E. Genome-wide analysis of mRNAs regulated by Drosha and Argonaute proteins in Drosophila melanogaster . Mol. Cell. Biol. 2006;26:2965–2975. doi: 10.1128/MCB.26.8.2965-2975.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider M.D., Najand N., Chaker S., Pare J.M., Haskins J., Hughes S.C., Hobman T.C., Locke J., Simmonds A.J. Gawky is a component of cytoplasmic mRNA processing bodies required for early Drosophila development. J. Cell Biol. 2006;174:349–358. doi: 10.1083/jcb.200512103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Till S., Lejeune E., Thermann R., Bortfeld M., Hothorn M., Enderle D., Heinrich C., Hentze M.W., Ladurner A.G. A conserved motif in Argonaute-interacting proteins mediates functional interactions through the Argonaute PIWI domain. Nat. Struct. Mol. Biol. 2007;14:897–903. doi: 10.1038/nsmb1302. [DOI] [PubMed] [Google Scholar]
- Wu L., Belasco J.G. Micro-RNA regulation of the mammalian lin-28 gene during neuronal differentiation of embryonal carcinoma cells. Mol. Cell. Biol. 2005;25:9198–9208. doi: 10.1128/MCB.25.21.9198-9208.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., Fan J., Belasco J.G. MicroRNAs direct rapid deadenylation of mRNA. Proc. Natl. Acad. Sci. 2006;103:4034–4039. doi: 10.1073/pnas.0510928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Ding L., Cheung T.H., Dong M.-Q., Chen J., Sewell A.K., Liu X., Yates J.R., Han M. Systematic identification of C. elegans miRISC proteins, miRNAs, and mRNA targets by their interactions with GW182 proteins AIN-1 and AIN-2. Mol. Cell. 2007;28:598–613. doi: 10.1016/j.molcel.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]